ABSTRACT
The influenza A virus M1 and M2 proteins play important roles in virus assembly and in the morphology of virus particles. Mutations in the distal cytoplasmic tail region of M2, and in particular a tyrosine-to-alanine mutation at residue 76 (Y76A), were essential for infectious virus production and filament formation while having limited effects on total virus particle budding. Using a novel selection method, mutations at seven different M1 amino acids (residue 73, 94, 135, 136, or 138 or a double mutation, 93/244) that are not found in circulating influenza virus strains or have not been previously identified to play a role in influenza A virus assembly were found to complement the lethal M2Y76A mutation. These M1 suppressor mutations restored infectious virus production in the presence of M2Y76A and mediated increased budding and filament formation even in the absence of M2. However, the efficiency of infectious virus replication was still dependent on the presence of the distal region of the M2 cytoplasmic tail. The data suggest that influenza A virus budding and genome incorporation can occur independently and provide further support for complementary roles of the M1 and M2 proteins in virus assembly.
IMPORTANCE Influenza virus particle assembly involves the careful coordination of various viral and host factors to optimally produce infectious virus particles. We have previously identified a mutation at position 76 of the influenza A virus M2 protein that drastically reduces infectious virus production and filament formation with minimal effects on virus budding. In this work, we identified suppressor mutations in the M1 protein which complement this lethal M2 mutation by increasing the efficiency with which virus particles bud from infected cells and restoring filament formation at the infected-cell surface. M2 distal cytoplasmic domain sequences were still required for optimal infectivity. This indicates that M1 and M2 can functionally replace each other in some, but not all, aspects of virus particle assembly.
KEYWORDS: M1, M2, assembly, influenza
INTRODUCTION
Influenza A virus (IAV), a member of the Orthomyxoviridae, is a respiratory pathogen. The genome consists of eight segmented negative-sense RNAs associated with a viral replicase complex (PA, PB1, and PB2) and multiple viral nucleoproteins (NPs) to form viral ribonucleoproteins (vRNPs). Newly assembled vRNPs may also contain matrix 1 (M1) and nuclear export protein (NEP). The lipid bilayers of influenza A virus particles contain three viral integral membrane proteins, hemagglutinin (HA), neuraminidase (NA), and matrix 2 (M2) (1).
The HA, NA, M1, and M2 proteins are important for influenza A virus assembly and budding (2). HA and NA cluster in virus budding sites at the plasma membrane and may induce membrane curvature (3–5). M1 is a central structural component of the virus particle, and electron microscopy shows that M1 proteins are present on the inner leaflet of the membrane of virus particles (6). M1 potentially interacts with the cytoplasmic tails of HA and NA (7–9). M1 has also been shown to directly bind vRNPs and the NEP to shuttle vRNP from the nucleus to the cytoplasm and may serve as a docking site for vRNPs packing into virions (10–12). The M1 protein is 252 amino acids (aa) in length and contains an N-terminal domain, a middle domain, and a C-terminal domain. Amino acid residues across all three domains have been shown to alter virus budding, virus particle morphology, and/or vRNP incorporation into virions (13–19). The structure of the N-terminal and middle domains of M1 (residues 1 to 164) contains two 4-helix bundles that are connected by a helical structure (20, 21). M1 crystal structures and computer modeling show that the M1-M1 interphase can have different orientations depending upon the pH (22, 23). This may indicate that the structure and oligomeric state of M1 vary at different stages of the virus life cycle.
M2 is a 97-amino-acid protein and forms homotetramers in cellular membranes. The ion channel activity of M2 is important for the release of vRNPs into the cytoplasm during virus entry (24). Another function of M2 is during virus assembly. Mutations or deletions in the M2 cytoplasmic domain alter virus particle morphology, decrease virus infectivity, and reduce the incorporation of NP and viral RNA into progeny virus particles (25–29). It is also thought that M2 is important for membrane scission that allows for release of virions from the cell surface (30–32). Studies have shown that M1 and M2 are located close to each other in the infected cells and directly interact in vitro (26, 28).
Previously, we used a scanning-alanine mutation strategy to identify a lethal tyrosine-to-alanine mutation at amino acid 76 of M2 (M2Y76A) in the influenza A virus A/Udorn/72 strain which also decreased the infectivity of the A/WSN/33 strain (33). The M2 mutation did not affect ion channel activity but led to reduced NP incorporation into virions and altered filament formation on virus-infected cells (33). To better investigate whether mutations in other viral proteins could complement the lethal M2Y76A mutation, blind passage of an A/Udorn/72 recombinant virus containing a premature stop codon in the M2 proteins (rUdorn-M2Stop) was conducted in an MDCK cell line stably expressing M2Y76A. Since M2 is not expressed by the viral genome in this system, mutations in other viral proteins are needed to complement the M2Y76A mutation. Eight viruses containing 7 different mutations in the M1 protein were identified. These mutations were located at the M1-M1 interphase and resulted in viruses with enhanced budding and filamentous particle formation. The data provide important insights into the role of M1 in influenza A virus assembly and on the cooperative roles of M1 and M2 in virus budding.
RESULTS
Isolation of IAV with suppressor mutations for M2Y76A.
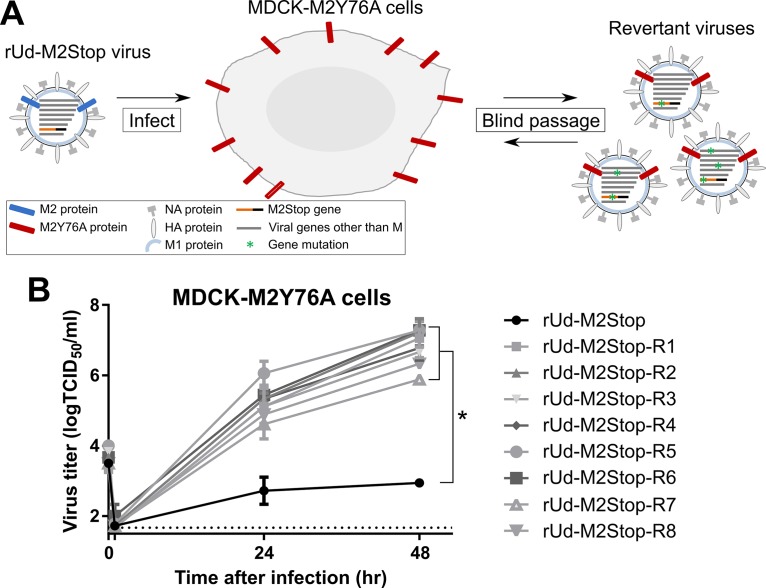
A tyrosine-to-alanine mutation at amino acid 76 of the influenza virus M2 protein was found to abolish infectious virus production and reduce virus particle assembly (33). A revertant virus which contained a serine-to-tyrosine mutation at amino acid 71 of the M2 protein had been isolated (33). To gain insight into other viral proteins which might be important in interacting with the M2 Y76 residue, we blind passaged an A/Udorn/72 H3N2 virus containing a premature truncation in the M2 open reading frame (ORF) (rUd-M2Stop) on MDCK cells expressing the Udorn M2Y76A protein (MDCK-M2Y76A). Using this selection, we would minimize the identification of suppressor mutations in the M2 protein—since it was now expressed under the control of a cellular promoter—and maximize the identification of mutations in other viral proteins that could complement the M2Y76A mutation (Fig. 1A). As expected, the rUd-M2Stop virus was unable to produce significant amounts of infectious virus particles in MDCK-M2Y76A cells. After two blind passages on MDCK-M2Y76A cells, a cytopathic effect was detected in the cultures. Plaque assays were performed on the infected-cell supernatants using MDCK-M2Y76A cells to isolate clonal populations of viruses. Eight plaques yielded viruses capable of producing infectious virus in MDCK-M2Y76A cells after infection at a low multiplicity of infection (MOI) (Fig. 1B).
