ABSTRACT
Several subtypes of avian influenza viruses (AIVs) are emerging as novel human pathogens, and the frequency of related infections has increased in recent years. Although neuraminidase (NA) inhibitors (NAIs) are the only class of antiviral drugs available for therapeutic intervention for AIV-infected patients, studies on NAI resistance among AIVs have been limited, and markers of resistance are poorly understood. Previously, we identified unique NAI resistance substitutions in AIVs of the N3, N7, and N9 NA subtypes. Here, we report profiles of NA substitutions that confer NAI resistance in AIVs of the N4, N5, N6, and N8 NA subtypes using gene-fragmented random mutagenesis. We generated libraries of mutant influenza viruses using reverse genetics (RG) and selected resistant variants in the presence of the NAIs oseltamivir carboxylate and zanamivir in MDCK cells. In addition, two substitutions, H274Y and R292K (N2 numbering), were introduced into each NA gene for comparison. We identified 37 amino acid substitutions within the NA gene, 16 of which (4 in N4, 4 in N5, 4 in N6, and 4 in N8) conferred resistance to NAIs (oseltamivir carboxylate, zanamivir, or peramivir) as determined using a fluorescence-based NA inhibition assay. Substitutions conferring NAI resistance were mainly categorized as either novel NA subtype specific (G/N147V/I, A246V, and I427L) or previously reported in other subtypes (E119A/D/V, Q136K, E276D, R292K, and R371K). Our results demonstrate that each NA subtype possesses unique NAI resistance markers, and knowledge of these substitutions in AIVs is important in facilitating antiviral susceptibility monitoring of NAI resistance in AIVs.
IMPORTANCE The frequency of human infections with avian influenza viruses (AIVs) has increased in recent years. Despite the availability of vaccines, neuraminidase inhibitors (NAIs), as the only available class of drugs for AIVs in humans, have been constantly used for treatment, leading to the inevitable emergence of drug-resistant variants. To screen for substitutions conferring NAI resistance in AIVs of N4, N5, N6, and N8 NA subtypes, random mutations within the target gene were generated, and resistant viruses were selected from mutant libraries in the presence of individual drugs. We identified 16 NA substitutions conferring NAI resistance in the tested AIV subtypes; some are novel and subtype specific, and others have been previously reported in other subtypes. Our findings will contribute to an increased and more comprehensive understanding of the mechanisms of NAI-induced inhibition of influenza virus and help lead to the development of drugs that bind to alternative interaction motifs.
KEYWORDS: avian influenza virus, neuraminidase inhibitor, resistance, random mutagenesis, N4, N5, N6, N8
INTRODUCTION
Wild waterfowl, a natural reservoir of influenza A viruses, can be infected with avian influenza viruses (AIVs) of 16 hemagglutinin (HA) and 9 neuraminidase (NA) subtypes (1). Influenza A viruses are known to have a strong host species barrier, and only a few subtypes (i.e., H1N1, H3N2, and H2N2) have been successfully established in humans (1–3). However, the recent increase in the number of humans infected with AIVs of different HA (e.g., H5, H6, H7, H9, and H10) and NA (e.g., N1, N2, N3, N6, N8, and N9) subtypes has elevated public health concerns, and no one can know for certain which strain, if any, will initiate the next pandemic. Therefore, effective measures, such as prophylaxis with influenza vaccines and antiviral drug treatment, are essential for mitigating the risk of a pandemic. The most effective control for human infection and spread of influenza virus is vaccine administration; however, the length of time it takes to prepare vaccine and its lack of efficacy for serologically distinct strains pose some limitations. Thus, antivirals are essential at the initial stage of a pandemic outbreak. To date, the neuraminidase inhibitors (NAIs) oseltamivir (OS), zanamivir (ZA), peramivir (PER), and laninamivir are the only available therapeutic options for control of influenza virus infections (2).
The mechanism of action of NAIs involves targeting NA, a surface glycoprotein that plays an important role in influenza virus replication by releasing the newly budding virions from the membranes of virus-infected cells, allowing them to spread into noninfected cells (4). The NA glycoprotein possesses pockets that consist of several residues with enzymatic activity (5). Its active sites are divided into catalytic and framework residues. Catalytic residues (R118, D151, R152, R224, E276, R292, R371, and Y406) are known to be highly conserved in influenza viruses and interact directly with the substrate, while the framework residues (E119, R156, W178, S179, D198, I222, E227, H274, E277, N294, and E425), which are also highly conserved, stabilize the catalytic residues. The enzymatic activity of the catalytic site cleaves two types of substrates on the host cell membrane, the α2-3- and α2-6-linked sialic acids, which bind to the viral HA surface glycoprotein during attachment, resulting in the release of offspring virions from host cells and subsequent spread throughout target organs (5). Thus, NA has been an attractive target for antiviral drug development. However, the increased use of NAIs and the error-prone nature of viral RNA polymerase result in the development of NAI-resistant influenza viruses. Molecular markers of NAI resistance were identified in the NA glycoprotein and have been intensively studied in human influenza viruses of the N1 and N2 subtypes (6–14). Although we previously profiled NAI resistance-associated substitutions in N3, N7, and N9 NA subtypes of AIVs that have caused human infections and identified novel and subtype-specific substitutions (15), there is still a lack of knowledge relating to the molecular basis of NAI resistance within the remaining AIV NA subtypes. In the current study, we profiled NA substitutions conferring resistance to NAIs in the N4, N5, N6, and N8 NA subtypes, which have recently been reported to infect humans and pose a public health risk, such as the recently prevalent highly pathogenic A(H5N6) and A(H5N8) AIVs (16–19). We identified novel, subtype-specific substitutions or amino acid substitutions that have been previously reported in other subtypes in the N4, N5, N6, and N8 subtypes using gene-fragmented random mutagenesis (GFRM). Furthermore, the replication efficiency in Madin-Darby canine kidney (MDCK) cells, plaque formation, and genetic stability of the recombinant viruses possessing the substitution(s) were also determined.
RESULTS
Screening for NAI resistance-associated markers in AIV of the N4 NA subtype.
To supplement the previously reported resistance database profile in AIVs of the N3, N7, and N9 NA subtypes (15), we screened for molecular markers that confer NAI resistance in the N4, N5, N6, and N8 AIV subtypes. To study the N4 AIV subtype, random mutations were introduced into the catalytic domain of the NA gene of A/EM/Korea/W140/2006 (H10N4) virus, followed by generation of a random mutant virus library in the background of PR8 virus (7+1; NA gene is from avian H10N4 virus and the remaining 7 genes are from PR8 virus) by reverse genetics (RG). The recombinant viruses were serially passaged in the presence of OS or ZA in MDCK cells. Sequence analysis of viruses passaged 3 or 4 times under drug pressure identified 4 substitutions via OS selection and 6 substitutions via ZA selection (Table 1). We subsequently generated recombinant viruses containing the identified substitutions by RG and performed an NA inhibition assay using 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid sodium salt hydrate (MUNANA) substrate. Of note, two recombinant viruses possessing one of the ZA-induced substitutions (E276D or I427L) hade reduced susceptibility to multiple NAIs. Virus with a single E276D substitution exhibited reduced inhibition by OS (10.6-fold 50% inhibitory concentration [IC50] increase) and PER (18.0-fold IC50 increase) and highly reduced inhibition by ZA (248.8-fold IC50 increase) compared to that of the N4 wild-type (WT) virus. Virus carrying the I427L substitution showed reduced inhibition by OS (18.8-fold IC50 increase) and ZA (75.5-fold IC50 increase) compared to the N4 WT virus. However, other mutations identified exhibited normal inhibition based on the antiviral susceptibility criteria defined by the World Health Organization (WHO) Antiviral Working Group (20), although virus with the double V114I-V116A substitution showed 8.6- to 9.9-fold-reduced susceptibility to ZA and OS. The H274Y and R292K substitutions, which are frequently identified in clinical samples and are known to have reduced susceptibility to OS in N1 and N2 subtypes (21, 22), were not identified in the initial screening. Thus, using site-directed mutagenesis, we created these substitutions to investigate if their alteration changed susceptibility to NAIs in the N4 AIV subtype. The H274Y substitution demonstrated highly reduced inhibition by OS (318.3-fold IC50 increase) and PER (114.9-fold IC50 increase), and of note, R292K showed highly reduced inhibition by all the NAIs tested (Table 1). Although the plaque size and infectivity of influenza viruses carrying E276D and I427L substitutions did not differ significantly from those of the parental N4 WT virus, both recombinant viruses showed decreased NA activity, and furthermore, the I427L mutant showed a tendency to return to the WT genotype (I427; 25%) after three serial passages in MDCK cells, indicating low genetic stability. The plaque size and NA activity of the H274Y mutant remained comparable to those of the N4 WT virus, whereas the R292K mutant showed significantly reduced plaque size and NA activity (Table 1).
