ABSTRACT
Selective autophagy and macroautophagy sequester specific organelles/substrates or bulk cytoplasm, respectively, inside autophagosomes as cargo for delivery to lysosomes. The mammalian ATG8 orthologues (MAP1LC3A/B/C and GABARAP/L1/L2) are ubiquitin (UB)-like proteins conjugated to the autophagosome membrane and are thought to facilitate cargo receptor recruitment, vesicle maturation, and lysosomal fusion. To elucidate the molecular functions of the ATG8 proteins, we engineered cells lacking genes for each subfamily as well as all six mammalian ATG8s. Loss of GABARAPs alone attenuates autophagic flux basally and in response to macroautophagic or selective autophagic stimuli, including parkin-dependent mitophagy, and cells lacking all ATG8 proteins accumulate cytoplasmic UB aggregates, which are resolved following ectopic expression of individual GABARAPs. Autophagosomes from cells lacking GABARAPs had reduced lysosomal content by quantitative proteomics, consistent with fusion defects, but accumulated regulators of late endosome (LE)/autophagosome maturation. Through interaction proteomics of proteins accumulating in GABARAP/L1/L2-deficient cells, we identified C18orf8/RMC1 as a new subunit of the CCZ1-MON1 RAB7 guanine exchange factor (GEF) that positively regulates RAB7 recruitment to LE/autophagosomes. This work defines unique roles for GABARAP and LC3 subfamilies in macroautophagy and selective autophagy and demonstrates how analysis of autophagic machinery in the absence of flux can identify new regulatory circuits.
KEYWORDS: macroautophagy, selective autophagy, ATG8, autophagosome maturation, mitophagy
INTRODUCTION
Catabolic recycling of cytoplasmic components via autophagy is a fundamental cellular mechanism for survival and sustainability under basal conditions and is used under stress to remove damaged proteins or organelles (1, 2). Sequential recruitment of conserved autophagy-related (ATG) proteins, in concert with VPS34/class III phosphatidylinositol 3-kinase-dependent generation of phosphatidylinositol-3-phosphate (PI3P), drives formation of a double-membrane autophagosome structure, the vehicle for autophagic cargo degradation via the lysosome (3, 4). The canonical autophagy pathway, as initially discovered in budding yeast, is composed of three major activities that converge to promote flux through the pathway: the ATG8 conjugation arm, the ULK1 kinase arm, and the VPS34 lipid kinase arm (3, 4). The conjugation arm is responsible for covalent attachment of the ubiquitin (UB)-like protein ATG8 to phosphatidylethanolamine (PE) on the growing phagophore membrane (5–7). This conjugation system uses ATG7 as an E1, ATG3 as an E2 and a complex of ATG12∼ATG5-ATG16 (where “∼” indicates an isopeptide bond) as an E3 on the surface of the growing phagophore for ATG8 conjugation to PE through its C-terminal Gly residue. While a single ATG8 protein is found in yeast, mammals contain 6 ATG8s in two subfamilies (MAP1LC3A/B/C and GABARAP/L1/L2), all of which are conjugated to PE. VPS34 generates PI3P on target membranes, which then serves to recruit PI3P-binding WIPI proteins (principally WIPI1 and WIPI2) to the phagophore, and subsequently recruit the ATG12∼ATG5-ATG16 complex to facilitate ATG8 conjugation to membranes (8, 9).
Macroautophagy supplies the cell with macromolecules for biosynthetic and metabolic processes through nonspecific capture of cytosol. This process can occur, albeit at a reduced rate, in the absence of the conjugation arm of the autophagy system (10, 11). In contrast, selective clearance of damaged organelles including mitochondria (mitophagy), intracellular pathogens (xenophagy), and protein aggregates (aggrephagy) is thought to occur in a manner that relies more heavily on ATG8 proteins to support cargo capture. Interaction proteomics of all 6 human ATG8 proteins revealed an extensive network of interacting proteins (12) including the conjugation machinery, components involved in vesicle fusion, consistent with a potential role in vesicle tethering (13), and UB-binding cargo adaptors such as p62 and OPTN that link particular types of ubiquitinated cargo with the autophagosomal machinery (14). Many of the interactions involve short hydrophobic sequence elements in ATG8-binding proteins referred to as LIR (LC3-interacting region) motifs. These sequences bind to a hydrophobic cavity on the surfaces of ATG8 proteins referred to as the LIR docking site (LDS) (15, 16). Indeed, interaction of proteins with the LDS is thought to underlie many aspects of ATG8 function.
While significant overlap exists in human ATG8-interacting proteins (12), there are also distinct interactions, and several scenarios have been reported where particular adaptors selectively engage ATG8 family members (14, 15, 17). Work in mammalian cells using RNA interference (RNAi) targeting all three members in each family revealed a distinction between the functions of mammalian LC3 and GABARAP subfamilies, where GABARAPs were required for late steps in maturation, while LC3s play a role in earlier steps of autophagosome formation (18). Consistent with this, GABARAPs were shown to specifically mediate maturation/fusion by recruiting the homotypic fusion and vacuole protein sorting (HOPS)-interacting lysosomal tether protein PLEKHM1 to autophagosomes and promoting phosphatidylinositol 4-phosphate (PI4P)-dependent lysosomal fusion (19, 20). This simple hierarchical picture, however, is made more complex by the report that GABARAPs facilitate upstream assembly and activation of the ULK1 complex, which is required for initiation of autophagy (21, 22). Moreover, in Caenorhabditis elegans, distinct roles for the ATG8 orthologues LGG-1 and LGG-2 in aggrephagy have been reported, although how these requirements correlate with early and late functions is less clear (23).
These studies support a model where functions of the single yeast ATG8 protein diversified into subfamilies of mammalian ATG8 orthologues. However, the precise molecular function and relative redundancy of each subfamily remain obscure. Further highlighting the complexity of ATG8 family members and conjugation machinery as regulators of autophagic flux, recent work revealed that cells deficient in the conjugation arm display a 60% decrease in the frequency of membrane closure and a 30-min delay in the breakdown of the inner autophagosomal membrane for autophagosomes that successfully undergo lysosome fusion (11). These findings, coupled with the division of ATG8 family function in mammalian cells, raise the following questions. (i) Are GABARAPs or LC3s required for selective forms of autophagy? (ii) Does attenuation of a single subfamily of ATG8s impact cargo recruitment or flux through the lysosome? (iii) Do individual family members play specific roles, as suggested by the apparent specific role of LC3C in xenophagy (17)? (iv) How do ATG8 subfamilies interface with lysosomal fusion machinery to drive lysosomal fusion and inner membrane degradation?
Here, we examined ATG8 family member functions through systematic genetic analysis via clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-based gene editing (24), in combination with assays measuring various forms of macroautophagic and selective autophagic flux. Deletion of the GABARAP subfamily is sufficient to attenuate basal and starvation-dependent autophagic flux, as well as depolarization and parkin-dependent mitophagy, indicating that LC3s at endogenous levels cannot fully support the essential functions of the GABARAP subfamily. Cells lacking GABARAPs, and to a greater extent cells lacking all ATG8s, accumulate UB-positive aggregates that are also positive for the UB-dependent cargo adaptor p62 and its regulatory kinase TBK1. p62-positive aggregates are absent upon expression of any single GABARAP family member, suggesting functional redundancy for GABARAP family members. Interestingly, differential p62 accumulation in cells lacking LC3 family members versus ATG8-deficient cells suggests that GABARAP subfamily members may be capable of functionally suppressing loss of LC3 family members under basal conditions. Consistent with impaired autophagic flux observed upon GABARAP subfamily depletion, quantitative autophagosome proteomics reveal accumulation of autophagic cargo receptors, including p62 (SQSTM1) and TAX1BP1, in GABARAP-deficient autophagosomes. Among the proteins enriched in autophagosomes from GABARAP-deficient cells was C18orf8, an unstudied protein containing N-terminal WD40 repeats and C-terminal α-helical elements. Interaction proteomics revealed C18orf8 as a member of the CCZ1-MON1 complex, which is known to function as a guanine exchange factor (GEF) for RAB7 during endocytic and autophagic flux to the lysosome. Functional analysis revealed C18orf8 as a positive regulator of CCZ1-MON1 function that is necessary for endosomal/autophagic flux and efficient RAB7 localization on CD63-positive vesicle membranes. We therefore refer to C18orf8 as RMC1 (regulator of MON1-CCZ1). This work, as well as the recent work of Nguyen et al. also examining the role of ATG8 family members in parkin-dependent mitophagy (25), reveals primary roles for GABARAP family members in selective forms of autophagy and suggests that LC3 family members are insufficient to support all steps in selective autophagy. This work also supports the use of autophagosomal proteomics from autophagy-defective cells in the discovery of new components of the system.
RESULTS
Construction of a tool kit for analysis of ATG8 function.
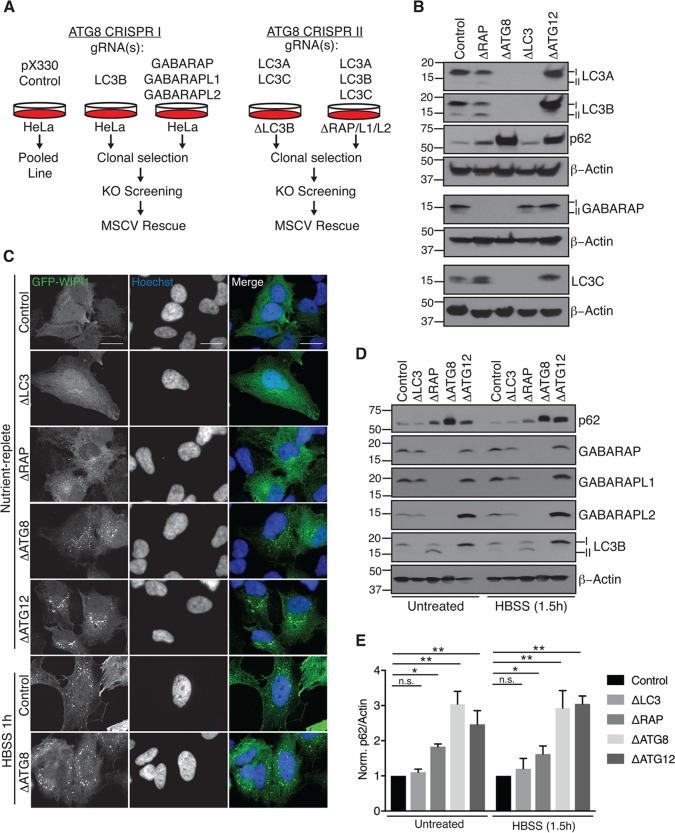
Previous work suggested that ATG8 proteins facilitate cargo recruitment to the autophagosome via cargo receptor interactions and drive lysosomal degradation of mature autophagosomes (18, 19, 26). To elucidate distinct roles of the mammalian ATG8s in these processes, we targeted each subfamily (LC3s and GABARAPs) and all six mammalian ATG8 homologues in HeLa cells via CRISPR/Cas9-mediated genome editing. Cells were cotransfected with plasmids encoding ATG8-specific guide RNAs (gRNAs) or empty pX330 vector as a nontargeting, CAS9-expressing control (Fig. 1A). Deletion of all LC3 and all GABARAP family proteins, referred to as ΔLC3 and ΔRAP, respectively, as well as ΔATG8, was confirmed via Western blotting (Fig. 1B and D), and in some cases, single deletions were also obtained (ΔLC3B, ΔGABARAP, ΔGABARAPL1, and ΔGABARAPL2), as described below. In parallel, we generated ΔATG12 cells, which lack the UB-like ATG12 protein required for conjugation of ATG8s to PE (Fig. 1B and D). Importantly, phagophore formation, as visualized by green fluorescent protein (GFP)-WIPI1 localization, remained intact in all ATG8 deletion cell lines (Fig. 1C). Consistent with previous work utilizing ATG conjugation machinery-deficient cells, ΔATG8s and ΔATG12 cell lines exhibit increased WIPI1 punctum formation basally in comparison to control cells (27). These cell lines provide a tool kit for systematic analysis of ATG8 function.