FIG 1.
Identification of suppressor mutations of IAV M2Y76A. (A) Schematic depicting the selection of IAV M2Y76A suppressor mutations. MDCK cells overexpressing M2Y76A were infected with rUd-M2Stop virus. After two blind passages, cytopathic effects were observed and viruses were isolated by plaque picking. (B) Low-MOI growth curve of plaque-purified viruses with suppressor mutations for M2Y76A. The dotted line indicates the limit of detection. The asterisk indicates a statistically significant difference compared to rUd-M2Stop-infected cells (P < 0.01 using two-way multiple-comparison ANOVA with Bonferroni posttest). Experiments were done three times, and representative data are shown. Error bars are standard deviations.
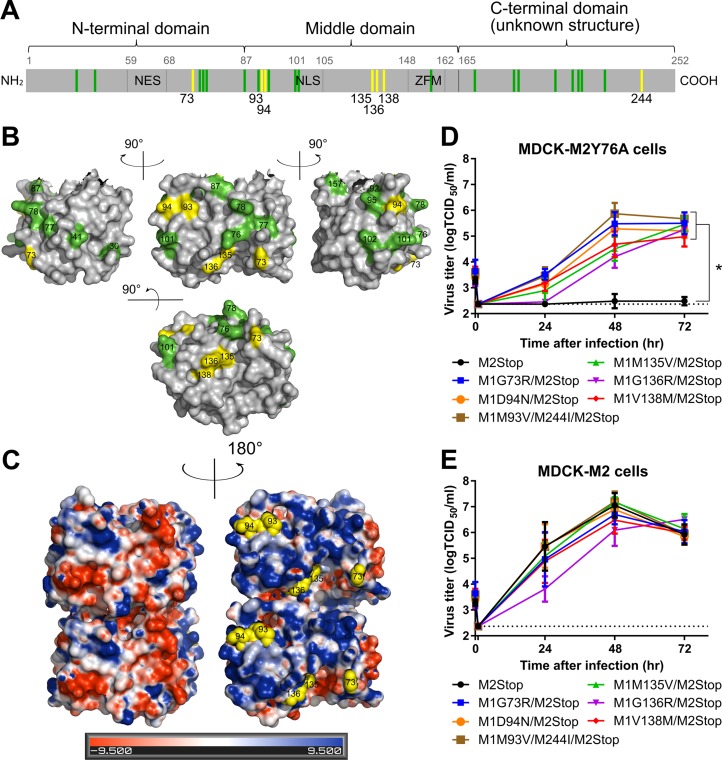
The M segment sequences of the 8 plaque-purified viruses were determined. All contained the two stop codons in the M2 ORF present in the parental rUd-M2Stop virus in addition to either one or two amino acid mutation(s) in the M1 ORF (Table 1). These M1 suppressor mutations are located at 7 distinct positions of the protein and are present in the N-terminal, middle, and C-terminal regions of the protein (yellow lines, Fig. 2A). When mapped onto the M1 X-ray diffraction crystal structure (yellow, Fig. 2B), they all fall onto one face of the monomer. None of these amino acid positions have been previously identified as playing a role in any M1 function, including a role in virus particle assembly, but they do fall close to a number of residues previously identified to play a role in influenza virus particle formation (green, Fig. 2A and B) (13–19). The M1 protein has both a positively charged and a negatively charged surface on opposite sides of the protein (21). The M1 suppressor mutations are all located on the positively charged surface of M1 (Fig. 2C). The positively charged side of M1 has been proposed to interact with the negatively charged surface of M1, the cell membrane, and vRNPs (21, 23). Residues 93 and 94 are located in M1 helix 6, which was previously shown to influence virion morphology (15). Residue 73 is within the region that connects the two 4-helix bundles. Residues 94 and 136 have been shown to be within the M1-M1 interface in several studies, and residues 73, 93, 135, and 138 are located close to the M1-M1 interface (22, 23).
TABLE 1.
M gene sequences present in rUd-M2Stop viruses which have the ability to produce infectious virus particles from MDCK-M2Y76A cells
| Revertant virus | M gene sequence |
|
|---|---|---|
| M1 ORF | M2 ORF | |
| R1 | G73R | Stop |
| R2 | M135V | Stop |
| R3 | V138M | Stop |
| R4 | G136R | Stop |
| R5 | M135T | Stop |
| R6 | M93V/M244I | Stop |
| R7 | G73R | Stop |
| R8 | D94N | Stop |
FIG 2.
M1 suppressor mutations complement the M2Y76A defect in virus replication. The M gene of viruses with suppressor mutations for M2Y76A was sequenced, and rUd-M2Stop viruses with M1 suppressor mutations were generated by reverse genetics. (A) The identified suppressor mutations in M1 protein (yellow) and residues known to affect virus morphology/assembly (green) are shown (13–19). The nuclear export sequence (NES), nuclear localization sequence (NLS), and the zinc finger motif (ZFM) are also indicated. (B) M1 suppressor mutations (yellow) and known residues that affect virus morphology (green) are shown on the M1 crystal structure (aa 1 to 164, Protein Data Bank [PDB] accession no. 1EA3 [21]) using PyMOL. (C) M1 suppressor mutations (yellow spheres) are located on the positively charged surface (blue) and not the negatively charged surface (red) of the M1 dimer neutral-pH structure (PDB accession no. 1EA3). Electrostatic surface potentials of M1 were generated by the PDB2PQR server and PyMOL APBS tools. (D and E) rUd-M2Stop virus and rUd-M2Stop viruses with M1 suppressor mutations were used to infect MDCK-M2Y76A cells (D) or MDCK-M2 cells (E) at an MOI of 0.001. Infectious virus titers in the cell supernatants were quantified using TCID50 assays. The dotted lines indicate the limits of detection. Asterisks indicate statistically significant differences compared to rUd-M2Stop-infected cells (P < 0.01 using two-way multiple-comparison ANOVA with Bonferroni posttest). Data were analyzed from three independent experiments. Error bars are standard deviations.
M1 suppressor mutations complement the M2Y76A defect in infectious virus production.
To determine whether a single mutation on M1 can complement the M2Y76A defect, recombinant viruses encoding each M1 suppressor mutation were rescued. Six viruses with M1 suppressor mutations (G73R, M135V, G136R, V138M, D94N, and M93V/M244I) were generated in the Ud-M2Stop background. MDCK-M2Y76A cells were infected with these recombinant M1 mutant/M2Stop viruses at a low MOI. As expected, the parental rUd-M2Stop virus produced virtually no infectious virus, but all recombinant viruses encoding M1 suppressor mutations produced infectious virus, though the amounts varied depending on the specific mutation (Fig. 2D). In MDCK-M2 cells, all the suppressor viruses grew to infectious virus titers that were comparable to those of rUd-M2Stop (Fig. 2E). All M1 suppressor viruses grew to higher titers in MDCK-M2 cells than in MDCK-M2Y76A cells, suggesting that the M1 suppressor mutations were not able to completely restore infectious virus production in the presence of M2Y76A. The data indicate that the M1 suppressor mutations alone can complement the M2Y76A defect in virus replication.
