ABSTRACT
To interrogate genes essential for cell growth, proliferation and survival in human cells, we carried out a genome-wide clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 screen in a B-cell lymphoma line using a custom extended-knockout (EKO) library of 278,754 single-guide RNAs (sgRNAs) that targeted 19,084 RefSeq genes, 20,852 alternatively spliced exons, and 3,872 hypothetical genes. A new statistical analysis tool called robust analytics and normalization for knockout screens (RANKS) identified 2,280 essential genes, 234 of which were unique. Individual essential genes were validated experimentally and linked to ribosome biogenesis and stress responses. Essential genes exhibited a bimodal distribution across 10 different cell lines, consistent with a continuous variation in essentiality as a function of cell type. Genes essential in more lines had more severe fitness defects and encoded the evolutionarily conserved structural cores of protein complexes, whereas genes essential in fewer lines formed context-specific modules and encoded subunits at the periphery of essential complexes. The essentiality of individual protein residues across the proteome correlated with evolutionary conservation, structural burial, modular domains, and protein interaction interfaces. Many alternatively spliced exons in essential genes were dispensable and were enriched for disordered regions. Fitness defects were observed for 44 newly evolved hypothetical reading frames. These results illuminate the contextual nature and evolution of essential gene functions in human cells.
KEYWORDS: CRISPR/Cas9, alternative splicing, gene essentiality, genetic screen, hypothetical gene, protein complex, proteome
INTRODUCTION
Essential genes underpin the genetic architecture and evolution of biological systems (1). In all species, essential genes are needed for survival and proliferation, while in multicellular organisms, additional essential genes function in different tissues at various stages of development. In the budding yeast Saccharomyces cerevisiae, only 1,114 of the ∼6,000 genes in the genome are essential for growth under nutrient-rich conditions (1). The nonessential nature of most genes suggests that the genetic landscape of the cell is shaped by redundant gene functions (2). Consistently, systematic screens in S. cerevisiae have uncovered more than 500,000 binary synthetic lethal interactions (3–5). In parallel, context-specific chemical screens have revealed that virtually every gene can be rendered essential under the appropriate condition (6). The prevalence of nonessential genes has been verified by systematic genetic analysis in other single-cell organisms, including Schizosaccharomyces pombe (7), Candida albicans (8), and Escherichia coli (9). In metazoans, the knockdown of gene function by RNA interference (RNAi) in the nematode worm Caenorhabditis elegans revealed that 1,170 genes are essential for development (10), while in the fruit fly Drosophila melanogaster at least 438 genes are required for cell proliferation in vitro (11). In the mouse Mus musculus, ∼25% of genes tested to date are required for embryonic viability (12). Essential genes tend to be highly conserved, to interact with one another in local modules, and to be highly connected in protein and genetic interaction networks (5, 13, 14). Although essentiality is often framed as an all-or-none binary phenotype, in reality the loss of gene function causes a spectrum of fitness defects that depend on developmental and environmental contexts. For example, in S. cerevisiae an additional ∼600 genes are required for optimal growth in rich medium (1), and in C. elegans most genes are required for fitness at the whole-organism level (15). The definition of essentiality is thus dependent on context and the experimental definition of fitness thresholds.
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (CAS) protein system directs the cleavage of specific DNA sequences in prokaryotes, where it serves as an adaptive immunity defense mechanism against infection by foreign phage DNA (16). The Cas9 endonuclease can be targeted toward a specific DNA sequence by a single-guide RNA (sgRNA) that contains a 20-nucleotide match to the locus of interest (17). The coexpression of Cas9 and an sgRNA leads to a blunt-end double-strand break (DSB) at a precisely specified position in the genome. Repair of the DSB by error-prone nonhomologous end joining (NHEJ) leads to random DNA insertions or deletions (indels) that cause frameshift mutations and an ersatz knockout of the target gene with high efficiency (18).
CRISPR/Cas9 technology has been recently adapted to perform large-scale functional gene knockout screens in human cells (19). A pooled library of 64,751 sgRNAs that targets 18,080 genes was used in screens against melanoma and stem cell lines (20, 21), while a library of 182,134 sgRNAs that targets 18,166 genes was used in screens against chronic myelogenous leukemia and Burkitt's lymphoma cell lines (22). A third library of 176,500 sgRNAs that targets 17,661 genes was used in screens against colon cancer, cervical cancer, glioblastoma, and hTERT-immortalized retinal epithelial cell lines (23). In parallel, genome-scale screens based on transposon-mediated gene trap technology have been performed in two haploid human cell lines (22, 24). Each of these genome-wide screens identified on the order of 1,500 to 2,200 essential genes that were enriched in functions for metabolism, DNA replication, transcription, splicing, and protein synthesis (22–24). Importantly, CRISPR/Cas9 and gene trap screens exhibit a strikingly high degree of overlap and overcome limitations of previous RNAi-based gene knockdown approaches (22–24).
Currently available human genome-wide CRISPR libraries target most well-characterized gene loci. Two reported libraries target the well-validated RefSeq gene collection (21, 22), while a third library targets confirmed protein-coding gene regions in the GenCode v17 assembly (23). To extend functional CRISPR/Cas9 screens to less-well-characterized regions of the genome at potential subgene resolution, we generated a high-complexity extended-knockout (EKO) library of 278,754 sgRNAs that targets 19,084 RefSeq genes, 20,852 unique alternative exons, and 3,872 hypothetical genes. The EKO library also includes 2,043 control sgRNAs with no match to the human genome to allow estimation of screen noise. We used the EKO library to identify essential genes in the NALM-6 human pre-B-cell lymphocytic leukemia line, including alternatively spliced exons and hypothetical genes not previously tested in CRISPR/Cas9 screens. Our analysis revealed the broad influence of Cas9-induced double-strand breaks on cell viability, the residue-level determinants of essential protein function, the prevalence of nonessential exons, and the evolution of new essential genes. Integration of our data with nine previous genome-wide CRISPR/Cas9 screens revealed a bimodal distribution of essential genes across cell lineages and a range of subunit essentiality across protein complexes, consistent with a continuous context-dependent variation in gene essentiality. High-resolution CRISPR/Cas9 genetic screens can thus uncover the organization and evolution of the essential human proteome.
RESULTS
Design and generation of the EKO library.
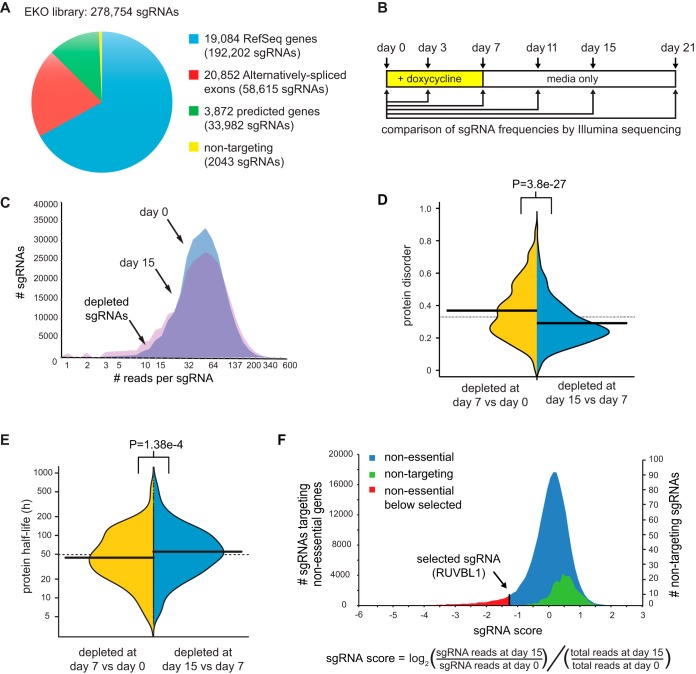
We generated a custom sgRNA library that targeted more genomic loci than previously published libraries, including additional RefSeq genes, alternatively spliced exons, and hypothetical protein-coding genes (Fig. 1A; see Table S1 in the supplemental material for all sgRNA sequences). The core set of the EKO library was comprised of 233,268 sgRNAs that target 19,084 RefSeq protein-coding genes and 17,139 alternatively spliced exons within these coding regions. An extended set of 43,443 sgRNAs within the EKO library was designed to target loci predicted to be potential protein-coding regions by AceView (25) or GenCode (26), as well as additional alternatively spliced exons predicted by AceView that we validated by independent transcriptome sequencing (RNA-seq) evidence (see the supplemental material). Each gene in the EKO library was targeted by approximately 10 sgRNAs and each alternative exon by 3 sgRNAs. The combined gene list represented in the EKO library contains almost 5,000 more candidate genes than any sgRNA library screened to date, including almost 1,000 additional RefSeq genes (20–23). To estimate the effect of noise in our screens, we also designed 2,043 sgRNAs that had no sequence match to the human genome.
FIG 1.
Genome-wide sgRNA library generation and screen for essential genes. (A) Composition of the EKO library. Note that some sgRNAs were used to evaluate both an alternative exon and the entire gene. (B) Experimental design for screen. A NALM-6 doxycycline-inducible Cas9 cell line was transduced with the EKO library at an MOI of ∼0.5, followed by sgRNA vector selection, population outgrowth, and determination of sgRNA frequencies at the indicated time points. (C) Number of sgRNA reads before doxycycline induction (day 0) superimposed on distributions after doxycycline induction (day 7) and after a further 8 days of outgrowth in medium only (day 15). A constant factor was used to center the two distributions. (D) Effect of protein intrinsic disorder on kinetics of sgRNA depletion. The average probability of disordered stretches over the entire protein as predicted by IUPred (76) for the 1,000 genes with most highly depleted sgRNAs at day 7 versus day 15 (two-tailed Wilcoxon test) is shown. (E) Effect of target protein half-life on kinetics of sgRNA depletion. The half-lives of mouse orthologs of the 1,000 most highly depleted genes after day 7 (n = 645) were compared to those after day 15 (n = 724). P value, two-tailed Wilcoxon test. (F) Distribution of log2 sgRNA frequency changes for day 0 versus day 15 for all previously described nonessential sgRNAs (22–24) compared to nontargeting sgRNAs in this study. An sgRNA targeting an essential gene in this study (RUVBL1) is shown for reference. Single-tailed P values for each sgRNA were calculated as the area under the curve of the control distribution to the left of the sgRNA score divided by the total area under the control curve.
Identification of cell fitness genes with the EKO library.