FIG 1.
Construction of a toolkit for analysis of ATG8 function. (A) Schematic for sequential CRISPR/Cas9-mediated genome editing to generate GABARAP family-deficient (ΔRAP), MAP1LC3 family-deficient (ΔLC3), or mammalian ATG8 family-deficient (ΔATG8) clonal cell lines. (B) Depletion of ATG8 proteins following gene editing. Denatured whole-cell extracts were subjected to SDS-PAGE and immunoblotted as indicated. (C) Phagophore formation remains intact in the absence of ATG8 proteins. Cells stably expressing GFP-WIPI1 were grown in nutrient-replete culture medium or HBSS as indicated, fixed, and imaged by spinning-disk confocal microscopy. Representative maximum-intensity projections of z-stacks are shown; scale bars represent 20 μm. (D) Elevated p62 expression in ΔRAP, ΔATG8, and ΔATG12 cells following starvation to induce autophagic flux. Cells were treated as indicated and subjected to immunoblot analysis as for panel B. (E) Plot of the ratio of p62 intensity to β-actin (loading) normalized to control cells for two independent biological experiments. Error bars represent standard deviation of the mean, and significance was determined by one-way analysis of variance (ANOVA) with post hoc testing (Dunnett's multiple-comparison test). *, P < 0.05; **, P < 0.001; n.s., not significant.
Impaired autophagic flux and accumulation of p62 in the absence of GABARAP proteins.
In order to characterize ATG8 mutant cells, we probed immunoblots from cells grown under nutrient-rich conditions with antibodies for p62, an autophagy receptor known to be degraded by the autophagy pathway that accumulates when there are defects in the pathway (14) (Fig. 1B and D). Interestingly, levels of p62 were unchanged in ΔLC3 cells, suggesting that LC3 proteins are not required for flux. In contrast, ΔATG8 cells, and to a lesser extent ΔRAP cells, displayed increased levels of p62 (a 3-fold increase in ΔATG8 and a 2-fold increase in ΔRAP). Moreover, in ΔATG8 cells, the levels of p62 were comparable to those seen in ΔATG12 cells, indicating reduced autophagic flux under nutrient-rich (basal) conditions (Fig. 1B, D, and E). Similar results were found after subjecting cells to starvation for 1.5 h in Hanks buffered saline solution (HBSS), indicating a requirement for GABARAPs in starvation-induced autophagic flux (Fig. 1D and E).
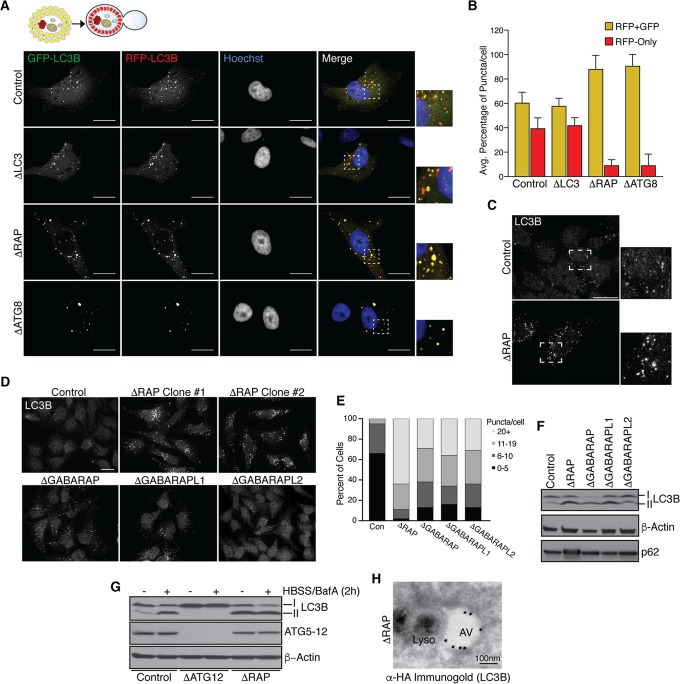
To examine the role of GABARAPs in autophagic flux, ΔLC3, ΔRAP, and ΔATG8 cells ectopically expressing near-endogenous levels of red fluorescent protein (RFP)-GFP-LC3B as a flux reporter were starved in HBSS for 1.5 h, followed by fixation and visualization of RFP-GFP (yellow) or RFP (red) puncta via confocal microscopy. Control cells displayed significant flux through the lysosome, as indicated by quenching of acid-sensitive GFP fluorescence in the lysosomal compartment (Fig. 2A and B). As expected based on p62 accumulation, both ΔRAP and ΔATG8 cells displayed a dramatic decrease in red puncta, consistent with reduced flux (Fig. 2A and B). In contrast, ΔLC3 cells expressing RFP-GFP-LC3B displayed flux rates similar to that seen in wild-type cells, indicating that ectopic expression of LC3B fails to accelerate flux in this system (Fig. 2A and B).
FIG 2.
Impaired autophagic flux and accumulation of p62 in the absence of GABARAPs. (A and B) Confocal microscopy analysis of RFP-GFP-LC3B flux following starvation (HBSS) for 1.5 h. Note accumulation of red (RFP-only) puncta in control and ΔLC3 cell lines. Scale bars represent 20 μm. Panel B depicts quantification of autophagic flux as analyzed in panel A; the average percentage of RFP-GFP and RFP-only puncta per cell was calculated for two pooled biological replicate experiments. Error bars represent the standard deviation of the mean. (C) Representative accumulation of basal LC3B puncta in ΔRAP cells as visualized by endogenous LC3B staining and confocal microscopy; the scale bar represents 20 μm. (D) Basal LC3B puncta accumulation, as visualized in panel C, with cells lacking individual GABARAP proteins or all three GABARAP proteins; the scale bar represents 20 μm. (E) Quantification of panel D. The number of LC3B puncta per cell was counted for each genotype and plotted according to the indicated classifications. (F) Immunoblot analysis of LC3-II accumulation in the absence of GABARAPs. (G) Loss of GABARAPs mimics LC3-II accumulation observed with bafilomycin A (BafA) treatment. Immunoblot analysis of LC3-II accumulation in control cells treated as indicated compared to that in ATG conjugation-deficient cells (ΔATG12) and ΔRAP cells is shown. (H) Impaired lysosomal fusion in ΔRAP cells. Immunogold staining for FLAG-HA-LC3B was performed, followed by TEM to visualize LC3B-positive autophagosomal structures and electron-dense lysosomes. The scale bar represents 100 nm.
Given reduced autophagic flux in ΔRAP cells, we next examined the phenotypes of cells lacking individual GABARAPs. In previous studies using RNAi to examine the roles of GABARAP proteins, all family members were depleted, making it difficult to deduce the relative roles of the 3 proteins (18). In addition, GABARAPs display ∼50% sequence identity, and the extent to which their functions overlap was unclear. Interestingly, we found increased endogenous LC3B puncta in cells lacking each individual GABARAP, suggesting that each family member contributes to overall flux (Fig. 2C to E). With each individual deletion, >60% of cells had 11 or more puncta (Fig. 2D and E). However, loss of all three GABARAPs resulted in a further increase in LC3B puncta, with more than 90% of cells having 11 or more puncta (Fig. 2C to E). This suggests that GABARAPs cumulatively support autophagic flux. This also resulted in an increase in the fraction of LC3B found in the LC3-II lipidated form, consistent with reduced flux (Fig. 2F). However, cells lacking individual GABARAPs did not display elevated p62 levels, indicating sufficient flux though the pathway (Fig. 2F). Immunoblot analysis revealed that LC3-II levels in ΔRAP cells under basal conditions were equivalent to those seen upon addition of bafilomycin A1 (BafA), an inhibitor of the vacuolar H+ ATPase (V-ATPase) and autophagic flux, to either wild-type or ΔRAP cells (Fig. 2G), thereby confirming that loss of GABARAPs impacts flux. Further supporting this notion, immunogold staining followed by transmission electron microscopy (TEM) analysis of FLAG-hemagglutinin (FLAG-HA)-tagged LC3B revealed electron-dense lysosomal structures juxtaposed to distinct LC3B-positive autophagosomal structures in ΔRAP cells (Fig. 2H). Given that loss of GABARAPs impairs Cherry-GFP-LC3B autophagic flux and that the LC3 immunogold signal was distinct from electron-dense lysosomal structures, we conclude that GABARAPs are critical mediators of productive lysosomal fusion with autophagosomes.
Accumulation of ubiquitinated protein aggregates in the absence of GABARAPs.
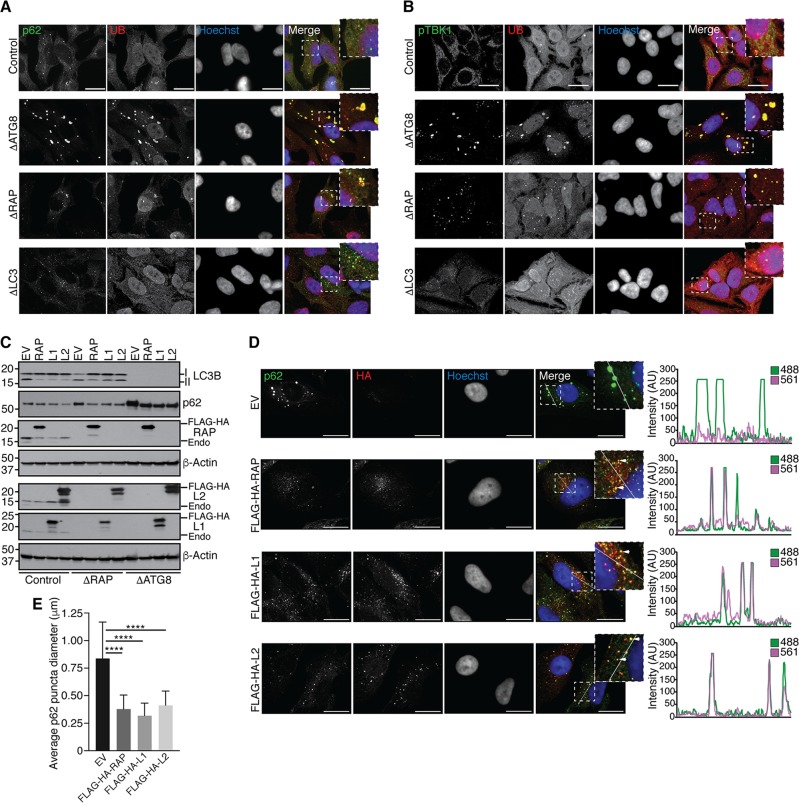
Since p62 accumulation in ΔATG8 cells exceeded that in ΔRAP cells, and because p62 is known to function in the autophagic degradation of UB aggregates, we examined whether ΔATG8 cells harbor an increased level of aggregated proteins. UB and p62 immunostaining in control cells was diffuse, as expected. In contrast, ΔATG8 cells and to a lesser extent ΔRAP cells accumulate p62 and UB-positive puncta under basal conditions, while UB-positive aggregates were not observed in ΔLC3 cells (Fig. 3A). Previous studies demonstrated that UB-positive autophagic cargo such as damaged mitochondria or intracellular pathogens also contain activated TBK1, as visualized using a phospho-specific S172 antibody (activating phosphorylation site) and via TBK1-dependent p62 phosphorylation (28–31). We found that anti-pTBK1 and -UB colocalize on structures in ΔATG8 cells and to a lesser extent in ΔRAP cells but were absent in ΔLC3 cells (Fig. 2B). Thus, like cells deficient in the conjugation apparatus, cells lacking GABARAPs are unable to efficiently remove ubiquitinated protein aggregates. The finding that ΔATG8 cells displayed larger and more numerous UB-positive puncta than ΔRAP cells (Fig. 3A) suggests that endogenous LC3 proteins are contributing to flux, whereas the absence of UB-positive puncta in ΔLC3 cells indicates that endogenous GABARAPs can perform all the necessary functions to promote flux.