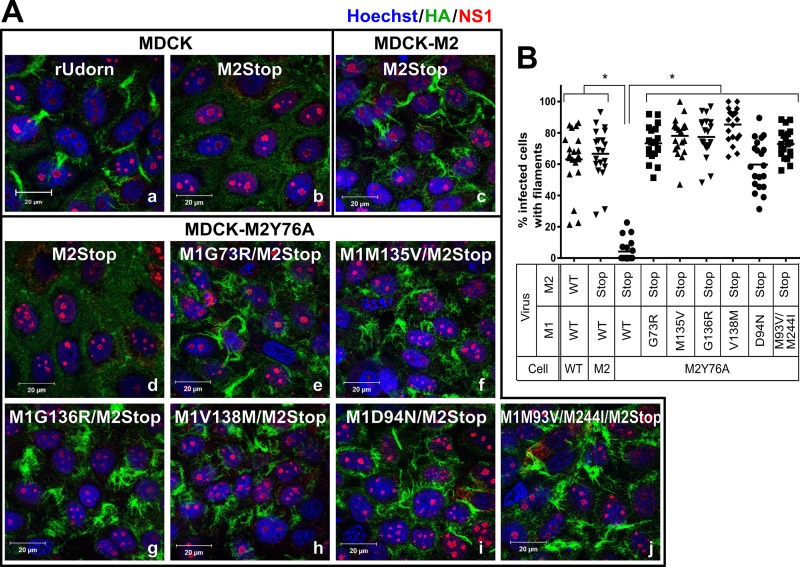
M1 suppressor mutations complement filament formation on the surface of MDCK-M2Y76A cells.
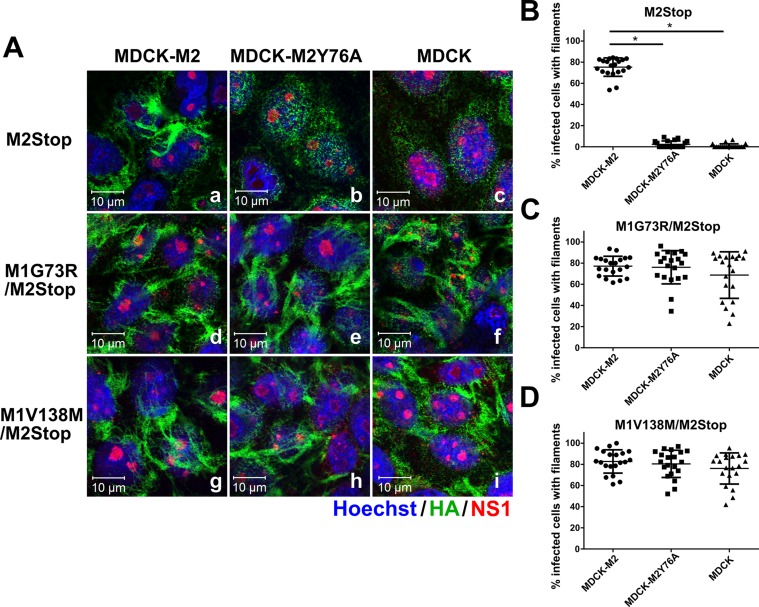
To determine whether M1 suppressor mutations complement the M2Y76A defect in filament formation, the recombinant viruses were used to infect confluent MDCK-M2Y76A cells and filament formation on the infected-cell surface was assessed 16 h postinfection (hpi) by immunostaining for the viral HA protein (Fig. 3A). The rUd-M2Stop virus produced filaments after infection of MDCK-M2 cells, but filaments were not detected after infection of MDCK or MDCK-M2Y76A cells (Fig. 3A-b to -d). All six recombinant viruses encoding M1 suppressor mutations produced clearly visible filamentous structures in 60 to 80% of infected M2Y76A cells (Fig. 3A-e to -j), which were similar to those found on rUdorn virus-infected MDCK cells (Fig. 3A-a) and M2Stop virus-infected MDCK-M2 cells (Fig. 3A-c and B). These data indicate that the M1 suppressor mutations could restore the M2Y76A defect on filament formation on the infected-cell surface.
FIG 3.
M1 suppressor mutations complement the M2Y76A defect in filamentous particle budding. (A) rUdorn and rUd-M2Stop viruses with the indicated M1 suppressor mutations were used to infect MDCK-M2Y76A cells at a high MOI. rUdorn-infected MDCK cells (wild type) were used as a positive control for filament formation. Surface HA staining by immunofluorescence assay with anti-HA antibody (green) at 16 hpi was used to detect filamentous structures. NS1 (red) is shown to identify all virus-infected cells, and nuclei are stained with Hoechst stain (blue). The images were taken with an LSM 510 confocal microscope with a 100× objective. Independent experiments were done three times, and representative images are shown. (B) The percentage of infected cells showing viral filaments was quantified by counting total infected cells and cells possessing filaments in 20 independent fields per sample. The data are from one experiment and are representative of three independent experiments. Asterisks indicate statistically significant differences compared to rUd-M2Stop-infected M2Y76A cells (P < 0.01, one-way ANOVA with Bonferroni posttest). WT, wild type.
M1 suppressor mutations enhance filament formation on the cell surface even in the absence of M2.
To better understand the mechanisms by which the M1 suppressor mutations restore infectious virus production, two M1 suppressor mutations, M1G73R and M1V138M, were selected for further characterization. These two mutations have robust phenotype changes and have different amino acid substitutions that are in different regions of M1 (Fig. 2B). To determine if the enhanced filament formation mediated by the M1 suppressor mutations required the presence of the M2 protein, recombinant viruses encoding M1 suppressor mutations were used to infect MDCK, MDCK-M2, and MDCK-M2Y76A cells and filament formation was assessed at 16 hpi (Fig. 4). Viruses with the M1 suppressor mutations produced filaments on the surface of MDCK-M2 cells that were indistinguishable from those seen in MDCK-M2Y76A cells (Fig. 4A-d, -e, -g, and -h). Interestingly, we observed that these M1 suppressor viruses could also produce filamentous structures in infected MDCK cells (Fig. 4A-f and -i), indicating that the increased filament production was independent of the expression of M2. As expected, the parental rUd-M2Stop virus was defective in filament formation in MDCK and MDCK-M2Y76A cells (Fig. 4A-b and -c). Quantitation of the cell surface filaments indicated that the M1 suppressor-encoding viruses were equally efficient at producing filaments in all three cell lines (Fig. 4C and D) while the parental rUd-M2Stop was able to produce filaments only in MDCK-M2 cells (Fig. 4B). The data suggested that the two M1 suppressor mutations can restore filament formation even in the absence of M2.
FIG 4.
M1 suppressor mutations also induce filamentous virus budding in the absence of M2. (A) MDCK, MDCK-M2Y76A, and MDCK-M2 cells were infected at a high MOI with rUd-M2Stop, rUd-M1G73R/M2Stop, and rUd-M1V138M/M2Stop. Filamentous particle budding at 16 hpi was analyzed by immunofluorescence assay. HA, NS1, and nucleus are shown in green, red, and blue, respectively. The images were taken with an LSM 510 confocal microscope with a 100× objective. Independent experiments were done three times, and representative images are shown. (B to D) The percentages of infected cells showing viral filaments per field of rUd-M2Stop (B), rUd-M1G73R/M2Stop (C), and rUd-M1V138M/M2Stop (D) viruses were quantified by counting 20 fields per sample (*, P < 0.01; one-way ANOVA with Bonferroni posttest compared to MDCK-M2 cells). The data shown are from one of three independent experiments, all of which had similar results.
M1 suppressor mutations increase virus budding, alter virion protein and vRNA composition, and increase infectious virus production.