We used the EKO library in a viability screen to assess fitness defects caused by loss of gene function in the human pre-B-cell acute lymphoblastic leukemia NALM-6 line, which is pseudodiploid and grows in suspension culture with a doubling time of approximately 24 h (27). A tightly regulated doxycycline-inducible Cas9 clone of NALM-6 for use in library screens was generated by random integration of a lentiviral construct. The EKO library was transduced at a low multiplicity of infection (MOI) and selected for lentiviral integration on blasticidin for 6 days, after which Cas9 expression was induced for 7 days with doxycycline, followed by outgrowth for up to 14 more days in the absence of doxycycline (Fig. 1B). Changes in sgRNA frequencies across the entire library were monitored by next-generation sequencing on an Illumina HiSeq 2000 at six different time points during the screen. The EKO library pool was well represented after blasticidin selection (i.e., the day zero time point for the screen), with 94% of all sgRNA read counts falling within a 10-fold range relative to one another (Fig. 1C; see Fig. S1A in the supplemental material). Little change in sgRNA frequency was observed after only 3 days of doxycycline induction (Fig. S1B), likely due to the lag in Cas9 induction, the kinetics of indel generation, and the time required for effective protein depletion. A progressively larger spread in sgRNA read frequencies was observed over the 21-day time course (Fig. S1C to F). We observed that genes with greater sgRNA depletion at earlier time points tended to encode proteins that were more disordered (Fig. 1D) and that had a shorter half-life (Fig. 1E). However, genes depleted by day 21 did not possess significantly longer half-lives than those depleted by day 15 (P > 0.05 by the Wilcoxon test) (data not shown). Genes scoring in the top 2,000 most-depleted genes at day 15 but not at day 7 or at day 21 were more likely to be validated by gene trap scores in HAP1 cells than those uniquely depleted on day 7 (P = 1.0e−91 by the Wilcoxon test) (Fig. S1G) or day 21 (P = 2.37e−6 by the Wilcoxon test) (Fig. S1H). In agreement with this finding, essential genes within specific functional classes would have been missed had we used the day 7 time point to identify essential genes (Fig. S1I). In light of these results, we chose to use the sgRNA read frequencies at day 15 for our subsequent analyses, with the day 0 frequencies serving as the reference time point. To further validate our screen data, we compared our results to those of previous CRISPR/Cas9 screens in 9 other cell lines (22, 23) and found that our screen had the best overall performance in identifying genes that were essential across multiple cell lines (see Fig. S2A in the supplemental material). We experimentally validated our pooled genome-wide screen data by recapitulating proliferation rate defects in single cell line knockouts for 10 essential genes in our screen and by examining the protein interaction contexts of 2 previously uncharacterized essential proteins (Fig. S2B to E) (see below).
Calculation of significant sgRNA depletion by RANKS.
To compute the relative level of depletion of each sgRNA, we developed a custom tool called robust analytics and normalization for knockout screens (RANKS), which enables statistical analysis of any pooled CRISPR/Cas9 library screen or short hairpin RNA (shRNA) screen. First, the log2 ratio of the sgRNA read frequency at the day 15 versus day zero time points, normalized by the total read count ratio, was used to quantify the relative abundance of each sgRNA (Fig. 1F). The average log2 ratio for all ∼10 sgRNAs that target each gene was used to generate a gene score (22), which reflected the potential fitness defect for each gene knockout. To account for experimental variation in sgRNA read counts within any given screen, instead of using the log2 ratio of the sgRNA itself, we used RANKS to estimate a P value based on how each log2 ratio compared to a selected set of nontargeting or other control sgRNAs. The resulting gene log P value score was then obtained from the average of the log P values for each of the 10 sgRNAs per gene. As applied to our data set, RANKS performed better than other, previously used methods (22, 23, 28) as judged by established correlates (see Fig. S3 and Table S2 in the supplemental material). Subsequent analyses were based on the RANKS statistical scores for each gene, exon, or hypothetical gene represented in the EKO library.
Genome-scale correlates of sgRNA depletion.
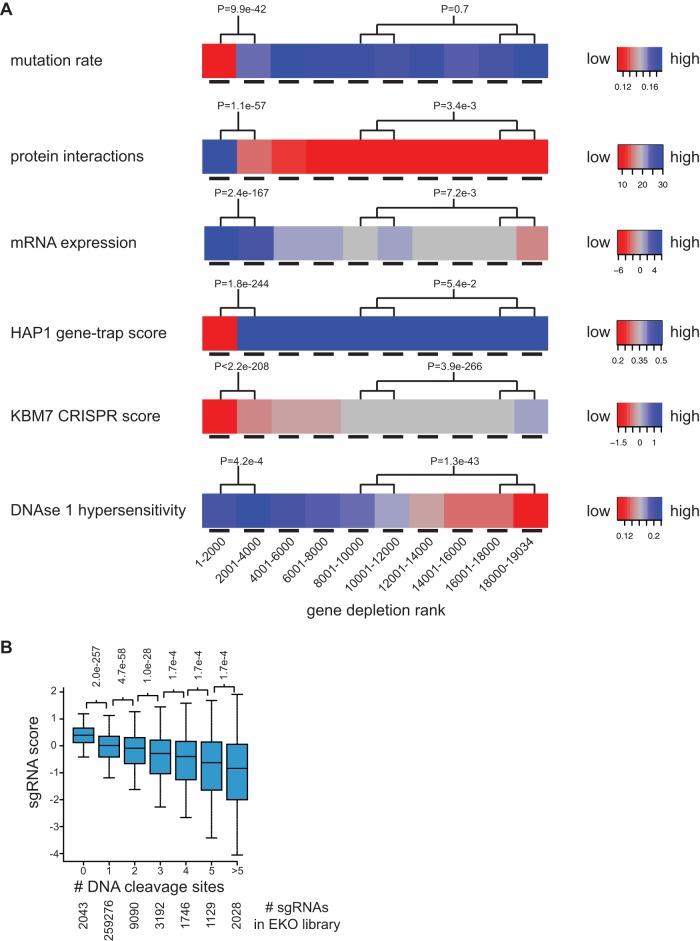
The results of the NALM-6 screen revealed that fitness scores correlated well with features known to be associated with essential genes. We examined correlates with mutation rate as estimated by protein sequence conservation across 46 vertebrate species (29), node degree in protein-protein interaction (PPI) networks (30), mRNA expression level in NALM-6 cells, essentiality detected by the gene trap method (24), essentiality detected by a whole-genome CRISPR/Cas9 screen in the near-haploid KBM7 cell line (22), and DNA accessibility as assessed by DNase I hypersensitivity peak density in naive B cells (Fig. 2A; see Fig. S4 in the supplemental material). For every feature, we observed a highly significant difference between the top-ranked 2,000 genes in our screen (1 to 2000) and the next 2,000 ranked genes (2001 to 4000), with P values of <0.05 (Wilcoxon test), such that each feature correlated strongly with gene essentiality. The strong agreement between depletion scores from an independent genome-wide CRISPR screen and our ranked gene list over the entire distribution confirmed the reproducibility of the CRISPR/Cas9 method.
FIG 2.
Features correlated with gene depletion. (A) RefSeq genes were ranked in bins of 2,000 genes from most to least depleted sgRNAs. The mutation rate was the fraction of aligned residues that differed from the human sequence across 45 vertebrate species in a 46-way Multi-Z whole-genome alignment. Protein interaction was the number of partners reported in the BioGRID database (3.4.133 release). mRNA expression was the log2 RNA-seq reads per million in the NALM-6 cell line. The HAP1 gene trap score was the ratio of sense to antisense intronic insertions (24). The KBM7 CRISPR score was the average log2 read frequency change (22). DNase I hypersensitivity was the number of read peaks per kbp in naive B cells from ENCODE. (B) Correlation of gene depletion with potential sgRNA target sites. The log2 read frequency fold change for day 0 to day 15 was binned by number of perfect matches or single-base mismatches (within the first 12 bases of the sgRNA) to the human genome.
For each feature, we also compared the union of the fourth and fifth bins (6001 to 10000) to the union of the last two bins (16001 to 19034). This comparison revealed that mutation rate and gene trap score differences flattened out after the first 2,000 to 4,000 genes (P values of >0.05 by the two-tailed Wilcoxon test), whereas the other features still correlated with gene rank in the latter bins to various degrees (PPI degree, P = 0.00335; mRNA expression, P = 0.0072; KBM7 score, P = 3.94e-266; DNase I hypersensitivity, P = 1.27e−46). The tight concordance between the NALM-6 and KBM7 scores over all bins was expected given that the same sgRNAs were used to target all RefSeq genes included in both libraries. The strong correlation between rank score in the NALM-6 screen and DNA accessibility suggested a potential nonspecific effect of sgRNA-directed DSB formation. The two additional significant correlations may be explained by the facts that mRNA expression level also correlated with DNA accessibility (Spearman's rank correlation coefficient = 0.34; P < 2.2e−16) and that protein interaction degree in turn correlated with gene expression (Pearson correlation coefficient = 0.22; P < 2.2e−16). These cross-correlations are likely explained by unrepaired Cas9-mediated cleavage events in a fraction of cells leading to a DNA damage-dependent growth arrest and depletion from the pool independent of indel formation. Highly accessible DNA is more likely to be cleaved by Cas9 (31), and sgRNAs with multiple potential cleavage sites in amplified regions cause a greater fitness defect (22, 32, 33), although the latter trend has not been observed in other screens (34). To test this idea on a genome-wide scale we compared the predicted number of sgRNA matches in the genome to the level of depletion in the pool and indeed found a strong overall correlation (Fig. 2B). For this same reason, nontargeting control sgRNAs in the EKO library were actually enriched compared to the rest of the targeting sgRNAs in the pool (Fig. 1F) and therefore were not an ideal control distribution. For subsequent analysis, we instead redefined the control set for RANKS as the 213,886 sgRNAs that targeted genes never reported as essential in any published screen, and we also removed all sgRNAs with more than two potential off-target cleavage sites and applied a fixed correction factor to sgRNAs with one or two potential off-target sites (see Fig. S5 and text in the supplemental material). This adjustment of the control sgRNA set allowed significance values to be reliably established but left the relative gene rank order virtually unchanged (r2 = 0.978).
A bimodal distribution of gene essentiality across cell lines.