TABLE 1.
Susceptibility to NAIs and viral properties caused by the selected NA substitutions in avian N4 influenza viruses
| Influenza virus NA amino acid change (N2 numbering) | Selection NAIa | NA activity (nM) ± SD | IC50 ± SDb (fold change)c |
Mean infectivity (log10 PFU/ml) ± SD | Mean plaque size (mm) ± SD | Genetic stabilityd (% read) | ||
|---|---|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | Peramivir | ||||||
| rgN4-WT | Not applicable | 47,981 ± 546 | 0.83 ± 0.002 (1.0) | 0.14 ± 0.001 (1.0) | 0.06 ± 0.001 (1.0) | 8.8 ± 0.0 | 0.78 ± 0.19 | Not applicable |
| rgN4-F132I | OS | 2,671 ± 79 | 1.55 ± 0.001 (1.9) | 0.46 ± 0.003 (3.4) | 0.13 ± 0.001 (2.2) | 7.5 ± 0.5 | 0.41 ± 0.14g | NDf |
| rgN4 S246F | ZA | 46,091 ± 1,753 | 0.85 ± 0.007 (1.0) | 0.51 ± 0.002 (3.8) | 0.09 ± 0.001 (1.6) | 8.6 ± 0.1 | 0.78 ± 0.24 | F (100) |
| rgN4 H274Y | Inducede | 44,508 ± 1,696 | 264.10 ± 0.006 (318.3) | 0.29 ± 0.001 (2.1) | 6.85 ± 0.002 (114.9) | 8.5 ± 0.1 | 0.92 ± 0.33 | Y (100) |
| rgN4 E276D | ZA | 5,469 ± 94 | 8.76 ± 0.001 (10.6) | 33.74 ± 0.006 (248.8) | 1.07 ± 0.001 (18.0) | 8.7 ± 0.1 | 0.73 ± 0.22 | D (100) |
| rgN4 R292K | Induced | 1,038 ± 25 | 438,650,000.21 ± 0.021 (52,862,135.4) | 63.09 ± 0.005 (465.3) | 1803 ± 0.047 (30251.7) | 8.0 ± 0.4 | 0.36 ± 0.11g | K (100) |
| rgN4 N342Y | ZA | 37,596 ± 8,599 | 0.71 ± 0.006 (0.9) | 0.13 ± 0.002 (1.0) | 0.07 ± 0.001 (1.2) | 8.6 ± 0.0 | 0.87 ± 0.27 | ND |
| rgN4 D379E | ZA | 8,854 ± 488 | 1.16 ± 0.004 (1.4) | 0.29 ± 0.001 (2.1) | 0.13 ± 0.002 (2.2) | 7.1 ± 0.3g | 0.65 ± 0.20 | ND |
| rgN4 V394K | ZA | 49,273 ± 1,788 | 1.18 ± 0.001 (1.4) | 0.20 ± 0.005 (1.5) | 0.09 ± 0.001 (1.4) | 8.3 ± 0.0 | 0.81 ± 0.23 | ND |
| rgN4 T414S | OS | 47,490 ± 883 | 0.91 ± 0.003 (1.1) | 0.16 ± 0.001 (1.1) | 0.08 ± 0.001 (1.3) | 9.1 ± 0.1 | 0.80 ± 0.24 | ND |
| rgN4 I427L | ZA | 27,707 ± 956 | 15.60 ± 0.002 (18.8) | 10.24 ± 0.007 (75.5) | 0.53 ± 0.001 (8.9) | 8.3 ± 0.0 | 0.70 ± 0.22 | L (75), I (25) |
| rgN4 V114I+V116A | OS | 33,845 ± 1,162 | 8.23 ± 0.001 (9.9) | 1.16 ± 0.001 (8.6) | 0.11 ± 0.001 (1.8) | 8.6 ± 0.0 | 0.82 ± 0.21 | I (100), A (100) |
| rgN4 S246F+N342Y | ZA | 47,123 ± 1,671 | 0.93 ± 0.001 (1.1) | 0.64 ± 0.001 (4.7) | 0.08 ± 0.001 (1.3) | 8.5 ± 0.0 | 0.74 ± 0.25 | F (100), Y (100) |
The NAI used in the selection of the corresponding mutations.
The values are means obtained from two independent experiments.
Fold change relative to the mean IC50s of the rgN4-WT virus.
Illumina sequencing was used to analyze the sequence variation.
This mutant was introduced for comparison.
ND, not determined due to insignificant alteration of NAI susceptibility of the indicated mutant virus.
P < 0.05 compared with the corresponding infectivity value or plaque size of WT virus.
Screening for NAI resistance-associated markers in AIV of the N5 NA subtype.
To study the N5 AIV subtype, we used A/EM/Korea/W69/2005 (H6N5) virus and generated virus libraries containing random mutations in the catalytic domain of the NA gene. Four and one amino acid substitutions were observed under the selective pressure of OS and ZA, respectively. Recombinant viruses possessing the identified NA substitutions were generated by RG to eliminate side effects that were potentially caused by a minor population of substitutions that might have occurred in other genes (Table 1). Of all the N5 substitutions, viruses with E119V and N147I substitutions showed reduced susceptibility to all the NAIs compared to the N5 WT virus. Of note, virus with the single E119V substitution showed reduced inhibition by OS (58.5-fold IC50 increase) and PER (63.9-fold IC50 increase) and highly reduced inhibition by ZA (736.6-fold IC50 increase) compared to that of the N5 WT virus. The single N147I substitution selected by ZA showed reduced inhibition by the drug (30.7-fold IC50 increase). For comparison, H274Y and R292K substitutions were additionally introduced into recombinant viruses of the N5 subtype. Viruses carrying both H274Y and R292K substitutions demonstrated highly reduced OS-mediated (1081.8- and 963.7-fold IC50 increases, respectively) and PER-mediated (311.7- and 283.7-fold IC50 increases, respectively) inhibition compared to that of the N5 WT virus but normal inhibition by ZA. Although recombinant viruses carrying either single E119V or double L91I-E119V substitutions formed significantly smaller plaques than the N5 WT virus, they maintained infectivity similar to that of the N5 WT virus in MDCK cells and showed high genetic stability after 3 sequential passages in MDCK cells (Table 2). Except for E119V, substitutions conferring NAI resistance, including N147I, H274Y, and R292K, showed poor genetic stability and reverted to their parental amino acid sequences after three serial passages in MDCK cells (Table 2).
TABLE 2.
Susceptibility to NAIs and viral properties caused by the selected NA substitutions in avian N5 influenza viruses
| Influenza virus NA amino acid change (N2 numbering) | Selection NAIa | NA activity (nM) ± SD | IC50 ± SDb (fold change)c |
Mean infectivity (log10 PFU/ml) ± SD | Mean plaque size (mm) ± SD | Genetic stabilityd (% read) | ||
|---|---|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | Peramivir | ||||||
| rgN5 WT | Not applicable | 44,311 ± 405 | 0.55 ± 0.002 (1.0) | 0.17 ± 0.001 (1.0) | 0.05 ± 0.001 (1.0) | 8.6 ± 0.1 | 0.79 ± 0.19 | Not applicable |
| rgN5 L91I | OS | 46,801 ± 913 | 0.59 ± 0.001 (1.1) | 0.20 ± 0.001 (1.1) | 0.05 ± 0.001 (1.1) | 8.8 ± 0.1 | 0.71 ± 0.22 | NDf |
| rgN5 E119V | OS | 13,527 ± 457 | 31.89 ± 0.009 (58.5) | 128.1 ± 0.021 (736.6) | 2.88 ± 0.001 (63.9) | 8.5 ± 0.1 | 0.57 ± 0.18g | V (100) |
| rgN5 N147I | ZA | 36,505 ± 798 | 0.56 ± 0.001 (1.0) | 5.34 ± 0.003 (30.7) | 0.15 ± 0.001 (3.3) | 8.83 ± 0.2 | 0.74 ± 0.21 | I (89), N(11) |
| rgN5 K187R | OS | 48,329 ± 278 | 0.87 ± 0.001 (1.6) | 0.27 ± 0.001 (1.5) | 0.05 ± 0.001 (1.0) | 8.64 ± 0.1 | 0.66 ± 0.14 | ND |
| rgN5 Q221H | OS | 48,749 ± 392 | 0.58 ± 0.001 (1.1) | 0.47 ± 0.001 (2.7) | 0.05 ± 0.001 (1.1) | 8.65 ± 0.1 | 0.71 ± 0.18 | ND |
| rgN5 H274Y | Inducede | 35,023 ± 79 | 589.80 ± 0.061 (1,081.8) | 0.47 ± 0.001 (2.7) | 14.04 ± 0.004 (311.7) | 7.5 ± 0.1g | 0.57 ± 0.10g | Y (98), H (2) |
| rgN5 R292K | Induced | 15,150 ± 545 | 525.40 ± 0.042 (963.7) | 0.68 ± 0.001 (3.9) | 12.78 ± 0.003 (283.7) | 8.4 ± 0.1 | 0.68 ± 0.16 | K (83), R (17) |
| rgN5 L91I+E119V | OS | 9,820 ± 49 | 23.58 ± 0.005 (43.3) | 96.61 ± 0.016 (555.5) | 3.9 ± 0.001 (86.5) | 8.34 ± 0.1 | 0.42 ± 0.13g | I (100), V (100) |
The NAI used in the selection of the corresponding mutations.