FIG 3.
Accumulation of ubiquitinated aggregates in the absence of GABARAPs. (A) Cells of the indicated genotypes were cultured in complete medium on glass coverslips, followed by fixation and immunostaining for endogenous p62 and ubiquitin (UB). Cells were visualized by confocal microscopy, and representative images observed in biological replicate experiments are shown as summed intensity projections of z-stacks. Scale bars represent 20 μm. (B) Immunostaining and confocal microscopy as for panel A, visualizing endogenous phosphorylated TBK1 (pTBK1) and UB. (C) Ectopic expression of GABARAPs reduces p62 accumulation. Control, ΔRAP, and ΔATG8 cells were transduced with retrovirus encoding empty vector (EV) (control), FLAG-HA-GABARAP (RAP), FLAG-HA-GABARAPL1 (L1), or FLAG-HA-GABARAPL2 (L2), followed by puromycin selection to generate stable cell lines. Cells were harvested for immunoblot analysis with the indicated antibodies. (D) Left panel, cells reconstituted with ectopic GABARAPs as for panel C were grown on glass coverslips, fixed, and stained for p62 and anti-HA to visualize tagged GABARAPs. Note the redistribution of p62 signal from large aggregates to GABARAP-positive punctate structures. Representative summed intensity projections of z-stacks are shown; scale bars represent 20 μm. Right panel, intensity profiles of a 20-μm line segment drawn across an individual z-section for each channel. Overlap of 488 (p62) and 561 (anti-HA) fluorescence intensity profiles (arbitrary units [AU]) indicates colocalization of p62 with GABARAPs. (E) Quantification of p62 punctum diameter as shown in panel D. Error bars represent standard deviation of the mean, and significance was determined by one-way ANOVA with post hoc testing (Dunnett's multiple-comparison test). ****, P < 0.0001.
To determine whether ectopic expression of GABARAP proteins could rescue aggregate accumulation in ΔATG8 cells or accumulated autophagic vesicles in ΔRAP cells, we transduced cells with control (empty vector) or GABARAP-encoding retroviruses and visualized LC3-II and p62 accumulation by immunoblotting (Fig. 3C). Expression of each GABARAP family protein reduced p62 levels in ΔRAP and ΔATG8 cells and reduced accumulation of large p62 aggregates in ΔATG8 cells (Fig. 3C to E). While a reduction in p62 aggregate size was observed upon expression of GABARAP proteins (Fig. 3E), immunostaining revealed a concomitant distribution of p62 signal into autophagic structures, as demonstrated by p62 and FLAG-HA-GABARAP colocalization (Fig. 3D). Our data suggest that expression of any individual GABARAP protein is sufficient to restore autophagic flux to remove UB aggregates that accumulate in the absence of GABARAP function, indicating functional redundancy in this form of selective autophagy.
GABARAP proteins facilitate clearance of damaged mitochondria via the parkin pathway.
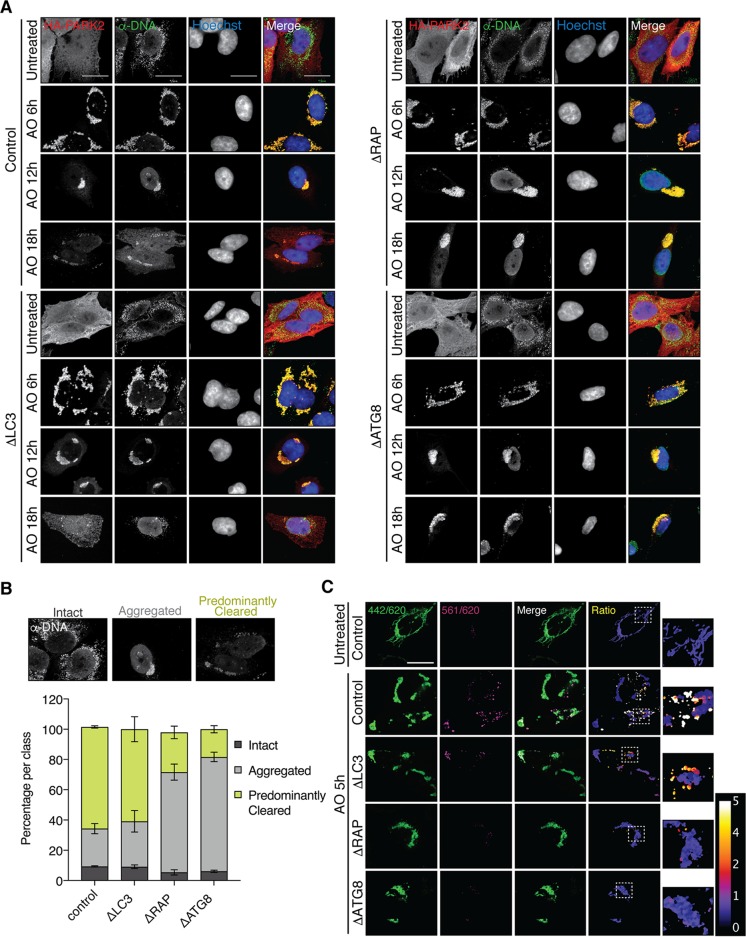
Parkin-dependent mitophagy is perhaps the best-understood form of selective autophagy (32). In this process, PINK1-dependent activation of parkin on the mitochondrial outer membrane leads to assembly of UB chains, which are then recognized by several UB-binding autophagy adaptors in HeLa cells, including OPTN, NDP52, and TAX1BP1, in a manner that is enhanced by TBK1-dependent phosphorylation of OPTN (28, 29, 33). To examine the roles of LC3 and GABARAP proteins in mitophagy, we retrovirally expressed FLAG-HA-parkin in wild-type, ΔLC3, ΔRAP, and ΔATG8 cells and examined the efficiency of mitophagy using anti-DNA to measure mitochondrial DNA content after depolarization with 10 μM antimycin A–5 μM oligomycin (AO). This assay has been used extensively to monitor mitochondrial abundance (33). While cells of each genotype displayed mitochondrial clustering at 6 h after AO treatment, as expected (34), a significant delay in mitochondrial clearance was observed in ΔRAP and ΔATG8 cells, consistent with attenuated autophagic flux in the absence of GABARAPs (Fig. 4A). Cells lacking GABARAPs and all six ATG8s displayed ∼25% and ∼20% cleared mitochondria, respectively, after 18 h of AO treatment, with a marked accumulation of aggregated mitochondria (∼70%) (Fig. 4B). In contrast, ΔLC3 and control cells displayed ∼60% cleared mitochondria with ∼25 to 30% aggregated mitochondria remaining (Fig. 4B). To examine mitophagic flux, we coexpressed the mitochondrion-targeted Keima fluorescent reporter (mtKeima), which shifts its excitation spectrum from 440 nm at neutral pH to 586 nm at the acidic pH found in the lysosome (29, 33, 35–37). Cells of each genotype were depolarized for 5 h and examined using ratiometric imaging of the mtKeima signal in live cells to determine mitophagy flux (Fig. 4C). Control and to a lesser extent ΔLC3 cells displayed high 561-nm/442-nm mtKeima ratios, consistent with significant flux through the mitophagy pathway. In contrast, ΔRAP and ΔATG8 cells displayed low acidic mtKeima signals. Taken together, these data indicate that ATG8s are not essential for mitochondrial clustering/aggregation following depolarization. However, GABARAPs are necessary for timely flux of damaged mitochondria through the lysosome.
FIG 4.
GABARAP proteins facilitate clearance of damaged mitochondria via the parkin pathway. (A) ΔATG8 cells exhibit delayed mitochondrial clearance. HeLa cells of each genotype stably expressing FLAG-HA-PARK2 were treated with antimycin A and oligomycin (AO) as indicated, followed by fixation and staining for anti-DNA to demarcate mitochondria and anti-HA to indicate PARK2 expression and analysis by confocal microscopy. Maximum-intensity projections of z-stacks are shown, and scale bars represent 20 μm. (B) Quantification of the 18-h time point as shown in panel A. PARK2-positive cells of each genotype were quantified by classification of mitochondria as intact, aggregated, or predominantly cleared. Error bars represent the standard deviation for two representative biological experiments; n > 30 cells per experiment analyzed. (C) Cells of the indicated genotypes stably expressing FLAG-HA-PARK2 and mtKeima were treated with AO for 5 h and analyzed for acidic (lysosomal) Keima signal using confocal microscopy. Representative ratiometric images were constructed by dividing the 561-nm excitation signal (acidic Keima) by the 442-nm excitation signal (neutral Keima). The color scale (arbitrary units [AU]) represents the pixel value for the 561 nm/442 nm ratio (orange/white; 4 to 5 AU indicates lysosomal mtKeima). The scale bar represents 20 μm.
Altered content of autophagic vesicles in ΔRAP cells revealed by quantitative proteomics.