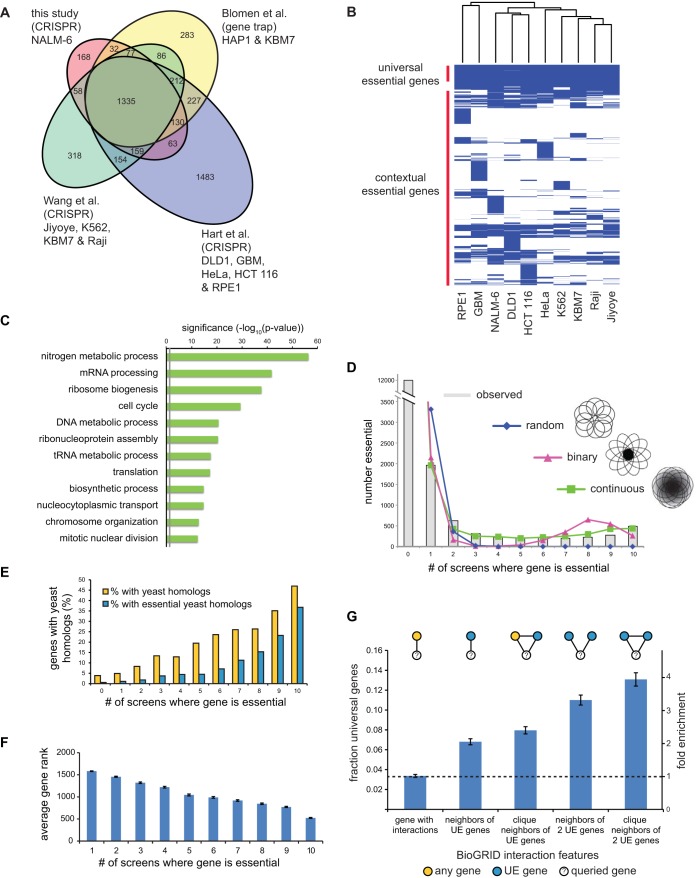
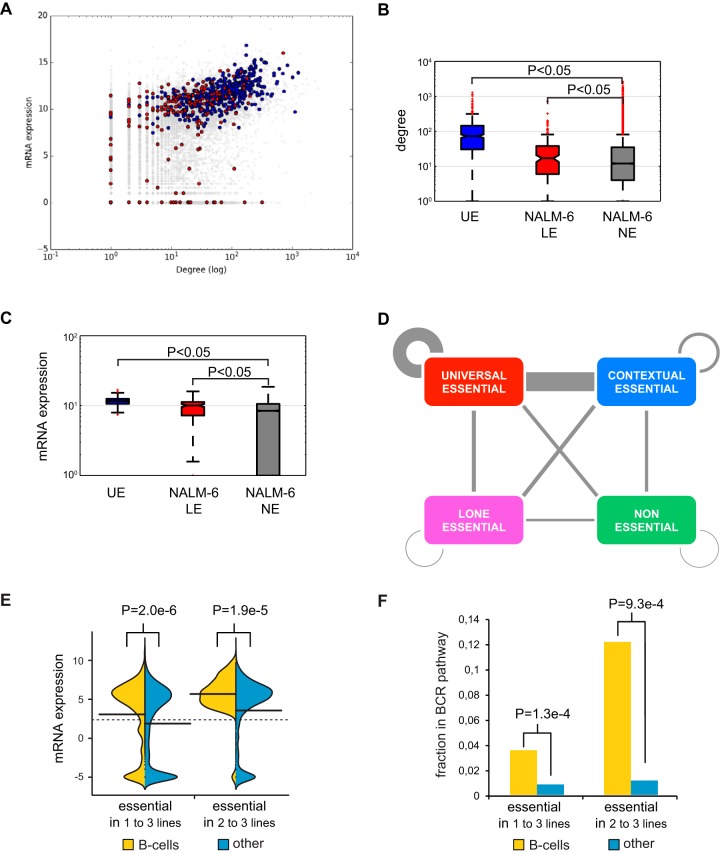
The EKO library screen identified a total of 2,280 fitness genes in NALM-6 cells below a false-discovery rate (FDR) of 0.05, which we generically termed essential genes (see Table S3 in the supplemental material). Of these essential genes, 269 were specific to the NALM-6 screen: 225 corresponded to well-validated RefSeq genes, including 19 RefSeq genes not assessed previously, and 44 hypothetical genes annotated only in AceView or GenCode. The set of essential RefSeq genes in NALM-6 cells overlapped strongly with three sets of essential genes previously identified using different libraries, cell lines, and methods (Fig. 3A) (22–24). For purposes of direct comparison between studies, we examined a set of 16,996 RefSeq genes shared between the EKO library and two published sgRNA libraries (22, 23). We identified 486 genes that were uniformly essential across our NALM-6 screen and 9 previous screens in different cell lines (22, 23), referred to here as universal essential (UE) genes (Fig. 3B and Table S3). This UE gene set was smaller than sets of previously reported common essential genes in previous screens (22–24) but was similarly enriched for processes required for cell proliferation and survival, including transcription, translation, energy metabolism, DNA replication, and cell division (Fig. 3C).
FIG 3.
Universal essential genes. (A) Overlap between genes defined as essential in this study and three other studies (22–24). The indicated overlap is between all essential genes identified in each study, i.e., an amalgam across all cell lines, in order to assess interlibrary reproducibility. (B) Clustergram of essential genes across 10 different cell lines from the CRISPR screens in panel A. (C) Biological processes enriched in UE genes. (D) Number of essential genes shared between cell lines. Experimentally observed essential genes are shown as a histogram. The indicated models to account for shared essential genes were fitted using maximum likelihood. See the text for details. (E) Fraction of essential genes with orthologous nonessential or essential yeast genes as a function of cell line number. (F) Average essentiality rank of each essential gene as a function of cell line number. (G) Fraction and relative enrichment of UE proteins in specific protein-protein interaction network motifs from BioGRID.
To investigate the nature of essential genes as a function of cell type, we plotted the number of shared essential genes across the 10 different cell lines. For clarity, we use the terms contextually essential (CE) to refer to genes essential in more than one cell line but fewer than in all cell lines and lone essential (LE) to designate any gene that is uniquely essential in a single line. As opposed to a simple monotonic decay in shared essential genes as a function of cell line number, we observed a bimodal distribution whereby the number of shared essential genes rapidly declined as the number of cell lines increased but then plateaued, with a slight peak at the maximum number of lines (Fig. 3D). To assess the potential effect of score threshold on the bimodal distribution, we examined the relative distribution of scores for essential versus nonessential genes as a function of cell line number (see Fig. S6A in the supplemental material). This analysis revealed that CE genes, especially those essential across many lines, often scored only slightly below the FDR threshold in other lines, consistent with genuine fitness defects that failed to reach significance in particular screens. However, it was also clear that many CE genes had no apparent fitness defect in particular cell lines, suggesting that bimodality was not merely driven by random effects in borderline-essential genes (Fig. S6A). Indeed, we observed that bimodality was preserved over a wide range of essentiality thresholds (Fig. S6B). Variable essentiality across cell types may also reflect the expression of partially redundant paralogs (22). Consistently, genes with one or more close paralogs (>30% protein sequence identity) tended to have significantly lower essentiality scores in NALM-6 cells (P = 3.6e−121 by the Wilcoxon test) and to be essential in fewer lines overall (P = 1.5e−121 by the Wilcoxon test) (Fig. S6C), but again this effect accounted for only a small fraction of the observed CE genes.
We assessed three different models that might help explain the observed bimodality of gene essentiality across cell lines. A random model represented the situation in which each gene was equally likely to be essential in any given cell line. A binary model corresponded to the scenario in which each gene was partitioned as either essential in all cell lines or not essential in any cell line, with an arbitrary constant for experimental noise. A continuous model represented the case in which each gene was assigned a specific probability of being essential in any given cell line (Fig. S6D and text in the supplemental material). The continuous model provided the best fit to the observed distribution, as it was the only model that accounted for the prevalence of essential genes in the medial fraction of cell lines (Fig. 3D). This result suggested that gene essentiality is far from an all-or-none effect across different human cell types.
Features of universal essential genes.
Model organism studies have shown that essential protein-coding genes are required for evolutionarily conserved processes in cell metabolism, macromolecular biosynthesis, proliferation, and survival. Consistently, and as reported previously (23, 24), we observed that the more cell lines in which a gene was essential, the higher the probability that this gene possessed a budding yeast ortholog and that the ortholog was also essential in yeast (Fig. 3E) (1, 35). We also examined the converse question of why almost half of the essential genes in yeast were not universally essential in human cell lines. Out of 444 essential yeast genes with a human ortholog tested in all 10 cell lines, 387 orthologs were essential in at least one cell line, with a tendency to be essential in multiple cell lines. For the remaining 57 yeast genes that appeared to be nonessential in humans, 40 of these had gene ontology (GO) terms linked to more specialized features of yeast biology. We also observed that as the number of cell lines in which a gene was essential increased, depletion of its sgRNAs from the library pool was greater (Fig. 3F), such that UE genes were associated with significantly greater fitness defects. Consistent with the enrichment for crucial cellular functions, the set of GO terms associated with essential genes became progressively more restricted as essentiality spread over more cell lines (see Fig. S7 in the supplemental material).
Proteins encoded by essential genes tend to cluster together within interaction networks in yeast (36), a feature also shown recently for human cells (22, 24). Using human protein interaction data from the BioGRID database (30), we observed that UE proteins tended to interact with each other more often than with random proteins (P < 0.05 by Fisher's exact test) and associated preferentially within maximally connected subnetworks, referred to as cliques (for clique n = 3, P < 0.05 by Fisher's exact test) (Fig. 3G). These results suggested that clusters of UE genes carry out a limited set of indispensable cellular functions.
Essentiality in human protein complexes.