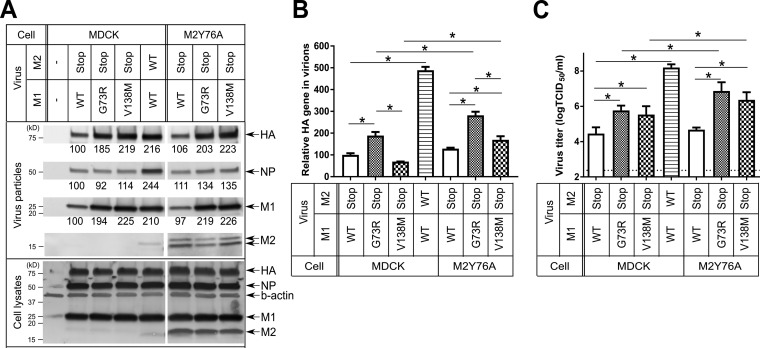
To determine the effect of the M1 suppressor mutations on the composition of released virions, Western blotting was performed on virions that were pelleted through a sucrose cushion (Fig. 5A). Equal levels of HA, NP, and M1 protein expression were detected in all virus-infected-cell lysates, indicating that there was no alteration in the amount of viral proteins produced (Fig. 5A). Similarly to our previous studies using the same monoclonal antibody to M2 (33), there were M2 doublets detected in virus particles but not in the cell lysates of MDCK-M2Y76A cells (Fig. 5A). To determine the relative difference in the amount of virus budding, the band intensity of each viral protein was measured and normalized to the protein levels found in rUd-M2Stop-infected MDCK cells (Fig. 5A). In MDCK cells, viruses encoding M1 suppressor mutations had a greater amount of released HA and M1 protein than did the parental rUd-M2Stop virus. The amount of NP protein released from M1 suppressor virus-infected cells was not increased, suggesting that not all viral structural proteins were preferentially packaged into M1 suppressor virus particles. Supernatants from rUdorn-infected cells showed increases in HA, M1, and NP proteins compared to rUd-M2Stop-infected cells, which reflects increased particle budding and infectivity in rUdorn-infected-cell supernatants. MDCK-M2Y76A cells showed a phenotype similar to that of MDCK cells with supernatants from M1 suppressor virus-infected cells showing increased HA and M1 proteins but not a comparable increase in NP protein. Taken together, the data suggest that the M1 suppressor mutations increase the overall budding of virions from infected cells even in the absence of M2, without a corresponding increase in NP incorporation.
FIG 5.
M1 suppressor mutations increase virus budding, alter virion protein composition, and enhance infectious virus production. The indicated viruses were used to infect MDCK and MDCK-M2Y76A cells at an MOI of 1.5. Virus particles and infected-cell lysates were collected at 16 hpi. (A) Virus particles partially purified from infected-cell supernatants and the infected-cell lysates were analyzed by Western blotting. Band intensity was quantified using ImageJ and normalized to the intensity of each protein of rUd-M2Stop-infected MDCK cells. Independent experiments were done two to three times, and representative Western blotting data are shown. (B) Viral HA RNA of virus particles was quantified using RT-qPCR with Udorn-HA-specific primers and probes. Relative HA gene in virions was calculated by normalizing the CT value of each sample to that from rUd-M2Stop-infected MDCK cells. (C) Infectious virus titers of virus particles were determined by TCID50 assays (*, P < 0.01; one-way ANOVA with Bonferroni posttest). Data were analyzed from two to three independent experiments. Error bars are standard deviations. Two independent experiments were performed, each in triplicate. The same set of samples was analyzed in all three panels. WT, wild type.
To determine the effect of the M1 suppressor mutations on viral RNA packaging, real-time reverse transcription-PCR (RT-PCR) targeting the HA gene segment was used to quantify viral RNA extracted from the partially purified virus particles. The relative amount of HA RNA of each sample was normalized to the HA RNA levels found in rUd-M2Stop-infected MDCK cells (Fig. 5B). In MDCK cells infected with the M1 suppressor viruses, there was no consistent increase in vRNA packaging, with the M1G73R virus showing increased HA vRNA packaging while the M1V138M virus had no change. This suggested that the M1 suppressor mutations, alone, had minimal effects on vRNA packaging, even though they were able to increase particle budding (Fig. 5A). After infection of MDCK-M2Y76A cells, both M1 suppressor viruses showed an increase in the amount of HA vRNA packaged compared to MDCK-M2Y76A cells infected with rUd-M2Stop. Both M1 suppressor viruses also packaged more HA vRNA in MDCK-M2Y76A cells than in MDCK cells, indicating that the M2 cytoplasmic tail was contributing to vRNA packaging (Fig. 5B). The vRNA packaging of M1G73R virus was higher than that of the M1V138M virus in both MDCK and MDCK-M2Y76A cells (Fig. 5B), which is consistent with a higher growth of M1G73R virus than M1V138M virus after low-multiplicity infection (Fig. 2D).
To determine the effect of the M1 suppressor mutations on infectious virus particle production, the infectious virus titers of the partially purified virus particles were determined by 50% tissue culture infective dose (TCID50) assay on MDCK-M2 cells (Fig. 5C). Compared to the parental virus, the M1 suppressor mutants produced about 10-fold more infectious particles in MDCK cells but approximately 100-fold more infectious particles in MDCK-M2Y76A cells (Fig. 5C). The increase in infectious virus titers seen in MDCK-M2Y76A cells is consistent with the increase in viral RNA packaging (Fig. 5B). This most likely reflects the requirement of the M2 ion channel activity, which is essential for infectious virus production (34), and the presence of the C terminus of M2, which is known to enhance vRNP incorporation into virus particles (25, 28). Combined with the data regarding filament formation (Fig. 4), these data indicate that the M1 suppressor mutants are likely able to increase virion budding in the absence of M2 but that the infectivity of those virus particles is dependent upon the presence of M2.
The M2 C-terminal tail affects M1 suppressor mutation efficiency in restoring virus replication.
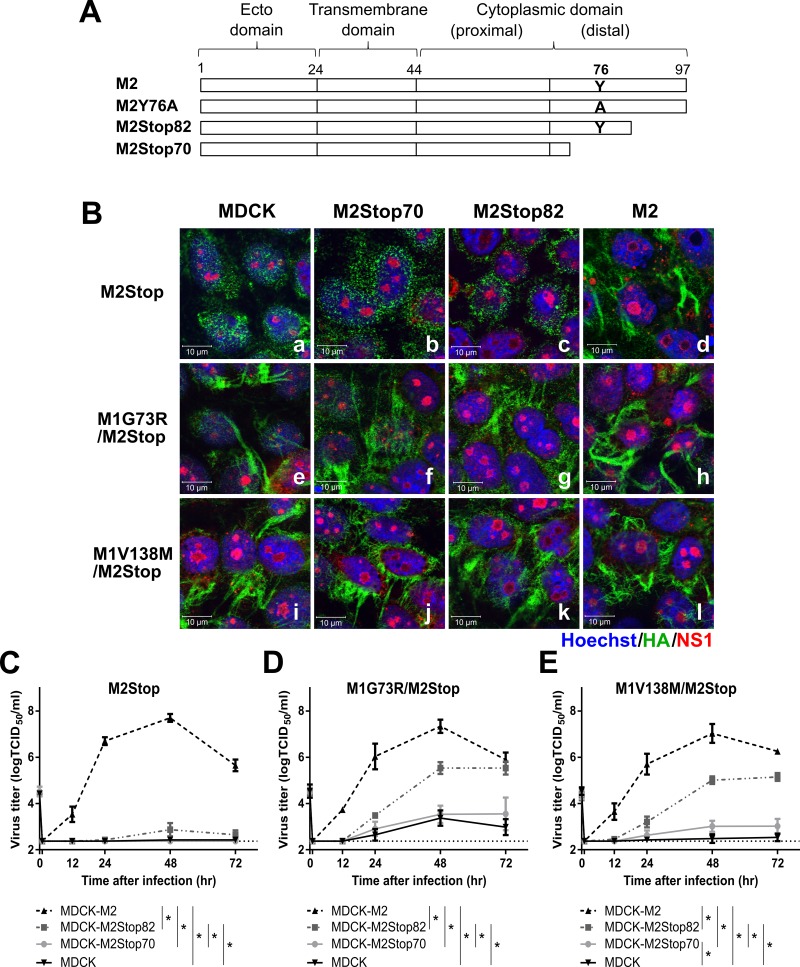
The membrane-distal region of the M2 cytoplasmic tail (amino acids [aa] 71 to 97) is important for the growth, assembly, and virion morphology of influenza A virus (25, 27). In order to determine whether the M1 suppressor mutations functionally replace the membrane-distal regions of M2 for infectious virus production, two previously established cell lines, MDCK cells expressing M2Stop82 (MDCK-M2Stop82) and MDCK cells expressing M2Stop70 (MDCK-M2Stop70) (Fig. 6A) (28), were infected with viruses encoding the M1 suppressor mutations. The two M2 truncations do not alter the ion channel activity of M2, but M2Stop70 deletes a region between amino acids 72 and 78 which has previously been shown to be critical for infectious virus production. Viruses encoding the M1 suppressor mutations complemented the defect of filament formation at the cell surface of the parental rUd-M2Stop virus on MDCK-M2Stop70 and MDCK-M2Stop82 cells as well as on MDCK cells (Fig. 6B-a, -b, -c, -e, -f, -g, -i, -j, and -k). The data support the results that M1 suppressor viruses can enhance filament formation independently of M2.