The values are means obtained from two independent experiments.
Fold change relative to the mean IC50s of the rgN5-WT virus.
Illumina sequencing was used to analyze the sequence variation.
The mutant was introduced for comparison.
ND, not determined due to insignificant alteration of NAI susceptibility of the indicated mutant virus.
P < 0.05 compared with the corresponding infectivity value or plaque size of WT virus.
Screening for NAI resistance-associated markers in AIV of the N6 NA subtype.
The N6 subtype of influenza viruses, in combination with highly pathogenic (HP) AIVs of the H5 subtype, have been recently reported to infect humans in China (16, 23). Although there have been no recent reports of NAI resistance in the N6 subtype (24), the fatality risk of A(H5N6) human cases has been found to be similar to or higher than those of HPAI A(H5N1) and A(H7N9) infections (17). To study the N6 AIV subtype, we used A/EM/Korea/W20/2005 (H4N6) virus and generated a random mutant virus library in the background of PR8 virus (7+1). Sequence analysis identified a total of 8 NA substitutions: 3 after OS selection and 5 after ZA selection from the mutant virus libraries (Table 3). Four substitutions (E119D, A246V, R292K, and R371K) reduced susceptibility to NAIs (Table 3). Except for R292K (selected by OS), the resistance-associated substitutions were selected by ZA. Viruses with R292K and R371K substitutions showed reduced or highly reduced inhibition by all the NAIs tested, and virus with E119D showed highly reduced inhibition by ZA and reduced inhibition by PER, indicating multi-NAI-resistant markers. The novel A246V substitution (selected by ZA) showed reduced inhibition by ZA (17.0-fold IC50 increase). For comparison, an induced H274Y substitution that generally has reduced susceptibility to OS in group 1 NA was also introduced in the N6 subtype (group 2 NA). However, the N6 virus possessing an H274Y substitution showed normal inhibition by all the NAIs tested. Viruses with R292K and A246V substitutions retained their infectivity and plaque formation capacity in MDCK cells and showed high genetic stability, while the E119D mutant formed smaller plaques and the R371K mutant showed poor genetic stability compared to the N6 WT virus.
TABLE 3.
Susceptibility to NAIs and viral properties caused by the selected NA substitutions in avian N6 influenza viruses
| Influenza virus NA amino acid change (N2 numbering) | Selection NAIa | NA activity (nM) ± SD | IC50 ± SDb (fold change)c |
Mean infectivity (log10 PFU/ml) ± SD | Mean plaque size (mm) ± SD | Genetic stabilityd (% read) | ||
|---|---|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | Peramivir | ||||||
| rgN6-WT | Not applicable | 36,653 ± 910 | 0.63 ± 0.001 (1.0) | 0.61 ± 0.001 (1.0) | 0.10 ± 0.002 (1.0) | 8.4 ± 0.1 | 0.75 ± 0.19 | Not applicable |
| rgN6 E119D | ZA | 10,593 ± 680 | 1.28 ± 0.001 (2.0) | 150.60 ± 0.038 (248.6) | 3.24 ± 0.001 (31.4) | 8.5 ± 0.1 | 0.53 ± 0.18g | D (100) |
| rgN6 R130M | ZA | 21,595 ± 329 | 0.44 ± 0.001 (0.7) | 1.07 ± 0.001 (1.8) | 0.14 ± 0.001 (1.3) | 7.8 ± 0.2g | 0.74 ± 0.19 | NDf |
| rgN6 P166T | ZA | 16,253 ± 322 | 0.50 ± 0.008 (0.8) | 1.12 ± 0.001 (1.9) | 0.18 ± 0.001 (1.7) | 7.9 ± 0.2g | 0.68 ± 0.16g | ND |
| rgN6 A246V | ZA | 8,830 ± 215 | 1.77 ± 0.002 (2.8) | 10.31 ± 0.009 (17.0) | 0.56 ± 0.001 (5.4) | 8.7 ± 0.2 | 0.92 ± 0.25 | Y (100) |
| rgN6 H274Y | Inducede | 22,647 ± 126 | 0.99 ± 0.001 (1.6) | 1.23 ± 0.001 (2.0) | 0.15 ± 0.001 (1.5) | 8.4 ± 0.1 | 0.62 ± 0.53g | ND |
| rgN6 R292K | OS | 12,078 ± 291 | 9,440 ± 0.031 (15,067.8) | 27.59 ± 0.004 (45.5) | 87.30 ± 0.011 (844.3) | 8.6 ± 0.2 | 0.82 ± 0.20 | K (100) |
| rgN6 R371K | ZA | 4,769 ± 59 | 14.59 ± 0.011 (23.3) | 31.93 ± 0.005 (52.7) | 1.22 ± 0.001 (11.8) | 8.8 ± 0.2 | 0.83 ± 0.23 | K (90), R (10) |
| rgN6 V444I | OS | 29,829 ± 257 | 0.36 ± 0.001 (0.6) | 0.96 ± 0.001 (1.6) | 0.12 ± 0.001 (1.2) | 8.4 ± 0.1 | 0.68 ± 0.22g | ND |
| rgN6 V302E+P347H | OS | 945 ± 43 | 0.4 ± 0.002 (0.6) | 1.32 ± 0.001 (2.2) | 0.22 ± 0.001 (2.2) | 6.0 ± 0.3g | 0.46 ± 0.15g | ND |
The NAI used in the selection of the corresponding mutations.
The values are means obtained from two independent experiments.
Fold change relative to the mean IC50s of the rgN6-WT virus.
Illumina sequencing was used to analyze the sequence variation.
The mutant was introduced for comparison.
ND, not determined due to insignificant alteration of NAI susceptibility of the indicated mutant virus.
P < 0.05 compared with the corresponding infectivity value or plaque size of WT virus.
Screening for NAI resistance-associated markers in AIV of the N8 NA subtype.
The influenza A(H10N8) viruses have recently emerged in humans in China and caused two deaths among three virus-infected patients (25). Furthermore, HPAI A(H5N8) virus has spread worldwide and has been causing large poultry outbreaks after reassortment with HPAI A(H5N1) virus (18), indicating pandemic potential of the viruses of N8 subtype. Therefore, screening for NAI resistance in the subtype is also important. To study the N8 AIV subtype, we used A/EM/Korea/W468/2015 (H5N8) virus and found three NA substitutions using OS selection and three using ZA selection (Table 4). Viruses with two substitutions (Q136K and G147V) showed reduced susceptibility to NAIs (Table 4). Although virus with the V116D substitution showed slightly decreased susceptibility to OS (4.8-fold IC50 increase) and ZA (7.0-fold IC50 increase) compared to the N8 WT virus, it still had normal inhibition. The Q136K substitution demonstrated highly reduced inhibition by ZA (263.4-fold IC50 increase) and PER (184.9-fold IC50 increase), and the novel G147V substitution showed reduced inhibition by ZA (24.9-fold IC50 increase) compared to the N8 WT virus. The additionally induced H274Y and R292K substitutions in the virus of N8 subtype conferred reduced susceptibility to multiple NAIs. Specifically, the H274Y mutant exhibited highly reduced inhibition by OS (425.1-fold IC50 increase) and PER (102.5-fold IC50 increase), and the R292K mutant showed highly reduced inhibition mediated by OS (50,636,335.7-fold IC50 increase) and PER (8,540.4-fold IC50 increase) and reduced ZA-mediated inhibition (92.9-fold IC50 increase). All the mutant viruses exhibited infectivities in MDCK cells similar to that of the N8 WT virus. However, viruses with H274Y and R292K substitutions formed smaller plaques than the N8 WT virus, and G147V virus was genetically unstable after three passages in MDCK cells (Table 4). The infectivity, plaque formation in MDCK cells, and genetic stability of Q136K virus were similar to those of the N8 WT virus.