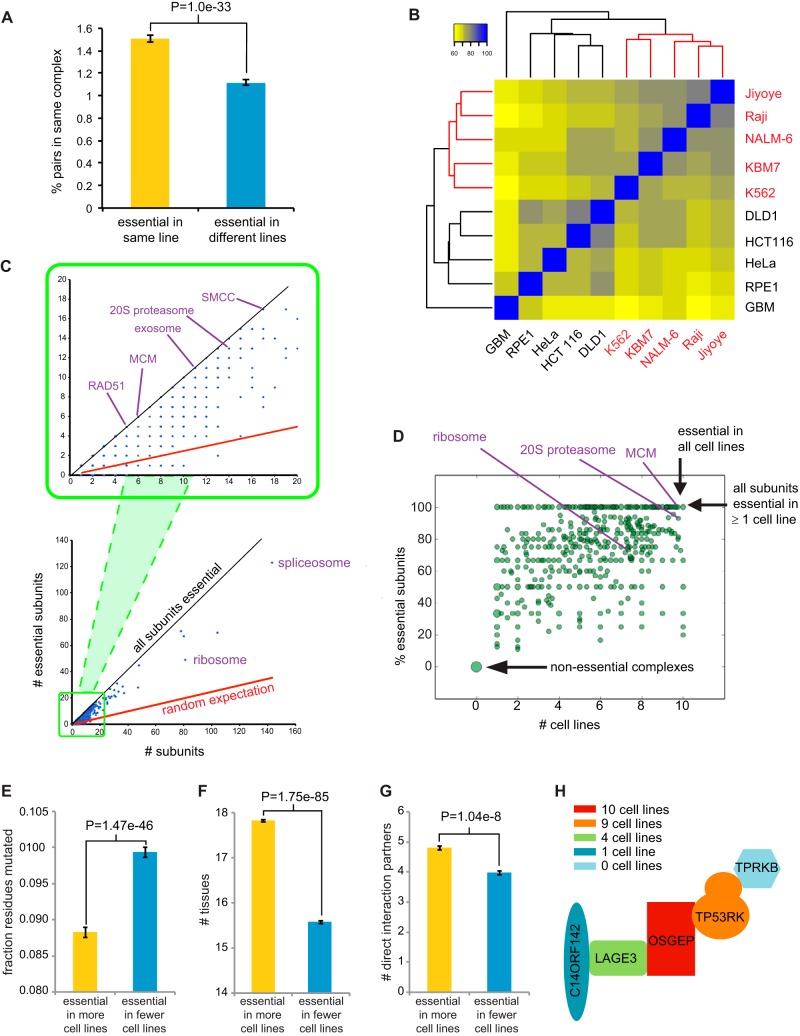
Essential genes tended to encode subunits of protein complexes, as shown previously (22–24), with an overall distribution similar to that of yeast essential genes (see Fig. S8A and B and Table S4 in the supplemental material). We assessed the propensity of CE genes to interact with each other and found that CE gene pairs essential in the same cell line were far more likely to encode subunits of the same protein complex than gene pairs that were essential in different cell lines (Fig. 4A). This result suggested that like UE genes, CE genes tended to encode essential modules in the proteome.
FIG 4.
Essential subunits of protein complexes. (A) Pairs of essential genes in the same cell line tend to encode subunits of the same protein complex compared to essential genes unique to different cell lines. All possible pairs of cell lines were sampled by randomly choosing two pairs of genes per sample, one in which each gene was essential in line 1 but not in line 2 and then one additional gene essential in line 2 but not line 1. The probability of the first and second genes belonging to the same complex and that of the first and third genes were estimated from 10 million trials. P value, Fisher's exact test. Self-interacting proteins were excluded. (B) Cell lines clustered as function of shared essential protein complexes. All protein complexes were taken from the CORUM database release 17.02.2012 (77) and are listed in Table S4 in the supplemental material. (C) Distribution of essential subunits in protein complexes. (D) Fraction of essential subunits as a function of cell line number. The dot size is proportional to the number of complexes. (E) Mean fraction of mutated residues across 45 vertebrate species for all pairs of proteins that are part of a common CORUM complex and essential in at least one cell line. P value, paired Wilcoxon rank sum test. (F) Expression of subunits of CORUM complexes across 30 different tissues types as function of essentiality. (G) Proximity of subunits with at least 4 mapped subunits in a common PDB structure as a function of essentiality. (H) Variable essentiality of subunits of the KEOPS complex (38).
We predicted that shared essential genes should strongly cluster cell lines by cell type identity, but we found that a number of cell lines did not segregate with similar lineages (Fig. S8C). However, when cell lines were clustered by shared essential complexes, defined as complexes with at least one essential subunit, the hematopoietic cell lines NALM-6, KBM7, Raji, and Jiyoye and the colon cell lines DLD1 and HCT116 were all precisely grouped together (Fig. 4B and S8C and D). The functions carried out by essential complexes thus correlate with cell type identity more closely than the complete spectrum of essential genes.
At the level of individual complexes, a small number were comprised entirely of subunits essential in one or more cell lines, such as for the highly conserved SRB- and MED-containing cofactor complex (SMCC), exosome, the Rad51 homologous recombination repair complex, and the DNA replicative helicase (MCM) complex (Fig. 4C). However, the vast majority of complexes contained a mixture of essential and nonessential (NE) subunits (Fig. 4D; Table S4). In order to assess whether the variation in essentiality between subunits of the same complex reflected the evolutionary history of the complex, we examined protein sequence conservation across 46 vertebrate species and found that subunits essential in more cell lines tended to be more conserved and expressed in more tissues than other subunits of the same complex (Fig. 4E and F). To test the notion that essentiality may reflect centrality in protein complex structure, we estimated the physical proximity of subunits for known complex structures in the Protein Data Bank (PDB) with at least four mapped subunits (37). Protein subunits that were essential in a greater number of cell lines tended to form more direct contacts with other subunits (Fig. 4G). As an example, the conserved KEOPS complex, that mediates an essential tRNA modification reaction (38), contained a core catalytic subunit (OSGEP, also called Kae1) that was essential in all lines, a tightly linked core subunit (TP53RK) essential in nine lines, and three auxiliary subunits essential in four (LAGE3), one (C14ORF142), and zero (TPRKB) lines (Fig. 4H). The core OSGEP-TP53RK subcomplex is flanked by the other subunits such that essentiality parallels structural centrality. The evolutionary plasticity of subunit essentiality is illustrated by the observations that OSGEP/Kae1 alone is sufficient for function in the mitochondrion, that three KEOPS subunits are essential in bacteria, and that all five KEOPS subunits are essential in yeast (38).
Many genes that are essential across multiple cell lines remain functionally uncharacterized for protein interactions. We used proximity-dependent biotin identification (BioID) and mass spectrometry to assess the protein interactions of two CE genes, UBALD1 (essential in NALM-6 and 2 other cell lines) and C19orf53 (essential in NALM-6 and 4 other cell lines). Duplicate BioID experiments with UBALD1 and C19orf53 as the bait proteins captured 49 and 183 statistically significant interactions, respectively (Fig. S2B; see Table S7 in the supplemental material). UBALD1, also known as FAM100A, contains a UBA-like domain and appears to be nonspecifically localized in the cell. The only reported interactions for UBALD1 are with the mitochondrial malonyl coenzyme A (malonyl-CoA) decarboxylase MLYCD, the 60S ribosomal subunit RPL9, and the RNA polymerase II (pol II) mediator subunit MED8, all identified in a pooled yeast two-hybrid screen (39). Of the 49 UBALD1 interactions we detected, 19 were previously annotated proteins implicated in stress response. C19orf53, also known as LYDG10 or HSPC023, is localized to the nucleus and interacts with the nucleoporin Nup133, the nuclear protein HNRNPU, the homeobox transcription factor POU5F1, and the mitochondrial ribosome component MRPL34 according to high-throughput studies curated in the BioGRID database (30). We found that C19orf53 interaction partners were enriched in NALM-6 essential genes (P = 4.5e−27 by Fisher's exact test) and in genes involved in rRNA biogenesis (P = 3.0e−40 by Fisher's exact test, FDR corrected). These results indirectly validate the essentiality of C19orf53 by interaction context and implicate this uncharacterized protein in the essential process of ribosomal biogenesis.
NALM-6-specific essential genes.
Of all the genes tested in common between our study and nine previous CRISPR/Cas9 screens (22, 23), 218 were uniquely essential to the NALM-6 screen. To ascertain that these LE genes were not merely due to screen noise, we generated single sgRNA-directed knockout cell populations for 4 different RefSeq genes that were uniquely essential to NALM-6 cells: the translation initiation factor gene EIF2AK1, the Fanconi anemia pathway gene FANCG, the mitotic spindle checkpoint gene MAD1L1, and the uncharacterized open reading frame (ORF) KIAA0141 (also called DELE). Each gene was targeted with two different sgRNAs in independent experiments, and cell proliferation effects were determined after 6 days of outgrowth on selective medium compared to those for two different control nontargeting sgRNAs. These experiments validated the essentiality of EIF2AK1, FANCG, MAD1L1, and KIAA0141 in the NALM-6 cell line (Fig. S2C).
The NALM-6 LE genes had more protein interaction partners than NE genes (Fig. 5A and B) and exhibited higher expression levels in NALM-6 cells (Fig. 5A and C), two defining features of essential genes identified in other cell lines and model organisms (22–24). However, LE proteins unique to NALM-6 cells were not as highly clustered in the protein interaction network as UE proteins (see Fig. S9A and B in the supplemental material) and had no more tendency to interact with UE proteins than NE proteins (Fig. S9C and D). Across all cell lines, while UE and CE proteins showed a strong propensity to interact, LE proteins were no more likely to interact with the UE core than NE proteins (Fig. 5D). These results suggested that LE proteins carry out a diversity of functions not strongly connected to the UE core, potentially as a consequence of synthetic lethal interactions with cell line-specific mutations.
FIG 5.
Cell-type-specific essential genes. (A) Expression level as a function of protein interaction degree for essential genes unique to the NALM-6 screen (red) versus UE genes (blue). (B) Average interaction degree for proteins encoded by UE genes, LE genes in NALM-6, and NE genes. (C) Expression levels of UE genes, LE genes in NALM-6, and NE genes. (D) Interactions between UE, CE, LE, and NE proteins. Counts were normalized by number of genes and interactions per class. (E) mRNA expression in NALM-6 cells of essential genes either unique to one or more B-cell-derived cell lines or unique to an equivalent number of other cell lines. (F) Participation in the B-cell receptor (BCR) pathway of essential genes either unique to one or more B-cell-derived cell lines or unique to an equivalent number of other cell lines.
The NALM-6 line bears an A146T mutation in NRAS (40), analogous to mutations that activate KRAS in some leukemias (41). As NALM-6 cells required the NRAS gene for optimal growth (Table S2), other NALM-6 specific essentials may be required to buffer the effects of oncogenic NRAS signaling. For example, two of the 11 components of cytochrome c oxidase (mitochondrial complex IV), COX6A1 and COX8A, were essential for survival exclusively in NALM-6 cells, as were two cytochrome c oxidase assembly factors, COA6 and COX16 (Table S3). Cytochrome c oxidase is known to be activated by oncogenic RAS and is required for survival of other cancer cell lines that bear an activated RAS allele (42).
