Abstract
Objective
To configure and validate a novel prostate disease nomogram providing prostate biopsy outcome probabilities from a prospective study correlating clinical indicators and diagnostic parameters among Filipino adult male with elevated serum total prostate specific antigen (PSA) level.
Methods
All men with an elevated serum total PSA underwent initial prostate biopsy at our institution from January 2011 to August 2014 were included. Clinical indicators, diagnostic parameters, which include PSA level and PSA-derivatives, were collected as predictive factors for biopsy outcome. Multiple logistic-regression analysis involving a backward elimination selection procedure was used to select independent predictors. A nomogram was developed to calculate the probability of the biopsy outcomes. External validation of the nomogram was performed using separate data set from another center for determination of sensitivity and specificity. A receiver-operating characteristic (ROC) curve was used to assess the accuracy in predicting differential biopsy outcome.
Results
Total of 552 patients was included. One hundred and ninety-one (34.6%) patients had benign prostatic hyperplasia, and 165 (29.9%) had chronic prostatitis. The remaining 196 (35.5%) patients had prostate adenocarcinoma. The significant independent variables used to predict biopsy outcome were age, family history of prostate cancer, prior antibiotic intake, PSA level, PSA-density, PSA-velocity, echogenic findings on ultrasound, and DRE status. The areas under the receiver-operating characteristic curve for prostate cancer using PSA alone and the nomogram were 0.688 and 0.804, respectively.
Conclusion
The nomogram configured based on routinely available clinical parameters, provides high predictive accuracy with good performance characteristics in predicting the prostate biopsy outcome such as presence of prostate cancer, high Gleason prostate cancer, benign prostatic hyperplasia, and chronic prostatitis.
Keywords: Nomogram, Prostate cancer, Benign prostatic hyperplasia, Prostatitis
1. Introduction
Recently, controversy has arisen regarding whether early detection of prostate cancer through prostate specific antigen (PSA) screening is actually beneficial or not [1]. This is so because it was known that most patients with indolent cancer may not die from it within 15 years [2]. Clinicians started to realize that the drawbacks of screening for prostate cancer with PSA alone may lead to overdetection and consequently may expose men to further intensive diagnostic screening and invasive management strategies affecting quality of life [3]. Thus, there is realization that PSA alone is not sufficient enough to justify the appropriateness of a full evaluation for prostate cancer (i.e., biopsy). There are several other clinical indicators, identified risk factors, and PSA derivatives that help increase differentiation between malignant and benign prostatic conditions, such as age, family history, race, body mass index (BMI), prior prostatitis, medications, abnormal digital rectal examination, heterogenic echo lesion on transrectal ultrasound, age-specific PSA, PSA density (PSAD), PSA velocity, and free PSA percentage [4], [5].
With the advent of evidence-based medicine, bias-free prediction models such as nomogram has started to emerge in aiding clinical decision making. A nomogram is able to quantify probability of the event of interest by multivariate analysis of combined contribution of identified risk factors and clinical parameters [6]. Currently, several existing models were developed to predict positive prostate biopsy among men undergoing evaluation for prostate cancer [7]; however, these models were only able to provide prostate cancer probability, and cannot differentiate probability for clinically significant prostate cancer versus differential benign conditions. Hence, the objective of this study is to configure and validate a novel prostate disease nomogram providing prostate biopsy outcome probabilities from a prospective study correlating clinical indicators among Filipino adult male with elevated PSA.
2. Material and methods
2.1. Data source
This is a cross-sectional study prospectively collected data from all patients who had their first transrectal ultrasound (TRUS) prostate biopsy at a tertiary medical center from January 2011–August 2014. The protocol of this study was reviewed and approved by the Institutional Scientific Review Board (ISRB), Institutional Ethics Review Board (IERB), and registered at www.Clinicaltrial.gov (Identifier: NCT01826617).
Two datasets were collected uniformly for the purpose of building a clinical care prostate biopsy database in two separate institutional prostate centers. The data acquired from the main center was used to generate the nomogram, while dataset from the other center with similar biopsy protocol was used as external validation data source. Included data for analysis for the purpose of nomogram development were Filipino patients, who have PSA-based indications for prostate biopsy (elevated serum total PSA level >4.0 ng/mL) and gave consent for inclusion of their data into the data bank. Dataset excluded were information from non-Filipino patients, incomplete data due to patients' refusal to provide required information, patients taking Finasteride or Dutasteride, PSA obtained outside the involved institutions, past history of prior biopsy, PSA greater than 40 ng/mL, equivocal biopsy results with no confirmatory immunohistopathologic staining (includes atypical small acinar proliferation and high grade prostatic intraepithelial neoplasia) or non-adenocarcinoma that Gleason score is not applicable (i.e., lymphoma).
