ABSTRACT
Enterovirus 71 (EV71) is the major causative agent of severe hand, foot, and mouth disease, which affects millions of young children in the Asia-Pacific region annually. In this study, we engineered a novel EV71 virus-like particle (VLP) that lacks VP4 (therefore designated VLPΔVP4) and investigated its structure, antigenicity, and vaccine potential. The cryo-electron microscopy (cryo-EM) structure of VLPΔVP4 was reconstructed to 3.71-Å resolution. Results from structural and biochemical analyses revealed that VLPΔVP4 resembles the end product of the viral uncoating process, the 80S empty capsid. VLPΔVP4 is able to elicit high-titer neutralizing antibodies and to fully protect mice against lethal viral challenge. Mechanistic studies showed that, at the cellular level, the anti-VLPΔVP4 sera exert neutralization effects at both pre- and postattachment stages by inhibiting both virus attachment and internalization, and at the molecular level, the antisera can block multiple interactions between EV71 and its key receptors. Our study gives a better understanding of EV71 capsid assembly and provides important information for the design and development of new-generation vaccines for EV71, and perhaps for other enteroviruses, as well.
IMPORTANCE Enterovirus 71 (EV71) infection may lead to severe hand, foot, and mouth disease, with significant morbidity and mortality. Knowledge regarding EV71 particle assembly remains limited. Here, we report the generation and characterization of a novel EV71 virus-like particle that lacks the VP4 capsid subunit protein. This particle, termed VLPΔVP4, structurally mimics the 80S empty capsid, which is the end stage of EV71 uncoating. We further show that VLPΔVP4 exhibits desirable immunogenicity and protective efficacy in proof-of-concept studies. In addition, the inhibitory mechanisms of the VLPΔVP4-induced antibodies are unraveled at both the cellular and molecular levels. Our work provides the first evidence of picornaviral particle assembly in the complete absence of VP4 and identifies VLPΔVP4 as an improved EV71 vaccine candidate with desirable traits. These findings not only enhance our understanding of particle assembly and uncoating of picornaviruses, but also provide important information for structure-guided vaccine design for EV71 and other enteroviruses.
KEYWORDS: cryo-EM, enterovirus 71, structure, uncoating, vaccine, virus-like particle
INTRODUCTION
Enterovirus 71 (EV71), a member of the genus Enterovirus of the family Picornaviridae, is the major causative agent of severe hand, foot, and mouth disease (HFMD) (1, 2). The genome of EV71 is a single-stranded positive-sense RNA that encodes the structural protein P1 and the nonstructural proteins P2 and P3. Cleavage of P1 by the viral protease 3CD results in the release of capsid proteins VP0 (VP4 plus VP2), VP3, and VP1 and, subsequently, the assembly of the viral capsid (1). Two native forms of viral particles have been observed for cell culture-derived EV71: one is the mature virion, which contains the viral RNA genome encapsulated within a capsid made of 60 copies each of VP4, VP2, VP3, and VP1, and the other is the natural empty particle (NEP), assembled from 60 copies each of VP0, VP3, and VP1 without viral-RNA encapsulation (3, 4). The NEP of EV71 is ∼4% larger than the mature virion (3, 4), which has not been observed in other enteroviruses. The capsid of EV71 adopts an icosahedral particle conformation with characteristic surface features, including a “mesa” at the 5-fold symmetry axis; a three-blade propeller-like feature surrounding the 3-fold axis; and a depression, called the “canyon,” surrounding each 5-fold axis (3, 4). VP1, VP2, and VP3 of EV71 are largely exposed on the outer surface of the capsid, while VP4, together with the N terminus of VP1, decorates the inner surface of the capsid shell of the mature virion (3, 4). Multiple receptors have been reported to mediate EV71 entry, including cell surface heparan sulfate, SCARB2, and PSGL-1 (5). Among them, heparan sulfate has been suggested to initiate EV71 attachment to the cell surface (6), SCARB2 has been identified as the functional receptor that mediates EV71 uncoating (7), and PSGL-1 has been suggested to facilitate EV71 infection of leukocytes (8).
For EV71 and several other picornaviruses, the mature virions may undergo serial structural alterations during the entry process, yielding at least two entry-related intermediate particles (9). Specifically, upon receptor binding and heat treatment, the mature virus transforms first to an expanded 135S uncoating intermediate (also termed the A particle), during which VP4 is extruded and the N terminus of VP1 is externalized (9, 10). Subsequently, the 135S A particle releases its RNA content, as well as VP4, yielding the 80S empty capsid (also termed the B particle in structural studies of rhinoviruses [11–13]), which is the end product of the uncoating process (9). The structures of both the 135S A particle and the uncoated 80S empty capsid of EV71 have been determined at relatively low resolutions (6.3 Å and 9.2 Å, respectively) by cryo-electron microscopy (cryo-EM) studies, showing that they are radially expanded compared to the mature virion (10). However, high-resolution structural details of the EV71 80S uncoated empty particle remain unavailable.
Inactivated whole-virus EV71 vaccines contain both the mature virion and NEP (3, 14), and both particle forms have been shown to be highly immunogenic and capable of eliciting protection in numerous preclinical and clinical trials (reviewed in reference 15). The two EV71 uncoating intermediates, the 135S A particle and the 80S empty capsid (10), are structurally distinct from the mature virion and NEP. Their vaccine potentials have not been assessed yet, due to the difficulty in generating sufficient amounts of the 135S A particle and the 80S empty capsid for immunization studies. The two particles are typically induced from the mature virion upon binding the receptor under acidic conditions or being heated in hypotonic buffers (10, 16, 17). These methods are difficult to scale up and cannot ensure complete particle transformations (10, 17).
Besides naturally occurring EV71 particles, recombinant virus-like particles (VLPs) have been produced and evaluated as an alternative EV71 vaccine candidate. Coexpression of full-length P1 and 3CD genes of EV71 in recombinant systems resulted in the cleavage of P1 into full-length VP0, VP1, and VP3 subunits; together, they assemble into full-length VLPs (termed VLPfull here) (18–21). VLPfull resembles the naturally occurring NEP in protein composition and overall structure (19, 22). Although VLPfull is highly immunogenic and protective in animal models (20, 23, 24), autocleavage of VP0 has been observed for unknown reasons in studies of EV71 (20, 24) and the related virus coxsackievirus A16 (CVA16) (25, 26). This autocleavage may occur during VLPfull assembly, purification, and storage and thus result in antigen inconsistency, which is an important issue that needs to be addressed prior to further product development.
In the present study, we created a novel EV71 VLP that lacks the VP4 protein (termed VLPΔVP4). We further investigated the structure, immunogenicity, protective efficacy, and working mechanism of VLPΔVP4. Our study revealed that VLPΔVP4 resembles the 80S empty capsid and represents an EV71 vaccine candidate superior to VLPfull, thus providing important information and a methodology for structural study and development of improved vaccines for EV71 and other enteroviruses.
RESULTS
Expression and assembly of VLPΔVP4.
Two plasmids and the corresponding recombinant baculoviruses were generated for expression of P1ΔVP4 and 3CD, respectively (Fig. 1A). The two resulting baculoviruses, IExBac-P1ΔVP4 and IExBac-3CD, were used to infect Sf9 insect cells individually or in combination. The infected cells were analyzed for protein expression by enzyme-linked immunosorbent assay (ELISA) and Western blotting using VP1-, VP0-, or VP3-specific antisera. The lysates from mock-infected or IEXBac-3CD-infected cells did not show positive signals in ELISA, whereas those from IEXBac-P1ΔVP4-infected or IEXBac-P1ΔVP4/IEXBac-3CD-coinfected cells exhibited significant reactivity with each of the three antisera (Fig. 1B). In agreement with the ELISA results, the IEXBac-P1ΔVP4-infected and IEXBac-P1ΔVP4/IEXBac-3CD-coinfected cells, but not the mock- or IEXBac-3CD-infected samples, produced positive bands on Western blots (Fig. 1C). However, the banding patterns were significantly different between the IEXBac-P1ΔVP4 and the IEXBac-P1ΔVP4/IEXBac-3CD samples. Specifically, all three antisera detected a predominant band of ∼90 kDa for the IEXBac-P1ΔVP4 samples, suggesting the presence of P1ΔVP4 protein in the unprocessed form, whereas for the coinfected samples, predominant bands of ∼34 kDa, ∼28 kDa, and ∼26 kDa were detected by the VP1-, VP0-, and VP3-specific antisera, respectively (Fig. 1C), indicating successful coexpression of P1ΔVP4 and 3CD and cleavage of P1ΔVP4 by 3CD into VP1, VP2, and VP3. Several weak bands were seen in the mock- and the IEXBac-3CD-infected samples when probed with the anti-VP1 or anti-VP0 antisera, reflecting minimal nonspecific binding. Some other minor bands were additionally observed for the IEXBac-P1ΔVP4 and the IEXBac-P1ΔVP4/IEXBac-3CD samples, likely representing partially cleaved P1ΔVP4 or degraded capsid subunit proteins.
FIG 1.
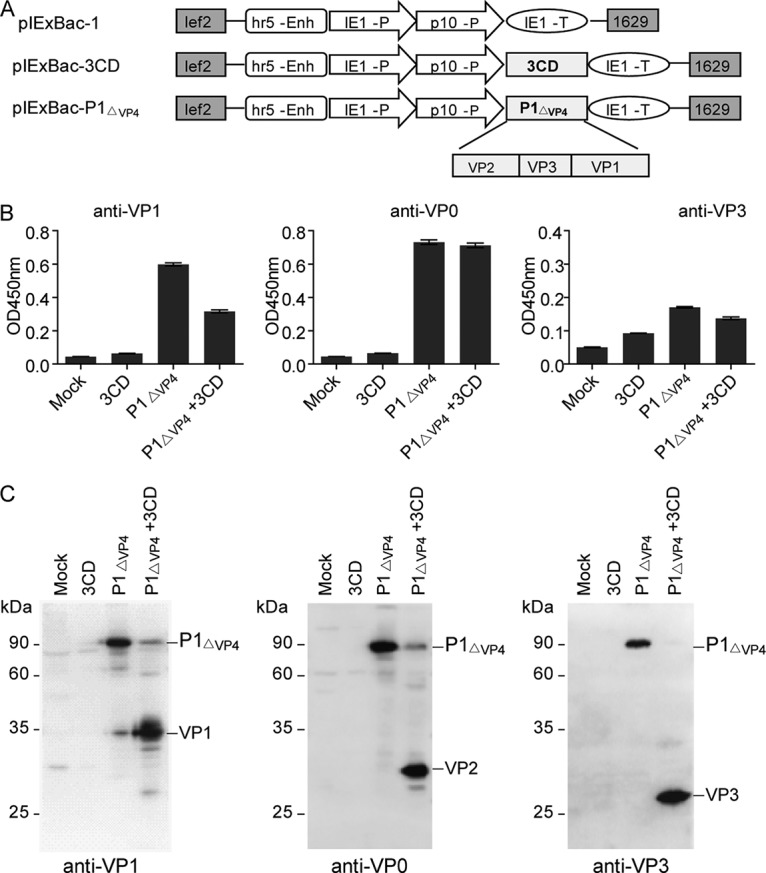
Coexpression of P1ΔVP4 and 3CD in Sf9 cells. (A) Schematic diagram of the constructs used in the present study. lef2 and 1629, the upstream and downstream elements for homologous recombination; hr5-Enh, the Autographa californica multiple nucleopolyhedrovirus (AcNPV) homologous region (hr) 5 enhancer; IE1-P, the IE1 immediate-early promoter; p10-P, p10 promoter; IE1-T, IE1 terminator. (B and C) ELISA (B) and Western blotting (C) of Sf9 cell lysates with polyclonal antibodies as indicated. Mock, uninfected Sf9 cells; 3CD, Sf9 cells infected by IEXBac-3CD; P1ΔVP4, Sf9 cells infected by IEXBac-P1ΔVP4; P1ΔVP4 + 3CD, Sf9 cells coinfected by IEXBac-P1ΔVP4 and IEXBac-3CD.