TABLE 4.
Susceptibility to NAIs and viral properties caused by the selected NA substitutions in avian N8 influenza viruses
| Influenza virus NA amino acid change (N2 numbering) | Selection NAIa | NA activity (nM) ± SD | IC50 ± SDb (fold change)c |
Mean infectivity (log10 PFU/ml) ± SD | Mean plaque size (mm) ± SD | Genetic stabilityd (% read) | ||
|---|---|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | Peramivir | ||||||
| rgN8-WT | Not applicable | 22,872 ± 349 | 0.65 ± 0.001 (1.0) | 0.28 ± 0.001 (1.0) | 0.05 ± 0.001 (1.0) | 8.0 ± 0.1 | 0.74 ± 0.21 | Not applicable |
| rgN8 V116D | OS, ZA | 7,482 ± 24 | 3.16 ± 0.002 (4.8) | 1.96 ± 0.001 (7.0) | 0.10 ± 0.001 (1.8) | 7.9 ± 0.1 | 0.50 ± 0.14g | D (100) |
| rgN8 Q136K | ZA | 5,760 ± 275 | 0.29 ± 0.001 (0.4) | 73.53 ± 0.029 (263.4) | 10.06 ± 0.011 (184.9) | 8.0 ± 0.1 | 0.68 ± 0.17 | K (100) |
| rgN8 G147V | ZA | 5,857 ± 116 | 1.29 ± 0.001 (2.0) | 6.94 ± 0.007 (24.9) | 0.27 ± 0.001 (4.9) | 7.7 ± 0.1 | 0.68 ± 0.18 | V (80), G (20) |
| rgN8 H274Y | Inducede | 13,397 ± 35 | 276.60 ± 0.036 (425.1) | 0.39 ± 0.001 (1.4) | 5.57 ± 0.003 (102.5) | 7.8 ± 0.1 | 0.40 ± 0.14g | Y (100) |
| rgN8 R292K | Induced | 1,366 ± 20 | 32,944,000.31 ± 0.095 (50,636,335.7) | 25.94 ± 0.008 (92.9) | 464.60 ± 0.092 (8540.4) | 7.6 ± 0.3 | 0.40 ± 0.11g | K (100) |
| rgN8 S367C | OS | 39,887 ± 510 | 1.02 ± 0.004 (1.6) | 0.21 ± 0.006 (0.8) | 0.05 ± 0.001 (0.9) | 8.2 ± 0.1 | 0.70 ± 0.20 | NDf |
| rgN8 N216D+I379V | OS | 34,828 ± 728 | 1.16 ± 0.003 (1.8) | 0.22 ± 0.002 (0.8) | 0.05 ± 0.001 (0.8) | 8.1 ± 0.1 | 0.87 ± 0.24 | ND |
The NAI used in the selection of the corresponding mutations.
The values are means obtained from two independent experiments.
Fold change relative to the mean IC50s of the rgN8-WT virus.
Illumina sequencing was used to analyze the sequence variation.
The mutant was introduced for comparison.
ND, not determined due to insignificant alteration of NAI susceptibility of the indicated mutant virus.
P < 0.05 compared with the corresponding infectivity value or plaque size of WT virus.
Analysis of molecular polymorphism and structural modeling of novel NAI-resistant substitutions.
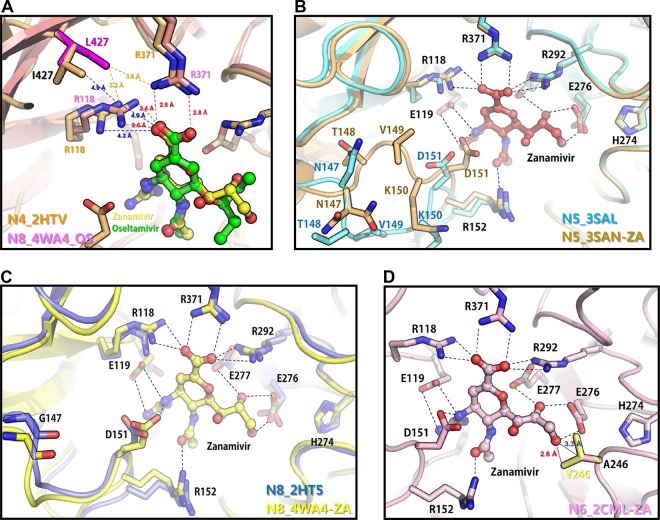
We identified novel NAI-resistant substitutions, specifically I427L in N4, N147I in N5, A246V in N6, and G147V in N8, via ZA selection and showed reduced ZA-mediated inhibition. Therefore, the molecular polymorphism and structural modeling of the substitutions interacting with ZA were further analyzed. I427 is highly conserved among influenza A viruses, and L427 has not been identified in surveillance studies, although about 15% of human A(H1N1) viruses possess V427 (Table 5). Due to the lack of OS/ZA-N4 complex structure, OS/ZA-N8 complex structure was adapted to predict the alteration of binding. The isoleucine-to-leucine change at residue 427 moves toward residue R118, which critically binds to OS/ZA, and this potentially results in reduced affinity for OS/ZA (Fig. 1A). Most influenza A viruses possess glycine at residue 147 in the NA protein, except avian N5 and human N2 subtypes, which predominantly possess N147 (Table 5). The identified changes in I147 and V147, which both showed reduced inhibition by ZA, were not observed in sequence data in the Influenza Research Database (IRD). Residue 147 is located at the 150 loop, and G147I and N147V substitutions may affect the interaction between D151 and ZA (Fig. 1B and C). While the N1 and N4 subtypes possess S246, the remaining NA subtypes predominantly have alanine at residue 246 (Table 5), and there was a small portion of V246 (0.01%) found in group 2 NAs in sequence data in the IRD, which potentially shows a reduction in susceptibility to ZA (Table 5). The A246V substitution potentially causes steric clashes with the inhibitor, reducing affinity for ZA (Fig. 1D).
TABLE 5.
NA sequence variations of the NAI resistance-associated substitutions identified in human and avian influenza viruses of N1 to N9 NA subtypes
| NA residuea | WT amino acid (subtype) | Amino acid substitution | NA subtype | Polymorphism(s) in database (frequency [%])b |
|||
|---|---|---|---|---|---|---|---|
| Avian | No. of sequences | Human | No. of sequences | ||||
| 147 | N/G (N5/N8) | I/V | N1 | G (99.76), R (0.24) | 2,110 | G (99.83), Q (0.11), R (0.06) | 8,065 |
| N4, N5, N8 | G (86.37), N (13.63)c | 3,258 | G (100) | 4 | |||
| N2 | G (98.78), D (0.65), S (0.58) | 2,776 | N (73.57), D (25.83), G (0.57), Y (0.02) | 8,809 | |||
| N3, N6, N7, N9 | G (99.94), R (0.03), T (0.01), Q (0.01) | 6,733 | G (99.12), Q (0.88) | 113 | |||
| 246 | A (N6) | F/V | N1 | S (99.76), N (0.19), G (0.05) | 2,111 | S (99.38), N (0.57), R (0.05) | 8,061 |
| N4, N5, N8 | A (89.1), S (10.81)d, T (0.09) | 3,276 | A (100) | 4 | |||
| N2 | A (99.96), S (0.04) | 2,775 | A (99.95), T (0.03), G (0.01) | 8,808 | |||
| N3, N6, N7, N9 | A (99.91), T (0.03), V (0.01), S (0.01), P (0.01), G (0.01) | 6,719 | A (100) | 125 | |||
| 427 | I (N4) | L | N1 | I (98.72), V (1.23), T (0.05) | 2,108 | I (85.11), V (14.87), M (0.01) | 8,061 |
| N4, N5, N8 | I (99.97), V (0.03) | 3,277 | I (100) | 4 | |||
| N2 | I (99.39), V (0.4), T (0.22) | 2,771 | I (99.93), V (0.05), M (0.01), T (0.01) | 8,805 | |||
| N3, N6, N7, N9 | I (97.4), T (2.51), M (0.07), V (0.01) | 6,692 | I (100) | 113 | |||
| 274 | H | Y | N1 | H (99.95), Y (0.047) | 2,112 | H (94.31), Y (5.672), N (0.012) | 8,056 |
| N4, N5, N8 | H (99.96), R (0.016), Q (0.016) | 6,176 | H (100) | 4 | |||
| N2 | H (100) | 2,775 | H (100) | 8,808 | |||
| N3, N6, N7, N9 | H (99.97), N (0.014), R (0.014) | 6,729 | H (100) | 125 | |||
Position numbers of amino acid substitutions are based on N2 numbering.