In addition to NALM-6, the Raji and Jiyoye cell lines, screened previously (22), are each derived from the B-cell lineage. Surprisingly, of the 351 genes uniquely essential to one or more of these B-cell lines (Table S2), only four genes were essential to all three lines: EBF1, CYB561A3, PAX5, and MANF. Two of these genes, PAX5 and EBF1 are key transcription factors that specify the B-cell lineage (43) and were identified previously as essential genes shared between the Raji and Jiyoye cell lines (22). MANF encodes mesencephalic astrocyte-derived neurotrophic factor and is expressed at high levels in secretory tissues such as the pancreas and B cells. MANF helps cells cope with high levels of protein folding stress in the endoplasmic reticulum (44) and also activates innate immune cells to facilitate tissue regeneration (45). CYB561A3 encodes a poorly characterized ascorbate-dependent cytochrome b561 family member implicated in transmembrane electron transfer and iron homeostasis (46), but its role in B-cell proliferation or survival is not characterized.
We also examined all of the genes that were essential only in B-cell lines and found that these tended to be more highly expressed in NALM-6 cells than genes essential in an equivalent number of other cell lines (Fig. 5E), consistent with the higher expression of genes involved in B-cell proliferation in NALM-6 cells. These essential genes were significantly more likely to participate in the B-cell receptor (BCR) signaling pathway (Fig. 5F), which is often hyperactivated in chronic lymphocytic leukemia (47, 48). Concordantly, disruption of this pathway reveals vulnerabilities specific to the B-cell lineage. For example, the BCR pathway components TSC2, PI3KCD, CD79B, and CD19 were essential in two of the three B-cell lines tested to date (Table S3).
Residue-level features predict phenotypic effects of sgRNA-directed in-frame mutations.
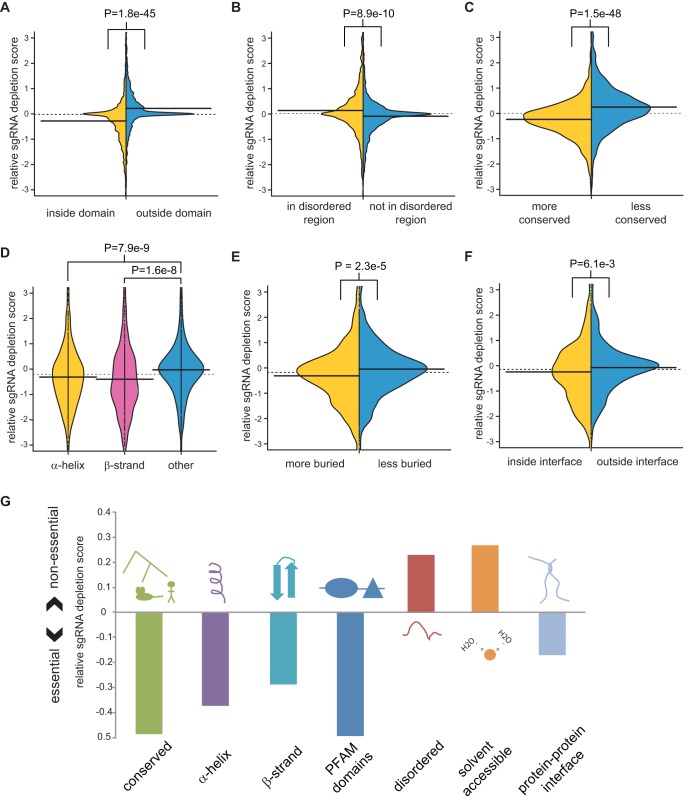
A fraction of indels introduced into genomic DNA following error-prone repair of a Cas9-mediated DSB will span a multiple of 3 bp such that the phenotypic effect will depend on the precise function of the mutated residue. For each sgRNA in the EKO library that targeted the 2,236 essential genes in NALM-6 cells, we identified the codon that would be subjected to Cas9 cleavage and hence the residue most likely to be affected by in-frame indels. We found that sgRNAs targeting predicted domain-coding regions were significantly more depleted from the pool than other sgRNAs targeting the same gene (P = 1.8e−45) (Fig. 6A). This result confirms a previous focused study on high-density sgRNA-mediated targeting of ∼200 genes (49) and generalizes the effect to the diverse classes of domains encoded by the entire genome. Based on the high degree of significance of this result, we asked whether other protein-level features would also correlate with sgRNA targeting sites. We found that sgRNAs targeting disordered protein regions were significantly less depleted than other sgRNAs targeting the same gene (Fig. 6B). The significance of this trend also held when the analysis was restricted to regions outside Pfam domains (P = 8.98e−6) (data not shown). Disruption of more conserved regions caused significantly more depletion than for less conserved regions of the same gene (Fig. 6C). Targeted regions that encoded α-helices or β-sheets were also significantly more depleted than other regions (Fig. 6D). sgRNAs targeting buried residues within a protein structure were more depleted than sgRNAs targeting accessible residues (Fig. 6E). Finally, sgRNAs targeting interfacial regions between two protein subunits were more depleted than noninterfacial regions of the same protein (Fig. 6F). Integration of each variable into a single linear multivariate model, together with the number of potential off-target cleavage sites and predicted sgRNA efficiencies, revealed that every variable was significantly and independently correlated with relative sgRNA depletion (Table 1). This result indicated that each residue-level feature contributed to the phenotypic effects of in-frame mutations (Fig. 6G).
FIG 6.
Correlation of sgRNA depletion with subgene- and protein-level features. The average log P values of sgRNAs linked to each feature were normalized to the gene average for all sgRNAs. (A) Overlap with Pfam domains. (B) Predicted protein disorder of >40% probability as determined by UIPred (76). (C) Protein-level conservation of a 30-residue window, either more conserved or less conserved than gene average. (D) Location of residues within an α-helix, an extended β-strand, or neither as annotated from PDB. (E) Residue burial, defined as the number of nonhydrogen atoms from nonadjacent residues present within 6 Å of the residue, for residues more or less buried than the average for the protein. (F) Residue proximity to a protein-protein interface. (G) Summary of protein-level features that influence sgRNA depletion from the library pool, as analyzed in panels A to F.
TABLE 1.
Multivariate Pearson correlation between sgRNA features and gene-normalized sgRNA depletion score
| sgRNA feature | P value |
|---|---|
| Overlapping domain | 0.0018 |
| Relative conservation | 1.33e−6 |
| Relative burial | 3.23e−6 |
| Overlapping α-helix | 8.27e−5 |
| Overlapping β-strand | 8.33e−6 |
| Overlapping PPI interface | 0.0034 |
| Overlapping protein disorder | 3.30e−5 |
Essentiality of alternatively spliced exons.
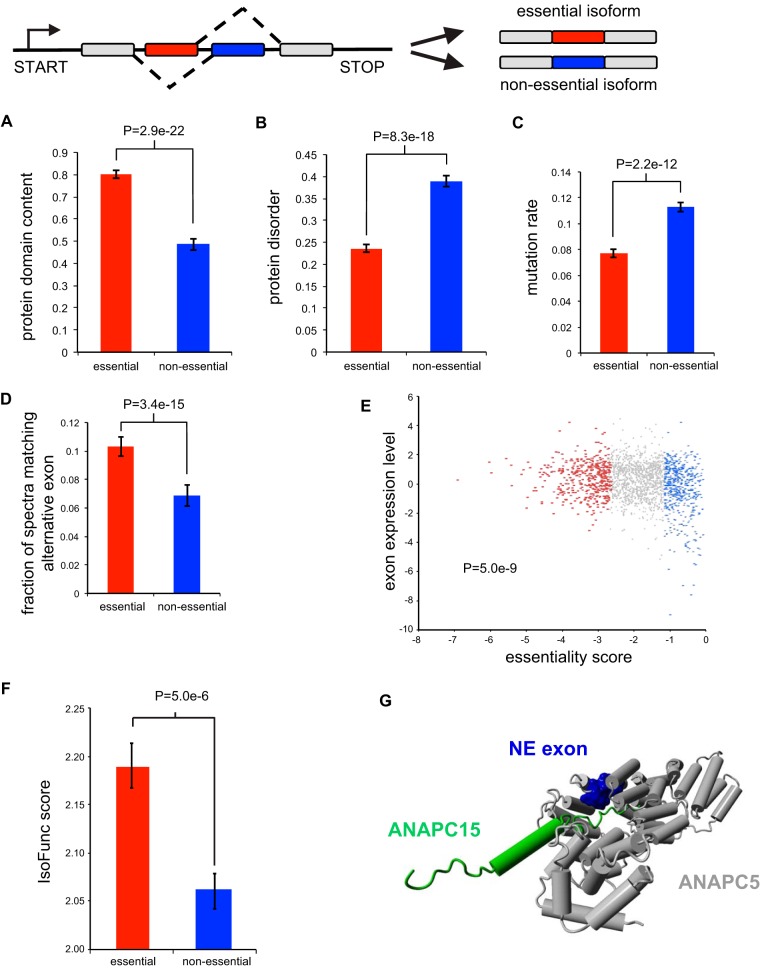
The EKO library was designed in part to target specific alternatively spliced exons, many of which are found in essential gene loci. From the depletion of sgRNAs targeting these exons, we were able to classify individual exons within essential genes as either essential or nonessential (see Table S5 in the supplemental material). We examined the effect of single sgRNA-directed knockouts of three different alternatively spliced exons that our screen identified as nonessential in otherwise essential genes: exon 5 of CSNK1A1, which encodes casein kinase 1α; exon 4 of MRPL43, which encodes a component of the large subunit of the mitochondrial ribosome; and exon 10 of ANAPC5, which encodes a subunit of the anaphase-promoting complex/cyclosome (APC/C). For each gene, two sgRNAs for the alternatively spliced exon and two sgRNAs for a constitutive exon were compared for effects on cell proliferation. We found that all sgRNAs that targeted the alternative exons affected proliferation considerably less than sgRNAs that targeted a constitutive exon from the same gene (Fig. S2D). The results confirmed that the EKO library was able to detect differential fitness effects for alternatively spliced exons.