FIG 6.
The efficiency of M1 suppressor-mediated restoration of IAV infectivity is enhanced by the C-terminal region of the M2 cytoplasmic tail. The indicated viruses were used to infect MDCK, MDCK-M2Stop70, MDCK-M2Stop82, and MDCK-M2 cells. (A) Diagram of M2 truncations and M2Y76A. (B) Filamentous particle budding after a high-MOI infection was analyzed by immunofluorescence assay using an LSM 510 confocal microscope and a 100× objective. HA, NS1, and nucleus are shown in green, red, and blue, respectively. Representative images are shown. (C to E) Virus growth curves of rUd-M2Stop (C), rUd-M1G73R/M2Stop (D), and rUd-M1V138M/M2Stop (E) at an MOI of 0.01 were quantified using TCID50 assays. The dotted lines indicate the limits of detection. Statistically significant differences between cell lines are indicated by an asterisk (P < 0.01, two-way ANOVA with Bonferroni posttest). Data were analyzed from 2 to 3 independent experiments. Error bars are standard deviations.
To test the requirement of the M2 cytoplasmic tail for M1 suppressor mutations to complement infectious virus production, a multicycle growth curve was performed with the M1 suppressor viruses on MDCK, MDCK-M2Stop70, MDCK-M2Stop82, and MDCK-M2 cells. While the parental rUd-M2Stop virus was able to productively replicate only in MDCK-M2 cells (Fig. 6C), the M1 suppressor viruses partially restored infectious virus production in MDCK-M2Stop82 cells (Fig. 6D and E), similar to the complementation in MDCK-M2Y76A cells (Fig. 2D). No significant amounts of infectious virus were detected in the supernatants of M1 suppressor virus-infected MDCK-M2Stop70 or MDCK cells (Fig. 6D and E), indicating that the M1 suppressor mutations enhanced filament formation at the surface of those cells (Fig. 6B) but that the released particles were not highly infectious. Of particular importance, the M1 suppressor viruses were not able to productively infect MDCK-M2Stop70 cells—which express an M2 with a functional ion channel activity and membrane scission domain—suggesting that M2 amino acids 71 to 82 are critical for efficient infectious virus particle formation (Fig. 6D and E) and that this function could not be complemented by the M1 suppressor mutations. Taken together, the M1 suppressor mutations enhance filament formation independently of M2, but the efficiency of infectious virus production requires M2 and specific regions of the M2 cytoplasmic tail.
DISCUSSION
M1 and virus assembly, morphology, and vRNP incorporation.
Mutations in the M1 or M2 proteins affect influenza A virus morphology. M1 and M2 have been shown to localize close to each other on the surface of infected cells and directly bind to each other in vitro (26, 28). Blocking M2 function with the M2 monoclonal antibody 14c2 allowed selection of revertant viruses with M1 mutations at residues 31 and 41, with the mutation at residue 41 altering virion morphology (17, 35). Swapping the M1 and/or M2 genes between filamentous and spherical influenza virus strains alters particle morphology (14, 28). All of these suggest that M1 and M2 may interact during the assembly of virus particles.
Using a novel M2 complementation and selection approach, M1 suppressor mutations which could complement a lethal M2Y76A mutation were identified and characterized for their ability to affect virus particle assembly. Since these M1 suppressor mutations increased virus budding in the absence of M2, it is likely that they function by increasing influenza virus budding in a manner that does not require an interaction with M2. Since the M1 suppressor mutations all mapped to a face of the M1 protein thought to be important for M1 oligomerization, we hypothesize that the mutations improved the oligomerization of M1, providing a driving force for increased budding and increased filamentous particle formation. While the recombinant viruses that were rescued clearly implicate the M1 suppressor mutations as driving the rescue of viruses encoding M2Y76A, it will be interesting to sequence all the other gene segments to see whether other amino acid changes have occurred during the selection of the M1 suppressor viruses.
The efficiency with which M1 suppressor mutations restored infectious virus production depended on the presence of the M2 protein and, specifically, the membrane-distal region of the cytoplasmic tail (aa 71 to 97). The M2 membrane-distal cytoplasmic tail is required for efficient influenza A virus assembly, vRNP incorporation, and infectious virus production (25, 26, 28, 33). While there was no increase in virus particle budding mediated by the M1 suppressor mutations in the presence of M2, virus infectivity increased significantly. This increased infectivity was not present with an M2 construct encoding the first 70 amino acids of the protein, M2Stop70, suggesting that those regions were not sufficient to increase virus particle infectivity. These data further support a role for M2 in infectious virus production in addition to its other roles in the virus life cycle (2). Our data are consistent with a model where the M1 suppressor mutations can become the driving factor for virus particle budding, with the distal region of the M2 cytoplasmic tail still being important for efficient packaging of NP and vRNPs into the budding particles. In its essence, the infectious virus assembly process can be divided into a particle budding stage and a genome incorporation stage, with the former being able to proceed in the absence of the latter.
We previously showed that an M2S71Y mutation can complement M2Y76A and restore virus replication and filamentous particle production as well as NP incorporation (33). In contrast, M1 suppressor mutations slightly enhanced NP incorporation in the infected M2Y76A cells but not in the infected cells lacking M2 (Fig. 5A). It is likely that proper vRNP incorporation requires the membrane-distal region (amino acids 71 to 82) of the M2 cytoplasmic tail (25–29, 33), and the M1 suppressor mutations are not able to completely complement this M2 function. The mechanisms involved in M2S71Y/M2Y76A reversion seem different from those mediating the M1 suppressor mutations' rescue of M2Y76A. It will be important to assess if the M1 suppressor mutations alter M1-M2 interactions directly or if the M1 suppressor effects are mediated through M1-M1 interactions or interactions with other viral proteins.
Roles for M1 and M2 in membrane scission.
The membrane-proximal region of the M2 cytoplasmic tail—an amphipathic helix (aa 45 to 62)—has been shown to be able to mediate membrane scission and subsequent virus release from cells (32, 36). This region is responsible for cholesterol binding which is thought to be necessary and sufficient for negative Gaussian curvature (NGC) generation to bend the membrane (37). Other studies suggest that this region has only modest effects on virus replication (38). However, other domains of M2 and other viral proteins cooperatively promote membrane curvature (37). In our study, M1 suppressor mutants enhanced virus budding even in the absence of M2, suggesting that M1 might be able to contribute significantly to membrane scission. The results support recent speculation (39) that M2 may not be the only factor required for influenza virus membrane scission, because the beads-on-string particles seen in viruses that do not encode M2 or encode mutations in the M2 membrane-proximal regions do not seem to contain a complete M1 layer (39). The M1 structure is composed of multiple amphipathic helices and has putative cholesterol binding domains, and M1 alone can induce membrane curvature (20, 21, 40, 41). Future experiments assessing the contribution of M1 and M2 to membrane scission in virus-infected cells are needed.