2.2. Clinical information
The clinical information gathered includes: (a) Identified risk factors (age, family history, race, BMI, prior prostatitis), (b) Clinical indicators of prostatic diseases (abnormal digital rectal examination, heterogenic echoic lesion on transrectal ultrasound), and (c) PSA (in ng/mL) and its derivatives (age-specific PSA classification was based from a previous race specific determination study [8], PSAD, PSA velocity). Basic demographic data such as patient's height and weight were measured using standard weighing scale (Detecto 439 Mechanical Scale with Height rod, Webb City, MO, USA). A urologist member of the team performed digital rectal exam (DRE) on all patients before or after the TRUS. Serum total PSA level was measured at the Institute of Pathology using “Siemens ADVIA Centaur Free PSA and PSA” (Seimens Healthcare Diagnostics, Sacramento, CA, USA) within 1 month before the prostate biopsy. Prior to prostate biopsy, the prostate was scanned using a biplanar 7.5 MHz probe (GE Medical Systems Kretz Ultrasound, Zipf, Austria). Prostate volume was measured by a radiology technician (member of the staff team) of the center using the transverse and sagittal planes with the standard equation of measurement for ellipsoid [width (w) × height (h) × length (l) × 0.523].
All prostate biopsies were uniformly-required 12 cores (extended scheme) or more prostate tissue strips under ultrasound guidance with Fr 18 25 cm biopsy device (Bard Urological, Covington, GA, USA). The technique was performed systematically to cover lateral and medial aspects of the apex, midgland, and base of the right and left prostate lobes. Two additional biopsies were obtained from suspected areas seen on ultrasound.
All acquired specimens were placed in a formalin-filled container and sent for histopathologic examination. They were all examined by at least two board-certified pathologists at the Institute of Pathology to determine the presence of inflammation (chronic prostatitis), other disease entity, or carcinoma (if positive for carcinoma, reading include grade using Gleason score, cancer length in biopsy specimen, percentage of cancer involvement). All the pathologists were blinded from the clinical indicators of the patients. In the event of inconclusive results, specimens were further subjected to immunohistostaining for a definitive conclusion. At least two pathologists were required to release the final report. Data collection and extraction used a pre-tested and standardized form. All collected forms were submitted to a third party clinical information management center for encoding and preliminary analysis of incidence and prevalence. Additional analyses were sent to a third party statistician for validation and reliability check.
2.3. Statistical analysis
Mean and SD were calculated for age, BMI, prostate volume, total PSA, PSA velocity and PSAD, count and percentage were calculated for abnormal age-specific PSA, family history, hypertension, diabetes, smoking, alcoholic beverage drinking, prior antibiotic intake for prostatitis, DRE findings, and TRUS echogenicity. Both univariate and multivariate logistic regression analysis were used to examine the association between predictive variables and biopsy outcomes (prostate cancer, high Gleason prostate cancer, chronic prostatitis, and BPH). Adjusted odds ratios and 95% confidence intervals (CI) were calculated. Using a backward model-selection procedure, the final model for prediction was conducted. Beta-coefficients of significant variables and slope for equation model were determined. Based on the final model, the estimated probability for each differential biopsy outcome were calculated, a nomogram was then developed into an electronic tool. In brief, the development method was that the regression coefficients representing the strengths of correlation were proportionately transferred to equation function in Excel program; upon which they can be linked to corresponding points and summed to a total estimated probability of a biopsy outcome. Receiver-operating characteristic (ROC) curves were used to assess the ability to discriminate as well as evaluate the corresponding accuracy by the different models (PSA level only vs. nomogram). The models were also externally validated using the validation set collected from another prostate center as described earlier. SPSS 17.0.1 for Windows (SPSS Inc., Chicago, IL, USA) and Microsoft Office Excel 2007 (Microsoft, Redmond, WA, USA) were used for all statistical analyses. All statistics considered significant were at the level of ≤0.05.
3. Results
A total of 552 patients were referred to our institution for initial prostate biopsy. Among the included eligible patients, 191 (34.6%) patients had benign prostatic hyperplasia (BPH) and 165 (29.9%) had prostatitis, particularly chronic type. The remaining 196 patients (35.5%) had prostate adenocarcinoma. Number of patients according to Gleason scoring classification of the prostate cancer ≥8, 7 and 6, were 82 (41.8%), 66 (33.7%), and 48 (24.5%), respectively.
The summary of patient characteristics, clinical parameters and PSA derivatives were described in Table 1. ANOVA test revealed a significant statistical difference in age, BMI, PSA level, PSAD, and PSA velocity between different group histopathologic outcomes. Prostate gland size was the only variable with no noted significant difference. With the Chi-square analysis of categorical parameters, a significant difference was noted on the variables of abnormal age-specific PSA, positive family history of prostate cancer, abnormal digital rectal exam, and heterogenic echogeneticity noted in TRUS.
Table 1.