Next, we determined whether the resulting VP1, VP2, and VP3 could assemble into VLPs. Lysates from the IEXBac-P1ΔVP4/IEXBac-3CD-coinfected cells were fractionated by sucrose gradient ultracentrifugation. SDS-PAGE analysis of the gradient fractions clearly showed three major protein bands of approximately 34, 28, and 26 kDa (Fig. 2A). These bands, which were rich in fractions 7, 8, and 9, were confirmed to be VP1, VP2, and VP3 by Western blotting (Fig. 2B) and ELISA (Fig. 2C) using specific antisera. The strongest signals were detected in fractions 7, 8, and 9 in all the assays (Fig. 2A to C), suggesting that the cosedimenting VP1, VP2, and VP3 coassemble into VLPs. The formation of VLPs was further confirmed by visualization of spherical particles with diameters of about 30 nm under electron microscopy (Fig. 2D). The resulting VLP does not contain VP4 protein; therefore, we refer to this novel VLP as VLPΔVP4 here.
FIG 2.
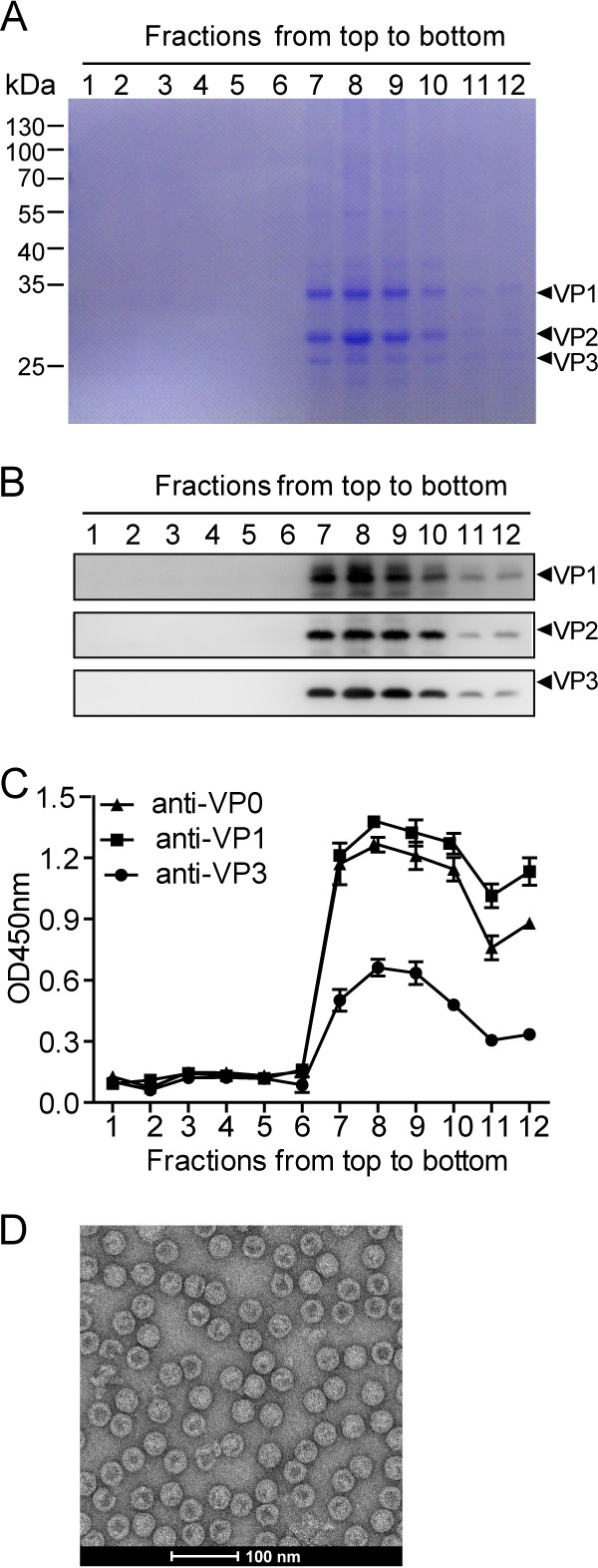
Assembly of VLPΔVP4. Cell lysate from IEXBac-P1ΔVP4- and IEXBac-3CD-coinfected Sf9 cells was layered onto 10 to 50% sucrose gradients for centrifugation. Twelve fractions were taken from top to bottom and then subjected to SDS-PAGE with Coomassie blue R-250 staining (A), Western blotting (B), or ELISA with antibodies as indicated (C). The OD450 values are means and standard deviations (SD) of three triplicate wells. (D) Electron microscopy of VLPΔVP4.
Cryo-EM structure of VLPΔVP4.
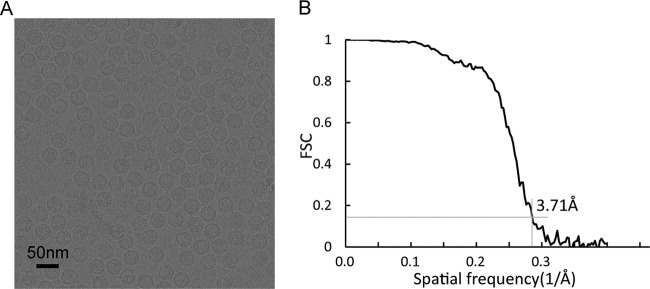
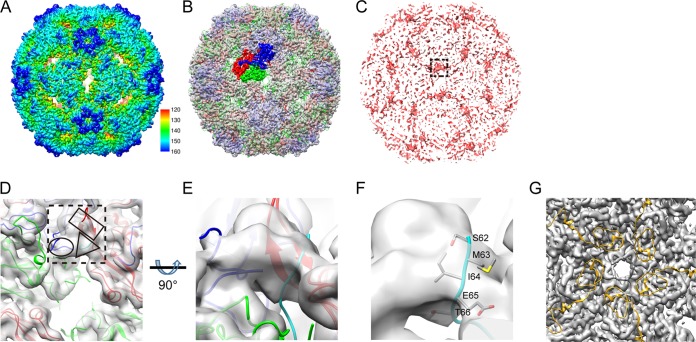
To better understand the structure of VLPΔVP4, we carried out cryo-EM reconstruction of the system. In representative micrographs, VLPΔVP4 appeared as spherical particles with a diameter of ∼30 nm (Fig. 3A). The cryo-EM structure of VLPΔVP4 was reconstructed to 3.71-Å resolution (Fig. 3B), revealing an icosahedral conformation. The structure displayed surface features characteristic of enteroviral capsids, such as a mesa at the 5-fold-symmetry axis, a three-blade propeller surrounding the 3-fold axis, and a canyon surrounding each 5-fold axis (Fig. 4A). VLPΔVP4 had a diameter of 334 Å, which is equivalent to that of the NEP and is about 4% larger than that of the mature virion (320 Å) (3).
FIG 3.
Cryo-EM data. (A) Representative cryo-EM image of EV71 VLPΔVP4. (B) Resolution evaluation of the cryo-EM reconstruction by FSC at 0.143 criterion.
FIG 4.
Cryo-EM reconstruction and analysis of VLPΔVP4. (A) Overall cryo-EM structure of VLPΔVP4 viewed along the 2-fold axis. The color bar labels the corresponding radius of the particle from the center of the sphere (in Å). (B) The crystal structure of the EV71 E particle (PDB ID 3VBU) was fitted into the cryo-EM density map of VLPΔVP4 (gray), with VP1, VP2, and VP3 structures within one asymmetric unit rendered in blue, green, and red, respectively. (C) Difference map (red) generated by subtracting the 5.2-Å VLPfull density map (EMDB accession no. EMD-2607) (19) from the low-pass-filtered VLPΔVP4 density map (to 6 Å). The difference map is shown in the same orientation as in panel A, and the most pronounced patch of density (near the 2-fold axis) is marked by the dashed square. (D) Enlarged top view of the low-pass-filtered VLPΔVP4 density map with the fitted crystal structure of the EV71 NEP (PDB ID 3VBU) in the equivalent location of the pronounced extra density, as indicated in the difference map (dashed square). Rectangle, ellipsoid, and triangle, possible densities of the VP3 GH loop, VP1 GH loop, and part of the VP1 N terminus, respectively. VP1, VP2, VP3, and a homology model of the VP1 N terminus are colored blue, green, red, and cyan, respectively. (E) Enlarged side view of the extra density from panel D. (F) Enlarged view of panel E at a slightly tilted angle to better visualize the side chains of the VP1 N terminus (residues 62 to 66), which are well embedded inside the density. (G) Enlarged view inside the VLPΔVP4 map along the 5-fold axis. We docked the EV71 mature virion crystal structure (PDB ID 3VBS) into the VLPΔVP4 cryo-EM density map (gray density) and rendered the VP4 crystal structure as a gold ribbon.
By comparison with the previously reported structures of various EV71 particles, including the mature virion (Protein Data Bank identifier [PDB ID] 3VBS), NEP (PDB ID 3VBU), and recombinant VLPfull (Electron Microscopy Data Bank [EMDB; http://www.emdatabank.org/] accession no. EMD-2607), we found that VLPΔVP4 was highly similar to the EV71 NEP and VLPfull in overall structure (Fig. 4B), with correlation coefficient (cc) values of 0.9275 and 0.8854, respectively (Table 1). In contrast, the similarity between VLPΔVP4 and the mature virion was much lower (cc = 0.3205) (Table 1), likely due to the expansion of VLPΔVP4 compared to the mature virion.
TABLE 1.