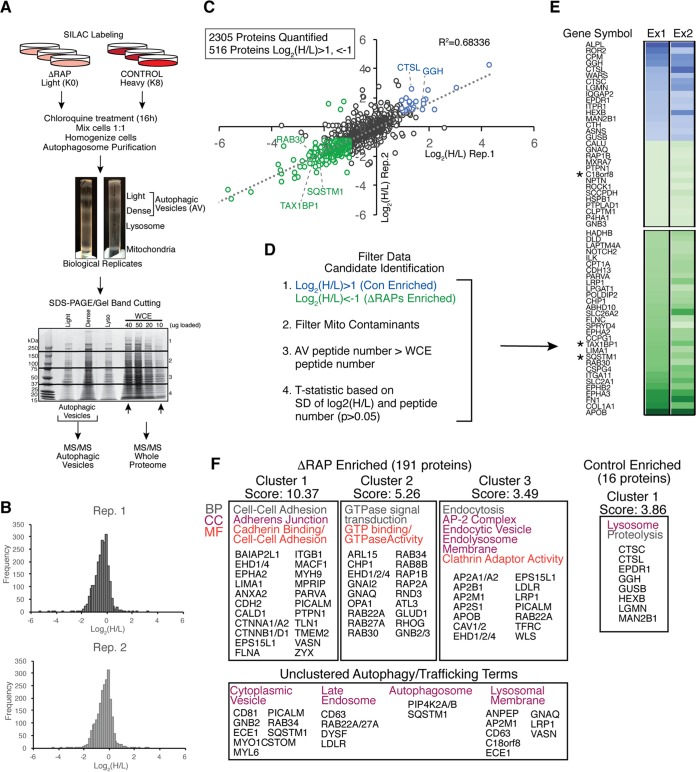
Previous studies demonstrated that autophagosomes can be generated in the absence of the ATG8 conjugation machinery and also suggested that late steps in the process, such as lysosomal fusion or degradation of the inner membrane after lysosomal engagement, require GABARAPs (10, 11). The inability of ΔRAP cells to promote autophagic turnover of ubiquitinated protein aggregates or damaged mitochondria suggested that autophagosomes from ΔRAP cells might accumulate pathway intermediates. To investigate this possibility, we used an approach we previously developed involving quantitative autophagosomal proteomics (38). Control cells labeled with heavy lysine (K8) or ΔRAP cells labeled with light lysine (K0) were treated for 16 h with 25 μM chloroquine to maximize accumulation of autophagosomes in biological duplicate experiments. Equal cell numbers were mixed, and extracts were subjected to density gradient fractionation prior to separation of autophagosomal vesicle (AV) fractions by SDS-PAGE (Fig. 5A). Digested peptides were liberated from gels and submitted for liquid chromatography-tandem mass spectrometry (LC-MS/MS), leading to the identification of 2,305 proteins in biological duplicate (Fig. 5C) (r2 = 0.68), of which 516 were changed in abundance by at least 2-fold in cells lacking GABARAPs (Fig. 5B and C; see Table S1 in the supplemental material). These proteins were then filtered based on previously described criteria (Fig. 5D) (38) to identify 197 candidate AV proteins that were significantly altered in abundance in ΔRAP cells (Fig. 5E and Table S1; see Materials and Methods for details).
FIG 5.
Altered content of autophagic vesicles in ΔRAP cells revealed by quantitative proteomics. (A) Control and ΔRAP cells were SILAC labeled and treated as indicated, followed by mixing of heavy and light labeled cells based on equal cell number. Autophagic vesicles were enriched via density gradient centrifugation, and samples were separated by SDS-PAGE for gel band cutting and in-gel Lys-C digest prior to MS/MS analysis. (B) Log2 (H/L) frequency distribution for two independent biological experiments as described for panel A. (C) Correlation of log2 (H/L) ratios for biological replicate experiments shown in panel B. A total of 2,305 proteins were quantified in both experiments, and 516 proteins passed the log2 (H/L) ratio cutoff of 1, −1 (2-fold change). Proteins significantly enriched in control cells and ΔRAP cells are shown in blue and green, respectively. (D) Filters applied to the biological replicate AV data set to enrich for novel autophagosome candidate proteins (see Materials and Methods for filtering details). (E) Log2 (H/L) ratio heat map for biological replicate experiments after filtering as described in panel D; the top and bottom 30 proteins are shown (see Table S1 in the supplemental material for the full data list). Asterisks indicate known autophagy receptors, as well as the previously uncharacterized protein C18orf8. (F) DAVID gene ontology (GO) analysis of the data set shown in panel E. Note enrichment of RAB GTPase proteins and endocytic machinery in ΔRAP cells, while lysosomal proteins are significantly enriched in control cells.
Consistent with GABARAPs facilitating late stages of autophagic flux, ΔRAP cells displayed accumulation of autophagosomal proteins relative to control counterparts, including TAX1BP1 and p62, in biological duplicate experiments (Fig. 5B, C, and E). We observed additional autophagy receptors significantly accumulating in ΔRAP cells, including CALCOCO2 (NDP52), NBR1, and NCOA4, along with LC3A and LC3B in the replicate with the deepest proteomic content (Table S1). Unbiased analysis of proteins accumulating in ΔRAP cells using gene ontology (GO) terms resulted in the identification of 5 clusters (P < 0.05) representing pathways linked with GTPases, endocytosis, the endolysosome membrane, and cell adhesion (Fig. 5F). The identification of numerous RABs in association with autophagosomes is interesting given the significant involvement of vesicle fusion in the autophagy process (39). We also analyzed proteins that were deenriched in ΔRAP cells relative to control cells (Fig. 5C, E, and F). This group of proteins included lysosomal proteins, including cathepsins, hydrolases, and proteases (CTSC, CTSL, GGH, GUSB, HEXB, LGMN, and MAN2B1) (Fig. 5E and F). Indeed, the only GO cluster identified for proteins deenriched in ΔRAP cells was a cluster centered on the lysosome (Fig. 5F). This result further supports the idea that ΔRAP cells are defective for productive lysosomal fusion (Fig. 2H).
Identification of C18orf8/RMC1 as a member of the CCZ1-MON1 GEF complex for RAB7.
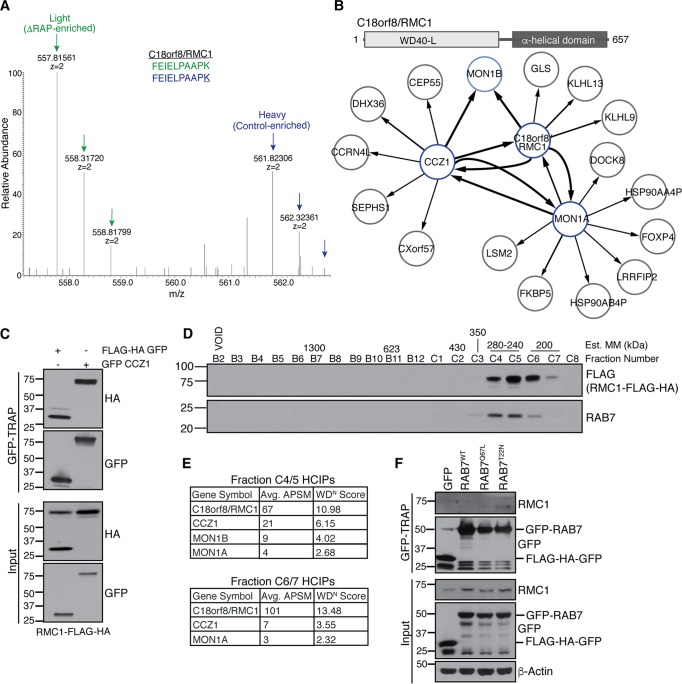
Among ΔRAP-enriched candidates was C18orf8 (Fig. 6A), a 70-kDa protein of unknown function previously reported to be lysosomal based on unbiased proteomics of purified lysosomes (40), which we have renamed RMC1 for reasons described below. To demonstrate the utility of our autophagosomal proteomics from ΔRAP cells to identify autophagy proteins, we further characterized RMC1. The structural prediction algorithms SwissModel and JPred identified a β-sheet-rich N-terminal region with a predicted WD40-like domain most similar to an aromatic amine dehydrogenase (PDB 2AGL), while the C-terminal half of the protein was exclusively helical (Fig. 6B, top panel), suggesting a possible scaffolding function. To identify RMC1-associated proteins, we performed affinity purification-mass spectrometry (AP-MS), followed by comparative proteomics software (CompPASS) analysis to identify high-confidence interacting proteins (HCIPs) associated with stably expressed HA-FLAG-tagged RMC1 (41). AP-MS of RMC1-FLAG-HA bait revealed several HCIPs, including MON1A, MON1B, and CCZ1, which were previously described as key components of the RAB7 GEF complex that is conserved in yeast and mammalian cells (Fig. 6B; see Table S2 in the supplemental material) (42–44). Multiple lines of evidence suggest the involvement of CCZ1-MON1 in autophagy. Initial studies in yeast demonstrated roles for CCZ1-MON1 in cytoplasm to vacuole targeting (Cvt) pathway and in autophagy, with evidence of a role at the stage of transfer vesicle-vacuole fusion (45, 46). Subsequent work identified CCZ1-MON1 as a GEF for RAB7 (42, 43). Moreover, CCZ1-MON1 promotes recruitment of RAB7 to PI3P-positive autophagosomes and is required for autophagosome-lysosome fusion in Drosophila (44).
FIG 6.
Identification of C18orf8/RMC1 as a member of the CCZ1-MON1 GEF complex for RAB7. (A) SILAC mass spectra of heavy (control) and light (ΔRAP-enriched) C18orf8/RMC1 peptides identified in autophagosomal proteomics; K represents the heavy-labeled lysine residue. (B) Top panel, predicted domain structure of RMC1. Bottom panel, Map of high-confidence interacting proteins (HCIPs) identified in reciprocal affinity purification-mass spectrometry (AP-MS) experiments with RMC1-FLAG-HA, FLAG-HA-CCZ1, or MON1A-MYC-FLAG bait proteins expressed in 293T cells. The line thickness correlates with the number of peptides observed in each interaction. (C) GFP-CCZ1 associates with RMC1-FLAG-HA. 293T cells stably expressing RMC1-FLAG-HA were transiently transfected with FLAG-HA-GFP control or GFP-CCZ1, followed by GFP-TRAP affinity purification. Note that RMC1-FLAG-HA associates with GFP-CCZ1 but not the FLAG-HA-GFP control. (D and E) Size exclusion chromatography and AP-MS of RMC1-containing complexes. 293T extracts stably expressing RMC1-FLAG-HA were separated by size exclusion chromatography; estimated molecular mass (MM) standards corresponding to fraction number are shown. Fractions containing RMC1 and RAB7 signals were subjected to FLAG AP-MS to identify complex members. (F) GFP-RAB7 associates with endogenous RMC1. 293T cells were transiently transfected with FLAG-HA-GFP control, GFP-RAB7WT, GFP-RAB7Q67L (constitutively active), or GFP-RAB7T22N (dominant negative), followed by GFP-TRAP affinity purification and immunoblot analysis as indicated.
In order to validate this interaction, we performed AP-MS on cells expressing HA-FLAG-CCZ1 or HA-FLAG-MON1A, which reciprocally associated with both RMC1 and each other (Fig. 6B). HA-FLAG-CCZ1 and RMC1 also associated with MON1B. We next performed two additional experiments to validate these interactions observed by AP-MS. First, we expressed GFP-CCZ1 (or FLAG-HA-GFP as a negative control) in HEK293T cells stably expressing RMC1-FLAG-HA and subjected cell extracts to affinity purification with GFP-TRAP resin. RMC1 was detected in GFP-CCZ1 complexes but not in control GFP affinity purifications (Fig. 6C). Second, extracts from 293T cells stably expressing RMC1-FLAG-HA were fractionated by size exclusion chromatography, and immunoblotting of fractions revealed RMC1 in a single major peak centered at ∼240 to 280 kDa (Fig. 6D). Fractions from ∼200 and ∼240 kDa were subjected to AP-MS, identifying peptides for RMC1, CCZ1, and MON1 proteins in both fractions (Fig. 6E). Although we did not detect RAB7 by mass spectrometry in this experiment, possibly due to its small size and limited tryptic fragments, RAB7 coeluted with RMC1 on gel filtration (Fig. 6D), consistent with a possible interaction. To examine this interaction further, we transiently transfected FLAG-HA-GFP control, GFP-RAB7WT (wild type), GFP-RAB7Q67L (a constitutively active mutant), or GFP-RAB7T22N (a GDP-bound dominant negative mutant) into 293T cells and identified an interaction between endogenous RMC1 and RAB7T22N, with minimal binding observed with RAB7WT or RAB7Q67L (Fig. 6F). Notably, previous work revealed an increased interaction between the MON1-CCZ1 GEF and GDP-locked RAB7T22N (43), consistent with increased RMC1 binding observed in our affinity purification (Fig. 6F). Taken together, these data suggest that RMC1 is a new component of the CCZ1-MON1 complex.