Parameters and comparison between prostate biopsy histopathologic results.
| Characteristics | Overall patient's summary (n = 552) | High risk PCa (n = 82) | Intermediate risk PCa (n = 66) | Low risk PCa (n = 48) | BPH (n = 191) | Prostatitis (n = 165) | p-Value |
|---|---|---|---|---|---|---|---|
| Age (year) a | 63.20 ± 8.23 | 68.0 ± 9.30 | 67.0 ± 7.72 | 62.0 ± 6.84 | 60.7 ± 7.06 | 62.0 ± 7.63 | <0.001 |
| BMI a | 25.80 ± 4.06 | 24.6 ± 4.57 | 25.8 ± 3.37 | 26.2 ± 3.96 | 26.2 ± 4.18 | 26.0 ± 4.31 | 0.038 |
| PSA levels (ng/mL) a | 9.38 ± 5.62 | 13.9 ± 6.75 | 11.4 ± 6.22 | 9.2 ± 4.30 | 7.5 ± 4.60 | 8.0 ± 4.43 | <0.001 |
| Prostate gland size by TRUS (g) a | 44.93 ± 18.60 | 44.3 ± 19.13 | 40.2 ± 17.56 | 44.8 ± 13.94 | 45.6 ± 18.02 | 45.8 ± 20.23 | 0.137 |
| PSAD a | 0.24 ± 0.18 | 0.4 ± 0.21 | 0.3 ± 0.22 | 0.2 ± 0.14 | 0.2 ± 0.15 | 0.2 ± 0.16 | <0.001 |
| PSA velocity a | 1.43 ± 1.32 | 2.8 ± 1.01 | 1.8 ± 0.88 | 1.4 ± 0.90 | 0.8 ± 0.70 | 1.3 ± 1.79 | <0.001 |
| Abnormal age specific PSA b | 379 (69) | 78 (95) | 54 (82) | 39 (81) | 100 (52) | 142 (86) | <0.001 |
| With family history of PCa b | 189 (34) | 69 (84) | 25 (38) | 14 (29) | 50 (26) | 53 (32) | <0.001 |
| With hypertension b | 282 (52) | 40 (49) | 32 (48) | 27 (56) | 95 (50) | 104 (63) | 0.830 |
| With diabetes mellitus b | 117 (21) | 16 (20) | 18 (27) | 12 (25) | 39 (20) | 50 (30) | 0.522 |
| Smoker b | 111 (20) | 15 (18) | 14 (21) | 9 (19) | 36 (19) | 48 (29) | 0.958 |
| Alcoholic beverage drinker b | 135 (25) | 18 (22) | 15 (23) | 11 (23) | 58 (30) | 45 (27) | 0.877 |
| With prior antibiotic drinker b | 133 (24) | 19 (23) | 13 (20) | 17 (35) | 61 (32) | 30 (18) | 0.246 |
| Abnormal digital rectal exam b | 103 (19) | 57 (70) | 18 (27) | 6 (13) | 13 (7) | 16 (10) | <0.001 |
| Heterogenic echogenecity on TRUS b | 151 (27) | 33 (40) | 11 (17) | 13 (27) | 24 (13) | 104 (63) | 0.013 |
Notes: aScale variables, values presented as mean ± SD; bCategorical variables, values presented as n(%).
PCa, prostate cancer; BMI, body mass index; PSA, prostate specific antigen; TRUS, transrectal ultrasound; PSAD, PSA density; BPH, benign prostatic hyperplasia.
The multivariate logistic regression analysis determined the variables included for equation in predicting probability of prostate cancer, high Gleason score (≥7) prostate cancer, BPH and chronic prostatitis were described in Table 2, Table 3, Table 4, Table 5, respectively. Most significant variables in predicting both prostate cancer and high Gleason prostate cancer were positive family history, abnormal DRE, and elevated PSA density. Significant predictors for BPH and chronic prostatitis were prior antibiotic intake for prostatitis, alcoholic beverage drinker, smoker, and heterogenous echoes in TRUS.
Table 2.
Multivariate analysis of factors associated with prostate cancer.
| Variables | Beta-coefficients | SE | Odds ratio | 95% CI |
p-Value | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Age (year) | 0.05 | 0.014 | 1.051 | 1.022 | 1.081 | <0.001 |
| Positive family history | 0.937 | 0.224 | 2.553 | 1.647 | 3.957 | <0.001 |
| PSA level (ng/mL) | 0.049 | 0.027 | 1.051 | 1.000 | 1.108 | 0.041 |
| PSA velocity (ng/mL/year) | 0.391 | 0.094 | 1.478 | 1.231 | 1.776 | <0.001 |
| Abnormal digital rectal exam | 1.556 | 0.304 | 4.739 | 2.612 | 8.597 | <0.001 |
| PSAD (ng/mL/g) | 1.557 | 0.786 | 4.746 | 1.016 | 22.159 | 0.048 |
| Hyperecho in TRUS | −0.578 | 0.26 | 0.561 | 0.337 | 0.934 | 0.026 |
| Constant (Slope) | −5.497 | 0.906 | ||||
PSA, prostate specific antigen; PSAD, PSA density; TRUS, transrectal ultrasound; CI, confidence interval.