M1 structure and a proposed function for the M1 suppressor mutations.
The surface charge potential of the M1 monomer shows that one side is positively charged and one side is negatively charged. During influenza virus assembly, M1 polymerizes underneath the plasma membrane and particularly at sites of virus budding. The positive face of M1 was proposed to electrostatically interact with the cytoplasmic side of the plasma membrane or the negatively charged face of M1 or perhaps with vRNPs (21, 23). The M1 suppressor mutations (aa 73, 93, 94, 135, 136, and 138) are located on a positively charged surface of M1 (Fig. 2C). Our data showed that at least two of the M1 suppressor mutations, aa 73 and 138, efficiently enhanced virus budding but seemed to have minimal effects on NP incorporation in the absence of M2, suggesting that these M1 suppressor mutations may be altering M1 oligomerization on the plasma membrane. Many of the M1 residues previously known to be important for virus morphology (aa 169, 183, 185, 198, 204, 207, and 209) are within the flexible C-terminal domain, which has been suggested to be important to maintain the integrity of the virion (42). All the other amino acids that affect morphology (aa 30, 41, 76, 77, 78, 87, 92, 95, 101, 102, and 157) are within the N-terminal and middle domains and are located on the surface of M1 (Fig. 2A and B) (13–19). Interestingly, none of these M1 residues are located on the negatively charged side of M1. While many of the M1 suppressor mutations lie in the positively charged face of the M1 protein, they do not consistently alter the charge of this region and can be rather conservative (M135V and V138M, for example), suggesting that the M1 suppressor mutations are not simply changing the overall charge in this region of the M1 protein.
The N-terminal and middle domains of M1 (aa 1 to 164) are composed of nine helices. M1 suppressor residues 93 and 94 and known residues that affect virus morphology (aa 92, 95, 101, and 102) are located in the amphipathic helix 6 (15). Residues 93 and 94 are located on the side that was suggested to interact with lipid raft membranes, and residues on the other side of the helix (R/K95, K98, and K102) have been shown to directly interact with vRNP (15, 40). M1 suppressor residues 94 and 136, residues in helix 6, and almost all other known residues within the N-terminal and middle domains that affect virus morphology were proposed to be involved in M1-M1 interactions under certain conditions (22, 23). M1 mutations (aa 87, 92, 101, and 157) that accumulated during repeated passage of a spherical A/PR8/1934 virus in guinea pigs resulted in filamentous particles (19). All of these residues are located at the M1-M1 interface, suggesting that M1 oligomerization is important for determination of influenza virus particle morphology.
Conservation of M1 suppressor mutation sequences and M1-M2 cooperativity in virus budding.
There is a high degree of conservation in natural isolates of influenza virus from animals and humans at the amino acids corresponding to the M1 suppressor mutations (43). This suggests a highly conserved function for these residues in the influenza virus life cycle. Influenza A virus assembly and budding require many viral proteins as well as host proteins to work together to induce membrane curvature, coordinate viral protein packaging, incorporate viral genomic RNA into budding sites, and release newly formed particles from the plasma membrane. Our study demonstrates that cooperativity between the M1 and M2 proteins is important for efficient assembly of infectious virus particles. Some assembly functions, such as membrane curvature, membrane scission, and particle budding, may be mediated by both proteins to different extents based on the specific sequences present in M1 and M2. However, efficient assembly of infectious virus particles requires unique functions ascribed to both the M1 and M2 proteins.
MATERIALS AND METHODS
Cells.
Madin-Darby canine kidney (MDCK) cells and human embryonic kidney cells (293T) cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma) with 10% fetal bovine serum (Gibco), 100 U/ml of penicillin, 100 μg/ml of streptomycin (Quality Biological), and 2 mM l-alanyl-l-glutamine (GlutaMAX; Gibco) at 37°C and 5% CO2. MDCK cells stably expressing the A/Udorn/72 M2, M2Y76A, M2Stop82, or M2Stop70 protein were generated as described previously (28, 33). These cells were maintained in medium containing 7.5 μg/ml of puromycin (Sigma) and 5 μM amantadine (Sigma) to maintain the M2 protein expression but inhibit its ion channel activity.
Viruses.
rUdorn (rUd), a recombinant virus based on sequences from the A/Udorn/72 (H3N2) virus, was propagated on MDCK cells. The rUd-M2Stop virus, containing two stop codons in the M2 ORF immediately after the region of overlap with the M1 ORF of the M segment (M2 aa 1 to 24), was described previously (28). The M segment coding region of plaque-purified viruses from Fig. 1 was sequenced to identify mutations that could complement the M2Y76A lethal mutation. M1 suppressor viruses were generated in the rUd-M2Stop backbone by reverse genetics. Briefly, site-directed mutagenesis was used to introduce the G73R, M135V, G136R, V138M, D94N, and M93V/M244I mutations into the pHH21 Udorn M1/M2Stop plasmid. The mutant M segment, along with other viral gene plasmids and 4 helper plasmids expressing PA, PB1, PB2, and NP proteins, was transfected into 293T cells and cocultured with MDCK-M2 cells. rUd-M2Stop and the M1 suppressor mutant viruses were plaque purified, sequenced, and expanded into working stocks by infecting MDCK-M2 cells at a multiplicity of infection (MOI) of 0.001 50% tissue culture infective doses (TCID50) per cell, and infectious virus titers were determined using MDCK-M2 cells.
Infection.
Low-multiplicity growth curves were performed in triplicate by infecting confluent cell monolayers in 24-well plates at an approximate MOI of 0.001 or 0.01 TCID50 units per cell. Virus was diluted in freshly made infection medium (IM; DMEM containing 2 mM l-alanyl-l-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin, 4 μg/ml N-acetyl trypsin, and 0.3% bovine serum albumin [BSA]). The cells were washed two times with phosphate-buffered saline (PBS) containing calcium and magnesium (100 μg/ml each; PBSplus), and virus inoculum was added and incubated with rocking for 1 h at room temperature. The inoculum was aspirated, and the cells were washed twice with PBSplus. Five hundred microliters of the medium was added per well, and the plates were incubated at 37°C in a 5% CO2 atmosphere. At the times indicated in the figures, infected-cell supernatants were removed and stored at −80°C and fresh infection medium was added. The amount of infectious virus in each sample was determined by TCID50 assays as previously described using MDCK-M2 cells.
Cells were infected for immunofluorescence microscopy as described previously (33). Cells were grown to confluence on 12-mm-diameter coverslips (Fisher Scientific) in 24-well plates and then incubated for an additional 2 days. An inoculum of 5 × 105 TCID50 of virus per well was added to PBSplus-washed cells and incubated with rocking for 1 h at room temperature, the inoculum was aspirated, and the cells were washed two times with PBSplus followed by the addition of IM. At 16 h after infection, cells were washed with PBSplus two times and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were then washed with PBS three times for further immunofluorescence staining.
For virus particle secretion analysis, cells grown in a T75 flask were incubated with viruses at an MOI of 1.5 by rocking for 1 h at room temperature. The inocula were removed, the cells were washed two times with PBSplus, 10 ml of IM without trypsin or bovine serum albumin (BSA) was added, and the flasks were incubated at 37°C in 5% CO2 for 16 h. Infected-cell supernatants were collected and centrifuged at 2,000 × g for 4 min to remove cell debris. Two hundred microliters of clarified supernatant was stored at −80°C for virus titration. The rest of the supernatant was loaded onto a 35% sucrose in PBS solution and centrifuged in a Beckman SW28 rotor at 118,000 × g for 1 h at 4°C to enrich virus particles. The pellet was resuspended in 150 μl of PBS and 50 μl of 4× SDS loading buffer (Bio-Rad) plus protease-phosphatase inhibitor (Halt; Thermo Scientific). For viral protein expression analysis, the infections were performed in one well of a 6-well plate. Infected cells were lysed in RIPA buffer (1% NP-40, 0.1% SDS, and 0.5% sodium deoxycholate in Tris-Cl, pH 7.4) with fresh Halt protease-phosphatase inhibitor. The lysates were homogenized by bath-ultrasonication (Misonix Sonicator Ultrasonic Processor XL) at intensity 3 for 30 s, and supernatants clarified by centrifugation at 12,000 × g for 10 min were quantified using the Pierce bicinchoninic acid (BCA) protein assay (Thermo Scientific). All samples were stored at −20°C for further Western blotting.
