ABSTRACT
Human astroviruses are recognized as a leading cause of viral diarrhea worldwide in children, immunocompromised patients, and the elderly. There are currently no vaccines available to prevent astrovirus infection; however, antibodies developed by healthy individuals during previous infection correlate with protection from reinfection, suggesting that an effective vaccine could be developed. In this study, we investigated the molecular mechanism by which several strains of human astrovirus serotype 2 (HAstV-2) are resistant to the potent HAstV-2-neutralizing monoclonal antibody PL-2 (MAb PL-2). Sequencing of the HAstV-2 capsid genes reveals mutations in the PL-2 epitope within the capsid's spike domain. To understand the molecular basis for resistance from MAb PL-2 neutralization, we determined the 1.35-Å-resolution crystal structure of the capsid spike from one of these HAstV-2 strains. Our structure reveals a dramatic conformational change in a loop within the PL-2 epitope due to a serine-to-proline mutation, locking the loop in a conformation that sterically blocks binding and neutralization by MAb PL-2. We show that mutation to serine permits loop flexibility and recovers MAb PL-2 binding. Importantly, we find that HAstV-2 capsid spike containing a serine in this loop is immunogenic and elicits antibodies that neutralize all HAstV-2 strains. Taken together, our results have broad implications for rational selection of vaccine strains that do not contain prolines in antigenic loops, so as to elicit antibodies against diverse loop conformations.
IMPORTANCE Human astroviruses (HAstVs) infect nearly every person in the world during childhood and cause diarrhea, vomiting, and fever. In this study, we investigated how several strains of HAstV are resistant to a virus-neutralizing monoclonal antibody. We determined the crystal structure of the capsid protein spike domain from one of these HAstV strains and found that a single amino acid mutation induces a structural change in a loop that is responsible for antibody binding. Our findings reveal how viruses can escape antibody neutralization and provide insight for the rational design of vaccines to elicit diverse antibodies that provide broader protection from infection.
KEYWORDS: astrovirus, neutralizing antibodies, structure-activity relationships, vaccines
INTRODUCTION
Astroviruses are a diverse family of small, nonenveloped, icosahedral positive-sense RNA viruses that infect a wide range of mammalian and avian species (1). In poultry, astroviruses have been associated with a large variety of disease manifestations, growth defects, and mortality (2). In humans, astroviruses are a leading cause of viral diarrhea in children, immunocompromised individuals, and the elderly (1, 3–5), accounting for 2 to 9% of all acute nonbacterial gastroenteritis in children worldwide (6). Human astroviruses (HAstVs) have also been associated with systemic infections and neurological complications such as encephalitis (7–9). There are three distinct phylogenetic clades of HAstV: canonical genotypes (HAstV-1 to -8) and noncanonical genotypes MLB1 to -3 and VA1 to -5 (6, 10, 11). HAstV-1 is the most prevalent serotype worldwide (3, 12), while the prevalence of HAstV-2, the subject of the present study, can vary widely (13).
Several studies demonstrate that the adaptive immune response plays a key role in controlling astrovirus infection and disease. Approximately 75% of people in the United States have developed antibodies against HastV by the age of 10 (14), indicating that an adaptive immune response is mounted in humans. Clinical studies of healthy adult volunteers infected with HAstV revealed that those that had anti-HAstV antibodies experienced little or no disease, whereas those that did not have anti-HAstV antibodies experienced more-severe diarrheal disease (15, 16). In another study, immunoglobulin therapy led to the recovery of an immunocompromised patient with a severe and persistent HAstV infection (17). More recently, astrovirus infectivity was tested in rag1 knockout mice (Rag1−/−), which lack mature B cells or T cells. These studies show that Rag1−/− mice infected with murine astrovirus had higher levels (by 2 log) of viral RNA than wild-type mice (18). Altogether, these studies support that adaptive immunity is key to the control of astrovirus infection. Furthermore, these studies suggest that a vaccine and therapeutic antibodies could be developed to prevent and/or treat HAstV infections.
Understanding the sites at which antibodies bind HAstV and understanding how the virus might resist antibody neutralization can inform the development of vaccines and antiviral therapeutics. Mature HAstV particles are comprised of a small RNA genome surrounded by an ∼35-nm T=3 icosahedral capsid protein shell projecting 30 knoblike spikes (19). Our lab and others have recently determined the crystal structure of the HAstV capsid core domain, which forms the icosahedral shell that encapsulates the viral RNA genome (20, 21). Our lab and others have also determined the crystal structure of the HAstV capsid spike domain, which forms the dimeric spike protrusions on the virus surface (20, 22, 23). While both the HAstV capsid core and spike domains are antigenic, the spike domain is ∼5-fold more antigenic (20). In addition, the capsid spike fragment binds all four of the previously described HAstV-neutralizing monoclonal antibodies (MAbs), including MAb PL-2, which neutralizes serotype HAstV-2 (24, 25).
We recently engineered the single-chain variable fragment (scFv) of the PL-2 antibody and used X-ray crystallography to determine the structure of the HAstV capsid spike/scFv PL-2 complex (23, 26). We found that the HAstV-2 capsid spike's loop 1 plays a major role in antibody PL-2 binding. We also provided evidence that the HAstV capsid spike is a receptor-binding domain and that antibody PL-2 blocks spike binding to human cells (23).