Table 3.
Multivariate analysis of factors associated with high Gleason score (7 and ≥8).
| Variables | Beta-coefficients | SE | Odds ratio | 95% CI |
p-Value | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Age (year) | 0.083 | 0.017 | 1.086 | 1.051 | 1.122 | <0.001 |
| Positive family history | 1.578 | 0.268 | 4.844 | 2.864 | 8.194 | <0.001 |
| Abnormal age specific PSA level | 1.084 | 0.362 | 2.955 | 1.452 | 6.013 | 0.003 |
| PSA velocity (ng/mL/year) | 0.327 | 0.093 | 1.387 | 1.155 | 1.666 | <0.001 |
| Abnormal digital rectal exam | 2.212 | 0.324 | 9.133 | 4.843 | 17.223 | <0.001 |
| PSAD (ng/mL/g) | 2.179 | 0.737 | 8.838 | 2.087 | 37.436 | 0.003 |
| Hyperecho in TRUS | −0.722 | 0.327 | 0.486 | 0.256 | 0.922 | 0.027 |
| Constant (Slope) | −9.260 | 1.178 | ||||
PSA, prostate specific antigen; PSAD, PSA density; TRUS, transrectal ultrasound; CI, confidence interval.
Table 4.
Multivariate analysis of factors associated with BPH on TRUS biopsy.
| Variables | Beta-coefficients | SE | Odds ratio | 95% CI |
p-Value | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Age (year) | −0.059 | 0.016 | 0.943 | 0.913 | 0.974 | <0.001 |
| Smoker | −0.720 | 0.325 | 0.487 | 0.258 | 0.920 | 0.027 |
| Alcoholic beverage drinker | 0.836 | 0.296 | 2.307 | 1.290 | 4.124 | 0.005 |
| Prior antibiotic intake | 0.693 | 0.252 | 2.00 | 1.221 | 3.274 | 0.006 |
| PSA level (ng/mL) | 0.072 | 0.036 | 1.075 | 1.001 | 1.153 | 0.046 |
| Abnormal age specific PSA level | −1.088 | 0.288 | 0.337 | 0.192 | 0.593 | <0.001 |
| PSA velocity (ng/mL/year) | −1.266 | 0.21 | 0.282 | 0.187 | 0.425 | <0.001 |
| PSAD (ng/mL/g) | −1.63 | 0.931 | 0.196 | 0.032 | 0.999 | 0.05 |
| Hyperecho echo in TRUS | −1.47 | 0.277 | 0.230 | 0.134 | 0.396 | <0.001 |
| Constant (Slope) | 4.919 | 1.019 | ||||
PSA, prostate specific antigen; PSAD, PSA density; TRUS, transrectal ultrasound; CI, confidence interval; PBH, benign prostatic hyperplasia.
Table 5.
Multivariate analysis of factors associated with chronic prostatitis on TRUS biopsy.
| Variables | Beta-coefficients | SE | Odds ratio | 95% CI |
p-Value | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| With hypertension | 0.535 | 0.268 | 1.708 | 1.010 | 2.888 | 0.046 |
| Smoker | 1.307 | 0.394 | 3.695 | 1.707 | 7.995 | 0.001 |
| Alcoholic beverage drinker | −1.002 | 0.381 | 0.367 | 0.174 | 0.774 | 0.008 |
| Prior antibiotic intake | 1.012 | 0.327 | 3.630 | 1.912 | 6.890 | 0.002 |
| PSA level (ng/mL) | −0.069 | 0.041 | 0.934 | 0.862 | 1.011 | 0.093 |
| Abnormal age specific PSA level | 0.880 | 0.331 | 2.412 | 1.262 | 4.610 | 0.008 |
| PSA velocity (ng/mL/year) | 0.636 | 0.217 | 1.888 | 1.235 | 2.887 | 0.003 |
| Hyperecho in TRUS | 2.061 | 0.319 | 7.854 | 4.199 | 14.689 | <0.001 |
| Constant (Slope) | −1.272 | 0.348 | ||||
PSA, prostate specific antigen; TRUS, transrectal ultrasound; CI, confidence interval.
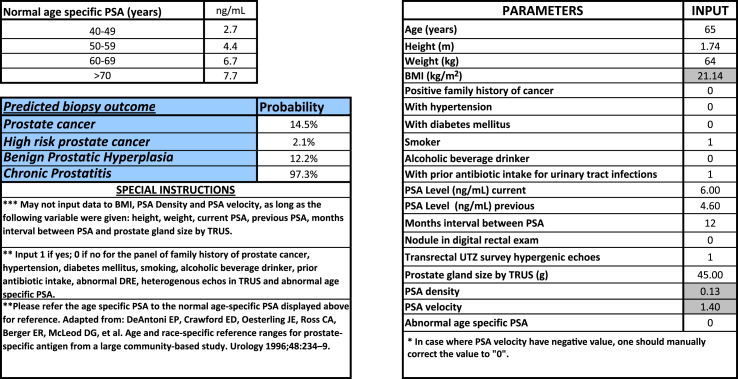
An electronic version of nomogram was constructed based on the determined significant associated variables and beta-coefficient values derived from multivariate logistic regression (Fig. 1). The parameters were requested in this electronic calculator, and corresponding probabilities of the biopsy outcome will be revealed.