TCID50 assay.
Assays were performed in confluent monolayers of MDCK-M2 cells grown in 96-well plates. The plates were washed with PBSplus two times, and 180 μl/well of IM with 2.5 μg/ml N-acetyl trypsin was added per well. Tenfold serial dilutions of all samples were made in infection medium. Twenty microliters of the diluted samples was added to each of 6 wells, incubated at 37°C for 5 days, fixed with the addition of 100 μl/well of 4% formaldehyde solution at room temperature for 4 h, and stained with naphthol blue-black at room temperature overnight. The cytopathic effect was scored visually, and 50% tissue culture infective dose was calculated as previously described (27).
Immunofluorescence microscopy.
Fixed cells on coverslips were incubated with blocking buffer (0.5% BSA and 3% donkey serum in PBS) for 1 h at room temperature. Cells were incubated with goat anti-HA (A/Aichi/2/1968 H3N2; 1:4,000 dilution) antibody (44) diluted in blocking reagent for 1 h at room temperature, washed twice in PBS, and then permeabilized with 0.2% Triton X in PBS for 10 min at room temperature. After washing with PBS, the cells were incubated with blocking buffer, followed by mouse anti-NS1 (1A7CL; 1:100 dilution) monoclonal antibody (45) diluted in blocking reagent for 1 h at room temperature. The cells were then incubated with the species-specific secondary antibodies coupled to Alexa Fluor 488 or 594 (Molecular Probes; 1:400 dilution) along with Hoechst 33258 (Life Technologies) for 1 h at room temperature. Coverslips were dried and mounted on slides using ProLong Gold antifade reagent (Invitrogen). Images from the bottom to just above the top of the cells (3.2 to 3.8 μm) were taken using z-stacks at 200-nm intervals with a 100× oil immersion objective on a Zeiss LSM 510 Meta confocal microscope. The images shown are one slice closest to the top of the cells (apical cell membrane) and linearly contrast enhanced to increase clarity. Images for quantifying filaments were obtained with a 63× oil immersion objective on a Zeiss Axio Imager M2 wide-field fluorescence microscope. Image stacks from the wide-field microscope were recorded at 200-nm intervals and subjected to iterative deconvolution using Volocity software (PerkinElmer). Deconvolved images were used to quantify filaments. Filament-positive cells are NS1-positive (infected) cells and contain short and/or long bundles of HA (green) filaments at the cell surface. Filament-negative cells are infected cells with HA (green) on the cell surface. The percentage of infected cells with filaments in each field was calculated. Twenty nonoverlapping fields of each sample were analyzed.
Western blotting.
Equal volumes of partially purified viral particles and equal amounts of cell lysate of each sample containing SDS loading buffer and 50 mM dithiothreitol (DTT) were boiled for 5 min, separated on a 4 to 15% precast Mini-Protean TGX gel (Bio-Rad), and transferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-FL; Millipore) using a tank transfer system (Mini Trans-Blot; Bio-Rad). The blots were blocked with 5% nonfat dry milk (Bio-Rad) in PBS with 0.05% Tween 20 at room temperature for 1 h. After blocking, the blots were incubated with primary antibodies at room temperature for 1 h, washed with PBS, incubated with secondary antibodies at room temperature for 1 h, washed with PBS, and then imaged with a FluorChem Q imager system (Alpha Innotech). All antibodies were diluted in the blocking buffer. The antibodies used in the study were goat anti-HA (A/Aichi/2/1968 H3N2; 1:2,000), rabbit anti-NP (GeneTex GTX125989; 1:1,000), rabbit anti-M1 (GeneTex GTX125928; 1:1,000), mouse anti-M2 (14c2; 1:500) (27), and mouse anti-β-actin (Abcam ab6276; 1:10,000) antibodies. The species-specific secondary antibodies coupled to Alexa Fluor 488, 594, or 647 (Molecular Probes; 1:1,000) were used to detect the primary antibodies.
Viral RNA quantification by RT-qPCR.
Equal volumes (140 μl) of cell supernatant of each sample from the high-MOI infections were used to extract viral RNA using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's protocol. Each vRNA sample was eluted in 60 μl of RNase-free water. Ten microliters of the vRNA solution was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen), an influenza A virus universal primer (5′-AGTAGAAACAAG-3′), and deoxynucleoside triphosphate (dNTP) (0.5 mM) at 65°C for 5 min and then immediately kept on ice for 2 min to denature RNA secondary structure. RNase inhibitor (Promega), DTT (5 mM; Invitrogen), 1× First-Strand buffer (Invitrogen), and the reverse transcriptase (400 U) were then added to a final 40-μl reaction mixture. The reaction mixture was incubated at 50°C for 30 min and inactivated at 70°C for 15 min. The viral cDNA was then quantified by quantitative PCR (qPCR) of the HA gene using TaqMan qPCR assays (Applied Biosystems) with F-primer (5′-GGACCCTCATTGTGATGGCTTTC-3′), R-primer (5′-CTAAGGGAGGCATAATCTGGCACAT-3′), and probe (5′-HEX [6-carboxy-2,4,4, 5,7, 7-hexachlorofluorescein]-ACGCAGCAA-ZEN-AGCTTTCAGCAACTGTTACC-Iowa Black FQ [IABkFQ]-3′; Integrated DNA Technologies) (46). The primers and probe were designed to be specific to the Udorn-HA gene segment. The qPCR mixture was 20 μl containing a final concentration of 1× TaqMan Gene Expression master mix (Applied Biosystems), 900 nM (each) primer, 250 nM probe, and 2 μl of the cDNA. The cycling parameters were 50°C for 2 min, 95°C for 10 min, and 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. Each sample was assayed in triplicate. The number of relative HA gene copies in each sample was determined by normalizing the threshold cycle (CT) value of each sample to the CT from rUd-M2Stop-infected MDCK cells.
Statistical analysis.
Low-multiplicity growth curves were compared using two-way analysis of variance (ANOVA) with the Bonferroni correction. The percentage of infected cells with filaments, vRNA incorporation, and infectious virus production from high-multiplicity infections were compared using one-way ANOVA with the Bonferroni correction. All statistical analyses and graphing were done with GraphPad Prism 6.
ACKNOWLEDGMENTS
We thank the members of the Pekosz laboratory, Sabra Klein, and members of the Klein laboratory for useful and critical discussions of the data.
The work was supported by the Shikani/El Hibri Prize for Discovery and Innovation (A.P.), a gift from the Eliasberg Family Foundation Inc. (A.P.), HHSN272201400007C (A.P.), and S10OD016374 (Zeiss LSM 510 confocal microscope).