We analyzed the 2,143 alternative exons within RefSeq-defined coding regions of essential genes in the NALM-6 screen, each of which was covered by at least 3 sgRNAs with ≥20 reads, to identify 462 exons with sgRNAs that were significantly depleted (FDR < 0.05). When we compared these essential alternative exons to nonessential exons (n = 592; FDR > 0.3) in essential genes, we found that the essential exons were more likely to overlap protein domains (Fig. 7A), less likely to contain long disordered regions (Fig. 7B), more conserved across 46 vertebrate species (Fig. 7C), and more highly expressed at both the protein (Fig. 7D) and mRNA (Fig. 7E) levels. Importantly, mRNA expression analysis showed that all 462 essential exons were expressed in NALM-6 cells. We also mapped the exons to full-length isoforms in the IsoFunct database, which assigns gene ontology functions to individual protein isoforms (50). Isoforms that contained essential exons were more likely to be functional (i.e., with higher IsoFunct scores) than isoforms with nonessential exons (Fig. 7F). The nonessential nature of particular exons may reflect structural features or protein interactions associated with the exon encoded region. For example, the anaphase-promoting complex/cyclosome (APC/C), contains a nonessential exon in the essential gene ANAPC5, which interacts with ANAPC15, itself a nonessential component of the complex (Fig. 7G) (51). These results suggested that the EKO library can effectively distinguish essential from nonessential alternatively spliced exons within essential gene loci and that many alternatively spliced exons of essential genes are nonessential.
FIG 7.
Alternatively spliced exons in essential genes. (A) Fraction of exons that overlap a Pfam domain (E value, <10−5) for essential exons (FDR < 0.05) and nonessential exons (FDR > 0.3) in essential genes. (B) Average IUPred predicted probability of exon residues belonging to a long disordered region. (C) Average fraction of residues mutated relative to human in 45 vertebrate species. (D) Fraction of matched mass spectra per alternative exon (FDR < 0.1) across a panel of human tissues. (E) Log2 RNA-seq read density in the exon normalized to gene average. (F) Average IsoFunc fold change, representing the likeliness that a specific isoform codes for a given GO function for isoforms of essential genes. (G) Nonessential alternative exon (blue) in the essential gene ANAPC5, within the anaphase-promoting complex/cyclosome, interacts with ANAPC15, a nonessential component (green) of the complex.
Genetic analysis of hypothetical genes.
The EKO library was also designed to target unvalidated hypothetical genes that are currently absent from the RefSeq database (35). We identified these hypothetical genes from the AceView and GenCode databases, which annotate loci on the basis of expressed sequence tag (EST) and/or RNA-seq evidence. Because many hypothetical genes may be expressed pseudogenes present in more than one copy in the genome, we excluded all sgRNAs with close mismatches to the genome in order to avoid depletion effects due to multiple cleavage events. When we considered only sgRNAs with a single potential cleavage site, we identified 44 essential genes (FDR < 0.05) that were absent from RefSeq (Table S3). To ascertain that hypothetical gene essentiality was not a consequence of screen noise, we generated single sgRNA-directed knockout cell populations for three different hypothetical genes that scored as essential in the NALM-6 screen: IGHV1-69, CROCCP2, and LOC100288778. We note that IGHV1-69 is expressed in chronic lymphocytic cell lines and encodes a predicted immunoglobulin heavy-chain variable region that appears to bind oxidation-specific epitopes (52). Each hypothetical gene was targeted with two different sgRNAs, and the effects on proliferation compared to those of two nontargeting control sgRNAs. These experiments validated the essentiality of IGHV1-69, CROCCP2, and LOC100288778 in NALM-6 cells (Fig. S2E).
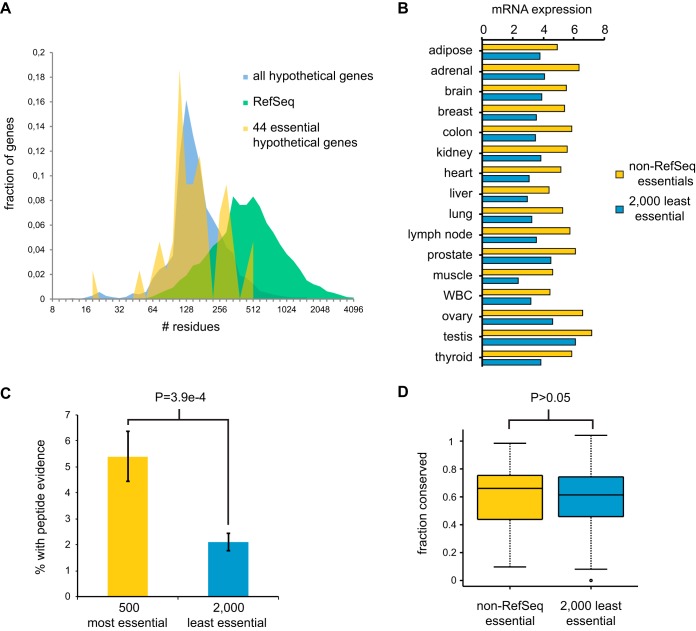
Similar to the case for most poorly annotated hypothetical genes, the essential hypothetical genes tended to encode short polypeptides, with a median length of 153 residues and a maximum length of 581 residues (Fig. 8A). We found that these essential hypothetical genes were more highly expressed across a range of tissues than the 2,000 hypothetical genes with the least depleted sgRNAs (Fig. 8B). We analyzed a comprehensive mass spectrometry-based proteomics data set that covers 73 tissues and body fluids (53) and found that the 500 hypothetical genes with the highest sgRNA depletion scores were more likely to have evidence of protein expression than the 2,000 genes with the lowest sgRNA depletion scores (Fig. 8C). We also note that of 37 small ORFs identified by peptide evidence in a recent mass spectrometry study, 20 corresponded to hypothetical genes in the EKO library, including all five small ORFs detected by two or more peptides (54). Of these 20 genes, two appeared to be essential in our screen (CROCCP2 and PPP1R35). Alignments across 46 vertebrate genomes revealed that the essential hypothetical genes were not more conserved than their nonessential counterparts (Fig. 8D), suggesting that the essential functions were acquired through recent evolution. These results suggest that at least a fraction of newly evolved uncharacterized hypothetical genes are likely to be expressed and perform important functions in human cells.
FIG 8.
Essentiality-correlated features of predicted hypothetical genes. (A) ORF length distribution of RefSeq genes and hypothetical ORFs from AceView or GenCode covered by the EKO library. The 44 hypothetical ORFs with significant (FDR < 0.05) essentiality scores in the NALM-6 screen are indicated separately. For genes with multiple transcripts, only the length of the longest predicted protein was considered. (B) Mean log2 (reads/kilobase) across 16 tissues from the Human BodyMap 2.0 for 44 essential and the 2,000 least essential hypothetical genes. (C) Fraction of genes with uniquely matched mass spectra at an FDR of <0.001 for the 500 most essential and the 2,000 least essential hypothetical genes. (D) Fraction of residues conserved across 46 vertebrate genomes for the 44 essential and the 2,000 least essential hypothetical genes.
DISCUSSION
Genome-wide collections of genetic reagents have allowed the identification of essential genes and insights into the functional architecture of model organisms. The first genome-wide CRISPR/Cas9 and gene trap screens have defined a draft map of essential genes across a variety of human cell types (19–24). Here, we have applied the high-complexity EKO library to define new essential features at the levels of protein residues, alternatively spliced exons, previously uncharacterized hypothetical coding regions, and protein complexes.
A continuum of gene essentiality.
We combined our screen data with two previously published CRISPR/Cas9 screen data sets to define a minimal set of 486 UE genes across 10 different cell lines of diverse origins. As opposed to a simple monotonic convergence on a core set of essential genes, we observed an unexpected bimodal distribution of essential genes as a function of the number of lines screened. This distribution was best explained by a continuous variation in the probability of gene essentiality as opposed to a simple all-or-none binary model. This continuous-probability model likely reflects genetic and/or epigenetic background effects, wherein different cell line contexts provide different levels of buffering against the loss of potentially essential genes. In this sense, the number of cell line backgrounds in which a gene is essential can be thought of as a form of genetic interaction degree, whereby the interaction occurs between any gene and a complex genetic background, as opposed to between two genes. In yeast, the strain background markedly affects gene essentiality due to high-order genetic interactions (55), such that the essentialome of a particular cell must be defined in the context of a precise set of genetic and/or epigenetic parameters.
The essentialome may be thought of as an onion with multiple layers that become progressively more context specific. As we show here, UE genes at the center of the onion make quantitatively greater fitness contributions than progressively more cell line-specific essential genes. The middle layers of the onion correspond to the trough in the bimodal distribution in which genes are essential in specific cell lines. The definition of essentiality is obviously influenced by the definition of experimental thresholds, but the continuum of essentiality is nevertheless apparent regardless of the specific threshold. Based on the current but limited analysis, we estimate that a consensus essentialome for any given 8 to 10 cell lines will encompass approximately 1,000 genes. This number, intriguingly, is close to the number of 1,114 essential genes required for yeast growth under optimal growth conditions (1).
It is important to note that this quantitative model of essentiality does not rule out the existence of a set of true UE genes. In fact, the continuum model predicts a smooth decline out to the maximal number of lines, in contrast to the sharp step increase observed experimentally at the maximum. This observation suggests the existence of a small set of UE genes that may be qualitatively distinct from the continuum of essentiality captured by the model. This set of essential functions likely corresponds to the structural and enzymatic core of the proteome, to which other essential functions have been added and subtracted through the course of evolution.
A continuum of essential subunits in protein complexes.
As in model organisms and as suggested by previous studies, the proteins encoded by UE genes tend to cluster together into modules in the protein-protein interaction network. This correlation between centrality and lethality is a robust feature of biological networks. Although originally posited to reflect the central location of essential proteins in network graphs (56), this topological argument has been overturned in favor of the idea that essential proteins perform their functions as complexes (14). This notion has in turn been supported by the claim that protein complexes exhibit a tendency to be composed of mainly essential or mainly nonessential subunits (36, 57), such that the interactions of essential protein subunits would naturally cluster together. A surprising outcome of our analysis across multiple different human cell lines is the extent to which subunit essentiality for any given complex varies between cell lines. Our analysis of structural data suggests that UE subunits form the functional and structural core of essential protein complexes, to which CE, LE, and NE subunits are appended. This observation probably reflects the facts that protein machines have acquired progressively more subunits through evolution and that more recently evolved subunits tend to lie outside the essential structural core (24, 58). This variable essentiality of protein subunits is consistent with the lethality-centrality relationship (14, 59) but suggests that network evolution may also help drive the observed correlation (60).
Cell line-specific essential genes and synthetic lethality.