Structural characteristics of VLPΔVP4-related particles
| Particle | Composition | ID | Similarity to VLPΔVP4 (cc value) |
|---|---|---|---|
| EV71 mature virion | VP1, VP2, VP3, VP4, RNA | 3VBS (PDB) | 0.3025 |
| EV71 NEP | VP0, VP1, VP3 | 3VBU (PDB) | 0.9275 |
| EV71 135S A particle | VP1, VP2, VP3, RNA | 5465 (EMDB) | NDa |
| EV71 80S empty capsid | VP1, VP2, VP3 | 5466 (EMDB) | ND |
| CVA16 135S-like particle | VP1, VP2, VP3, RNA | 4JGY (PDB) | 0.8112 |
| EV71 VLPfull | VP0, VP1, VP3 | 2607 (EMDB) | 0.8854 |
| EV71 VLPfull | VP0, VP1, VP3 | 4YVS (PDB) | 0.8851 |
| EV71 VLPΔVP4 | VP1, VP2, VP3 | 6702 (EMDB) | 1 |
ND, not determined.
To compare the structural differences between VLPΔVP4 and VLPfull, we calculated a difference map between the VLPΔVP4 and VLPfull (EMDB accession no. EMD-2607) cryo-EM structures (Fig. 4C). The difference map revealed a piece of pronounced density located at quasi-3-fold axes (boxed in Fig. 4C), indicating this was the density visible in our VLPΔVP4 map but missing in the VLPfull map. To further verify the identity of this pronounced density, we fitted the crystal structure of the EV71 NEP (PDB ID 3VBU) into the low-pass-filtered VLPΔVP4 density map, which was used to generate the difference map and could better render the pronounced extra density missing in VLPfull (Fig. 4D; the location of the pronounced density found in the difference map is indicated by the dashed box). We found that both the VP1 GH loop (residues 208 to 222) and the VP3 GH loop (residues 170 to 190) partly went through this density (Fig. 4D, ellipsoid and rectangle, respectively). Interestingly, there is an extra piece of density (Fig. 4D, triangle) that no model of the crystal structure of the EV71 NEP was able to fit in. The 9.2-Å cryo-EM density map of the EV71 80S empty capsid reported previously (10) suggested that its VP1 N terminus was externalized. We therefore speculated that the extra density in our difference map might be related to the externalized N terminus of VP1. As the only available structure in the expanded uncoating intermediate form of HFMD-related enteroviruses with an externalized VP1 N terminus is the crystal structure of the CVA16 135S-like particle (PDB ID 4JGY) (27), we built a homology model of the EV71 VP1 protein by using the structure of the CVA16 135S-like particle as the template and fitted the model into the low-pass-filtered map. As shown in Fig. 4E and F, the extruded N-terminal residues 62 to 66 of VP1 (in cyan) was indeed found to go through this extra density from inside to outside the VLP capsid (also shown in Fig. 4D, triangle).
To verify whether VP4 is present in VLPΔVP4, we docked the EV71 mature virion X-ray structure (PDB ID 3VBS), which is the only EV71 structure resolving the VP4 protein, into our VLPΔVP4 EM density map. We found that our VLPΔVP4 map had no electron densities presumably corresponding to the VP4 protein (Fig. 4G).
VLPΔVP4 is antigenically distinct from VLPfull.
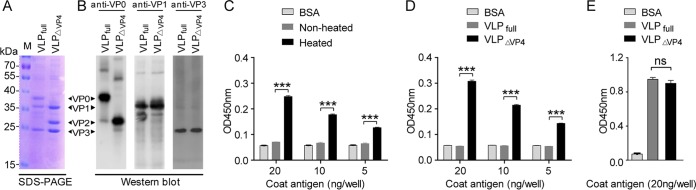
We then determined the antigenic properties of VLPΔVP4 in parallel with VLPfull, a VLP consisting of VP0, VP3, and VP1 (20). Purified VLPΔVP4 and VLPfull were first analyzed for their protein compositions and their reactivities with capsid subunit-specific antibodies. As shown in Fig. 5A and B, both VLPΔVP4 and VLPfull produced an ∼34-kDa band and an ∼26-kDa band that corresponded to VP1 and VP3, respectively; for VLPΔVP4, a predominant band of ∼28 kDa, representing VP2, was evident, whereas a band of ∼38 kDa, corresponding to VP0, was observed for VLPfull but not for VLPΔVP4. Some very minor bands were also observed on the Western blot (Fig. 5B), probably representing trace amounts of oligomeric proteins or degraded forms of the capsid subunit proteins. It is worth noting that the ∼28-kDa band (the position of VP2) was also detected in the VLPfull sample by the VP0-specific antisera (Fig. 5B), indicating that a proportion of VP0 may undergo autocleavage to yield VP2 and VP4. The above-mentioned results indicate that VLPΔVP4 consists of VP2, VP3, and VP1 and is distinct from VLPfull in protein composition.
FIG 5.
Antigenic comparison between VLPΔVP4 and VLPfull. (A) SDS-PAGE analysis of purified VLPΔVP4 and VLPfull. (B) Western blot analysis of purified VLPΔVP4 and VLPfull. M, protein marker. (C) Reactivity of the mature virion and the mixture of 135S A particles and 80S empty capsid with the VP1-N75-specific antiserum in ELISA. (D) Reactivity of VLPΔVP4 and VLPfull with the VP1-N75-specific antiserum in ELISA. (E) Reactivity of VLPΔVP4 and VLPfull with GH loop-specific monoclonal antibody D5 in ELISA. The data are mean OD450 values and SD of triplicate wells. Statistical significances were determined by one-way ANOVA using GraphPad Prism version 5. ns, P ≥ 0.05; ***, P < 0.001.
Our cryo-EM reconstruction showed that the VP1 N terminus of VLPΔVP4 is externalized (Fig. 4D to F). To verify this structural feature, we generated a polyclonal antibody specific for the first 75 amino acids of VP1 and used the antibody to examine the exposure of N-terminal VP1. As expected, the antibody was capable of recognizing the mixed 135S A particle and 80S empty capsid (generated by heating mature virions in hypotonic buffer, as described previously [10]), whereas it did not recognize the mature virion, for which the N terminus of VP1 was at the inner surface (3) (Fig. 5C), thus validating the use of the antibody for detection of the externalized VP1 N terminus. We then compared the reactivities of VLPΔVP4 and VLPfull with the N-terminal VP1-specific antibody. As shown in Fig. 5D, VLPfull, whose VP1 N terminus is at the inner face (19), did not react with the antibody; in contrast, VLPΔVP4 exhibited strong reactivity, indicating the externalization of the VP1 N terminus of VLPΔVP4.
We also determined the reactivity of VLPΔVP4 with a monoclonal antibody, D5, which recognizes the exposed VP1 GH loop (28), an immunodominant neutralization epitope (20). As shown in Fig. 5E, both VLPΔVP4 and VLPfull reacted strongly with D5, indicating that VLPΔVP4 retains the antigenic property of the VP1 GH loop.
The antibody profile induced by immunization with VLPΔVP4 was different from that with VLPfull.
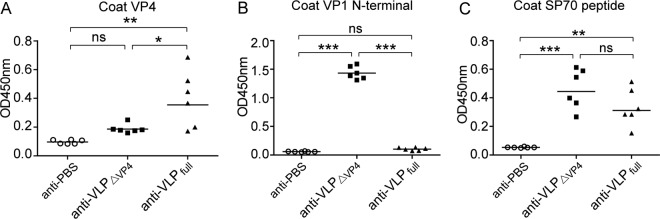
The absence of VP4 and externalization of the VP1 N terminus may provide VLPΔVP4 with immunogenic properties distinct from those of VLPfull. To verify this hypothesis, we immunized three groups of mice with VLPΔVP4, VLPfull, and phosphate-buffered saline (PBS) (as a control) and compared the resulting antisera for reactivity with recombinant VP4 and VP1-N75 (comprising N-terminal residues 1 to 75 of VP1) proteins. As shown in Fig. 6A and B, the control anti-PBS serum did not react with either VP4 or VP1-N75, and the anti-VLPΔVP4 serum did not react significantly with VP4 but did react strongly with VP1-N75; on the other hand, anti-VLPfull reacted significantly with VP4 but not with VP1-N75. We also compared the three groups of antisera for reactivity with the SP70 peptide (residues 208 to 222 of VP1, representing the VP1 GH loop). The SP70-binding activities of the anti-VLPΔVP4 sera appeared to be comparable to those of the anti-VLPfull sera (Fig. 6C), consistent with the antigenicity result showing that the VP1 GH loop epitopes were well retained on both VLPΔVP4 and VLPfull (Fig. 5E). The above-mentioned data demonstrate that antibodies targeting the VP4 or VP1 N terminus were distinctly elicited by VLPΔVP4 and VLPfull.
FIG 6.
Binding specificities of sera from VLP-immunized mice. (A) VP4-binding activities of individual antisera. (B) VP1-N75-binding activities of individual antisera. (C) SP70 peptide-binding activities of individual antisera. Each symbol represents an individual mouse, and the horizontal lines indicate the geometric mean values of the groups. Statistical significances were determined by one-way ANOVA using GraphPad Prism version 5. ns, P ≥ 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Protective efficacy of VLPΔVP4.
We compared the neutralization potencies of the anti-VLPΔVP4 and anti-VLPfull sera. When tested against the homologous EV71 strain G082, the neutralization titers of the anti-VLPΔVP4 sera appeared to be slightly higher than, but not statistically different from, those of the anti-VLPfull sera (Fig. 7A), with geometric mean titers (GMTs) of 3,649 and 2,896, respectively. The cross-neutralization potential of pooled antisera was examined using a panel of EV71 or CVA16 strains. Pooled antisera from the VLPΔVP4 and VLPfull groups, but not the PBS group, neutralized all EV71 strains tested, with titers ranging from 1,024 to 4,096, but not the CVA16 strains (Table 2). The overall neutralization patterns for anti-VLPΔVP4 and anti-VLPfull sera were nearly identical (Table 2). These results indicate that VLPΔVP4 is as potent as VLPfull in inducing neutralization antibodies.
FIG 7.