Figure 1.
Electronic nomogram calculator for predicting prostate biopsy outcome.
The nomogram accuracies in predicting the described conditions were evaluated by comparing the predicted and actual probabilities of conditions in the validation set. The validation set consists of a total of 66 patients from another center. Table 6 describes the comparability of the group characteristics and proportion of biopsy outcome conditions with the main group population.
Table 6.
Patient characteristics between the main group population and validation set population.
| Overall patient's summary (n = 552) | External validation patient's summary (n = 66) | p-Value | |
|---|---|---|---|
| Agea | 63.20 ± 8.23 | 64.32 ± 7.84 | 0.14 |
| BMIa | 25.80 ± 4.06 | 25.98 ± 4.83 | 0.39 |
| PSA levels (ng/mL)a | 9.38 ± 5.62 | 9.64 ± 6.12 | 0.74 |
| Prostate gland size by TRUS (g)a | 44.93 ± 18.60 | 52.75 ± 20.86 | <0.001 |
| PSADa | 0.24 ± 0.18 | 0.20 ± 0.17 | 0.96 |
| PSA velocitya | 1.43 ± 1.32 | 1.55 ± 1.48 | 0.53 |
| Abnormal age specific PSAb | 379 (69) | 44 (67) | |
| With family history of PCab | 189 (34) | 20 (30) | |
| With hypertensionb | 282 (52) | 26 (39) | |
| With diabetes mellitusb | 117 (21) | 12 (18) | |
| Smokerb | 111 (20) | 16 (24) | |
| Alcoholic beverage drinkerb | 135 (25) | 19 (29) | |
| With prior antibiotic drinkerb | 133 (24) | 12 (18) | |
| Abnormal digital rectal examb | 103 (19) | 12 (18) | |
| Heterogenic echogenecity on TRUSb | 151 (27) | 11 (17) | |
| PCab | 196 (36) | 22 (33) | |
| High GS PCab | 82 (15) | 10 (15) | |
| BPHb | 191 (35) | 30 (45) | |
| Chronic prostatitisb | 165 (30) | 25 (38) |
Notes: aScale varibles, values presented as mean ± SD; bCategorical variables, values presented as n (%).
PSA, prostate specific antigen; PSAD, PSA density; TRUS, transrectal ultrasound; PCa, prostate cancer; GS,Gleason score; BPH, benign prostatic hyperplasia.
The cut-off values set for each biopsy outcome and their corresponding sensitivity, specificity, and positive and negative likelihood ratios were described in Table 7. The recommended threshold for prostate biopsy in predicted prostate cancer probability is 46% with 73% sensitivity and 80% specificity; while the threshold for biopsy in predicted high Gleason cancer probability is 28% with 90% sensitivity and 77% specificity.
Table 7.
The cut-off values set for predicting each biopsy outcome and their corresponding sensitivity, specificity, positive and negative likelihood ratios.
| Prostate biopsy outcome | Novel nomogram cut-off value (%) | Sensitivity (%) | Specificity (%) | Positive likelihood ratio | Negative likelihood ratio |
|---|---|---|---|---|---|
| PCa | 46 | 73.0 | 80.0 | 3.6500 | 0.3375 |
| High risk PCa | 28 | 90.0 | 77.0 | 3.9130 | 0.1299 |
| Chronic prostatitis | 38 | 76.0 | 50.0 | 1.5200 | 0.4800 |
| BPH | 38 | 83.0 | 86.0 | 5.9200 | 0.1970 |
PCa, prostate cancer; BPH, benign prostatic hyperplasia.
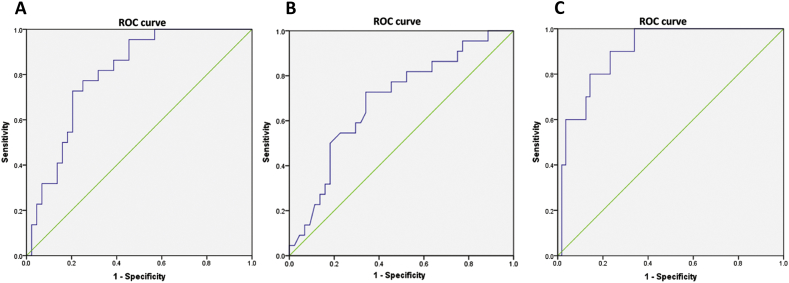
As described in Table 8, the accuracy of diagnosing prostate cancer by the developed nomogram-based cut-off was better than solely relying on PSA level either at the internationally accepted cut-off of 4 ng/mL or the locally determined cut-off for Filipino populations at 6.7 ng/mL. Likewise, the ROC determined area under the curve (AUC) for the nomogram in diagnosing prostate cancer was high at 0.804 which is better than the PSA level based AUC of 0.688 (Fig. 2A and B). The novel nomogram was even better in diagnosing high Gleason prostate cancer with the AUC of 0.902 (Fig. 2C).