Data were obtained from the Influenza Research Database.
N is 100% in the N5 subtype (350/350).
S is 99.55% in the N4 subtype (225/226).
FIG 1.
Interactions between NAI resistance-associated amino acid substitutions and inhibitors. (A) Locations of oseltamivir and zanamivir resistance-associated substitutions found in N4 NA. The structure of the I427L mutant was generated from the N4 WT structure (Protein Data Bank [PDB] entry 2HTV) using PyMOL. Superimposition of active sites of N4 WT, the N4 I427L mutant, and the N8 WT-oseltamivir complex (PDB entry 4WA4) shows the positional difference of R118 and the alteration of the interaction distance between R118 and inhibitors. (B) Superimposition of the active sites of N5 (PDB entry 3SAL) and an N5-zanamivir complex (PDB entry 3SAN) showing the different conformations of the 150 loop, including N147. (C) Superimposition of the active sites of N8 (PDB entry 2HT5) and N8-zanamivir (PDB entry 4WA4) showing the similar conformations of the 150 loop, including G147. (D) Location of a zanamivir resistance substitution found in N6-NA. The structure of the A246V mutant was generated from the N6 WT structure (PDB entry 2CML) by using PyMOL. Superimposition of active sites of N6 WT and the N6 A246V mutant shows the predicted interaction distance between V246 and zanamivir. The residues critical for the interaction between the NA protein and NAIs are shown.
DISCUSSION
As a continuation of our previous molecular-based resistance screening, which identified unique NA substitutions that confer reduced NAI susceptibility in AIVs of the N3, N7, and N9 subtypes (15), we report here the identification of 16 novel or previously reported NAI resistance-associated substitutions in AIVs of the N4, N5, N6, and N8 NA subtypes, with biosafety protocols conducted in the previous study maintained (15). These subtypes were already known to have resulted in human infections or represent a potential infection risk to humans. Furthermore, 13 of the substitutions conferred reduced inhibition by the multiple NAIs currently available as influenza treatments (i.e., OS, ZA, and PER). In addition, we also evaluated the replication efficiency, plaque formation, and genetic stability of the recombinant viruses containing the NA substitutions associated with NAI resistance.
The substitutions identified in avian N4, N5, N6, and N8 NA proteins belong to 2 categories: (i) novel subtype-specific substitutions (G/N147V/I, A246V, and I427L) and (ii) substitutions previously reported in viruses of other NA subtypes (E119A/D/G/V, Q136K, E276D, R292K, and R371K). The novel G/N147 substitution is located in the 150 loop (residues 147 to 152), which forms a portion of the enzyme active site. The 150 loop forms a cavity that binds to NAIs with similar affinities in N1 and N2 subtypes (26, 27). Structural analysis revealed that residue 147 plays an essential role in the conformation of the 150 cavity (26). The majority of AIVs possess G at residue 147 of the NA protein, while AIVs of the N5 subtype and human viruses of the N2 subtype mainly have N at this position (Table 5), resulting in an extended form of the 150 cavity (26). The G→N change at residue 147, conferring NAI resistance, was not reported previously in clinical samples after NAI treatment (28, 29). Although the G147R substitution found in the N9 subtype conferred NAI susceptibility similar to that of the parental virus (15), the novel G147V in virus of the N5 subtype and N147I in virus of the N8 subtype have reduced inhibition by ZA (Tables 2 and 4). However, the essential role of G/N147 in the formation of the 150 cavity could potentially lead to genetic instability of the change and the possibility of return to the intrinsic amino acid (Tables 2 and 4). This suggests that occurrence of the resistant phenotype in viruses of the N5 and N8 subtypes carrying substitutions at residue 147 is unlikely.
The novel A246V substitution in the N6 subtype was shown to reduce ZA-mediated susceptibility. The N1 and N4 subtypes possess serine at position 246, while other NA subtypes have an alanine at the same position (Table 5). S/A246, together with R224, I222, and/or E276, forms a hydrophobic side pocket adjacent to the NA active site (30–32). This side pocket is known to accommodate the glycerol side chains of OS and ZA (31). Thus, changes at residue 246 may cause disturbance of the hydrophobic side pocket (Fig. 1D) and result in reduced binding affinity between the side pocket and inhibitors, as was observed previously (15, 32–34). Previously, S246N/G and A246T substitutions were detected in association with NAI resistance in viruses of the N1 and N9 subtypes, respectively (15, 32, 35). The degree of inhibitor binding of residue 246 was higher for S246 of the N1 subtype than for A246 of the N2 subtype (35). Thus, resistance substitutions associated with S246 of the N1 subtype are predominantly detected compared with A246 of the other subtypes (35). However, we have found that an A246T substitution in the N9 subtype and an A246V substitution in the N6 subtype reduced ZA-mediated viral susceptibility. In addition to the novel A246V substitution in the current study, S246F was also observed in N4 subtype viruses; however, susceptibility to ZA was only modestly reduced (3.8-fold IC50 increase) compared to that of the N4 WT virus. Thus, investigating whether the S246N substitution in virus of the N4 subtype could possibly confer resistance to NAIs could be an interesting topic for further study.
The I427 and R371 residues in the N1 subtype play an important role in anchoring most of the NAIs (36). It was shown that the I→Q and I→T changes at residue 427 result in reduced inhibition by multiple NAIs (36, 37). In previous structural-analysis studies, a hypothesis that the substitutions at the I427 residue induce a conformational change in the side chain of the R371 residue or the R118 residue, interfering with inhibitor binding, was tested (36). Thus, the novel I427L substitution in the N4 subtype identified in the current study may potentially impact the structural composition; R118 may disrupt viral interactions with inhibitors (Fig. 1A), although the precise mechanism should be further determined by structural crystallography. In addition, corresponding to the I427T/Q substitution, I427L also reduced susceptibility to multiple NAIs (OS and ZA), which implies its importance as a molecular marker for multi-NAI resistance. Although further studies are needed, the NAI resistance-associated substitutions are subtype specific. These identified novel resistance substitutions were not observed in a previous study (37). Although the reason for this is still unclear, as proposed in our previous study (37), the novel NAI resistance substitutions may be specific to AIVs of the N4, N5, N6, or N8 subtype. Additionally, it was observed that all the novel substitutions were selected by ZA, which has been less frequently prescribed than OS.
We identified NAI resistance-associated amino acid changes that have been reported in influenza A viruses of N1, N2, N3, N7, and N9 subtypes and influenza B viruses. Changes at residue E119 associated with NAI resistance in both group 1 and 2 NAs have been reported. E119V/D substitutions have been detected in patients infected with influenza viruses of the N1 or N2 subtype and treated with OS and/or ZA (10, 11, 38). More importantly, the D/V substitutions at residue 119 showed reduced to highly reduced inhibition by multiple NAIs in the N1 and N2 subtypes. E119V and E119D substitutions found in AIVs of the N5 (group 1 NA) and N6 (group 2 NA) subtypes, respectively, showed reduced to highly reduced inhibition by multiple NAIs. Previous studies have shown that the E119V mutation is mainly selected in the group 2 NAs because this substitution in group 1 NAs leads to a significant decrease in viral fitness (39, 40). Although more tests should be done to more precisely evaluate viral fitness, based on our initial experiments, viral infectivity and genetic stability are retained in N5 subtype viruses (group 1 NA) with the E119V substitution.