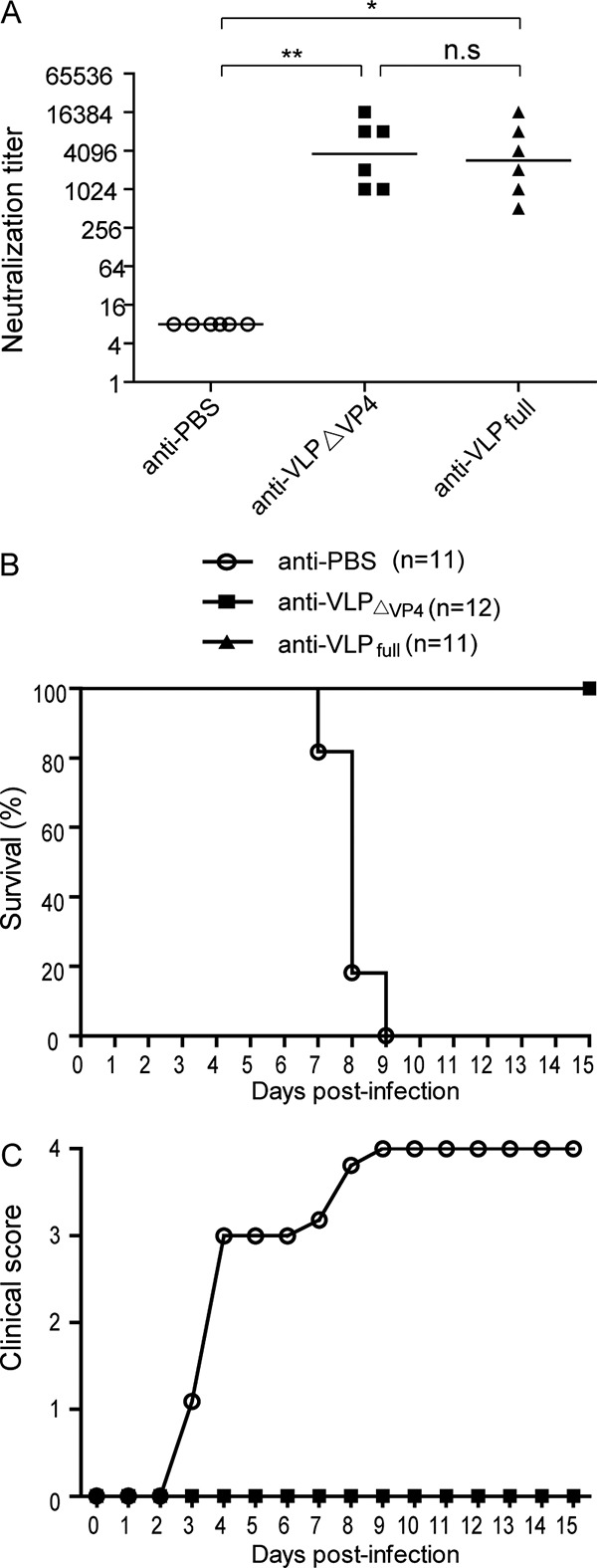
VLPΔVP4-elicited neutralizing antibodies protected mice against lethal viral challenge. (A) In vitro neutralization titers of sera from VLPΔVP4-immunized mice. Groups of female ICR mice were i.p. immunized with VLPΔVP4, VLPfull, or PBS control at week 0 and week 2. At week 4, sera were collected and subjected to neutralization tests against EV71 G082. Each symbol represents an individual mouse, and the horizontal lines indicate the geometric mean values of the groups. (B and C) In vivo challenge experiment. The mice in the VLPΔVP4, VLPfull, and PBS control groups were mated with naive male ICR mice. Neonatal mice born to the immunized dams were i.p. challenged with EV71 MAV-W at 5 days old. All the mice were monitored daily for survival (B) and clinical score (C) for a period of 15 days.
TABLE 2.
Neutralization of pooled antisera against a panel of EV71 and CVA16 strainsa
| Pooled antisera | Neutralization titer against: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| EV71 |
CVA16 |
||||||||
| BrCr | G081 | G082 | FY091 | FY092 | SZ98 | MAV-W | G08 | SZ05 | |
| Anti-PBS | <32 | <32 | <32 | <32 | <32 | <32 | <32 | <32 | <32 |
| Anti-VLPΔVP4 | 1,024 | 1,024 | 4,096 | 4,096 | 4,096 | 4,096 | 1,024 | <32 | <32 |
| Anti-VLPfull | 1,024 | 1,024 | 4,096 | 8,192 | 4,096 | 2,048 | 1,024 | <32 | <32 |
The lowest serum dilution tested was 1:32.
We then compared the protective efficacies of VLPΔVP4 and VLPfull in an established mouse model of EV71 infection (29, 30). Wild-type adult mice are usually resistant to EV71 infection, whereas wild-type neonatal mice inoculated with mouse-adapted EV71 may develop severe clinical manifestations and eventually die (31). Therefore, female ICR mice immunized with VLPΔVP4, VLPfull, or the PBS control were mated with naive male ICR mice. The neonatal mice at an age of 5 days were intraperitoneally (i.p.) challenged with a mouse-adapted EV71 strain and subsequently monitored for 15 days. The mice born to PBS-immunized dams rapidly developed clinical signs, including limb paralysis, and all of them died within 9 days postchallenge. In contrast, all of the mice born to VLPΔVP4- or VLPfull-immunized dams remained healthy without any clinical signs during the whole observation period (Fig. 7B and C). These data demonstrate that VLPΔVP4 immunization can completely protect mice against lethal viral challenge.
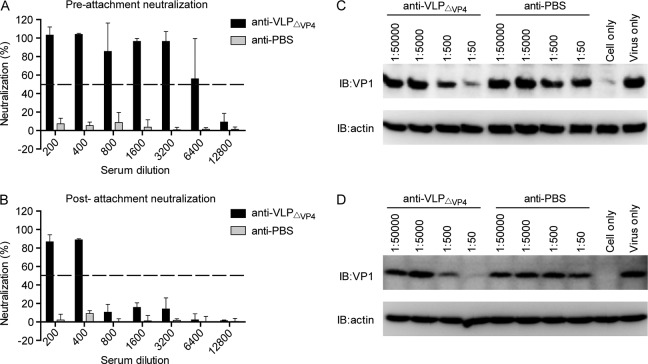
Anti-VLPΔVP4 sera inhibited EV71 infection at both pre-and postattachment stages.
To explore the mode of action of anti-VLPΔVP4 sera, we first determined the neutralization activity of anti-VLPΔVP4 sera at both pre- and postattachment stages. As shown in Fig. 8A and B, the control (anti-PBS) serum did not show any neutralization effect even at the lowest dilution (1:200) when applied at either the pre- or postattachment stage, whereas anti-VLPΔVP4 serum exhibited potent inhibitory effects at both stages, with the highest serum dilutions that conferred more than 50% neutralization being 6,400 and 400, respectively. These results suggest that anti-VLPΔVP4 serum exerted its neutralization function mainly by preventing virus attachment.
FIG 8.
Anti-VLPΔVP4 sera inhibit EV71 attachment to susceptible cells. (A and B) Antisera from an individual mouse of each group were pooled and tested for inhibitory effects by preattachment neutralization assay (A) and postattachment neutralization assay (B). The data are means and SD of three triplicate wells. (C and D) Blockade of EV71 attachment to susceptible cells by anti-VLPΔVP4 sera. EV71 was incubated with serially diluted anti-VLPΔVP4 sera as indicated at 37°C for 1 h. Then, the mixtures were added to RD (C) or Vero (D) cells and incubated at 4°C for another 1 h to allow virus attachment. After washing, cell-bound virus was detected by Western blotting with anti-VP1 polyclonal antibody. β-Actin was also detected, serving as the loading control. IB, immunoblotting.
Anti-VLPΔVP4 pretreatment blocks EV71 attachment to susceptible cells.
Next, we examined whether anti-VLPΔVP4 serum could indeed inhibit EV71 attachment to susceptible cells. Pretreatment with the anti-VLPΔVP4 serum, but not the anti-PBS serum, reduced the amounts of EV71 attached to either RD (Fig. 8C) or Vero (Fig. 8D) cells in an antiserum dose-dependent manner, indicating that anti-VLPΔVP4 serum could specifically block EV71 attachment to susceptible cells.
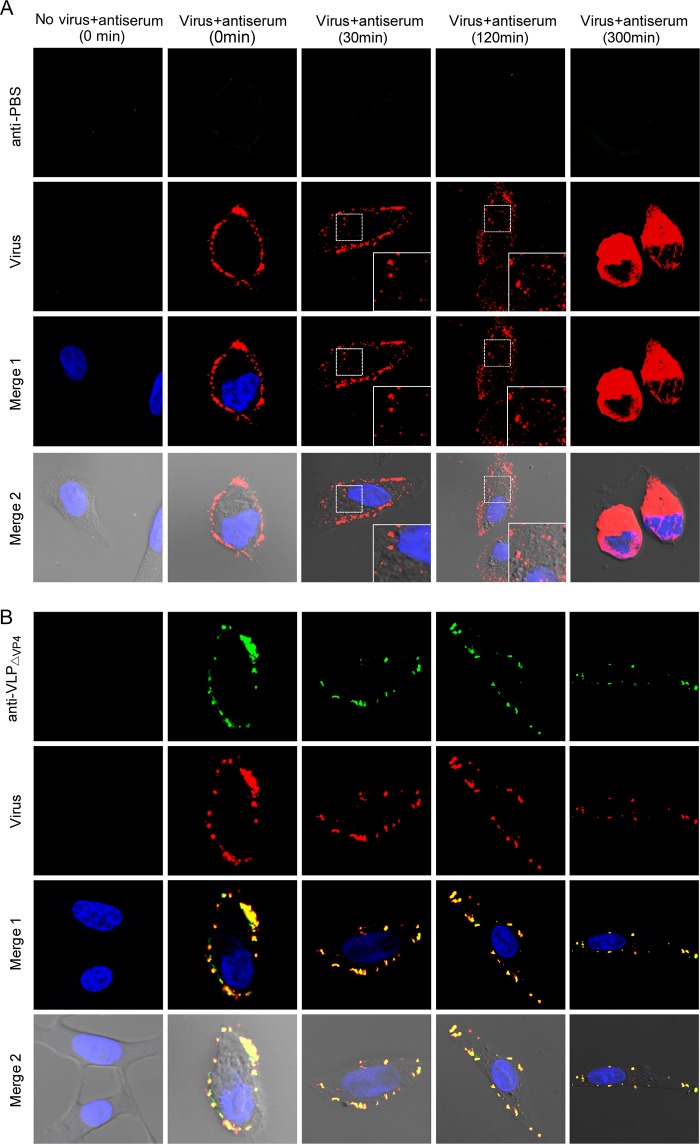
Anti-VLPΔVP4 serum inhibited EV71 internalization at the postattachment stage.
We further determined whether anti-VLPΔVP4 serum could inhibit EV71 internalization, an essential entry step following viral attachment. Vero cells were preattached to the EV71 mature virion and then treated with anti-VLPΔVP4 or anti-PBS serum as described in Materials and Methods. The locations of the virus and the antibodies on the cells at different time points after incubation at 37°C were examined by confocal microscopy. For the anti-PBS-treated sample, the signals of virus (red) were evident at the cell periphery prior to 37°C incubation (0 min) and were then found in the cytoplasm at 30 min after 37°C incubation, indicating rapid internalization of EV71; viral signals were also evident in the cytoplasm at 120 min and were even more significant at 300 min after 37°C incubation (Fig. 9A). In contrast, for the anti-VLPΔVP4-treated samples, the signals of virus (red) colocalized with those of antibodies (green) at the cell periphery regardless of incubation times at 37°C (Fig. 9B), indicating that the anti-VLPΔVP4 antibody-bound EV71 viruses were arrested at the cell periphery. These results demonstrate that anti-VLPΔVP4 treatment can inhibit EV71 internalization at the postattachment stage.