C18orf8/RMC1 is required for endolysosomal size control and autophagic flux.
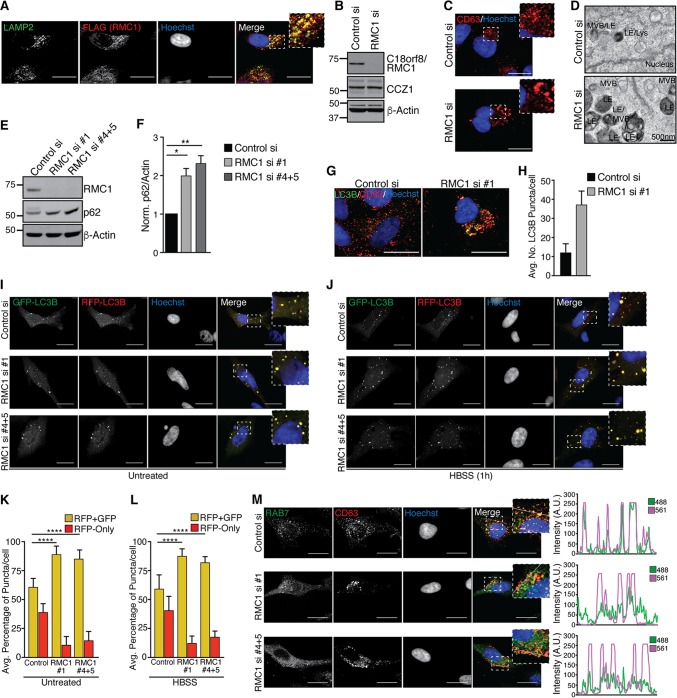
Given that RAB7 localizes to late endosomes, we used confocal analysis in combination with immunofluorescence to examine RMC1 subcellular localization. Antibodies that detect endogenous RMC1 by immunofluorescence are not available. Therefore, we expressed RMC1-FLAG-HA and found that it largely colocalized with LAMP2, a late endosomal marker (Fig. 7A). In order to examine the function of RMC1 in endosomal or CCZ1-MON1 action, we initially attempted to use CRISPR/CAS9 to delete the gene but were unsuccessful in identifying surviving clones despite extensive gene editing, suggesting that loss of expression is detrimental to cell viability. Therefore, we employed a small interfering RNA (siRNA) pool to deplete RMC1 transiently in HeLa cells (Fig. 7B). Using anti-CD63 to monitor late endosome/lysosome structures, we noted significantly enlarged structures, possibly suggesting a defect in endosomal clearance or maturation (Fig. 7C). Consistent with this notion, we observed accumulation of enlarged dense late endosome/lysosome structures by TEM analysis of thin sections of cells depleted of RMC1 (Fig. 7D).
FIG 7.
C18orf8/RMC1 is required for endolysosomal size control and autophagic flux. (A) HeLa cells stably expressing RMC1-FLAG-HA were grown on glass coverslips, fixed, and immunostained as indicated. (B) HeLa cells were transfected with control or RMC1 siRNA pools; at 72 h posttransfection, cells were harvested for immunoblot analysis with the indicated antibodies. (C) Cells treated with siRNAs as for panel B were grown on glass coverslips, fixed, and immunostained with the late endosomal marker anti-CD63; maximum-intensity projections of z-stacks are shown, and scale bars represent 20 μm. (D) Cells treated with siRNAs as for panel B were fixed and analyzed by TEM. (E) HeLa cells were treated with individual or combined siRNAs as indicated for 72 h, followed by immunoblot analysis to evaluate depletion of RMC1 and accumulation of p62 protein. (F) Quantification of p62 levels shown in panel E for two biological replicate experiments. Error bars represent standard deviation, and significance was determined by one-way ANOVA with post hoc testing (Dunnett's multiple-comparison test). *, P < 0.05; **, P < 0.01. (G) HeLa cells were grown on glass coverslips, fixed, and immunostained with anti-CD63 and LC3B; representative maximum-intensity projections are shown, and scale bars represent 20 μm. (H) Quantification of basal LC3B punctum number in cells stained as for panel G. Data represent two pooled biological experiments, and error bars represent standard deviation. (I and J) HeLa cells treated with control or RMC1 siRNAs (as for panel E) were treated with Cell Light RFP-GFP-LC3B for 16 h, followed by incubation in nutrient-replete DMEM (I) or starvation medium (HBSS) (J) for 1 h. Autophagic flux was monitored by confocal microscopy; single z-sections are shown for each channel, and scale bars represent 20 μm. (K and L) Quantification of average percentage of RFP-GFP-positive and RFP-only puncta per cell as shown in panels I and J. Data represent two pooled biological experiments, error bars represent standard deviation of the mean, and significance was determined by one-way ANOVA with post hoc testing (Dunnett's multiple-comparison test). ****, P < 0.0001. (M) Left panel, HeLa cells were grown on glass coverslips in nutrient-replete medium, fixed, and stained with endogenous antibodies against RAB7 and CD63. Individual representative z-sections for each channel are shown, and scale bars represent 20 μm. Right panel, intensity profiles of a 20-μm line segment drawn across the z-section shown for each channel. Overlap of 488 (RAB7) and 561 (CD63) fluorescence intensity profiles (arbitrary units [AU]) indicates colocalization of RAB7 with the late endosomal marker CD63.
Given the previously defined role for CCZ1-MON1 in late vesicle fusion events during autophagy and the link between RMC1 and late endosome/lysosomal disturbance when depleted, we asked whether basal autophagic flux in HeLa cells required RMC1. HeLa cells were transfected for 72 h with an individual siRNA targeting RMC1 (si1) or a combination of two individual siRNAs (si4 and si5) that resulted in greater than 80% depletion of endogenous RMC1 (Fig. 7E). RMC1 depletion led to accumulation of p62, compared with that in control siRNA-treated cells, consistent with impaired basal autophagic flux in the absence of RMC1 (Fig. 7E and F). Consistently, HeLa cells treated with RMC1 siRNAs harbored an increased number of endogenous LC3B-positive puncta as assayed by immunofluorescence compared to control siRNA-treated cells under basal conditions (Fig. 7G and H).
In order to directly examine a requirement for RMC1 in basal and starvation-induced autophagic flux, we employed the autophagic flux probe RFP-GFP-LC3B. HeLa cells were transfected with control or RMC1 siRNAs, and after 60 h, cells were induced to express RFP-GFP-LC3B under either basal (nutrient-replete) conditions or starvation (1 h in HBSS) conditions, followed by confocal imaging and image analysis (Fig. 7I to L). Under basal conditions, control siRNA cells displayed ∼60% RFP/GFP-positive (yellow) puncta and ∼40% RFP-positive puncta, indicating significant flux, and depletion of RMC1 with independent siRNAs reduced the percentage of RFP-positive puncta to ∼10%, consistent with a role for RMC1 in basal flux (Fig. 7K). Likewise, under starvation conditions, which display a larger overall flux, independent siRNAs targeting RMC1 also reduced flux to ∼10 to 15% with a concomitant increase in the percentage of RFP-GFP-positive puncta (Fig. 7L). Taken together, these data indicate that RMC1 is necessary for efficient autophagic flux under both basal and starvation conditions.
Previous work revealed that the MON1/CCZ1 GEF complex is required for RAB7 localization to PI3P-positive endosome/autophagosome membranes in Drosophila (44) and for Ypt7 localization on the vacuole in yeast (47). We hypothesized that if RMC1 positively regulated MON1/CCZ1 function, loss of RMC1 would be anticipated to block recruitment of RAB7 to late endosomes. To examine this question, HeLa cells were treated with control or independent RMC1 siRNAs, followed by fixation and immunostaining with endogenous RAB7 antibodies and anti-CD63 as a marker of the late endosomal compartment. Substantial CD63 and RAB7 colocalization in puncta was observed in control siRNA-treated cells, and image analysis of single confocal z-sections revealed extensive coincidence for the two antibody signals (Fig. 7M). In contrast, in cells treated with independent RMC1 siRNAs, RAB7 adopted a more diffuse cytosolic localization pattern, including a dramatic decrease in colocalization with CD63-positive puncta as measured using image analysis of individual confocal z-sections. Taken together, these data support a role for RMC1 in promoting the activity and localization of the MON1-CCZ1 complex for RAB7 GEF activity required for its assembly on late endosomes.
DISCUSSION
While mammalian ATG8 proteins have been shown to function centrally in autophagic cargo receptor engagement and vesicle maturation/lysosomal fusion, understanding of their individual molecular functions and absolute requirement for selective autophagic processes has remained limited. ATG8-dependent cargo recruitment has been highlighted in multiple selective autophagy pathways, including mitophagy, xenophagy, and aggrephagy (14). During parkin-dependent mitophagy, autophagy receptors OPTN, NDP52, p62, and TAX1BP1 are posttranslationally regulated by TBK1, thereby enhancing receptor association with UB chains on damaged mitochondria and recruitment of ATG8-positive autophagic membranes (28–30, 48). Similarly, p62 and NBR1 have been implicated in ATG8-dependent clearance of ubiquitinated protein aggregates in mammalian cells (49–51). Degradation of such cargo is critical following environmental insult or genetic alterations, as defects in cargo recognition or autophagic flux underlie a number of proteinopathies, including neurodegenerative disease (52). Cargo degradation relies on efficient flux through the lysosome, and previous work suggested that ATG8s, particularly the GABARAP subfamily, are mediators of this process (18–20). While ATG8 conjugation-independent alternative autophagy has been documented in murine cells and embryonic tissues, recent work revealed that autophagic flux is delayed in the absence of conjugation machinery due to inefficient autophagosomal closure and reduced rates of autophagosome inner membrane degradation (10, 11). In this study, we demonstrate that GABARAPs are key drivers of autophagic flux through the lysosome following starvation and selective autophagic stimuli. During preparation of this article, an additional study demonstrated that ATG8-deficient cells exhibit impaired autophagic flux following starvation-induced autophagy and mitochondrial depolarization, consistent with our findings (25).