Only 4 essential genes were uniquely shared between the three B-cell-derived Raji, Jiyoye, and NALM-6 lines, and only 35 uniquely essential genes were shared between any two of the three lines. Similarly, a recent study identified only five essential genes shared between five acute myeloid leukemia (AML) cell lines and 66 genes shared between at least three of the five lines (34). These results suggest that genetic and epigenetic variation between lines may dominate the cell line-specific essentialome. Indeed, the majority of essential genes identified in human genome-wide screens to date are unique to single cell lines. In comparisons of wild-type and laboratory yeast strains, which exhibit sequence variation similar to that between any two human individuals (61), strain-specific essential genes have a complex contextual basis due to multiple undefined genetic modifiers (55). It seems likely that many cell-type-specific essential genes will reflect synthetic lethal interactions associated with the unique spectrum of cancer-associated mutations in any given cancer cell line. If on average each mutation yields 20 synthetic lethal interactions (5, 24), only 10 cell line-specific loss-of-function mutations would be needed to account for a specific essentiality profile of 200 LE genes. As an example, several components of cytochrome c oxidase and its assembly factors were specifically essential for survival in NALM-6 cells, potentially as a consequence of oncogenic RAS signaling (42), which is known to cooperatively trigger senescence in conjunction with mitochondrial defects (62). From a therapeutic perspective, the variable subunit essentiality of protein complexes suggests that a window of genetic sensitivity may exist for essential functions that are partially compromised in cancer cells (2, 63).
Evolution of new functions by alternative splicing and de novo gene formation.
Recent RNA-seq and proteomics studies have illuminated the magnitude of alternative splicing in mammals and the attendant proteomic and phenotypic diversity generated by this mechanism (64). Different splice isoforms can have radically different functions, such as, for example, pro- or antiapoptotic splice variants of BCL2 (65). Limited systematic studies on dozens to hundreds of isoforms suggest that the exon composition can often dramatically alter protein interaction profiles (66, 67). Our genome-wide screen with the EKO library suggests that a large fraction of alternatively spliced exons in essential genes are not required for cell survival. This result suggests that alternative splicing has evolved as a means to diversify protein structure and interactions without compromising essential functions. Our data also show that essential exons in general tend to encode structured protein domains that are more highly conserved and highly expressed, whereas nonessential exons tend to encode intrinsically disordered regions. Nonessential exons likely represent the first step toward the evolution of new functions, some of which may be then destined to become essential.
In contrast to the generation of new genes by duplication (68), the de novo appearance of new protein-coding genes is a poorly understood but nevertheless important evolutionary mechanism. For example, in mammals, 1,828 known genes are unique to the primate lineage and 3,111 to rodents (69). Protogenes are genetic loci that produce very short proteins unique to each species, and it has been suggested that a large pool of such potential genes exists in yeast (70). If a protogene assumes a beneficial function, it will be subjected to selective pressure and coevolve across related species, such that even young genes can rapidly acquire essential functions (71). However, the detection of such protogenes is hampered by the absence of sequence conservation and short length, which often preclude detection by mass spectrometry (72). As shown here, CRISPR/Cas9-based screens allow the systematic functional analysis of hypothetical human genes. We detected an essential function for 44 hypothetical genes that was consistent with mRNA and protein expression data (53, 54). These essential hypothetical gene functions were also evident across 36 different EKO library screens under various chemical stress conditions (unpublished data). Although we cannot exclude the possibly that some of these loci are noncoding RNA genes or pseudogenes, it is probable that these short reading frames represent newly evolved human genes. The biochemical interrogation of the corresponding proteins should help identify the functions of these new candidate genes. Although we restricted our analysis to currently annotated hypothetical loci, it will be feasible to systematically query all short open reading frames in the human genome with dedicated sgRNA libraries.
High-resolution genetic analysis of the human genome.
Genome-wide CRISPR/Cas9-based screens have ushered in a new era of systems genetics in human cells (19). The EKO library demonstrates the capacity of high-resolution CRISPR/Cas9 screens to define gene essentiality across scales in the human proteome. Analogous deep-coverage sgRNA libraries and variant CRISPR/Cas9 strategies also enable the precise mapping of regulatory and noncoding regions in the human genome (73). Systematic functional analysis of these vast unexplored regions of the genome and proteome will provide insights into biological mechanisms and the evolution of phenotypic complexity in human cells.
MATERIALS AND METHODS
Genome-wide sgRNA library.
A set of 181,130 sgRNA sequences that target most RefSeq genes (∼10 sgRNAs per gene) was as reported previously (74). Similar design rules were used to target additional genes from the latest RefSeq, AceView, and GenCode releases (∼10 sgRNAs per gene), as well as 20,852 alternatively spliced exons derived from 8,744 genes (∼3 sgRNA per exon). A set of 2,043 nontargeting sgRNA sequences with no detectable match to the human genome was randomly generated. The complete set of 278,754 sgRNAs was divided into three sublibrary pools of 92,918 sgRNAs for array-based oligonucleotide synthesis as 60-mers (Custom Array), each containing 3 or 4 sgRNAs per gene, 1 sgRNA per alternative exon, and 681 nontargeting sgRNAs. Each pool was amplified by PCR, cloned by Gibson assembly into the pLX-sgRNA plasmid (74), expanded in plasmid format, and converted to a lentiviral pool by transfection into 293T cells (see Table S6 and text in the supplemental material).
Screen for essential genes.
A doxycycline-inducible Cas9 clonal cell line of NALM-6 was generated by infection with a Cas9-FLAG lentiviral construct and puromycin selection, followed by fluorescence-activated cell sorting (FACS) and immunoblotting with an anti-FLAG antibody to select a tightly regulated clone. The inducible Cas9 cell line was infected with pooled lentivirus libraries at an MOI of 0.5 and 500 cells per sgRNA. After 6 days of blasticidin selection, 140 million cells were induced with doxycycline for 7 days, followed by periods of outgrowth in the absence of doxycycline. sgRNA sequences were recovered by PCR of genomic DNA, reamplified with Illumina adapters, and sequenced on an Illumina HiSeq 2000 instrument.
Scoring of gene essentiality.
A RANKS score was calculated for every gene targeted by ≥4 sgRNAs represented by ≥20 reads in one sample. A single-tailed P value for each sgRNA was obtained by comparing its ratio to that of the control sgRNAs. The RANKS score was determined as the average loge P value of the sgRNAs targeting the gene or exon. Gene/exon P values were generated by comparing the RANKS score to that of a control distribution of an equal number of control sgRNAs for 5 million samples with FDR correction (75). The 2,043 nontargeting sgRNAs were used as controls in calculating the original RANKS score for scoring method comparisons and the binned rank analyses. To control for nonspecific effects of Cas9 cleavage, the entire EKO library was used as a control after removing sgRNAs that targeted previously documented essential genes (22–24) and sgRNAs with ≥2 predicted off-target cleavage sites. A fixed correction factor was applied to sgRNAs with 1 or 2 predicted off-target cleavage sites. For the identification of essential non-RefSeq genes, only sgRNAs with unique matches to the genome were used to avoid potential confounding cleavage events at paralogous loci.
Further details of the experimental, computational, and statistical methods are provided in the supplemental material. All materials are available upon request, and code for RANKS may be obtained at https://github.com/JCHuntington/RANKS.
Accession number(s).
Raw mass spectrometry data have been submitted to the MassIVE database under accession number MSV000081460.
Supplementary Material
ACKNOWLEDGMENTS
We thank Driss Boudeffa, Bobby-Joe Breitkreutz, Manon Lord, Jennifer Huber, Philippe Daoust, and Alfredo Staffa for technical support and Traver Hart, Jason Moffat, Luisa Izzi, and other members of the Tyers laboratory for helpful discussions.
J.C.-H. was supported by a Canadian Institutes of Health Research (CIHR) postdoctoral fellowship, K.G.B. was supported by Cole Foundation and Fonds de recherche du Québec Santé studentships, Y.X. was supported by a Canada Research Chair in Computational and Systems Biology, and M.T. was supported by a Canada Research Chair in Systems and Synthetic Biology. This work was funded by grants from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2014-03892 to Y.X.), the CIHR (MOP-126129, MOP-366608, and PJT 152962 to M.T.), the Canadian Cancer Society Research Institute (703906 to M.T.), and the National Institutes of Health (R01OD010929 to M.T.) and by an award from the Ministère de l'Enseignement Supérieur, de la Recherche, de la Science et de la Technologie du Québec through Génome Québec to M.T.
We declare no conflicts of interest.
T.B. and J.C.-H. designed the EKO library, T.B. built the EKO library and performed the screen with assistance from K.G.B., J.C.-H. developed the RANKS algorithm and performed all statistical analyses, A.C.-A. and E.C. performed protein interaction analyses, and T.B., J.C.-H., A.C.-A., B.R., Y.X., and M.T. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00302-17.