FIG 9.
Anti-VLPΔVP4 sera inhibit EV71 internalization. Vero cells were preattached to EV71 and then treated with anti-PBS (A) or anti-VLPΔVP4 (B) serum at 4°C for 1 h. After incubation at 37°C for 0, 30, 120, or 300 min, as indicated, the cells were fixed for immunofluorescence staining. Localization of virus and antibodies was visualized by confocal microscopy. The green signal represents EV71, which was detected by rabbit-anti-EV71 VLP serum; the red signal represents antibodies in anti-VLPΔVP4 serum; the blue signal represents nuclei stained by DAPI (4′,6-diamidino-2-phenylindole). Merge 1 represents a merge of green, red, and blue channels. Merge 2 represents a merge of green, red, blue, and white field channels. The insets are enlarged views of the boxed areas.
Anti-VLPΔVP4 serum interfered with multiple interactions between EV71 and its key receptors.
To understand the molecular basis of anti-VLPΔVP4-mediated neutralization, we examined whether anti-VLPΔVP4 could disrupt the interactions between EV71 and its key receptors, including heparan sulfate (6), SCARB2 (7), and PSGL-1 (8). As shown in Fig. 10A, preincubation with anti-VLPΔVP4 reduced the amounts of EV71 pulled down by heparin-conjugated agarose beads in an antiserum dose-dependent manner, whereas anti-PBS preincubation had no inhibitory effect regardless of the antiserum dose. The binding of EV71 to SCARB2-immobilized agarose beads was also reduced by pretreatment with anti-VLPΔVP4, but not with the control serum, in an antiserum dose-dependent manner (Fig. 10B). Similarly, the ability of EV71 to bind PSGL-1 in vitro was also impaired following anti-VLPΔVP4 pretreatment (Fig. 10C). These results demonstrated that anti-VLPΔVP4 serum can interfere with multiple EV71-receptor interactions.
FIG 10.
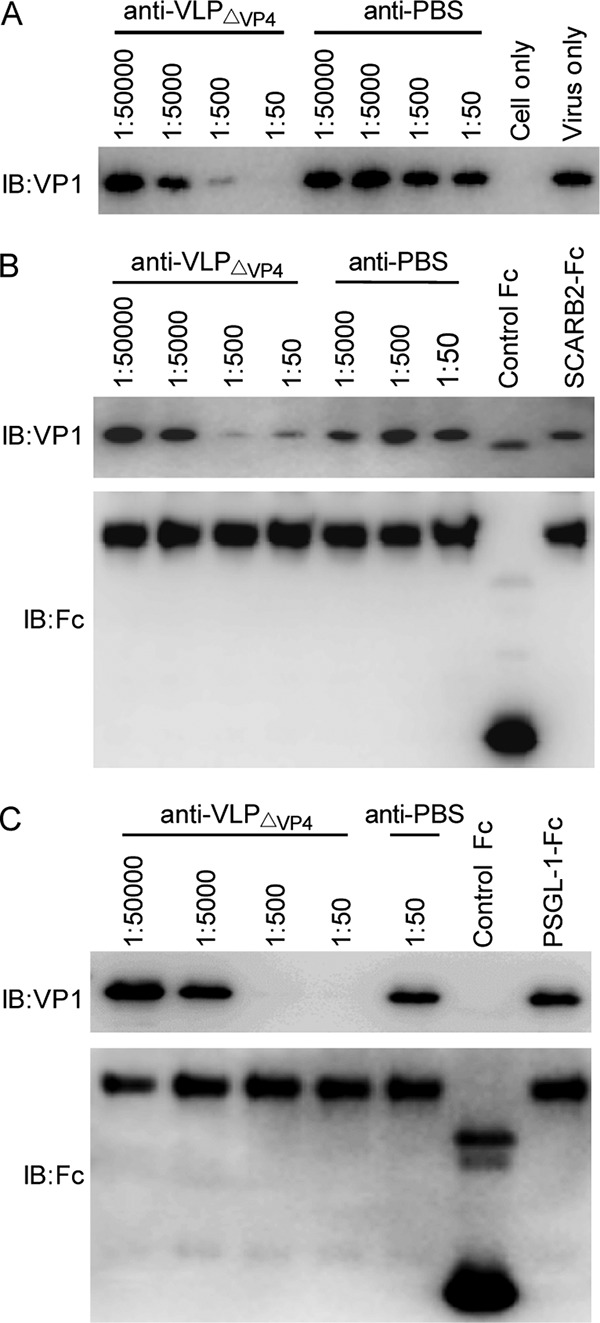
Anti-VLPΔVP4 sera block EV71 binding to its key receptors. EV71 was incubated with serially diluted anti-VLPΔVP4 or anti-PBS at 37°C for 1 h. The mixtures were then added to heparin-immobilized (A), SCARB2-Fc-immobilized (B), or PSGL-1–Fc-immobilized (C) agarose beads and incubated at 37°C for 2 h. After washing, agarose-bound virus was released by SDS-PAGE sampling buffer and analyzed by Western blotting with anti-VP1 polyclonal antibody. For SCARB2-Fc and PSGL-1–Fc pulldown assays, the SCARB2-Fc and PSGL-1–Fc were also detected using an anti-Fc polyclonal antibody as controls for protein immobilization.
DISCUSSION
In the present study, we demonstrated that a designed EV71 VLP that lacks the VP4 protein, VLPΔVP4, can be readily produced in the baculovirus-insect cell expression system. To our knowledge, this is the first evidence showing picornaviral particle assembly in the complete absence of VP4. In general, myristoylation of VP4/VP0 is required for the assembly of infectious virions or noninfectious subviral particles of picornaviruses, such as poliovirus (PV) and foot-and-mouth disease virus (FMDV) (32–37). It has been proposed that, for PV, VP4 myristoylation facilitates the interactions between the 5S protomer subunits required to form stable 14S pentamers (36). This proposition was supported by the results from a study on FMDV, which showed that an Escherichia coli-expressed VP4-truncated P1 precursor of FMDV could be processed by exogenous 3C protease in vitro but the resulting subunit proteins failed to form pentamers (35). However, contrasting observations have been made in hepatitis A virus (HAV), which also belongs to the family Picornaviridae. Specifically, Tesar et al. reported that VP4/VP0 of HAV was not myristoylated and that mutation of a potential myristoylation site in VP4/VP0 did not affect viral infectivity (38). Together, these findings and our results suggest that there may be a VP4 myristoylation-independent mechanism of capsid assembly for some picornaviruses.
During the uncoating process of picornaviruses, the mature virion undergoes a series of changes in conformation and composition, yielding the 135S uncoating intermediate and the 80S uncoated empty particle (9, 39). Although structures of mature virions of many picornaviruses, including EV71, have been determined at atomic resolutions, the structural information on their uncoated form, 80S empty capsids, remains limited (12), due partially to the difficulty in generating the large quantities of high-quality protein preparations required for crystallization. In previous structural studies, uncoated 80S empty capsids were generated by exposing mature virions and/or A particles to high temperatures and/or low pH (10–13, 40, 41). Such treatments often resulted in incomplete virions or A particle-to-80S particle conversion (10–13, 40–42) and in some cases even generated a mixture of distinct forms of 80S particles (40). A three-dimensional (3D) structure of the 80S empty capsid of EV71 has recently been reconstructed at 9.2 Å by cryo-EM single-particle analysis (10); however, high-resolution structural information for this particle is still lacking. The VLPΔVP4 developed in the present study resembles the EV71 80S empty capsid in critical features, such as the protein composition (VP1 plus VP2 plus VP3), the lack of viral RNA, the particle expansion relative to the mature virion, and the enlarged 2-fold channel. Therefore, VLPΔVP4 can be used to represent the EV71 80S empty capsid in structural studies. Indeed, our 3.71-Å VLPΔVP4 map agrees well with the 9.2-Å map of the 80S uncoated empty particle reported previously (10). Moreover, owing to the higher resolution of our cryo-EM map, we were able to observe the externalization of the VP1 N terminus at the base of the canyon near the quasi-3-fold axis (Fig. 4E and F), a characteristic of uncoating intermediates, which could not be identified in the 80S empty-capsid map at lower resolution (10). This observation indicates that the VP1 N terminus externalization can occur spontaneously in the absence of VP4, suggesting that VP4 may have constrained movement of the N-terminal VP1 in the interior of naturally occurring mature virions and NEPs. Therefore, during the mature-virion–to–A particle transition, the extrusion of VP4 must occur first to release the constraint, allowing the VP1 N terminus externalization to proceed. By following our protocol, homogeneous VLPΔVP4 can be easily produced and purified, thus providing high-quality materials for crystallography experiments. We believe that our VLPΔVP4 strategy can be applied to other picornaviruses, as well, for structural study of their 80S uncoated empty particles, ultimately leading to better understanding of the uncoating process of picornaviruses as a whole.
In the present study, we demonstrated that, as a mimic of the EV71 80S empty capsid, VLPΔVP4 is highly immunogenic and confers protection against EV71 challenge in mice. These data are in sharp contrast to the results from studies on other picornaviruses. Specifically, it was reported that the 80S empty particles of PV (generated by heating mature virions) failed to evoke neutralizing antibodies (43). This loss of vaccine potency was associated with antigenic changes in the 80S empty particles (43, 44). A subsequent study demonstrated that the conformation of neutralization epitopes was different between the PV mature virion and the 80S empty particle (45). In the case of FMDV, there is thus far no evidence showing the existence of uncoating intermediates, including the 80S empty capsid, during natural viral infection. Rather, upon heating or low-pH treatment, the 146S mature virion dissociated into 12S pentamers (46). As expected, the FMDV 12S pentamer was much less competent at inducing neutralizing antibodies than the 146S mature virus (47). Hence, it is surprising that our VLPΔVP4, which resembles the 80S empty capsid, retained protective immunogenicity. Two factors may have contributed to the vaccine potency of VLPΔVP4: (i) the particulate nature, as it is well known that VLPs are more immunogenic than the corresponding soluble proteins, owing to their ability to efficiently interact with antigen-presenting cells, to display epitopes at high density, and to provide T-cell help (reviewed in references 48 and 49); (ii) the preservation of important neutralizing epitopes with proper conformations on VLPΔVP4. The SP70 epitope located at the GH loop of VP1 is a predominant neutralizing epitope for EV71 (50) and is recognized by broadly neutralizing monoclonal antibodies (28, 51). The ability of the SP70 epitope to induce neutralizing antibodies did not appear to be strictly conformation dependent, as SP70-targeting antibodies recognized synthetic peptide and denatured VP1 protein (28, 52) and bound both the mature virion and VLPfull (51). Moreover, the SP70 epitope displayed on chimeric (hepatitis B core antigen-based) VLPs could elicit neutralizing antibodies that protected mice against EV71 challenge (53). In this study, VLPΔVP4 strongly reacted with the SP70-targeting neutralizing monoclonal antibody D5 (Fig. 5E), suggesting that the protective antigenicity may have been well maintained.