REFERENCES
- 1.Bouvier NM, Palese P. 2008. The biology of influenza viruses. Vaccine 26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda M, Leser GP, Russell CJ, Lamb RA. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc Natl Acad Sci U S A 100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leser GP, Lamb RA. 2005. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 5.Chen BJ, Leser GP, Morita E, Lamb RA. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol 81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. 2010. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci U S A 107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali A, Avalos RT, Ponimaskin E, Nayak DP. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J Virol 74:8709–8719. doi: 10.1128/JVI.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Pekosz A, Lamb RA. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J Virol 74:4634–4644. doi: 10.1128/JVI.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin H, Leser GP, Zhang J, Lamb RA. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J 16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. 2007. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J Gen Virol 88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao S, Liu X, Yu M, Li J, Jia X, Bi Y, Sun L, Gao GF, Liu W. 2012. A nuclear export signal in the matrix protein of influenza A virus is required for efficient virus replication. J Virol 86:4883–4891. doi: 10.1128/JVI.06586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Z, Liu T, Offringa DP, McInnis J, Levandowski RA. 1999. Association of influenza virus matrix protein with ribonucleoproteins. J Virol 73:7467–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das SC, Watanabe S, Hatta M, Noda T, Neumann G, Ozawa M, Kawaoka Y. 2012. The highly conserved arginine residues at positions 76 through 78 of influenza A virus matrix protein M1 play an important role in viral replication by affecting the intracellular localization of M1. J Virol 86:1522–1530. doi: 10.1128/JVI.06230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elleman CJ, Barclay WS. 2004. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology 321:144–153. doi: 10.1016/j.virol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Burleigh LM, Calder LJ, Skehel JJ, Steinhauer DA. 2005. Influenza A viruses with mutations in the M1 helix six domain display a wide variety of morphological phenotypes. J Virol 79:1262–1270. doi: 10.1128/JVI.79.2.1262-1270.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bialas KM, Desmet EA, Takimoto T. 2012. Specific residues in the 2009 H1N1 swine-origin influenza matrix protein influence virion morphology and efficiency of viral spread in vitro. PLoS One 7:e50595. doi: 10.1371/journal.pone.0050595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts PC, Lamb RA, Compans RW. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240:127–137. doi: 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Wang Z, Fan GZ, Wang J, Gao S, Li Y, Sun L, Yin CC, Liu WJ. 2015. Two polar residues within C-terminal domain of M1 are critical for the formation of influenza A virions. Cell Microbiol 17:1583–1593. doi: 10.1111/cmi.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seladi-Schulman J, Steel J, Lowen AC. 2013. Spherical influenza viruses have a fitness advantage in embryonated eggs, while filament-producing strains are selected in vivo. J Virol 87:13343–13353. doi: 10.1128/JVI.02004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sha B, Luo M. 1997. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat Struct Biol 4:239–244. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- 21.Arzt S, Baudin F, Barge A, Timmins P, Burmeister WP, Ruigrok RW. 2001. Combined results from solution studies on intact influenza virus M1 protein and from a new crystal form of its N-terminal domain show that M1 is an elongated monomer. Virology 279:439–446. doi: 10.1006/viro.2000.0727. [DOI] [PubMed] [Google Scholar]
- 22.Safo MK, Musayev FN, Mosier PD, Zhou Q, Xie H, Desai UR. 2014. Crystal structures of influenza A virus matrix protein M1: variations on a theme. PLoS One 9:e109510. doi: 10.1371/journal.pone.0109510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris A, Forouhar F, Qiu S, Sha B, Luo M. 2001. The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology 289:34–44. doi: 10.1006/viro.2001.1119. [DOI] [PubMed] [Google Scholar]
- 24.Edinger TO, Pohl MO, Stertz S. 2014. Entry of influenza A virus: host factors and antiviral targets. J Gen Virol 95:263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 25.Iwatsuki-Horimoto K, Horimoto T, Noda T, Kiso M, Maeda J, Watanabe S, Muramoto Y, Fujii K, Kawaoka Y. 2006. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J Virol 80:5233–5240. doi: 10.1128/JVI.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BJ, Leser GP, Jackson D, Lamb RA. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol 82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCown MF, Pekosz A. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J Virol 79:3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCown MF, Pekosz A. 2006. Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol 80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. 2010. Influenza virus M2 ion channel protein is necessary for filamentous virion formation. J Virol 84:5078–5088. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruce EA, Medcalf L, Crump CM, Noton SL, Stuart AD, Wise HM, Elton D, Bowers K, Digard P. 2009. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology 390:268–278. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grantham ML, Stewart SM, Lalime EN, Pekosz A. 2010. Tyrosines in the influenza A virus M2 protein cytoplasmic tail are critical for production of infectious virus particles. J Virol 84:8765–8776. doi: 10.1128/JVI.00853-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. 2002. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J Virol 76:1391–1399. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zebedee SL, Lamb RA. 1989. Growth restriction of influenza A virus by M2 protein antibody is genetically linked to the M1 protein. Proc Natl Acad Sci U S A 86:1061–1065. doi: 10.1073/pnas.86.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts KL, Leser GP, Ma C, Lamb RA. 2013. The amphipathic helix of influenza A virus M2 protein is required for filamentous bud formation and scission of filamentous and spherical particles. J Virol 87:9973–9982. doi: 10.1128/JVI.01363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt NW, Mishra A, Wang J, DeGrado WF, Wong GC. 2013. Influenza virus A M2 protein generates negative Gaussian membrane curvature necessary for budding and scission. J Am Chem Soc 135:13710–13719. doi: 10.1021/ja400146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart SM, Pekosz A. 2011. Mutations in the membrane-proximal region of the influenza A virus M2 protein cytoplasmic tail have modest effects on virus replication. J Virol 85:12179–12187. doi: 10.1128/JVI.05970-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chlanda P, Zimmerberg J. 2016. Protein-lipid interactions critical to replication of the influenza A virus. FEBS Lett 590:1940–1954. doi: 10.1002/1873-3468.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsfasman T, Kost V, Markushin S, Lotte V, Koptiaeva I, Bogacheva E, Baratova L, Radyukhin V. 2015. Amphipathic alpha-helices and putative cholesterol binding domains of the influenza virus matrix M1 protein are crucial for virion structure organisation. Virus Res 210:114–118. doi: 10.1016/j.virusres.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 41.Saletti D, Radzimanowski J, Effantin G, Midtvedt D, Mangenot S, Weissenhorn W, Bassereau P, Bally M. 2017. The matrix protein M1 from influenza C virus induces tubular membrane invaginations in an in vitro cell membrane model. Sci Rep 7:40801. doi: 10.1038/srep40801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shtykova EV, Baratova LA, Fedorova NV, Radyukhin VA, Ksenofontov AL, Volkov VV, Shishkov AV, Dolgov AA, Shilova LA, Batishchev OV, Jeffries CM, Svergun DI. 2013. Structural analysis of influenza A virus matrix protein M1 and its self-assemblies at low pH. PLoS One 8:e82431. doi: 10.1371/journal.pone.0082431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosier PD, Chiang MJ, Lin Z, Gao Y, Althufairi B, Zhou Q, Musayev F, Safo MK, Xie H, Desai UR. 2016. Broad spectrum anti-influenza agents by inhibiting self-association of matrix protein 1. Sci Rep 6:32340. doi: 10.1038/srep32340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pekosz A, Newby C, Bose PS, Lutz A. 2009. Sialic acid recognition is a key determinant of influenza A virus tropism in murine trachea epithelial cell cultures. Virology 386:61–67. doi: 10.1016/j.virol.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lalime EN, Pekosz A. 2014. The R35 residue of the influenza A virus NS1 protein has minimal effects on nuclear localization but alters virus replication through disrupting protein dimerization. Virology 458-459:33–42. doi: 10.1016/j.virol.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee HK, Lee CK, Loh TP, Tang JW, Chiu L, Tambyah PA, Sethi SK, Koay ES. 2010. Diagnostic testing for pandemic influenza in Singapore: a novel dual-gene quantitative real-time RT-PCR for the detection of influenza A/H1N1/2009. J Mol Diagn 12:636–643. doi: 10.2353/jmoldx.2010.100010. [DOI] [PMC free article] [PubMed] [Google Scholar]