REFERENCES
- 1.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 2.Hartman JLt, Garvik B, Hartwell L. 2001. Principles for the buffering of genetic variation. Science 291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 3.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 4.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, et al. 2010. The genetic landscape of a cell. Science 327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, Pelechano V, Styles EB, Billmann M, van Leeuwen J, van Dyk N, Lin ZY, Kuzmin E, Nelson J, Piotrowski JS, Srikumar T, Bahr S, Chen Y, Deshpande R, Kurat CF, Li SC, Li Z, Usaj MM, Okada H, Pascoe N, San Luis BJ, Sharifpoor S, Shuteriqi E, Simpkins SW, Snider J, Suresh HG, Tan Y, Zhu H, Malod-Dognin N, Janjic V, Przulj N, Troyanskaya OG, Stagljar I, Xia T, Ohya Y, Gingras AC, Raught B, Boutros M, Steinmetz LM, Moore CL, Rosebrock AP, et al. 2016. A global genetic interaction network maps a wiring diagram of cellular function. Science 353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, Han S, Jeffery L, Baek ST, Lee H, Shim YS, Lee M, Kim L, Heo KS, Noh EJ, Lee AR, Jang YJ, Chung KS, Choi SJ, Park JY, Park Y, Kim HM, Park SK, Park HJ, Kang EJ, Kim HB, Kang HS, Park HM, Kim K, Song K, Song KB, Nurse P, Hoe KL. 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, Martel N, Veronneau S, Lemieux S, Kauffman S, Becker J, Storms R, Boone C, Bussey H. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 9.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 11.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N, Heidelberg Fly Array C. 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu CW, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LM, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, et al. 2016. High-throughput discovery of novel developmental phenotypes. Nature 537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsh AE, Fraser HB. 2001. Protein dispensability and rate of evolution. Nature 411:1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- 14.Zotenko E, Mestre J, O'Leary DP, Przytycka TM. 2008. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput Biol 4:e1000140. doi: 10.1371/journal.pcbi.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramani AK, Chuluunbaatar T, Verster AJ, Na H, Vu V, Pelte N, Wannissorn N, Jiao A, Fraser AG. 2012. The majority of animal genes are required for wild-type fitness. Cell 148:792–802. doi: 10.1016/j.cell.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Samson JE, Magadan AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 17.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shalem O, Sanjana NE, Zhang F. 2015. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, Marceau C, Janssen H, Carette JE, Bennett KL, Colinge J, Superti-Furga G, Brummelkamp TR. 2015. Gene essentiality and synthetic lethality in haploid human cells. Science 350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 25.Thierry-Mieg D, Thierry-Mieg J. 2006. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol 7(Suppl 1):S12.1–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurwitz R, Hozier J, LeBien T, Minowada J, Gajl-Peczalska K, Kubonishi I, Kersey J. 1979. Characterization of a leukemic cell line of the pre-B phenotype. Int J Cancer 23:174–180. doi: 10.1002/ijc.2910230206. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, Haussler D, Miller W. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O'Donnell L, Reguly T, Nixon J, Ramage L, Winter A, Sellam A, Chang C, Hirschman J, Theesfeld C, Rust J, Livstone MS, Dolinski K, Tyers M. 2015. The BioGRID interaction database: 2015 update. Nucleic Acids Res 43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chari R, Mali P, Moosburner M, Church GM. 2015. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods 12:823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, Kost-Alimova M, Gill S, Xu H, Ali LD, Jiang G, Pantel S, Lee Y, Goodale A, Cherniack AD, Oh C, Kryukov G, Cowley GS, Garraway LA, Stegmaier K, Roberts CW, Golub TR, Meyerson M, Root DE, Tsherniak A, Hahn WC. 2016. Genomic copy number dictates a gene-independent cell response to CRISPR-Cas9 targeting. Cancer Discov 6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munoz DM, Cassiani PJ, Li L, Billy E, Korn JM, Jones MD, Golji J, Ruddy DA, Yu K, McAllister G, DeWeck A, Abramowski D, Wan J, Shirley MD, Neshat SY, Rakiec D, de Beaumont R, Weber O, Kauffmann A, McDonald ER 3rd, Keen N, Hofmann F, Sellers WR, Schmelzle T, Stegmeier F, Schlabach MR. 2016. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov 6:900–913. doi: 10.1158/2159-8290.CD-16-0178. [DOI] [PubMed] [Google Scholar]
- 34.Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, Mupo A, Grinkevich V, Li M, Mazan M, Gozdecka M, Ohnishi S, Cooper J, Patel M, McKerrell T, Chen B, Domingues AF, Gallipoli P, Teichmann S, Ponstingl H, McDermott U, Saez-Rodriguez J, Huntly BJ, Iorio F, Pina C, Vassiliou GS, Yusa K. 2016. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep 17:1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NCBI Resource Coordinators. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hart GT, Lee I, Marcotte ER. 2007. A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality. BMC Bioinformatics 8:236. doi: 10.1186/1471-2105-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose PW, Prlic A, Bi C, Bluhm WF, Christie CH, Dutta S, Green RK, Goodsell DS, Westbrook JD, Woo J, Young J, Zardecki C, Berman HM, Bourne PE, Burley SK. 2015. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res 43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan LC, Maisonneuve P, Szilard RK, Lambert JP, Ng TF, Manczyk N, Huang H, Laister R, Caudy AA, Gingras AC, Durocher D, Sicheri F. 2016. Proteomic analysis of the human KEOPS complex identifies C14ORF142 as a core subunit homologous to yeast Gon7. Nucleic Acids Res. doi: 10.1093/nar/gkw1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. 2005. A human protein-protein interaction network: a resource for annotating the proteome. Cell 122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 40.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, Wooster R. 2004. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer 91:355–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyner JW, Erickson H, Deininger MW, Willis SG, Eide CA, Levine RL, Heinrich MC, Gattermann N, Gilliland DG, Druker BJ, Loriaux MM. 2009. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood 113:1749–1755. doi: 10.1182/blood-2008-04-152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Telang S, Nelson KK, Siow DL, Yalcin A, Thornburg JM, Imbert-Fernandez Y, Klarer AC, Farghaly H, Clem BF, Eaton JW, Chesney J. 2012. Cytochrome c oxidase is activated by the oncoprotein Ras and is required for A549 lung adenocarcinoma growth. Mol Cancer 11:60. doi: 10.1186/1476-4598-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Somasundaram R, Prasad MA, Ungerback J, Sigvardsson M. 2015. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood 126:144–152. doi: 10.1182/blood-2014-12-575688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindahl M, Saarma M, Lindholm P. 2017. Unconventional neurotrophic factors CDNF and MANF: Structure, physiological functions and therapeutic potential. Neurobiol Dis 97:90–102. doi: 10.1016/j.nbd.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Neves J, Zhu J, Sousa-Victor P, Konjikusic M, Riley R, Chew S, Qi Y, Jasper H, Lamba DA. 2016. Immune modulation by MANF promotes tissue repair and regenerative success in the retina. Science 353:aaf3646. doi: 10.1126/science.aaf3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asard H, Barbaro R, Trost P, Berczi A. 2013. Cytochromes b561: ascorbate-mediated trans-membrane electron transport. Antioxid Redox Signal 19:1026–1035. doi: 10.1089/ars.2012.5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D'Eustachio P, Stein L. 2011. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res 39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seda V, Mraz M. 2015. B-cell receptor signalling and its crosstalk with other pathways in normal and malignant cells. Eur J Haematol 94:193–205. doi: 10.1111/ejh.12427. [DOI] [PubMed] [Google Scholar]
- 49.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panwar B, Menon R, Eksi R, Li HD, Omenn GS, Guan Y. 2016. Genome-wide functional annotation of human protein-coding splice variants using multiple instance learning. J Proteome Res 15:1747–1753. doi: 10.1021/acs.jproteome.5b00883. [DOI] [PubMed] [Google Scholar]
- 51.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. 2015. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Que X, Widhopf GF II, Amir S, Hartvigsen K, Hansen LF, Woelkers D, Tsimikas S, Binder CJ, Kipps TJ, Witztum JL. 2013. IGHV1-69-encoded antibodies expressed in chronic lymphocytic leukemia react with malondialdehyde-acetaldehyde adduct, an immunodominant oxidation-specific epitope. PLoS One 8:e65203. doi: 10.1371/journal.pone.0065203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B. 2014. Mass-spectrometry-based draft of the human proteome. Nature 509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 54.Ma J, Diedrich JK, Jungreis I, Donaldson C, Vaughan J, Kellis M, Yates JR 3rd, Saghatelian A. 2016. Improved identification and analysis of small open reading frame encoded polypeptides. Anal Chem 88:3967–3975. doi: 10.1021/acs.analchem.6b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, Bernstein DA, Rolfe PA, Heisler LE, Chin B, Nislow C, Giaever G, Phillips PC, Fink GR, Gifford DK, Boone C. 2010. Genotype to phenotype: a complex problem. Science 328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. 2001. Lethality and centrality in protein networks. Nature 411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 57.Ryan CJ, Krogan NJ, Cunningham P, Cagney G. 2013. All or nothing: protein complexes flip essentiality between distantly related eukaryotes. Genome Biol Evol 5:1049–1059. doi: 10.1093/gbe/evt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim PM, Lu LJ, Xia Y, Gerstein MB. 2006. Relating three-dimensional structures to protein networks provides evolutionary insights. Science 314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 59.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. 2002. Evolutionary rate in the protein interaction network. Science 296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 60.Coulombe-Huntington J, Xia Y. 2017. Network centrality analysis in fungi reveals complex regulation of lost and gained genes. PLoS One 12:e0169459. doi: 10.1371/journal.pone.0169459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Engel SR, Weng S, Binkley G, Paskov K, Song G, Cherry JM. 2016. From one to many: expanding the Saccharomyces cerevisiae reference genome panel. Database (Oxford). doi: 10.1093/database/baw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura M, Ohsawa S, Igaki T. 2014. Mitochondrial defects trigger proliferation of neighbouring cells via a senescence-associated secretory phenotype in Drosophila. Nat Commun 5:5264. doi: 10.1038/ncomms6264. [DOI] [PubMed] [Google Scholar]
- 63.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. 1997. Integrating genetic approaches into the discovery of anticancer drugs. Science 278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 64.Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 65.Kontos CK, Scorilas A. 2012. Molecular cloning of novel alternatively spliced variants of BCL2L12, a new member of the BCL2 gene family, and their expression analysis in cancer cells. Gene 505:153–166. doi: 10.1016/j.gene.2012.04.084. [DOI] [PubMed] [Google Scholar]
- 66.Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O'Hanlon D, Kim PM, Wrana JL, Blencowe BJ. 2012. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell 46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 67.Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray RR, Spirohn K, Begg BE, Duran-Frigola M, MacWilliams A, Pevzner SJ, Zhong Q, Trigg SA, Tam S, Ghamsari L, Sahni N, Yi S, Rodriguez MD, Balcha D, Tan G, Costanzo M, Andrews B, Boone C, Zhou XJ, Salehi-Ashtiani K, Charloteaux B, Chen AA, Calderwood MA, Aloy P, Roth FP, Hill DE, Iakoucheva LM, Xia Y, Vidal M. 2016. Widespread expansion of protein interaction capabilities by alternative splicing. Cell 164:805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohno S. 1970. Evolution by gene duplication. Springer, Berlin, Germany. [Google Scholar]
- 69.Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M. 2010. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol 8:e1000494. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, Brar GA, Weissman JS, Regev A, Thierry-Mieg N, Cusick ME, Vidal M. 2012. Proto-genes and de novo gene birth. Nature 487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen S, Zhang YE, Long M. 2010. New genes in Drosophila quickly become essential. Science 330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bruford EA, Lane L, Harrow J. 2015. Devising a consensus framework for validation of novel human coding loci. J Proteome Res 14:4945–4948. doi: 10.1021/acs.jproteome.5b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wright JB, Sanjana NE. 2016. CRISPR screens to discover functional noncoding elements. Trends Genet 32:526–529. doi: 10.1016/j.tig.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. [Google Scholar]
- 76.Dosztanyi Z, Csizmok V, Tompa P, Simon I. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 77.Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. 2010. CORUM: the comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Res 38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.