Our data support a model wherein GABARAPs are critical facilitators of autophagic flux. First, autophagosomal structures accumulate in the absence of GABARAPs, and LC3B overexpression in the context of ATG8 deletion fails to rescue flux in starved cells (Fig. 2A). This suggests that the demand for autophagic cargo clearance exceeds the capacity of the nonselective or ATG-independent autophagy arm of the pathway. This could potentially be a reflection of cargo size/loss of recruitment mechanisms that allow the assembly of autophagosomes proximal to large cargo. Second, cells lacking LC3 family members exhibit a UB aggregate profile similar to that of control HeLa cells, while GABARAP family-deficient cells accumulate p62-, UB-, and pTBK1-positive structures that are not present when any of the three individual GABARAP proteins are expressed (Fig. 3D). While ATG conjugation-independent autophagy may be capable of clearing small protein aggregates, it has been shown that large aggregates or aggresomes are targeted by a selective autophagy program, dependent in part on conjugation machinery (53). This is consistent with our observation that ATG8-deficient cells do not effectively degrade aggregated proteins. A strong reliance on GABARAP-dependent flux was also observed with parkin-dependent mitophagy, as also reported recently (25) (Fig. 4B). Although endogenous LC3s are recruited to damaged mitochondria to form the autophagosome, their expression alone is not sufficient for lysosomal degradation (25, 54).
Molecular determinants of autophagosome-lysosome tethering and fusion have been described (19, 55–57); however, the precise GABARAP-dependent interaction necessary for this process remains unclear. Using quantitative proteomics, we found that a cohort of lysosomal proteins was significantly enriched in control cells compared to ΔRAP counterparts, further supporting our conclusion that lysosomal tethering or fusion is impaired in the absence of GABARAP function. A recent study suggested that GABARAP promotes fusion via recruitment of PI4K2A and subsequent PI4P generation (20). Interestingly, we observed PI4K2A in both control and ΔRAP autophagic vesicles {nonsignificant log2 (heavy/light) [log2 (H/L)] for enrichment in either genotype} (see Table S1 in the supplemental material), suggesting that LC3s may play a compensatory role in PI4K2A recruitment.
The homotypic fusion and vacuole protein sorting (HOPS) tethering complex has also been shown to play a key role in autophagosome-lysosome fusion (19, 58). At the molecular level, bridging of ATG8, RAB7, and HOPS at the autophagosome-lysosome junction by PLEKHM1 drives the fusion event (19). While we did not observe PLEKHM1 peptides in our mass spectrometry analysis, RAB7A, along with HOPS complex members VPS39, VPS11, and VPS18, are found on both control and ΔRAP autophagic vesicles, albeit that the HOPS members were found in only the deeper of the two proteomic experiments. Similarly, SNARE machinery, including VAMP7, VAMP8, and SNAP-29, is identified in both genotypes (nonenriched). Syntaxin 17 (STX17) localizes to the outer membranes of mature autophagosomes, forming a complex with SNAP-29 and VAMP8 that drives lysosomal fusion (57). We did not observe STX17 peptides by mass spectrometry, but ectopically expressed full-length STX17 localizes to autophagosomes in both control and GABARAP family-depleted cells, although overexpression of this form is thought to function in a dominant negative fashion (data not shown). Further studies are required to identify key fusion factors or modification events that are dependent upon GABARAPs.
From a proteomics standpoint, ΔRAP cells serve as a valuable tool for discovery of associated or integral autophagosomal proteins. Our quantitative autophagic vesicle analysis identified a novel member of the RAB7 GEF complex, C18orf8/RMC1. The MON1/CCZ1 GEF components are recruited to the membrane by the early endosome marker RAB5 and PI3P, and GEF activity facilitates RAB7 recruitment to the membrane concomitant with exchange of GDP for GTP (43, 59). Recent structural analysis revealed that MON1/CCZ1 exists as a heterotetramer, and importantly, PI3P alone was insufficient for GEF function, suggesting that the GEF complex likely requires additional factors to coordinate its membrane recruitment (60). However, the stoichiometry of RMC1 within the heterotetrameric MON1-CCZ1 complex remains to be determined. Given the predicted structure of RMC1, we speculate that its N-terminal β-propeller may function analogously to the WIPI proteins to bind phosphoinositides at the membrane, while the C-terminal α-helical structure serves as a scaffold for CCZ1-MON1 recruitment, thereby bridging the membrane and GEF complex. Consistent with this notion, ectopically expressed RMC1 localizes on endolysosomal membranes (Fig. 7A), and loss of RMC1 impairs both recruitment of RAB7 to endosomes and autophagic flux (Fig. 7C and I to M). In the long term, full reconstitution of the mammalian GEF complex in the context of vesicle fusion will be necessary to understand the precise function of RMC1 in RAB7 activation. Collectively, our data indicate that GABARAP proteins, along with RAB7 activation via the MON1/CCZ1/RMC1 GEF complex, facilitate autophagic flux, and this study provides a framework for assessing mammalian ATG8 family function.
MATERIALS AND METHODS
Antibodies and reagents.
The following antibodies were used in this study for immunoblot analysis: LC3B (2775), LC3C (clone D3O6P 14736), GABARAP (13733), GABARAPL1 (26632), GABARAPL2 (14256), p62 (8025), ATG12 (4180), and RAB7 (9367) (Cell Signaling Technology); β-actin (sc-69879) and PCNA (sc-56) (Santa Cruz); HA.11 mouse monoclonal antibody (MAb) (901501) (BioLegend); M2 FLAG mouse MAb (F1804) (Sigma); GFP mouse MAb (11814460001) (Roche); C18orf8 mouse MAb (TA504561) (Origene); and C18orf8 (ab137276) and CCZ1 (ab175065) (Abcam). LC3A-, GABARAP-, GABARAPL1-, and GABARAPL2-specific antibodies used for Western blotting were gifts from Takeda Oncology. Secondary antibodies for immunoblotting were horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (H+L) or anti-mouse IgG (W4011 and W4021; Promega). For immunofluorescence staining, the following antibodies were used: LC3B rabbit MAb (3868), p62 rabbit MAb (7695), and p-S172 TBK1 rabbit MAb (5483) (Cell Signaling Technology); ubiquitin (FK2 clone, 04-263) and anti-DNA (CBL186) (EMD Millipore); M2 FLAG (F1804) (Sigma); LAMP2 mouse MAb H4B4 (ab25631) and CD63 (ab59479) (Abcam); and RAB7 (9367) (Cell Signaling Technology). Alexa Fluor 488–anti-rabbit or Alexa Fluor 594–anti-mouse secondary antibodies were used for immunofluorescence (Life Technologies). For nuclear staining, cells were incubated with Hoechst dye following antibody incubations. The following chemicals were used in this study: bafilomycin A1 (Sigma; 50 nM), antimycin A (Sigma; 10 μM), oligomycin (Sigma; 5μM), and chloroquine (Sigma; 25 μM).
Mammalian cell culture and plasmid/siRNA transfections.
HeLa and 293T cells were obtained from the American Type Culture Collection (ATCC) and tested for mycoplasma contamination. Cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (HyClone) at 37°C with 5% CO2. For stable cell line generation, plasmid DNA (retroviral pMSCV or lentiviral pHAGE Gateway compatible backbone with gene inserted) along with viral helper constructs (retroviral MSCV-vsvg or MSCV-gag/pol or lentiviral psPAX2 or pMD2) were diluted in Opti-MEM (Gibco) and combined with 1 μg/μl polyethylenimine (PEI) (Polysciences Inc.) in a PEI/DNA concentration ratio of 3:1. The transfection mixture was added to 293T cells, followed by incubation for 12 to 16 h. Fresh DMEM was added to the cells, followed by a 24-hour incubation to allow virus production. Viral supernatant was collected, filtered, and added to recipient HeLa or 293T cells along with 8 μg/ml Polybrene for infection. Stable cell lines were established by puromycin selection (2 μg/ml) for 48 to 72 h. For plasmid transient-transfection experiments, Lipofectamine 3000 transfection reagent (Life Technologies) was used for transfection of HeLa cells according to manufacturer's specifications, and PEI (as described above) was used for transient transfection of 293T cells. For small interfering RNA (siRNA) transfection, Lipofectamine RNAiMax was used to transfect indicated siRNA (20 nM) into HeLa cells. Transfected cells were analyzed at 60 to 72 h after siRNA delivery. The following siRNAs (all purchased from Dharmacon) were used in this study: ON-TARGETplus nontargeting control siRNA pool (D-001810-10-05), ON-TARGETplus human C18orf8 siRNA pool (L-016601-00-0005), siGENOME nontargeting control siRNA (D-001210-01-05), siGENOME human C18orf8 siRNA1 (si1) (D-016601-01-0002; GGAGAUGCCUCAUAAAUUU), siGENOME human C18orf8 siRNA4 (D-016601-04-0002; GCGCCUAAGUCAACUAAAC), and ON-TARGETplus human C18orf8 siRNA5 (J-016601-05-002; GGAGAUGCCUCAUAAAUUU). For cells treated simultaneously with si4 and si5, a 10 nM final concentration of each siRNA was delivered to cells.
cDNA expression constructs.
pMXs-IP-GFP-WIPI1, utilized for visualization of phagophore formation, was a gift from Noboru Mizushima (Addgene plasmid 38272) (27). pMSCV-Flag-HA-ATG8 retroviral constructs and the pHAGE-FLAG-HA-PARK2 construct were generated as previously described (12, 61). pIND(Spl)-mtKeima was a gift from Atsushi Miyawaki (37); the mtKeima sequence with stop codon included was cloned into our pHAGE-C-terminal FLAG-HA lentiviral vector via Gateway cloning, and a stop codon prevents expression of the FLAG-HA tag. C18orf8 and CCZ1 cDNAs were purchased from Origene and subjected to Gateway cloning into the pHAGE-C-terminal-FLAG-HA vector (C18orf8), pHAGE-N-terminal-FLAG-HA, or pHAGE-GFP vector (CCZ1). GFP-RAB7WT, GFP-RAB7Q67L, and GFP-RAB7T22N (dominant negative) were gifts from Qing Zhong (Addgene plasmids 28047, 28049, and 28048, respectively) (62). For transient RFP-GFP-LC3B expression, HeLa cells were transduced with Cell Light Premo autophagy tandem sensor BacMAM reagent (P36239; Life Technologies) according to the manufacturer's instructions for 16 h.
CRISPR/Cas9-mediated gene editing.
Gene editing for creation of mammalian ATG8-depleted cells was performed as described previously (24, 63). Briefly, 20-nucleotide gRNAs for the mammalian ATG8 family members were cloned into pX330 or pX459 CRISPR/Cas9 vectors; pX330-U6-Chimeric_BB-CBh-hSpCas9 and pSpCas9(BB)-2A-Puro (PX459) V2.0 were gifts from Feng Zhang (Addgene plasmids 42230 and 62988). The following gRNAs were utilized: LC3A (GTCAAGCAGCGGCGGAGCTT), LC3B (GTCAAGCAGCGCCGCACCTT), LC3C (GCTTGAAGGGTCTGACGCTT), GABARAP (GTCGAGAAGCGCCGCTCTGA), GABARAPL1 (GTCCGCTCCTCTACACTCAC), GABARAPL2 (GCGAGCGAAATATCCCGACA), and ATG12 (GCTCCGGGGTGGTTGTTTCT). Two hundred nanograms of pX330 vector containing gRNA was cotransfected with 200 ng of pHAGE-N-FLAG-HA-GFP vector (GFP expression vector with puromycin cassette) to allow for puromycin selection of transfected cells (1 μg/ml; 24-hour pulse performed at 48 h posttransfection). Alternatively, the pX459 vector with puromycin resistance was used. Following puromycin selection, single-cell clones were plated in 96-well format by limiting dilution and cultured to allow single colonies to grow out. Clones were expanded to 12-well plates, and samples of the resulting clones were screened via Western blotting with two independent gene-specific antibodies (see “Antibodies and reagents” above) and the Surveyor mutation detection kit (IDT, 706020) (24) to detect ATG8-edited and -depleted clones. The resulting ATG8-depleted cell lines were rescued by reexpression of near-endogenous levels of ectopic ATG8 expression using our MSCV-driven N-terminal FLAG-HA tagging retroviral vector (12).