VLPΔVP4 differs from VLPfull in particle composition and antigenic properties; however, VLPΔVP4 may represent a vaccine platform superior to VLPfull for developing recombinant vaccines for EV71 and other related enteroviruses for several reasons. First, VLPΔVP4 eliminates the potential for antigen inconsistency caused by the autocleavage of VP0 into VP4 and VP2, which has been observed for VLPfull derived from EV71 (20, 24) and the closely related CVA16 (25, 26), and therefore, quality control of antigen production for VLPΔVP4 should be simpler than that for VLPfull. Second, VLPΔVP4 appeared to elicit slightly higher neutralizing antibody titers than did VLPfull, likely due to the induction of antibodies against the VP1 N terminus, which was externalized in VLPΔVP4 but not in VLPfull, as previous studies have shown that antibodies to the VP1 N terminus of EV71 or other enteroviruses are neutralizing (54, 55). Third, VLPΔVP4 does not contain VP4 and therefore eliminates the potential for antibody-dependent enhancement (ADE) of infection mediated by anti-VP4 antibodies reported for several enteroviruses (56–58). The ADE phenomena have also been observed for EV71 (59–62), although it is unclear whether VP4-targeting antibodies contributed to the observed enhancement of infection. Finally, the externalized VP1 N terminus of VLPΔVP4 is immunogenic and may be inserted or replaced with multiple heterologous T- and/or B-cell epitopes to induce antibodies with more potent and broader neutralization ability or multifunctional T cells. In the latter case, VLPΔVP4 may also be used as a novel vaccine vector for epitope presentation.
Despite the fact that EV71 VLPs have been shown to induce neutralizing antibodies that protect mice against virus challenge (20, 23, 24), the mechanisms by which VLP-elicited antibodies exert inhibition are not entirely clear. In the present study, we demonstrated that anti-VLPΔVP4 is able not only to inhibit EV71 attachment in a cell-type-independent manner, but also to inhibit EV71 internalization at the postattachment stage. At the molecular level, we showed that (i) anti-VLPΔVP4 sera can interfere with EV71 binding to heparan sulfate, an attachment receptor for EV71 (6); (ii) they can interfere with EV71 binding to SCARB2, which mediates EV71 internalization and uncoating (16, 63); and (iii) they can also inhibit EV71 binding to PSGL-1, another identified receptor that facilitates EV71 infection of leukocytes (8). Taken together, these data suggest that blocking the interactions between EV71 and its key receptors is the main mechanism by which anti-VLPΔVP4 mediates neutralization of EV71 infection.
In summary, EV71 VLPΔVP4 mimicking the 80S uncoated empty particle represents an excellent EV71 vaccine platform. Our study not only elucidates the structural basis and protective mechanism of the EV71 VLPΔVP4 vaccine, but also establishes a methodology for VLPΔVP4 production that can be extended to other enteroviruses, as well, thus providing important information for vaccine development and structural study of enteroviruses.
MATERIALS AND METHODS
Cells and viruses.
RD (American Type Culture Collection [ATCC] strain CCL-136), Vero (ATCC strain CRL-1586), and Sf9 (ATCC strain CRL-1711) cells were grown as described previously (20, 25). Seven EV71 strains—EV71 BrCr (ATCC strain VR-1775), EV71 G081, EV71 G082, EV71 FY09-2, EV71 SZ98, and EV71 MAV-W—and two CVA16 strains, CVA16 G08 and CVA16 SZ05, were described previously (21). The EV71 mature virion was prepared as previously described (53). The EV71 135S A particle and 80S empty particle were prepared by heating mature virions in hypotonic buffer according to a previously published method (10), and the resultant sample was a mixture of two kinds of particles. The virus titers were determined by a microtitration method using RD cells and are expressed as the 50% tissue culture infectious dose (TCID50) (20).
Peptides, recombinant proteins, and antibodies.
The VP1-derived peptides, including the SP70 peptide and a control peptide, were synthesized in a previous study (20). VP4 and the N terminus of VP1 (amino acids 1 to 75, termed VP1-N75) were expressed as glutathione S-transferase (GST) fusion proteins in E. coli BL21. Then, the GST-tagged fusion proteins were purified from lysates of the transformed E. coli cells by using a glutathione-resin column (GE Healthcare) according to the manufacturer's instructions. The insect cell-derived full EV71 VLP (termed VLPfull) was generated as described previously (20). Rabbit polyclonal antibody against VLPfull was generated in house. Rabbit polyclonal antibodies against VP0 and VP1 of EV71 and guinea pig polyclonal antibody against VP3 of CVA16, which cross-react strongly with VP3 of EV71, were previously generated (64). A monoclonal antibody, D5, targeting the VP1 GH loop was previously described (28). Mouse polyclonal antibody against VP1-N75 was generated in house by immunizing BALB/c mice three times with purified VP1-N75 protein. The antigen formulation and immunization procedures were similar to those in a previous study (64) except that the antigen dose for each immunization was 75 μg per mouse.
Vector construction.
The construction of expression vectors and generation of recombinant baculovirus were performed as previously described (20) with some modifications. Briefly, the VP4-truncated P1 fragment (termed P1ΔVP4) was amplified with the primers forward, 5′-AATCCATGGGTTCGCAGGTGTCT-3′, and reverse, 5′-AATCCATGGGATCCCCATCCGCTGAGGCG-3′ (underlined sequences, cleavage sites of Nco I), and inserted into the plasmid pIExBac-1 to make pIExBac-P1ΔVP4. pIExBac-P1ΔVP4 was used to generate the baculovirus (termed IExBac-P1ΔVP4), as previously described (20). The pIExBac-3CD plasmid and the corresponding baculovirus, IExBac-3CD, used in this study were described previously (20).
Expression and verification of VLPΔVP4.
Sf9 cells were infected separately by IExBac-P1ΔVP4 at a multiplicity of infection (MOI) of 3 and IExBac-3CD at an MOI of 1 or coinfected by both of the baculoviruses at MOI of 3 and 1, respectively. After 72 h, infected cells were harvested for determination of protein expression. Cell lysates were prepared and used to coat 96-well ELISA plates or loaded onto 12% polyacrylamide gels. ELISA and Western blot analysis were carried out with polyclonal antibodies to VP0, VP1, or VP3, as previously described (20).
Sucrose gradient sedimentation was performed as previously described (20) with some modifications. Briefly, cell lysates from IExBac-P1ΔVP4- and IExBac-3CD-coinfected Sf9 cells were layered onto a 20% sucrose cushion and centrifuged at 27,000 rpm (131,000 × g) for 5 h, and the pellet was resuspended in PBS and layered onto a 10 to 50% sucrose gradient for ultracentrifugation. Then, 12 fractions were harvested from top to bottom. The samples were loaded onto 12% polyacrylamide gels for Coomassie blue R-250 staining, transferred onto polyvinylidene difluoride (PVDF) membranes for Western blot analysis, or used to coat 96-well ELISA plates for ELISA analysis. The sucrose gradient fractions with the strongest signals of capsid proteins in Western blot assays were pooled and subjected to further purification as previously described (20). The final VLPΔVP4 sample was quantified by Bradford assay and stored at −80°C until use.
Negative staining and electron microscopy of VLPΔVP4 were performed as previously described (53).
Cryo-EM imaging and reconstruction.
The cryo-EM imaging and reconstruction of VLPΔVP4 were performed as described previously (51) with some modifications. Images were recorded on a Gatan K2 Summit direct electron detector operated in superresolution counting mode with a pixel size of 0.665 Å. Each frame was exposed for 0.2 s, with an accumulation time of 7.6 s for each movie, leading to a total accumulated dose of 38 electrons (e−)/Å2 on the specimen. All the images were automatically collected using SerialEM software (65). To correct the drift and beam-induced motion, the frames in each movie were aligned to generate a single micrograph using Motioncorr 2.0. Then, particles were semimanually boxed using the e2boxer.py program from the EMAN2.1 package (66). A total of 21,362 particles were boxed out from 486 images. These particles were subjected to 2D and 3D classification using the software RELION 1.3 (67), and 7,887 cleaned up particles without inclusion of genome materials were selected. Then, the RELION project was converted to an EMAN project by using the EMAN 2.1 e2reliontoeman.py program. Contrast transfer function (CTF) fitting was automatically performed using the fitctf2.py program in the Jspr software package (68) and then visually validated and adjusted using the EMAN 1.9 ctfit program (69). Then, the 3D reconstruction was performed using Jspr (70), which coordinates the further refinement of defocus, magnification, astigmatism, and anisotropic magnification (71). The map resolution was assessed at a Fourier shell correlation (FSC) cutoff of 0.143.
Structural analysis.
UCSF Chimera software (72) was used to visualize the VLPΔVP4 density map, to measure the surface diameter, and to compare the structural similarities of VLPΔVP4 and related particles, including the EV71 mature virion (PDB ID 3VBS) (3), tbe EV71 NEP (PDB ID 3VBU) (3), EV71 VLPfull (EMDB accession no. EMD-2607) (19), and the CVA16 135S-like particle (PDB ID 4JGY) (27). The cc values for these comparisons were calculated using the Fit in Map module in UCSF Chimera. The difference map between VLPfull and VLPΔVP4 was calculated by subtracting the VLPfull density (5.2 Å) (EMDB accession no. EMD-2607) from our VLPΔVP4 density map, which was low- pass filtered into 6 Å and normalized to the VLPfull map. The homologous model of EV71 VP1 was built through the online SWISS-MODEL server (73).
Antigenic characterizations.
The VLPΔVP4 and VLPfull samples were loaded onto 12% polyacrylamide gels for protein separation and then transferred onto PVDF membranes for Western blot analysis using individual polyclonal antibodies against VP0, VP1, or VP3. To determine the reactivity of VLPΔVP4 or VLPfull with a VP1 N-terminus-specific antibody, ELISA plates were coated with protein samples, followed by detection with a polyclonal antibody against VP1-N75.
Mouse immunization and antibody measurement.