Although the Q136 residue is located outside the catalytic and framework residues, Q136K/L/R substitutions have been reported to confer NAI resistance, implying an important role for binding to NAIs. Previous studies have shown that mutations confer reduced susceptibility to ZA, PER, and/or laninamivir but do not affect OS susceptibility (8, 41). We detected a Q136K substitution in N8 subtype viruses and demonstrated highly reduced inhibition by ZA and PER, as was shown previously in other NA subtypes (including both group 1 and 2 NAs) (8, 13), suggesting that the Q136K substitution is subtype independent. The E276D substitution has been previously reported in group 1 and 2 NAs showing reduced inhibition by ZA (15, 42, 43). Although the E276D mutation showed little effect on the NAI susceptibility of viruses of the N1 subtype (group 1 NAs) (42, 43), E276D-possessing N4 viruses (group 1 NAs) showed reduced and highly reduced inhibition by all the NAIs tested, indicating multi-NAI resistance. The R371K substitution was identified in an influenza B virus-infected patient and caused resistance to OS, ZA, and PER (14). Recombinant A(H3N2) and B viruses carrying an R371K substitution showed reduced susceptibility to OS, ZA, and PER (14, 42, 44). Although it is not confirmed whether there is NA group or subtype specificity of the R371K substitution, we detected the substitution only in N6 subtype viruses (group 2 NAs). The R292 residue is located at the catalytic sites of the NA enzyme and binds to the NAIs (22). Due to the structural difference between group 1 and 2 NAs, the reduced susceptibility to NAIs caused by the R292K substitution has been detected in group 2 NAs and generally showed highly reduced inhibition by multiple NAIs (11, 28, 45–47). In this study, the R292K substitution was selected by OS only in N6 subtype viruses (group 2 NAs), although the introduction of this substitution into N4, N5, and N8 subtypes (group 1 NAs) also reduced inhibition by NAIs. It is possible that viral fitness properties of group 1 NA viruses possessing R292K substitution were significantly decreased, as evidenced by reduced plaque formation and genetic stability compared to the N6 virus (group 2 NA). The H274Y substitution has been frequently detected in N1 subtype viruses with high levels of OS resistance (48, 49). For comparison, we introduced the H274Y substitution into viruses of the N4, N5, N6, and N8 subtypes, although it was not selected in our cell culture experiments. Corresponding to the findings, no cases of H274Y change have been reported in subtypes other than N1 via single nucleotide polymorphism (SNP) analysis. The artificially induced H274Y substitution in all group 1 NAs conferred reduced to highly reduced inhibition by OS and PER, similar to the results in the N1 subtype in previous studies (46). Interestingly, an H274Y substitution in some group 1 NAs (specifically N5 and N8) conferred reduced plaque size or genetic stability, suggesting that it is an N1 subtype-specific substitution. On the other hand, although the V116A substitution was reported in association with OS and ZA resistance in H5N1 virus (33), the V116A substitution selected by OS in the N4 subtype and V116D selection by both OS and ZA in the N8 subtype showed normal inhibition by all the NAIs but had slightly reduced susceptibility to OS and ZA (ranging from 4.8- to 9.9-fold). This suggests that changes at this residue may cause reduced susceptibility to OS and ZA in group 1 NA viruses, although the levels of resistance are low.
The balance between HA and NA proteins during the spread of influenza virus is critical (50, 51), and a substitution(s) in HA elicited simultaneously with the change(s) in the NA protein may function as a permissive substitution(s) to maintain viral fitness properties. Thus, the use of a parental combination of HA and NA genes may help us to understand the precise mechanism of molecular-based NAI resistance. However, despite the limitations of this study, the profiling of the molecular-based resistant substitutions in AIVs of all NA subtypes, together with the subtypes reported previously (N3, N7, and N9), will further contribute to an increased and comprehensive understanding of mechanisms of influenza virus inhibition by NAIs and will help lead to the development of drugs that bind to alternative interaction motifs. Importantly, resistance profiling will also contribute to clinical management, especially by helping to guide the prescription of more appropriate or alternative NAIs for patients infected by NAI-resistant strains of influenza virus in the absence of other classes of anti-influenza virus drugs.
MATERIALS AND METHODS
Ethics statement.
The experimental protocol was approved by the Institutional Biosafety Committee of Chungbuk National University, Cheongju, Republic of Korea (16-RDM-028). All experiments with NAI-resistant viruses were performed in an enhanced biosafety level 3 facility approved by the Korea Center for Disease Control and Prevention (K-CDC).
Cells, viruses, plasmids, and neuraminidase inhibitors.
MDCK cells (American Type Culture Collection [ATCC], Manassas, VA) were maintained at 37°C in 5% CO2 in Eagle's minimal essential medium (EMEM) (Lonza, Allendale, NJ) containing 5% fetal bovine serum (FBS) and vitamins (Gibco) and used for virus titration and passages for resistance selection. Human embryonic kidney 293T (HEK-293T cells (ATCC, Manassas, VA) were maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Gibco, USA) containing 10% FBS and 1% antibiotics (penicillin-streptomycin; Gibco-Invitrogen) and used for virus rescue by reverse genetics. A/Environment/Korea/W140/2006 (H10N4), A/Environment/Korea/W69/2005 (H6N5), A/Environment/Korea/W20/2005 (H4N6), and A/Environment/Korea/W468/2015 (H5N8) were isolated from wild-bird fecal samples in South Korea. The viruses were propagated in specific-pathogen-free (SPF) 10-day-old embryonated chicken eggs and stored at −80°C until use. Viral RNA was isolated and used to clone NA genes, including N4, N5, N6, and N8, into a plasmid (pHW2000, kindly provided by Robert G. Webster, St. Jude Children's Research Hospital) for GFRM and random and single mutant virus generation using reverse genetics. Other gene plasmids (PB2, PB1, PA, NP, HA, M, and NS genes) were from A/Puerto Rico/8/1934 (H1N1; PR8) virus and were used as genetic background. The NAIs, oseltamivir carboxylate [ethyl (3R,4R,5S)-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylate) and zanamivir (2,4-dideoxy-2,3-didehydro-4-guanidineosialic acid), were obtained from Hoffmann-La Roche (Nutley, NJ), and peramivir [(1S,2S,3R,4R,1S)-3-(1-acetylamino-2-ethyl) butyl-4-(aminoimino)-methylamino-2-hydroxycyclopentane-1-carboxylic acid] was obtained from BioCryst Pharmaceuticals, Inc. (Birmingham, AL). The compounds were dissolved in dimethyl sulfoxide, and 5 mM aliquots were stored at −20°C until use.
Gene-fragmented random mutagenesis and generation of random mutant virus libraries.
The catalytic domain of the NA gene was divided into six regions (with sizes ranging from 110 to 470 nucleotides) based on the distribution of catalytic and framework residues with small overlapping areas using specific primers (the list of primers will be provided upon request). To generate a random mutant plasmid library, random mutations were introduced into the six individual regions of each NA gene (N4, N5, N6, and N8) using a GeneMorph II EZClone domain mutagenesis kit (Stratagene) according to the manufacturer's instructions. Briefly, 2 μg of each NA plasmid (N4, N5, N6, and N8) having random mutations or wild type and 1 μg of seven remaining plasmids (PR8) were cotransfected in cocultured 293T and MDCK cells (3:1 ratio) using cell transfection reagent (Mirus) in Opti-MEM medium (Gibco). After an 18-h incubation, the supernatants were replaced with 1 ml of Opti-MEM containing 1% antibiotics. At 48 h posttransfection, 1 ml of Opti-MEM with 0.2% l-tosylamide 2-phenylethyl chloromethyl ketone (TPCK) trypsin was added. At 4 days after transfection, 1 ml of supernatants was passaged to select resistant variants in the presence of OS or ZA, and the remaining supernatants were used for plaque assay and/or for determination of the 50% tissue culture infectious dose (TCID50) to confirm virus rescue. The distribution of random mutations was confirmed by sequencing 36 individual plasmid clones.
Screening for NAI resistance substitutions.
The random mutant virus libraries were subsequently passaged 3 or 4 times in MDCK cells with increased concentrations of inhibitors, OS or ZA (5 to 40 μM). After the passages, the NA genes of all the passaged viruses grown in the presence of inhibitors were sequenced and analyzed to identify any mutations relative to the parental NA gene. The screening procedure was performed at least three times in all the subtypes studied. All the substitutions identified in each NA subtype were introduced into the corresponding subtypes using site-directed mutagenesis (Stratagene), and mutagenized viruses were prepared according to the general protocol for rescuing influenza viruses by reverse genetics (52). The NA genes of all the recombinant viruses were verified to determine whether additional mutations were present.
Plaque assay.
MDCK cells were used to determine the infectivity of recombinant viruses (the number of PFU per milliliter). MDCK cells (in 6-well plates) were washed twice with PBS, and the cells were infected with 1 ml of viruses and incubated at 37°C in a CO2 incubator for 1 h for virus adsorption to the cells, and then the supernatants were removed and the 0.7% agarose (Lonza)-containing infection medium with penicillin (1,000 units/ml), streptomycin (1,000 μg/ml), vitamin solution, 0.5% bovine serum albumin, and 0.1% trypsin was added to the wells. Following incubation at 37°C with 5% CO2 for 48 h, the plates were chilled at 4°C for 10 min, and the 0.7% agarose overlay was removed and stained with a 1% (wt/vol) crystal violet solution containing 10% formaldehyde. After washing, the plates were dried, the appropriate wells containing 20 to 100 plaques were selected, and ImageJ software was used to measure individual plaque sizes (ImageJ 1.46r; National Institutes of Health).