Immunoblot analysis.
For immunoblotting experiments, cell extracts were prepared in denaturing urea lysis buffer (8 M urea, 50 mM Tris [pH 8.2], and 75 mM NaCl, supplemented with protease inhibitors [Roche]) or nondenaturing lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 1% [vol/vol] NP-40, supplemented with protease inhibitors and 1 mM dithiothreitol [DTT]). Extracts were incubated on ice for 20 min to allow lysis and cleared by centrifugation at 16,000 × g for 10 min. Cell extracts (20 to 40 μg) were separated via SDS-PAGE using Tris-glycine gels (Bio-Rad); gels were transferred to nitrocellulose membranes (Bio-Rad), followed by immunoblot analysis as indicated. For LC3C Western blotting, equal numbers of cells were lysed in 1× SDS sample buffer (Bio-Rad) and boiled for 5 min, followed by SDS-PAGE separation on 4 to 12% Bis-Tris gels in morpholineethanesulfonic acid (MES) buffer. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes, followed by incubation with primary antibody overnight. For RMC1 immunoblots, cells were lysed in nondenaturing lysis buffer; extracts were separated on 4 to 12% Bis-Tris gels in morpholinepropanesulfonic acid (MOPS) buffer to resolve the 75-kDa protein and transferred to PVDF membranes.
Immunofluorescence and confocal microscopy.
Cells of each genotype were grown on glass coverslips, treated as indicated, and fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) (pH 7.4). For endogenous LC3B staining, cells were fixed with 100% methanol for 10 min at −20°C. All cells were permeabilized and blocked with 5% normal goat serum (Cell Signaling Technology) in PBS containing 0.3% Triton X-100 (Sigma). Cells were subjected to immunostaining with the indicated antibodies diluted according to manufacturer's recommendations in PBS containing 1% bovine serum albumin (BSA) (Sigma) and 0.1% Triton X-100. Following washing with PBS, cells were subjected to secondary antibody staining using Alexa Fluor 488/594-conjugated secondary antibodies (Life Technologies) for 45 min at room temperature. Following immunostaining (or fixation of fluorescent proteins such as GFP, Cherry, and RFP), coverslips were stained with Hoechst dye, washed, and mounted on glass slides using Prolong Gold antifade reagent (Life Technologies). Images were acquired using a Yokogawa CSU-X1 spinning-disk confocal microscope with Borealis modification on a Nikon Ti-E inverted microscope using an X100 Plan ApoNA 1.4 objective lens. Images were collected with a Hamamatsu ORCA-AG cooled charge-coupled device (CCD) camera controlled by MetaMorph7 software (Molecular Devices). Fluorophores were excited using a Spectral Applied Research LMM-5 laser merge module with acousto-optic tunable filter (AOTF)-controlled solid-state lasers (488 nm and 561 nm). A Lumencor Sola fluorescence light source was used for imaging Hoechst staining. Z-series optical sections were collected with a step size of 0.2 μm, using the internal Nikon Ti-E focus motor, and stacked using MetaMorph to construct maximum- or summed-intensity projections where indicated. Images taken for each genotype in an experiment were acquired with matched laser intensities, and the resulting images were adjusted in MetaMorph to display matched intensities in each channel across the various genotypes. For RFP-GFP-LC3 analysis, puncta in the 488-nm (GFP) and 561-nm (RFP) channels were identified using Fiji (Image J; https://fiji.sc), and the average percentages of RFP-GFP-coinciding puncta and RFP-only puncta were calculated and plotted in GraphPad Prism7, where error bars represent the standard deviation. For analysis of colocalization of p62 and HA-GABARAPs (Fig. 3D) or RAB7 and CD63 (Fig. 7M), a 20-μm line segment was drawn across a single z-section and copied to each channel in Fiji. The plot profile for each channel was determined and exported as an Excel file; intensity profiles (shown in arbitrary units) are plotted for the 488-nm (p62 or RAB7) and 561-nm (HA or CD63) channels for each genotype. For Fig. 3, the diameter of p62 puncta (calculated in micrometers) within each cell was determined after thresholding to visualize puncta from background staining intensity using Fiji.
Live confocal microscopy.
For ratiometric imaging experiments, mtKeima was transduced into cells stably expressing FLAG-HA-PARK2 via lentivirus, followed by 1 μg/ml puromycin (Sigma) selection to enrich for mtKeima-expressing cells. Cells were either untreated or depolarized with 5 μM oligomycin and 10 μM antimycin A (Sigma) (referred to as AO treatment) in FluoroBrite DMEM (Thermo Fisher Scientific) supplemented with 10% FBS and 1% l-glutamine for periods of time as indicated. Following the desired treatment duration, live cells were analyzed using spinning-disk confocal microscopy using a 100×/Plan Apo 1.4-numerical-aperture objective lens, as described for immunofluorescence experiments, within a 37°C stage-top incubator with CO2 (Okolab). mtKeima measurements were conducted using dual excitation wavelengths of 442 nm (neutral) and 561 nm (acidic) and a single emission wavelength of 620 nm (dichroic). Single z-slices of each wavelength were adjusted for equal intensity across all genotypes for a given time point, followed by thresholding to include mitochondrial signals found in both channels and calculation of the 561/442 ratio using Fiji (64). Images are presented using the fire lookup table (ranging from dark blue/purple for a neutral Keima signal to orange/yellow/white for Keima signals in the acidic lysosomal compartment).
Electron microscopy.
Control and C18orf8/RMC1 knockdown HeLa cells were grown to 60% confluence in 6-cm culture dishes and fixed with 2% paraformaldehyde (PFA)–2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 5 min at room temperature. Fixed samples were submitted to the Harvard Medical School Electron Microscopy Facility and subsequently processed for imaging using a Tecnai G2 Spirit BioTwin transmission electron microscope. For immunogold labeling, the Tokayasu method was used. Cells expressing MSCV-FLAG-HA-LC3B were grown to ∼75% confluence, rinsed with PBS, and gently trypsinized. The cell suspension was layered on top of a cushion of 4% PFA–0.1% glutaraldehyde (in 0.1 M sodium phosphate buffer, pH 7.4) and pelleted by centrifugation for 3 min at 775 × g. The pellets were resuspended in fresh 4% PFA–0.1% glutaraldehyde and fixed for 2 h at room temperature. The fixative was replaced with PBS, and samples were submitted to the Electron Microscopy Facility, where pellets were infiltrated with 2.3 M sucrose in PBS (containing 0.2 M glycine) for 15 min. Samples frozen in liquid nitrogen were sectioned and transferred to Formvar-carbon-coated copper grids. Grids were floated on drops of 1% BSA to block nonspecific labeling, followed by incubation in anti-HA antibody, washing with PBS, and labeling with protein A-gold. Contrasting/embedding of the labeled grids was carried out on ice in 0.3% uranyl acetate in 2% methyl cellulose, grids were examined in the Tecnai G2 Spirit BioTWIN transmission electron microscope, and images were recorded with an AMT 2k CCD camera.
Quantitative autophagosome proteomics.
Control or ΔRAP cells were grown in heavy (K8) or light (K0) SILAC (stable isotope labeling with amino acids in cell culture) DMEM, respectively, and an equal number of cells was mixed prior to cell homogenization and Histodenz gradient centrifugation as described previously (38). Enriched autophagic vesicles or control cell pellets were lysed in 2% SDS–50 mM Tris (pH 7.5), and proteins were separated by SDS-PAGE. Gel bands were excised and subjected to in-gel Lys-C digestion (Wako). The resulting peptides were desalted using C18 Stage tips and analyzed using an Orbitrap Fusion mass spectrometer coupled to a Proxeon EASY-nLC 1000 liquid chromatography (LC) pump (Thermo Fisher Scientific). Approximately 2 μg of peptides was fractionated on a 100-μm-inner-diameter microcapillary column packed with ∼20 cm of Accucore C18 resin (2.6 μm, 150 Å; Thermo Fisher Scientific). Peptides were separated using a 2-hour gradient of 5 to 29% acetonitrile in 0.125% formic acid with a flow rate of ∼450 nl/min. The scan sequence began with an Orbitrap MS1 spectrum with the following parameters: resolution, 120,000; scan range, 400 to 1,400 Th; automatic gain control (AGC) target, 2 × 105; maximum injection time, 100 ms; and centroid spectrum data type. We selected the top 20 precursors for MS2 analysis, which consisted of high-energy collision dissociation (HCD) with the following parameters: resolution, 15,000; AGC, 1 × 104; maximum injection time, 150 ms; isolation window, 0.7 Th; normalized collision energy (NCE), 35; and centroid spectrum data type. In addition, unassigned and singly charged species were excluded from MS2 analysis, and dynamic exclusion was set to 60 s. For data analysis, mass spectra were processed using a Sequest-based in-house software pipeline. MS spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included all entries from the human UniProt database (10 August 2011), which was concatenated with a reverse database composed of all protein sequences in reverse order. Searches were performed using a 50-ppm precursor ion tolerance. The product ion tolerance was set to 0.9 Th. Oxidation of methionine residues (+15.99492 Da) was set as a variable modification. Peptide spectral matches (PSMs) were altered to a 1% false-discovery rate (FDR) (65, 66). PSM filtering was performed using a linear discriminant analysis, as described previously (67), while considering the following parameters: XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy. Peptide spectral matches were identified, quantified, collapsed to a 1% FDR, and then further collapsed to a final protein-level FDR of 1%. Furthermore, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides. Four gel band regions were cut per sample (individual lanes) of the SDS-polyacrylamide gel; these 4 regions were processed separately, and each is an independent MS sample (to decrease complexity of peptides in each run). Protein assembly was utilized to group the 4 runs of a given sample together for batched analysis and calculation of log2 (heavy/light) ratios. The resulting data were filtered for SILAC-quantified proteins based on the summed signal-to-noise ratio for heavy and light peptides; contaminant peptides identified in the search were removed at this step. An intensity cutoff of 10 was applied for summed heavy plus light channels, and each individual channel had an intensity cutoff of 5 (to avoid identifying proteins with an intensity of 0 in either the light or heavy channel, i.e., proteins found only in one condition or the other). Proteins with calculated log2 (H/L) ratios of >1 (control enriched) and <−1 (ΔRAP enriched) were considered significantly enriched (first filter applied in identifying candidate autophagosomal proteins). The following additional filters were then applied to create a high-confidence autophagosomal candidate list. For the second filter, mitochondrial contaminants generated during fragmentation of the organelle during homogenization of cells were removed by filtering proteins that had UniProt designations as only mitochondrial matrix or mitochondrial inner membrane. For the third filter, the peptide number for a given protein in the AV fraction was compared to a matched whole-cell extract (WCE) sample to determine whether the protein is “enriched” in the AV fraction (38), and proteins whose peptide number was greater in the WCE sample, possibly indicating a contaminating protein in the AV preparation, were filtered. For the fourth filter, a T-statistic was calculated based on the standard deviation of the log2 (H/L) and peptide number (38). Because this filter will remove any proteins identified as a single peptide, proteins identified with >1 peptide in at least one of the biological replicate experiments were retained in the analysis. Following application of the above filters, gene ontology (GO) analysis was performed on control and ΔRAP-enriched proteins using DAVID version 6.8 (68, 69), with medium classification stringency for the CC, BP, and MF terms.