For a single injection dose, 1 μg of VLPΔVP4 or VLPfull was adsorbed to 0.5 mg of aluminum hydroxide (Alhydrogel; InvivoGen, USA). Groups of six female ICR mice (8 weeks old) were injected i.p. with each dose of formulated antigen at weeks 0 and 2. A control group of mice were injected with the formulated PBS. Blood samples were collected at week 4 for determination of antibody responses and neutralization titers.
For determination of antibody responses, ELISA plates were coated with recombinant VP4 protein (200 ng/well), VP1-N75 (200 ng/well), or VP1-derived SP70 peptide (200 ng/well). Individual serum samples were diluted at 1:50, 1:100, and 1:100 and analyzed for reactivity with VP4, VP1-N75, or SP70 peptide, respectively. All the incubation steps and measurement of absorbance (optical density at 45 nm [OD450]) were performed as previously described (20).
For determination of neutralization titers of serum samples against a panel of EV71 and CVA16 strains, a standard microtitration assay was performed as previously described (53).
Maternal immunization/protection assay.
Three groups of 8-week-old female ICR mice were immunized as described above. The immunized mice were allowed to mate with naive male ICR mice 6 weeks after the last immunization. The neonatal mice born to the immunized dams in each group were challenged on day 5 after birth by i.p. injection with 2.1 × 104 TCID50 of EV71 MAV-W and then monitored daily for survival and clinical scores for 15 days. Clinical scores were graded as follows: 0, healthy; 1, reduced mobility; 2, limb weakness; 3, paralysis; 4, death.
The mouse experiments described above were approved by the Institutional Animal Care and Use Committee at the Institut Pasteur of Shanghai and carried out in accordance with the regulations in the Guide for the Care and Use of Laboratory Animals issued by the Ministry of Science and Technology of the People's Republic of China.
Pre- and postattachment neutralization assays.
Both the pre-and postattachment neutralization assays were performed as previously described (28) except that the antibodies used in this study were serially diluted pooled antisera for each group. Cell viabilities were measured using a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay, and neutralization efficiencies were calculated as previously described (28).
Attachment inhibition assay.
The attachment inhibition assay was performed as described previously with minor modifications (20): pooled antisera were diluted in 400 μl Dulbecco's modified Eagle's medium (DMEM) containing 4.8 × 107 TCID50 of EV71 G082.
Immunofluorescence assay.
The localization of virus and antibodies within cells was analyzed by immunofluorescence staining and confocal microscopy analysis according to a previously established protocol (28) with some minor modifications. Briefly, Vero cells were preattached to 9 × 106 TCID50 of mature virions and then incubated with the indicated antiserum samples at 4°C. After washing away unbound antibodies, the cells were immediately fixed using 4% paraformaldehyde; in some cases, prior to fixation, the cells were moved to a 37°C incubator for a period of time (e.g., 30, 120, or 300 min) to allow viral internalization to proceed. The fixed cells were then subjected to immunofluorescence staining and subsequent imaging.
Pulldown assays.
Heparin-, SCARB2-, and PSGL-1 pulldown assays were carried out according to previously established protocols (28) with minor modifications. Specifically, for heparin pulldown assays, serially diluted antisera were mixed with 4.8 × 107 TCID50 of EV71 G082 in 400 μl DMEM and then incubated at 37°C for 1 h; next, the virus-antiserum mixtures were incubated with heparin-agarose beads (Sigma) at 4°C for 2 h. After washing, the virus-bound beads were subjected to SDS-PAGE and Western blot analysis as described above.
Statistics.
Statistical significances were determined by one-way analysis of variance (ANOVA) using GraphPad Prism version 5.
Accession number(s).
The cryo-EM map for EV71 VLPΔVP4 has been deposited in the Electron Microscopy Data Bank (accession no. EMD-6702).
ACKNOWLEDGMENTS
We thank the Electron Microscopy Facility and the Data Base and Computation Facility at the National Centre for Protein Science Shanghai (NCPSS) for their assistance with EM instrument management, data storage, and parallel computing.
This work was supported by grants from the TOTAL Foundation, the National Key R&D Program of China(2017YFA0503503), CAS Pilot Strategic Science and Technology Projects B (XDB08030201), the National Natural Science Foundation of China (31370930, 31670754, and 31500153), the Science and Technology Commission of Shanghai Municipality (13431900600 and 15XD1524900), the CAS-Shanghai Science Research Center (CAS-SSRC-YH-2015-01), the STS Program of the Chinese Academy of Sciences (KFJ-EW-STS-098), and the State Key Laboratory of Molecular Biology.
REFERENCES
- 1.Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. 2010. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis 10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 2.Xing W, Liao Q, Viboud C, Zhang J, Sun J, Wu JT, Chang Z, Liu F, Fang VJ, Zheng Y, Cowling BJ, Varma JK, Farrar JJ, Leung GM, Yu H. 2014. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect Dis 14:308–318. doi: 10.1016/S1473-3099(13)70342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol 19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. 2012. Crystal structure of human enterovirus 71. Science 336:1274. doi: 10.1126/science.1218713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Rossmann MG. 2014. The cellular receptor for enterovirus 71. Protein Cell 5:655–657. doi: 10.1007/s13238-014-0092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan CW, Poh CL, Sam IC, Chan YF. 2013. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J Virol 87:611–620. doi: 10.1128/JVI.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, Koike S. 2009. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat Med 15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- 8.Nishimura Y, Shimojima M, Tano Y, Miyamura T, Wakita T, Shimizu H. 2009. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat Med 15:794–797. doi: 10.1038/nm.1961. [DOI] [PubMed] [Google Scholar]
- 9.Bergelson JM, Coyne CB. 2013. Picornavirus entry. Adv Exp Med Biol 790:24–41. doi: 10.1007/978-1-4614-7651-1_2. [DOI] [PubMed] [Google Scholar]
- 10.Shingler KL, Yoder JL, Carnegie MS, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2013. The enterovirus 71 A-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS Pathog 9:e1003240. doi: 10.1371/journal.ppat.1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korant BD, Lonbergh K, Stasny JT, Noble J. 1972. Naturally occurring and artificially produced components of 3 rhinoviruses. Virology 48:71. doi: 10.1016/0042-6822(72)90115-8. [DOI] [PubMed] [Google Scholar]
- 12.Garriga D, Pickl-Herk A, Luque D, Wruss J, Caston JR, Blaas D, Verdaguer N. 2012. Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid. PLoS Pathog 8:e1002473. doi: 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickl-Herk A, Luque D, Vives-Adrian L, Querol-Audi J, Garriga D, Trus BL, Verdaguer N, Blaas D, Caston JR. 2013. Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle. Proc Natl Acad Sci U S A 110:20063–20068. doi: 10.1073/pnas.1312128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu CC, Guo MS, Lin FH, Hsiao KN, Chang KH, Chou AH, Wang YC, Chen YC, Yang CS, Chong PC. 2011. Purification and characterization of enterovirus 71 viral particles produced from Vero cells grown in a serum-free microcarrier bioreactor system. PLoS One 6:e20005. doi: 10.1371/journal.pone.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JX, Mao QY, Liang ZL, Ji H, Zhu FC. 2014. Development of enterovirus 71 vaccines: from the lab bench to phase III clinical trials. Expert Rev Vaccines 13:609–618. doi: 10.1586/14760584.2014.897617. [DOI] [PubMed] [Google Scholar]
- 16.Yamayoshi S, Ohka S, Fujii K, Koike S. 2013. Functional comparison of SCARB2 and PSGL1 as receptors for enterovirus 71. J Virol 87:3335–3347. doi: 10.1128/JVI.02070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P, Song Z, Qi Y, Feng X, Xu N, Sun Y, Wu X, Yao X, Mao Q, Li X, Dong W, Wan X, Huang N, Shen X, Liang Z, Li W. 2012. Molecular determinants of enterovirus 71 viral entry: cleft around GLN-172 on VP1 protein interacts with variable region on scavenge receptor B 2. J Biol Chem 287:6406–6420. doi: 10.1074/jbc.M111.301622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung YC, Huang JH, Lai CW, Sheng HC, Shih SR, Ho MS, Hu YC. 2006. Expression, purification and characterization of enterovirus-71 virus-like particles. World J Gastroenterol 12:921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong M, Zhu H, Zhou J, Yang C, Feng J, Huang X, Ji G, Xu H, Zhu P. 2014. Cryo-electron microscopy study of insect cell-expressed enterovirus 71 and coxsackievirus a16 virus-like particles provides a structural basis for vaccine development. J Virol 88:6444–6452. doi: 10.1128/JVI.00200-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ku Z, Ye X, Huang X, Cai Y, Liu Q, Li Y, Su Z, Huang Z. 2013. Neutralizing antibodies induced by recombinant virus-like particles of enterovirus 71 genotype C4 inhibit infection at pre- and post-attachment steps. PLoS One 8:e57601. doi: 10.1371/journal.pone.0057601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C, Ku ZQ, Liu QW, Wang XL, Chen T, Ye XH, Li DP, Jin X, Huang Z. 2015. High-yield production of recombinant virus-like particles of enterovirus 71 in Pichia pastoris and their protective efficacy against oral viral challenge in mice. Vaccine 33:2335–2341. doi: 10.1016/j.vaccine.2015.03.034. [DOI] [PubMed] [Google Scholar]
- 22.Lyu K, He YL, Li HY, Chen R. 2015. Crystal structures of yeast-produced enterovirus 71 and enterovirus 71/coxsackievirus A16 chimeric virus-like particles provide the structural basis for novel vaccine design against hand-foot-and-mouth disease. J Virol 89:6196–6208. doi: 10.1128/JVI.00422-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin YL, Yu CI, Hu YC, Tsai TJ, Kuo YC, Chi WK, Lin AN, Chiang BL. 2012. Enterovirus type 71 neutralizing antibodies in the serum of macaque monkeys immunized with EV71 virus-like particles. Vaccine 30:1305–1312. doi: 10.1016/j.vaccine.2011.12.081. [DOI] [PubMed] [Google Scholar]
- 24.Li HY, Han JF, Qin CF, Chen R. 2013. Virus-like particles for enterovirus 71 produced from Saccharomyces cerevisiae potently elicits protective immune responses in mice. Vaccine 31:3281–3287. doi: 10.1016/j.vaccine.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, Yan K, Feng Y, Huang X, Ku Z, Cai Y, Liu F, Shi J, Huang Z. 2012. A virus-like particle vaccine for coxsackievirus A16 potently elicits neutralizing antibodies that protect mice against lethal challenge. Vaccine 30:6642–6648. doi: 10.1016/j.vaccine.2012.08.071. [DOI] [PubMed] [Google Scholar]