Table 8.
The accuracy comparison of parameters used in diagnosing prostate cancer.
| Parameter | Sensitivity (%) | Specificity (%) | Positive likelihood ratio | Negative likelihood ratio |
|---|---|---|---|---|
| PSA level 4.0 ng/mL | 96.3 | 10.4 | 1.0748 | 0.3558 |
| PSA level 6.7 ng/mL | 80.0 | 54.2 | 1.7000 | 0.4200 |
| Novel nomogram cut-off at 46% | 73.0 | 80.0 | 3.6500 | 0.3380 |
PSA, prostate specific antigen.
Figure 2.
Receiver operating characteristic curves for the nomogram and PSA level (ng/mL) in predicting prostate cancer and for the nomogram in predicting high Gleason prostate cancer. (A): Area under the curve (AUC), 0.804; SE = 0.53; p-value < 0.001; 95% confidence interval (CI), 0.70-0.908. (B): AUC, 0.688; SE = 0.069; p-value = 0.013; 95% CI, 0.55-0.823. (C): AUC, 0.902; SE = 0.43; p-value < 0.001; 95% CI, 0.818-0.986.
4. Discussion
For the past few decades, widespread use of PSA screening has increased early detection of prostate cancer, but particularly that of the indolent prostate cancer (Gleason score <7 or cancer volume <5 mL) [9]. Issues have emerged regarding harms brought about by PSA-indicated prostate biopsy because since increased serum PSA level is not specific for prostate cancer. Because of low specificity of the PSA level in the detection of prostate cancer, patients suspected of having this disease using PSA screening usually receive an unnecessary biopsy which is an invasive procedure with accompanying potential complications [10].
It is apparent that the elevation of PSA can be caused by various non-malignant conditions of the prostate such as BPH and prostatitis [11]. Discriminating prostate cancer from benign condition, particularly in the intermediate PSA range of 4–10 ng/mL is difficult [12]. In our dataset, among patients with established PSA-based indications for biopsy (such as elevated PSA or abnormal age-specific PSA level), only 35.5% actually have prostate cancer and 65% have benign conditions (BPH and chronic prostatitis). In fact, only 15% of the patients were with high Gleason Score prostate cancer that will benefit from further management. Therefore, there is a pressing need to pursue an optimal approach in reducing prostate cancer over-diagnosis and over-treatment as well as lessening their associated morbidities.
The traditional cut-off for an abnormal PSA level in the major screening studies has been 4.0 ng/mL though the sensitivity has been reported as being between 67% and 80% with a low specificity of only 20%–30% [13], [14]. When the PSA level of 4 ng/mL was used in our validation dataset, the sensitivity was 96%, however with a severely low specificity of 10.4%. There were several previous local studies that also suggest increasing PSA level cut-off between 6.8 and 8.0 ng/mL specified for Filipino population. However, due to their small sample size and inadequate method of statistical interpolation, increasing the PSA level threshold was not widely adapted [15], [16]. With the available dataset at hand, this study was able to generate a better PSA level cut-off specific for Filipino population from the ROC derived new cut-off value of 6.7 ng/mL, which compared to traditional cut-off of 4 ng/mL, has better specificity of 54% while not compromising the sensitivity at 80%.
Several attempts were made in applying PSA derivatives and PSA composites in increasing the accuracy of prostate cancer detection. Specifically, the percent free PSA was being mentioned to have highest accuracy (sensitivity 70.8% and specificity 67.4%) [14]. However, in the local setting, percent free PSA determination was considered expensive given that both free PSA and total PSA must be requested at one instance to determine the percent free PSA. In this study, it is not included as an additional parameter due to its low utilitization among urologist, leading to insufficient data for inference. In the development of this nomogram, the readily available PSA derivatives of PSA density and PSA velocity were included instead of percent free PSA. Studies have shown that PSA density and PSA velocity when used in combination improves the sensitivity to 85.8% and specificity 45.3%. In the multivariate analysis both parameters significantly contributes in the predictive equation for both prostate cancer and high Gleason score prostate cancer.
A reliable nomogram that can accurately predict and differentiate the presence of aggressive prostate cancer from indolent or benign conditions would be useful for urologist and patients to make decisions. In the current dataset for nomogram development, multivariate analysis also depicted that presence of family history is a predominant variable predicting biopsy outcome of prostate cancer and high Gleason score prostate cancer. This finding was consistent to a recent study by Saarimäki et al. [17], who postulated that positive family history is particularly a strong indicator for presence or development of high Gleason prostate cancer among Finnish males who underwent screening [16].