Affinity purification and proteomic analysis.
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 1% [vol/vol] NP-40, supplemented with protease inhibitors [Roche] and 1 mM DTT) for 20 min on ice to obtain whole-cell extracts. The lysates were cleared for 15 min at 4°C at 16,000 × g and diluted with lysis buffer without DTT to reduce the DTT concentration to 0.1 mM prior to affinity purification. For GFP-TRAP affinity purifications, ∼0.5 mg of extract expressing GFP-tagged proteins was incubated with 10 μl solid GFP-TRAP magnetic agarose resin (Chromotek) for 1 h at 4°C, followed by 4 1-min washes with ice-cold lysis buffer without DTT. Following washes, beads were resuspended in 1× SDS loading buffer (Bio-Rad) and boiled for 2 min, and eluted proteins were separated by SDS-PAGE (Tris-glycine gels; Invitrogen) prior to immunoblotting. Affinity purification-mass spectrometry (AP-MS) and CompPASS analysis were performed as previously described (12, 41). Briefly, ∼1 × 107 293T cells were lysed in lysis buffer, incubated, cleared, and diluted to reduce the DTT concentration as described above. The resulting extracts were incubated with M2 FLAG magnetic beads (Sigma) for 2 h at 4°C, followed by 4 1-min washes with ice-cold lysis buffer plus protease inhibitors (without DTT), 2 washes with ice-cold PBS (pH 7.2) to remove detergent, and elution with 3× FLAG peptide (0.25 mg/ml). Eluted proteins were reduced and alkylated, followed by trichloroacetic acid (TCA) precipitation and digestion with sequencing-grade trypsin (Promega). The resulting peptides were desalted on C18 Stage tips and analyzed using a Velos LTQ mass spectrometer (Thermo Fisher Scientific), followed by CompPASS analysis to identify high-confidence interacting proteins (HCIPs) (12, 41). For size exclusion chromatography/AP-MS analysis, cells were lysed in lysis buffer as described above. Size exclusion chromatography (gel filtration) of whole-cell extracts was performed using a Superose 6 column, collecting 0.5-ml fractions. The fast protein liquid chromatography (FPLC) gel filtration process was repeated 6 times, pooling each fraction obtained from replicate runs (3-ml total elution for each fraction) for subsequent Western blotting and M2-FLAG (Sigma) affinity purification. Proteins were eluted from beads using 3× FLAG peptide, and samples were prepared and analyzed using a Velos LTQ mass spectrometer (Thermo Fisher Scientific) as described above for CompPASS analysis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jennifer Waters, Talley Lambert, and Anna Jost of the Nikon Imaging Center (NIC) at Harvard Medical School for imaging support and technical assistance, Hunter Elliott and David Richmond of the Imaging and Data Analysis Core (IDAC) for data analysis support, Maria Ericsson and the HMS Electron Microscopy Facility for EM/immunogold support, Joseph Mancias for technical support with autophagosome isolation/proteomics and gel filtration analysis, Christian Munch for maintaining the Velos LTQ and running CompPASS samples on the instrument, Alban Ordureau for cloning of mtKeima into the pHAGE lentiviral expression backbone, and Takeda Oncology for the gift of ATG8 antibodies.
This work was supported by research grants to J.W.H. from the NIH (R37-NS083524 and RO1-GM095567) and an NIH grant (K01DK098285) to J.A.P. L.P.V. was supported by a Damon Runyon postdoctoral fellowship.
L.P.V. and J.W.H. conceived the study and prepared the manuscript. L.P.V. performed the experiments. J.A.P. provided proteomics support. E.L.H. provided proteomics data analysis support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00392-17.
REFERENCES
- 1.Yang Z, Klionsky DJ. 2010. Eaten alive: a history of macroautophagy. Nat Cell Biol 12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G, Marino G, Levine B. 2010. Autophagy and the integrated stress response. Mol Cell 40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ktistakis NT, Tooze SA. 2016. Digesting the expanding mechanisms of autophagy. Trends Cell Biol 26:624–635. doi: 10.1016/j.tcb.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Yoshimori T, Ohsumi Y. 2011. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. 1998. A protein conjugation system essential for autophagy. Nature 395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 6.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 7.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. 2014. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell 55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakoh-Nakatogawa M, Matoba K, Asai E, Kirisako H, Ishii J, Noda NN, Inagaki F, Nakatogawa H, Ohsumi Y. 2013. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol 20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- 10.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. 2009. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 11.Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N. 2016. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354:1036–1041. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 12.Behrends C, Sowa ME, Gygi SP, Harper JW. 2010. Network organization of the human autophagy system. Nature 466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakatogawa H, Ichimura Y, Ohsumi Y. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 14.Khaminets A, Behl C, Dikic I. 2016. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol 26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Rogov V, Dotsch V, Johansen T, Kirkin V. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Stolz A, Ernst A, Dikic I. 2014. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 17.von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein A, Bloor S, Rutherford TJ, Freund SM, Komander D, Randow F. 2012. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell 48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. 2010. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, Kirchof A, Gatti E, Helfrich MH, Wakatsuki S, Behrends C, Pierre P, Dikic I. 2015. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Sun HQ, Zhu X, Zhang L, Albanesi J, Levine B, Yin H. 2015. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc Natl Acad Sci U S A 112:7015–7020. doi: 10.1073/pnas.1507263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, Olsvik H, Overvatn A, Kirkin V, Johansen T. 2012. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem 287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joachim J, Jefferies HB, Razi M, Frith D, Snijders AP, Chakravarty P, Judith D, Tooze SA. 2015. Activation of ULK kinase and autophagy by GABARAP trafficking from the centrosome is regulated by WAC and GM130. Mol Cell 60:899–913. doi: 10.1016/j.molcel.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu F, Watanabe Y, Guo XY, Qi X, Wang P, Zhao HY, Wang Z, Fujioka Y, Zhang H, Ren JQ, Fang TC, Shen YX, Feng W, Hu JJ, Noda NN, Zhang H. 2015. Structural basis of the differential function of the two C. elegans Atg8 homologs, LGG-1 and LGG-2, in autophagy. Mol Cell 60:914–929. doi: 10.1016/j.molcel.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 24.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M. 2016. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol 215:857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaid S, Brandts CH, Serve H, Dikic I. 2013. Ubiquitination and selective autophagy. Cell Death Differ 20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itakura E, Mizushima N. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. 2015. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell 60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, Youle RJ, Dikic I. 2016. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A 113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thurston TL, Boyle KB, Allen M, Ravenhill BJ, Karpiyevich M, Bloor S, Kaul A, Noad J, Foeglein A, Matthews SA, Komander D, Bycroft M, Randow F. 2016. Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy. EMBO J 35:1779–1792. doi: 10.15252/embj.201694491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickrell AM, Youle RJ. 2015. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron 85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. 2010. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, Finkel T. 2015. Measuring in vivo mitophagy. Mol Cell 60:685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. 2014. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 37.Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. 2011. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol 18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 38.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. 2014. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szatmari Z, Sass M. 2014. The autophagic roles of Rab small GTPases and their upstream regulators: a review. Autophagy 10:1154–1166. doi: 10.4161/auto.29395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroder B, Wrocklage C, Pan C, Jager R, Kosters B, Schafer H, Elsasser HP, Mann M, Hasilik A. 2007. Integral and associated lysosomal membrane proteins. Traffic 8:1676–1686. doi: 10.1111/j.1600-0854.2007.00643.x. [DOI] [PubMed] [Google Scholar]
- 41.Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nordmann M, Cabrera M, Perz A, Brocker C, Ostrowicz C, Engelbrecht-Vandre S, Ungermann C. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol 20:1654–1659. doi: 10.1016/j.cub.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Cabrera M, Nordmann M, Perz A, Schmedt D, Gerondopoulos A, Barr F, Piehler J, Engelbrecht-Vandre S, Ungermann C. 2014. The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J Cell Sci 127:1043–1051. doi: 10.1242/jcs.140921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hegedus K, Takats S, Boda A, Jipa A, Nagy P, Varga K, Kovacs AL, Juhasz G. 2016. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol Biol Cell 27:3132–3142. doi: 10.1091/mbc.E16-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang CW, Stromhaug PE, Shima J, Klionsky DJ. 2002. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J Biol Chem 277:47917–47927. doi: 10.1074/jbc.M208191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meiling-Wesse K, Barth H, Thumm M. 2002. Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett 526:71–76. doi: 10.1016/S0014-5793(02)03119-8. [DOI] [PubMed] [Google Scholar]
- 47.Cabrera M, Ungermann C. 2013. Guanine nucleotide exchange factors (GEFs) have a critical but not exclusive role in organelle localization of Rab GTPases. J Biol Chem 288:28704–28712. doi: 10.1074/jbc.M113.488213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto G, Shimogori T, Hattori N, Nukina N. 2015. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet 24:4429–4442. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- 49.Kirkin V, Lamark T, Johansen T, Dikic I. 2009. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 5:732–733. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- 50.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. 2011. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell 44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 52.Menzies FM, Fleming A, Rubinsztein DC. 2015. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci 16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 53.Wong E, Bejarano E, Rakshit M, Lee K, Hanson HH, Zaarur N, Phillips GR, Sherman MY, Cuervo AM. 2012. Molecular determinants of selective clearance of protein inclusions by autophagy. Nat Commun 3:1240. doi: 10.1038/ncomms2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stolz A, Putyrski M, Kutle I, Huber J, Wang C, Major V, Sidhu SS, Youle RJ, Rogov VV, Dotsch V, Ernst A, Dikic I. 2017. Fluorescence-based ATG8 sensors monitor localization and function of LC3/GABARAP proteins. EMBO J 36:549–564. doi: 10.15252/embj.201695063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Miao G, Xue X, Guo X, Yuan C, Wang Z, Zhang G, Chen Y, Feng D, Hu J, Zhang H. 2016. The Vici syndrome protein EPG5 is a Rab7 effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol Cell 63:781–795. doi: 10.1016/j.molcel.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 56.Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, Pfuetzner RA, Brunger AT, Zhong Q. 2015. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520:563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Itakura E, Kishi-Itakura C, Mizushima N. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 58.Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. 2014. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell 25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. 2010. Identification of the switch in early-to-late endosome transition. Cell 141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 60.Kiontke S, Langemeyer L, Kuhlee A, Schuback S, Raunser S, Ungermann C, Kummel D. 2017. Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat Commun 8:14034. doi: 10.1038/ncomms14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. 2013. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. 2010. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci U S A 107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elias JE, Gygi SP. 2010. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol 604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elias JE, Gygi SP. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 67.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 69.Huang da W, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.