NA activity and inhibition assays.
A fluorometric assay was used to quantify levels of NA activity in recombinant viruses (53). We measured the NA activity with the fluorogenic substrate MUNANA (Sigma-Aldrich, Inc., St. Louis, MO; 0 to 5,000 mM). Briefly, each recombinant virus was diluted to the appropriate concentration based on NA activity and standardized to 10 μM 4-methylumbelliferone (MUSS). The incubation was then continued for 1 h with the MUNANA substrate and NAIs, and stop solution was added to measure the value. The NAI concentration (OS, ZA, and peramivir ranging from 5 × 10−7 μM to 50 μM) that inhibited 50% of the NA activity (IC50) was calculated from the dose-response curve using GraphPad (La Jolla, CA) Prism 5.0 software. The susceptibilities of recombinant viruses to NAIs were categorized according to the criteria recommended by the WHO Antiviral Working Group and based on the fold change in the IC50 compared with that of the susceptible virus: normal inhibition, <10-fold; reduced inhibition, 10- to 100-fold; highly reduced inhibition, >100-fold (20, 54).
Deep sequencing of NA genes of resistant variants.
Viral RNA was extracted using an RNeasy minikit (Qiagen), and cDNA was synthesized using a Moloney murine leukemia virus (MMLV) reverse transcriptase kit (Enzynomics). PCR of the influenza virus NA gene was performed using gene-specific primers. PCR products were extracted from 0.8% agarose and purified using a DNA purification kit (Cosmogenetech). The libraries were prepared using Illumina's Nextera XT DNA sample preparation kit according to the protocol. The libraries were sequenced using Illumina's MiSeq platform. Sequencing reads were then demultiplexed, quality trimmed, and filtered using CLC Genomics Workbench 7 (CLC Bio). The reads were aligned to the wild-type virus sequences, and the mapped reads were put through the quality-based variant detection pipeline. The variants were considered if they met the predefined quality scores and were present in both forward and reverse reads at equal ratios. In addition, the minimum variant read frequency was set at 5%, and variants had to be supported by a minimum of 10 reads.
ACKNOWLEDGMENTS
This work was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant numbers HI15C2888 and HI16C1032), and by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning (NRF-2015R1C1A2A01054406 to Y.H.B.).
We declare no conflict of interest.
REFERENCES
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webster RG, Govorkova EA. 2014. Continuing challenges in influenza. Ann N Y Acad Sci 1323:115–139. doi: 10.1111/nyas.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reperant LA, Kuiken T, Osterhaus AD. 2012. Influenza viruses: from birds to humans. Hum Vaccin Immunother 8:7–16. doi: 10.4161/hv.8.1.18672. [DOI] [PubMed] [Google Scholar]
- 4.Kim CU, Lew W, Williams MA, Wu H, Zhang L, Chen X, Escarpe PA, Mendel DB, Laver WG, Stevens RC. 1998. Structure-activity relationship studies of novel carbocyclic influenza neuraminidase inhibitors. J Med Chem 41:2451–2460. doi: 10.1021/jm980162u. [DOI] [PubMed] [Google Scholar]
- 5.Colman PM. 1994. Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci 3:1687–1696. doi: 10.1002/pro.5560031007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe C, Greenwald I, Chen L. 2010. Pandemic (H1N1) 2009 and oseltamivir resistance in hematology/oncology patients. Emerg Infect Dis 16:1809. doi: 10.3201/eid1611.101053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore C, Galiano M, Lackenby A, Abdelrahman T, Barnes R, Evans MR, Fegan C, Froude S, Hastings M, Knapper S. 2011. Evidence of person-to-person transmission of oseltamivir-resistant pandemic influenza A (H1N1) 2009 virus in a hematology unit. J Infect Dis 203:18–24. doi: 10.1093/infdis/jiq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurt AC, Holien JK, Parker M, Kelso A, Barr IG. 2009. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J Virol 83:10366–10373. doi: 10.1128/JVI.01200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le QM, Kiso M, Someya K, Sakai YT, Nguyen TH, Nguyen KH, Pham ND, Nguyen HH, Yamada S, Muramoto Y. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. doi: 10.1038/4371108a. [DOI] [PubMed] [Google Scholar]
- 10.Baz M, Abed Y, McDonald J, Boivin G. 2006. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis 43:1555–1561. doi: 10.1086/508777. [DOI] [PubMed] [Google Scholar]
- 11.Mishin VP, Hayden FG, Gubareva LV. 2005. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemother 49:4515–4520. doi: 10.1128/AAC.49.11.4515-4520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, Zhang X, Yen HL, Shi B, Li T, Guan W, Xu L, Liu Y, Wang S, Tian D, Zhu Z, He J, Huang K, Chen H, Zheng L, Li X, Ping J, Kang B, Xi X, Zha L, Li Y, Zhang Z, Peiris M, Yuan Z. 2013. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 381:2273–2279. doi: 10.1016/S0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- 13.Dapat C, Suzuki Y, Saito R, Kyaw Y, Myint YY, Lin N, Oo HN, Oo KY, Win N, Naito M. 2010. Rare influenza A (H3N2) variants with reduced sensitivity to antiviral drugs. Emerg Infect Dis 16:493. doi: 10.3201/eid1603.091321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheu TG, Deyde VM, Okomo-Adhiambo M, Garten RJ, Xu X, Bright RA, Butler EN, Wallis TR, Klimov AI, Gubareva LV. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother 52:3284–3292. doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song M-S, Marathe BM, Kumar G, Wong S-S, Rubrum A, Zanin M, Choi Y-K, Webster RG, Govorkova EA, Webby RJ. 2015. Unique determinants of neuraminidase inhibitor resistance among N3, N7, and N9 avian influenza viruses. J Virol 89:10891–10900. doi: 10.1128/JVI.01514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Chen M, Huang Y, Zhu W, Yang L, Gao L, Li X, Bi F, Huang C, Kang N. 2017. Human infections with novel reassortant H5N6 avian influenza viruses in China. Emerg Microbes Infect 6:e50. doi: 10.1038/emi.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang H, Wu P, Uyeki TM, He J, Deng Z, Xu W, Lv Q, Zhang J, Wu Y, Tsang TK. 2017. Preliminary epidemiologic assessment of human infections with highly pathogenic avian influenza A (H5N6) virus, China. Clin Infect Dis 65:383–388. doi: 10.1093/cid/cix334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Shesheny R, Barman S, Feeroz M, Hasan M, Jones-Engel L, Franks J, Turner J, Seiler P, Walker D, Friedman K. 2017. Genesis of influenza A (H5N8) viruses. Emerg Infect Dis 23:1368–1371. doi: 10.3201/eid2308.170143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y-I, Pascua PNQ, Kwon H-I, Lim G-J, Kim E-H, Yoon S-W, Park S-J, Kim SM, Choi E-J, Si Y-J, Lee O-J, Shim W-S, Kim S-W, Mo I-P, Bae Y, Lim YT, Sung MH, Kim C-J, Webby RJ, Webster RG, Choi YK. 2014. Pathobiological features of a novel, highly pathogenic avian influenza A(H5N8) virus. Emerg Microbes Infect 3:e75. doi: 10.1038/emi.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO. 2012. Meetings of the WHO working group on surveillance of influenza antiviral susceptibility—Geneva, November 2011 and June 2012. Wkly Epidemiol Rec 87:369–374. [PubMed] [Google Scholar]
- 21.van der Vries E, Jonges M, Herfst S, Maaskant J, Van der Linden A, Guldemeester J, Aron G, Bestebroer T, Koopmans M, Meijer A. 2010. Evaluation of a rapid molecular algorithm for detection of pandemic influenza A (H1N1) 2009 virus and screening for a key oseltamivir resistance (H275Y) substitution in neuraminidase. J Clin Virol 47:34–37. doi: 10.1016/j.jcv.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ives J, Carr J, Roberts N, Tai C, Mendel D, Kelly L, Lambkin R, Oxford J. 2000. An oseltamivir treatment-selected influenza A: N2 virus with a R292K mutation in the neuraminidase gene has reduced infectivity in vivo. J Clin Virol 18:251–269. doi: 10.1016/S1386-6532(00)00111-6. [DOI] [Google Scholar]