This study was the first study that was able to predict and differentiate prostate biopsy outcome into high Gleason score prostate cancer and differential benign conditions such as BPH and chronic prostatitis. Likewise, this is the first to take into account several established risk factors, clinical parameters and PSA derivatives into the modeling of a nomogram. In this study, the adequacy of sample size of 552 and the ideal proportion of the prostate biopsy outcome into 1∶1∶1 (prostate cancer∶BPH∶chronic prostatitis) was suitable for developing a bias-free nomogram. The concordance of TRUS prostate biopsy histopathology and final prostatectomy histopathology was also well studied in our institution. According to a recent study by Bascuna et al. [18], an 80% concordance rate was established, which makes the biopsy reading in our institution an excellent standard as surrogate for prostatectomy histopathologic outcome.
The main advantage of this electronic nomogram tool is that clinicians can assess risk of prostate cancer versus benign conditions in an individual. On the basis of this differentiation, evaluation and management decisions will be facilitated. For instance, if the nomogram predicts a low chance for having aggressive prostate cancer for an older patient with a high PSA level, and points to a chronic prostatitis (Fig. 1), it would be reasonable for the patient to forego a biopsy. The exact probability cut-off for undergoing or foregoing a biopsy would be left with the treating physician and patient. This shared decision-making should be individualized. Although a list of sensitivities and specificities has been provided based on various probability cut-offs for each biopsy outcome, as illustrated in Fig. 2, the AUC 0.804 and 0.902 of this nomogram in diagnosing prostate cancer and high Gleason prostate cancer is way better than utilizing the PSA level alone (either the standard 4.0 ng/mL or 6.7 ng/mL specific for Filipino population); likewise, it has better accuracy than other Asian-based nomogram with AUC ranging from 0.72 to 0.88 [19], [20], [21], [22], [23], [24].
This study reflected a center-based screening population. Nonetheless, it was able to describe an external validation for its accuracy, albeit limited to only one other center. It is imperative for other centers to confirm and validate the present findings. A multi-institutional study is strongly recommended. Still, the instrument that was formulated in the study contributed important information for urologists and patients in assessing an individual's risk for prostate cancer and provided aid on how to further proceed after such an assessment. The tool is made available to the general public for free.
5. Conclusion
This study configured and validated a novel nomogram intended for Filipino males with elevated serum total PSA level. The nomogram based on routinely available clinical parameters and PSA derivatives provides high predictive accuracy with good performance characteristics in predicting the prostate biopsy outcome such as presence of prostate cancer, high Gleason score prostate cancer, BPH, and chronic prostatitis.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgment
The study was sponsored by the Institution where the study was conducted; likewise, it was also awarded with research grant from the Philippine Urological Association, Inc.
Footnotes
Peer review under responsibility of Chinese Urological Association and SMMU.
References
- 1.Barry M.J. Screening for prostate cancer — the controversy that refuses to die. N Engl J Med. 2009;360:1351–1354. doi: 10.1056/NEJMe0901166. [DOI] [PubMed] [Google Scholar]
- 2.Bangma C.H., Roemeling S., Schröder F.H. Overdiagnosis and overtreatment of early detected prostate cancer. World J Urol. 2007;25:3–9. doi: 10.1007/s00345-007-0145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen C., Kattan M.W. Formalized prediction of clinically significant prostate cancer: is it possible? Asian J Androl. 2012;14:349–354. doi: 10.1038/aja.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abouassaly R., Thompson I.M., Plats E.A., Klein E.A. Epidemiology, etiology, and prevention of prostate cancer. In: Wein A.J., editor. 10th ed. vol. 3. Saunders; Philadelphia: 2012. pp. 2704–2725. (Campbell-Walsh urology). [Google Scholar]
- 5.Kamoi K., Babaian R.J. Advances in the application of prostate specific antigen in the detection of early-stage prostate cancer. Semin Oncol. 1999;26:140–149. [PubMed] [Google Scholar]
- 6.Chun F.K.H., Karakiewicz P.I., Briganti A., Walz J., Kattan M.W., Huland H. A critical appraisal of logistic regression-based nomogram, artificial neural networks, classifications and regression-tree models, look-up tables and risk-group stratification models for prostate cancer. BJU Int. 2007;99:794–800. doi: 10.1111/j.1464-410X.2006.06694.x. [DOI] [PubMed] [Google Scholar]