- 26.Zhao H, Li HY, Han JF, Deng YQ, Li YX, Zhu SY, He YL, Qin ED, Chen R, Qin CF. 2013. Virus-like particles produced in Saccharomyces cerevisiae elicit protective immunity against Coxsackievirus A16 in mice. Appl Microbiol Biotechnol 97:10445–10452. doi: 10.1007/s00253-013-5257-3. [DOI] [PubMed] [Google Scholar]
- 27.Ren JS, Wang XX, Hu ZY, Gao Q, Sun Y, Li XM, Porta C, Walter TS, Gilbert RJ, Zhao YG, Axford D, Williams M, McAuley K, Rowlands DJ, Yin WD, Wang JZ, Stuart DI, Rao ZH, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun 4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ku Z, Ye X, Shi J, Wang X, Liu Q, Huang Z. 2015. Single neutralizing monoclonal antibodies targeting the VP1 GH loop of enterovirus 71 inhibit both virus attachment and internalization during viral entry. J Virol 89:12084–12095. doi: 10.1128/JVI.02189-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai Y, Ku Z, Liu Q, Leng Q, Huang Z. 2014. A combination vaccine comprising of inactivated enterovirus 71 and coxsackievirus A16 elicits balanced protective immunity against both viruses. Vaccine 32:2406–2412. doi: 10.1016/j.vaccine.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Ku Z, Liu Q, Ye X, Cai Y, Wang X, Shi J, Li D, Jin X, An W, Huang Z. 2014. A virus-like particle based bivalent vaccine confers dual protection against enterovirus 71 and coxsackievirus A16 infections in mice. Vaccine 32:4296–4303. doi: 10.1016/j.vaccine.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 31.Wang YF, Yu CK. 2014. Animal models of enterovirus 71 infection: applications and limitations. J Biomed Sci 21:31. doi: 10.1186/1423-0127-21-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marc D, Girard M, Vanderwerf S. 1991. A Gly1 to Ala substitution in poliovirus capsid protein Vp0 blocks its myristoylation and prevents viral assembly. J Gen Virol 72:1151–1157. doi: 10.1099/0022-1317-72-5-1151. [DOI] [PubMed] [Google Scholar]
- 33.Moscufo N, Simons J, Chow M. 1991. Myristoylation is important at multiple stages in poliovirus assembly. J Virol 65:2372–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abrams CC, King AMQ, Belsham GJ. 1995. Assembly of foot-and-mouth-disease virus empty capsids synthesized by a vaccinia virus expression system. J Gen Virol 76:3089–3098. doi: 10.1099/0022-1317-76-12-3089. [DOI] [PubMed] [Google Scholar]
- 35.Goodwin S, Tuthill TJ, Arias A, Killington RA, Rowlands DJ. 2009. Foot-and-mouth disease virus assembly: processing of recombinant capsid precursor by exogenous protease induces self-assembly of pentamers in vitro in a myristoylation-dependent manner. J Virol 83:11275–11282. doi: 10.1128/JVI.01263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansardi DC, Porter DC, Morrow CD. 1992. Myristylation of poliovirus capsid precursor-p1 is required for assembly of subviral particles. J Virol 66:4556–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang P, Liu Y, Ma HC, Paul AV, Wimmer E. 2014. Picornavirus morphogenesis. Microbiol Mol Biol Rev 78:418–437. doi: 10.1128/MMBR.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tesar M, Jia XY, Summers DF, Ehrenfeld E. 1993. Analysis of a potential myristoylation site in hepatitis-A virus capsid protein Vp4. Virology 194:616–626. doi: 10.1006/viro.1993.1301. [DOI] [PubMed] [Google Scholar]
- 39.Hogle JM. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu Rev Microbiol 56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levy HC, Bostina M, Filman DJ, Hogle JM. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J Virol 84:4426–4441. doi: 10.1128/JVI.02393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lonberg-Holm K, Korant BD. 1972. Early interaction of rhinoviruses with host cells. J Virol 9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bostina M, Levy H, Filman DJ, Hogle JM. 2011. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J Virol 85:776–783. doi: 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Bouvier GL. 1955. The modification of poliovirus antigens by heat and ultraviolet light. Lancet ii:1013–1016. [DOI] [PubMed] [Google Scholar]
- 44.Ferguson M, Minor PD, Magrath DI, Qui YH, Spitz M, Schild GC. 1984. Neutralization epitopes on poliovirus type-3 particles: an analysis using monoclonal-antibodies. J Gen Virol 65:197–201. doi: 10.1099/0022-1317-65-1-197. [DOI] [PubMed] [Google Scholar]
- 45.Ferguson M, Minor PD. 1990. Differences in conformation of type-3 poliovirus antigenic sites on noninfectious empty particles and infectious virus. J Gen Virol 71:1271–1274. doi: 10.1099/0022-1317-71-6-1271. [DOI] [PubMed] [Google Scholar]
- 46.Vasquez C, Denoya CD, Latorre JL, Palma EL. 1979. Structure of foot-and-mouth-disease virus capsid. Virology 97:195–200. doi: 10.1016/0042-6822(79)90387-8. [DOI] [PubMed] [Google Scholar]
- 47.Rao MG, Butchaiah G, Sen AK. 1994. Antibody-Response to 146s particle, 12s protein subunit and isolated vp1 polypeptide of foot-and-mouth-disease virus type Asia-1. Vet Microbiol 39:135–143. doi: 10.1016/0378-1135(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 48.Noad R, Roy P. 2003. Virus-like particles as immunogens. Trends Microbiol 11:438–444. doi: 10.1016/S0966-842X(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 49.Chackerian B. 2007. Virus-like particles: flexible platforms for vaccine development. Expert Rev Vaccines 6:381–390. doi: 10.1586/14760584.6.3.381. [DOI] [PubMed] [Google Scholar]
- 50.Foo DG, Alonso S, Phoon MC, Ramachandran NP, Chow VT, Poh CL. 2007. Identification of neutralizing linear epitopes from the VP1 capsid protein of Enterovirus 71 using synthetic peptides. Virus Res 125:61–68. doi: 10.1016/j.virusres.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 51.Ye XH, Fan C, Ku ZQ, Zuo T, Kong LL, Zhang C, Shi JP, Liu QW, Chen T, Zhang YY, Jiang W, Zhang LQ, Huang Z, Cong Y. 2016. Structural basis for recognition of human enterovirus 71 by a bivalent broadly neutralizing monoclonal antibody. PLoS Pathog 12:e1005454. doi: 10.1371/journal.ppat.1005454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ku Z, Shi J, Liu Q, Huang Z. 2012. Development of murine monoclonal antibodies with potent neutralization effects on enterovirus 71. J Virol Methods 186:193–197. doi: 10.1016/j.jviromet.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 53.Ye X, Ku Z, Liu Q, Wang X, Shi J, Zhang Y, Kong L, Cong Y, Huang Z. 2014. Chimeric virus-like particle vaccines displaying conserved enterovirus 71 epitopes elicit protective neutralizing antibodies in mice through divergent mechanisms. J Virol 88:72–81. doi: 10.1128/JVI.01848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim XF, Jia Q, Chow VTK, Kwang J. 2013. Characterization of a novel monoclonal antibody reactive against the N-terminal region of Enterovirus 71 VP1 capsid protein. J Virol Methods 188:76–82. doi: 10.1016/j.jviromet.2012.11.038. [DOI] [PubMed] [Google Scholar]
- 55.Li QH, Yafal AG, Lee YMH, Hogle J, Chow M. 1994. Poliovirus neutralization by antibodies to internal epitopes of Vp4 and Vp1 results from reversible exposure of these sequences at physiological temperature. J Virol 68:3965–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauter P, Lobert PE, Lucas B, Varela-Calvino R, Alm G, Wattre P, Hober D. 2007. Role of the capsid protein VP4 in the plasma-dependent enhancement of the Coxsackievirus B4E2-infection of human peripheral blood cells. Virus Res 125:183–190. doi: 10.1016/j.virusres.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Sauter P, Chehadeh W, Lobert PE, Lazrek M, Goffard A, Soumillon M, Caloone D, Vantyghem MC, Weill J, Fajardy I, Alm G, Lucas B, Hober D. 2008. A part of the VP4 capsid protein exhibited by coxsackievirus B4 E2 is the target of antibodies contained in plasma from patients with type 1 diabetes. J Med Virol 80:866–878. doi: 10.1002/jmv.21171. [DOI] [PubMed] [Google Scholar]
- 58.Chehadeh W, Lobert PE, Sauter P, Goffard A, Lucas B, Weill J, Vantyghem MC, Alm G, Pigny P, Hober D. 2005. Viral protein VP4 is a target of human antibodies enhancing coxsackievirus B4- and B3-induced synthesis of alpha interferon. J Virol 79:13882–13891. doi: 10.1128/JVI.79.22.13882-13891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang SM, Chen IC, Su LY, Huang KJ, Lei HY, Liu CC. 2010. Enterovirus 71 infection of monocytes with antibody-dependent enhancement. Clin Vaccine Immunol 17:1517–1523. doi: 10.1128/CVI.00108-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen IC, Wang SM, Yu CK, Liu CC. 2013. Subneutralizing antibodies to enterovirus 71 induce antibody-dependent enhancement of infection in newborn mice. Med Microbiol Immunol 202:259–265. doi: 10.1007/s00430-013-0289-y. [DOI] [PubMed] [Google Scholar]
- 61.Han JF, Cao RY, Deng YQ, Tian X, Jiang T, Qin ED, Qin CF. 2011. Antibody dependent enhancement infection of enterovirus 71 in vitro and in vivo. Virol J 8:106. doi: 10.1186/1743-422X-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao RY, Dong DY, Liu RJ, Han JF, Wang GC, Zhao H, Li XF, Deng YQ, Zhu SY, Wang XY, Lin F, Zhang FJ, Chen W, Qin ED, Qin CF. 2013. Human IgG subclasses against enterovirus type 71: neutralization versus antibody dependent enhancement of infection. PLoS One 8:e64024. doi: 10.1371/journal.pone.0064024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin YW, Lin HY, Tsou YL, Chitra E, Hsiao KN, Shao HY, Liu CC, Sia C, Chong P, Chow YH. 2012. Human SCARB2-mediated entry and endocytosis of EV71. PLoS One 7:e30507. doi: 10.1371/journal.pone.0030507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Q, Huang X, Ku Z, Wang T, Liu F, Cai Y, Li D, Leng Q, Huang Z. 2013. Characterization of enterovirus 71 capsids using subunit protein-specific polyclonal antibodies. J Virol Methods 187:127–131. doi: 10.1016/j.jviromet.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 65.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 66.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 67.Scheres SHW. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang W, Guo F, Liu Z. 2012. A graph theory method for determination of cryo-EM image focuses. J Struct Biol 180:343–351. doi: 10.1016/j.jsb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 70.Guo F, Jiang W. 2014. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol 1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z, Guo F, Wang F, Li TC, Jiang W. 2016. 2.9 Angstrom resolution Cryo-EM 3D reconstruction of close-packed virus particles. Structure 24:319–328. doi: 10.1016/j.str.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 73.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]