- 23.Bi Y, Mei K, Shi W, Liu D, Yu X, Gao Z, Zhao L, Gao GF, Chen J, Chen Q. 2015. Two novel reassortants of avian influenza A (H5N6) virus in China. J Gen Virol 96:975–981. doi: 10.1099/vir.0.000056. [DOI] [PubMed] [Google Scholar]
- 24.Du Y, Chen M, Yang J, Jia Y, Han S, Holmes EC, Cui J. 2017. Molecular evolution and emergence of H5N6 avian influenza virus in central China. J Virol 91:e00143-. doi: 10.1128/JVI.00143-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu M, Li X, Yuan H, Zhou J, Wu J, Bo H, Xia W, Xiong Y, Yang L, Gao R. 2015. Genetic diversity of avian influenza A (H10N8) virus in live poultry markets and its association with human infections in China. Sci Rep 5:7632. doi: 10.1038/srep07632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Qi J, Liu Y, Vavricka CJ, Wu Y, Li Q, Gao GF. 2011. Influenza A virus N5 neuraminidase has an extended 150-cavity. J Virol 85:8431–8435. doi: 10.1128/JVI.00638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Qin G, Gao F, Liu Y, Vavricka CJ, Qi J, Jiang H, Yu K, Gao GF. 2013. Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci Rep 3:1551. doi: 10.1038/srep01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samson M, Pizzorno A, Abed Y, Boivin G. 2013. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res 98:174–185. doi: 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Orozovic G, Orozovic K, Lennerstrand J, Olsen B. 2011. Detection of resistance mutations to antivirals oseltamivir and zanamivir in avian influenza A viruses isolated from wild birds. PLoS One 6:e16028. doi: 10.1371/journal.pone.0016028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoll V, Stewart KD, Maring CJ, Muchmore S, Giranda V, Gu Y-Y, Wang G, Chen Y, Sun M, Zhao C. 2003. Influenza neuraminidase inhibitors: structure-based design of a novel inhibitor series. Biochemistry 42:718–727. doi: 10.1021/bi0205449. [DOI] [PubMed] [Google Scholar]
- 31.Eshaghi A, Shalhoub S, Rosenfeld P, Li A, Higgins RR, Stogios PJ, Savchenko A, Bastien N, Li Y, Rotstein C. 2014. Multiple influenza A (H3N2) mutations conferring resistance to neuraminidase inhibitors in a bone marrow transplant recipient. Antimicrob Agents Chemother 58:7188–7197. doi: 10.1128/AAC.03667-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurt A, Lee R, Leang S, Cui L, Deng Y, Phuah S, Caldwell N, Freeman K, Komadina N, Smith D. 2011. Increased detection in Australia and Singapore of a novel influenza A (H1N1) 2009 variant with reduced oseltamivir and zanamivir sensitivity due to a S247N neuraminidase mutation. Euro Surveill 16:19884. [PubMed] [Google Scholar]
- 33.Boltz DA, Douangngeun B, Phommachanh P, Sinthasak S, Mondry R, Obert C, Seiler P, Keating R, Suzuki Y, Hiramatsu H. 2010. Emergence of H5N1 avian influenza viruses with reduced sensitivity to neuraminidase inhibitors and novel reassortants in Lao People's Democratic Republic. J Gen Virol 91:949–959. doi: 10.1099/vir.0.017459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abed Y, Baz M, Boivin G. 2009. A novel neuraminidase deletion mutation conferring resistance to oseltamivir in clinical influenza A/H3N2 virus. J Infect Dis 199:180–183. doi: 10.1086/595736. [DOI] [PubMed] [Google Scholar]
- 35.Prachanronarong KL, Ozen A, Thayer KM, Yilmaz LS, Zeldovich KB, Bolon DN, Kowalik TF, Jensen JD, Finberg RW, Wang JP. 2016. Molecular basis for differential patterns of drug resistance in influenza N1 and N2 neuraminidase. J Chem Theory Comput 12:6098–6108. doi: 10.1021/acs.jctc.6b00703. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann A, Schade D, Kirchmair J, Clement B, Sauerbrei A, Schmidtke M. 2016. Platform for determining the inhibition profile of neuraminidase inhibitors in an influenza virus N1 background. J Virol Methods 237:192–199. doi: 10.1016/j.jviromet.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 37.Hurt AC, Chotpitayasunondh T, Cox NJ, Daniels R, Fry AM, Gubareva LV, Hayden FG, Hui DS, Hungnes O, Lackenby A. 2012. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis 12:240–248. doi: 10.1016/S1473-3099(11)70318-8. [DOI] [PubMed] [Google Scholar]
- 38.Abed Y, Baz M, Boivin G. 2006. Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir Ther 11:971. [PubMed] [Google Scholar]
- 39.Zürcher T, Yates PJ, Daly J, Sahasrabudhe A, Walters M, Dash L, Tisdale M, McKimm-Breschkin JL. 2006. Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J Antimicrob Chemother 58:723–732. doi: 10.1093/jac/dkl321. [DOI] [PubMed] [Google Scholar]
- 40.Abed Y, Goyette N, Boivin G. 2005. Generation and characterization of recombinant influenza A (H1N1) viruses harboring amantadine resistance mutations. Antimicrob Agents Chemother 49:556–559. doi: 10.1128/AAC.49.2.556-559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuraminidase Inhibitor Flu Treatment Investigator Group, Little K, Leang S-K, Butler J, Baas C, Harrower B, Mosse J, Barr IG, Hurt AC. 2015. Zanamivir-resistant influenza viruses with Q136K or Q136R neuraminidase residue mutations can arise during MDCK cell culture creating challenges for antiviral susceptibility monitoring. Euro Surveill 20. doi: 10.2807/1560-7917.ES.2015.20.45.30060. [DOI] [PubMed] [Google Scholar]
- 42.Yen H-L, Hoffmann E, Taylor G, Scholtissek C, Monto AS, Webster RG, Govorkova EA. 2006. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J Virol 80:8787–8795. doi: 10.1128/JVI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho H-T, Hurt AC, Mosse J, Barr I. 2007. Neuraminidase inhibitor drug susceptibility differs between influenza N1 and N2 neuraminidase following mutagenesis of two conserved residues. Antivir Res 76:263–266. doi: 10.1016/j.antiviral.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Burnham AJ, Baranovich T, Marathe BM, Armstrong J, Webster RG, Govorkova EA. 2014. Fitness costs for Influenza B viruses carrying neuraminidase inhibitor-resistant substitutions: underscoring the importance of E119A and H274Y. Antimicrob Agents Chemother 58:2718–2730. doi: 10.1128/AAC.02628-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaymard A, Charles-Dufant A, Sabatier M, Cortay J-C, Frobert E, Picard C, Casalegno J-S, Rosa-Calatrava M, Ferraris O, Valette M, Ottmann M, Lina B, Escuret V. 2016. Impact on antiviral resistance of E119V, I222L and R292K substitutions in influenza A viruses bearing a group 2 neuraminidase (N2, N3, N6, N7 and N9). J Antimicrob Chemother 71:3036–3045. doi: 10.1093/jac/dkw275. [DOI] [PubMed] [Google Scholar]
- 46.Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 47.Varghese JN, Smith PW, Sollis SL, Blick TJ, Sahasrabudhe A, McKimm-Breschkin JL, Colman PM. 1998. Drug design against a shifting target: a structural basis for resistance to inhibitors in a variant of influenza virus neuraminidase. Structure 6:735–746. doi: 10.1016/S0969-2126(98)00075-6. [DOI] [PubMed] [Google Scholar]
- 48.Ives JAL, Carr JA, Mendel DB, Tai CY, Lambkin R, Kelly L, Oxford JS, Hayden FG, Roberts NA. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antivir Res 55:307–317. doi: 10.1016/S0166-3542(02)00053-0. [DOI] [PubMed] [Google Scholar]
- 49.Hurt AC, Holien JK, Parker MW, Barr IG. 2009. Oseltamivir resistance and the H274Y neuraminidase mutation in seasonal, pandemic and highly pathogenic influenza viruses. Drugs 69:2523–2531. doi: 10.2165/11531450-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 50.Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 51.Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol 74:6015–6020. doi: 10.1128/JVI.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potier M, Mameli L, Belisle M, Dallaire L, Melancon S. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-DN-acetylneuraminate) substrate. Anal Biochem 94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 54.Okomo-Adhiambo M, Nguyen HT, Abd Elal A, Sleeman K, Fry AM, Gubareva LV. 2014. Drug susceptibility surveillance of influenza viruses circulating in the United States in 2011-2012: application of the WHO Antiviral Working Group criteria. Influenza Other Respir Viruses 8:258–265. doi: 10.1111/irv.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]