- 7.Karakiewicz P.I., Benayoun S., Kattan M.W., Perrotte P., Valiquette L., Scardino P.T. Development and validation of a nomogram predicting the outcome of prostate biopsy based on patient age, digital rectal examination and serum prostate specific antigen. J Urol. 2011;173:1930–1934. doi: 10.1097/01.ju.0000158039.94467.5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeAntoni E.P., Crawford E.D., Oesterling J.E., Ross C.A., Berger E.R., McLeod D.G. Age and race-specific reference ranges for prostate-specific antigen from a large community-based study. Urology. 1996;48:234–239. doi: 10.1016/s0090-4295(96)00091-x. [DOI] [PubMed] [Google Scholar]
- 9.Welch H.G., Albertsen P.C. Prostate cancer diagnosis and treatment after the introduction of prostate specific antigen screening: 1986–2005. J Natl Cancer Inst. 2010;101:1325–1329. doi: 10.1093/jnci/djp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf A.M., Wender R.C., Etzioni R.B., Thompson I.M., D'Amico A.V., Volk R.J. American Cancer Society guideline for the early detection of prostate cancer: update 2010. CA Cancer J Clin. 2010;60:70–98. doi: 10.3322/caac.20066. [DOI] [PubMed] [Google Scholar]
- 11.Loeb S., Carter H.B. Early detection, diagnosis, and staging of prostate cancer. In: Wein A.J., editor. 10th ed. vol. 3. Saunders; Philadelphia: 2012. p. 2764. (Campbell-Walsh urology). [Google Scholar]
- 12.Catalona W.J., Partin A.W., Slawin K.M., Brawer M.K., Flanigan R.C., Patel A. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: a prospective multicenter clinical trial. JAMA. 1998;27:1542–1547. doi: 10.1001/jama.279.19.1542. [DOI] [PubMed] [Google Scholar]
- 13.Shariat S.F., Scardino P.T., Lilja H. Screening for prostate cancer: an update. Can J Urol. 2008;15:4363–4374. [PMC free article] [PubMed] [Google Scholar]
- 14.Murray N.P., Reyes E., Orellana N., Fuentealba C., Dueñas R. A comparative performance analysis of total PSA, percentage free PSA, PSA velocity, and PSA density versus the detection of primary circulating prostate cells in predicting initial prostate biopsy findings in Chilean men. Biomed Res Int. 2014;2014:676572. doi: 10.1155/2014/676572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitero V.G., Bolong D.T., Letran J.L. Systematic 12-core biopsy protocol versis 6-core biopsy protocol: the value of six additional laterally directed cores in improving prostate cancer detection. Philipp J Urol. 2002;12:1–5. [Google Scholar]
- 16.Petero V.G., Letran J.L. Prostate specific antigen density of the transition zone enhances specificity of prostate specific antigen in the detection of prostate cancer in patients with intermediate PSA levels. Philipp J Urol. 2003;13:15–19. [Google Scholar]
- 17.Saarimäki L., Tammela T.L., Määttänen L., Taari K., Kujala P.M., Raitanen J. Family history in the finnish prostate cancer screening trial. Int J Cancer. 2015;136:2172–2177. doi: 10.1002/ijc.29243. [DOI] [PubMed] [Google Scholar]
- 18.Bascuna R., Luna S.L., Serrano D.P., Morales M.L. Concordance rate of Gleason score between transrectal ultrasound-guided needle biopsy and radical prostatectomy specimens among Filipino men with prostate cancer done in a Tertiary hospital from January 2008–September 2012. St Luke’s J Med. 2015 [in press]. Abstract presented in USANZ 2013 in Australia. [Google Scholar]
- 19.Ahn J.H., Lee J.Z., Chung M.K., Ha H.K. Nomogram for prediction of prostate cancer with serum prostate specific antigen less than 10 ng/mL. J Korean Med Sci. 2014;29:338–342. doi: 10.3346/jkms.2014.29.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakami S., Numao N., Okubo Y., Koga F., Yamamoto S., Saito K. Development, validation, and head-to-head comparison of logistic regression-based nomograms and artificial neural network models predicting prostate cancer on initial extended biopsy. Eur Urol. 2008;54:601–611. doi: 10.1016/j.eururo.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Jeong I.G., Lim J.H., Hwang S.S., Kim S.C., You D., Hong J.H. Nomogram using transrectal ultrasound-derived information predicting the detection of high grade prostate cancer on initial biopsy. Prostate Int. 2013;1:69–75. doi: 10.12954/PI.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J.Y., Yoon S., Park M.S., Cho D.Y., Park H.S., Moon du G. Initial biopsy outcome prediction in Korean patients-comparison of a noble web-based Korean prostate cancer risk calculator versus prostate-specific antigen testing. J Korean Med Sci. 2011;26:85–91. doi: 10.3346/jkms.2011.26.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang P., Chen H., Uhlman M., Lin Y.R., Deng X.R., Wang B. A nomogram based on age, prostatespecific antigen level, prostate volume and digital rectal examination for predicting risk of prostate cancer. Asian J Androl. 2013;15:129–133. doi: 10.1038/aja.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo S.C., Hung S.H., Wang H.Y., Chien C.C., Lu C.L., Lin H.J. Chinese nomogram to predict probability of positive initial prostate biopsy: a study in Taiwan region. Asian J Androl. 2013;15:780–784. doi: 10.1038/aja.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]