In the present study, we investigated the molecular mechanism by which several strains of HAstV-2 are resistant to neutralization by MAb PL-2. We find that a single point mutation in the capsid spike loop 1 of these HAstV-2 strains is responsible for resistance to antibody neutralization. Structural studies reveal how this mutation prevents antibody binding. Altogether, these studies have broad implications for a rational design of vaccines and therapeutic antibodies.
RESULTS
HAstV-2 strain resistance to antibody PL-2 neutralization.
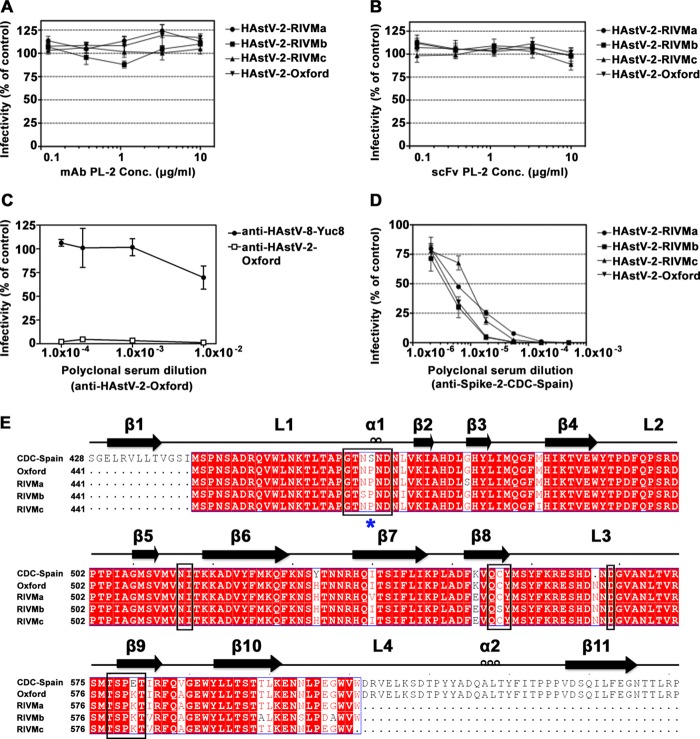
Monoclonal antibody PL-2 (MAb PL-2) was reported to neutralize infectivity of HAstV-2 strain CDC-Spain (HAstV-2-CDC-Spain), and our recent structural and functional studies suggested that MAb PL-2 may neutralize HAstV-2 by blocking a receptor-binding site on the capsid surface (23, 24). With the intention to directly investigate the mechanism of MAb PL-2 neutralization, we began our studies by aiming to confirm that MAb PL-2 neutralizes HAstV-2 infection in cell culture. HAstV-2 strain Oxford (HAstV-2-Oxford) was preincubated with increasing concentrations of MAb PL-2 or scFv PL-2 and then added to Caco-2 cells. To our surprise, no neutralization was observed by either MAb PL-2 or scFv PL-2 (Fig. 1A and B). Similar results were observed with three other HAstV-2 strains tested (HAstV-2-RIVMa, -RIVMb, and -RIVMc). In comparison, complete neutralization of HAstV-2-Oxford was obtained with polyclonal antibodies raised against HAstV-2-Oxford virus, and even partial neutralization was obtained with polyclonal antibodies raised against HAstV-8-Yuc8 (Fig. 1C). Unfortunately, HAstV-2-CDC-Spain, which was originally used to produce MAb PL-2, was no longer available in the several labs that we queried.
FIG 1.
Antibody neutralization of HAstV-2 strains and alignment of HAstV-2 capsid spike sequences. (A and B) Infectivity of indicated HAstV-2 strains preincubated with MAb PL-2 (A) and with scFv PL-2 (B). (C) Infectivity of HAstV-2-Oxford preincubated with anti-HAstV-2-Oxford or anti-HAstV-8-Yuc8 polyclonal sera. (D) Infectivity of indicated HAstV-2 strains preincubated with anti-Spike-2-CDC-Spain polyclonal sera. All infectivity experiments were performed in biological triplicates, and the error bars represent the standard errors of the means. (E) Sequence alignment of Spike-2-CDC-Spain, Spike-2-Oxford, and Spike-2-RIVMa-c. Conserved, similar, and nonconserved amino acids are colored red, pink, and white, respectively. Alignments and mapping of conservation onto the structure were performed with the online ENDscript server. Black boxes highlight amino acids in the antibody PL-2 epitope. A blue star indicates the location of Ser463 or Pro463.
Sequence of HAstV-2 strains.
We used reverse transcription (RT)-PCR to determine the sequence of the capsid spike from each of the four HAstV-2 strains (strains Oxford, RIVMa, RIVMb, and RIVMc). We then aligned these sequences to the reported capsid protein sequence of HAstV-2-CDC-Spain (Fig. 1E). Focusing on the amino acids within the MAb PL-2 epitope, we observed two amino acids that are mutated in all four HAstV-2 strains: Ser463Pro and Glu580Lys. Ser463 falls in the middle of the PL-2 epitope in loop 1 of the capsid spike domain, whereas Glu580 resides at the edge of the epitope (23).
Structure of Spike-2-Oxford.
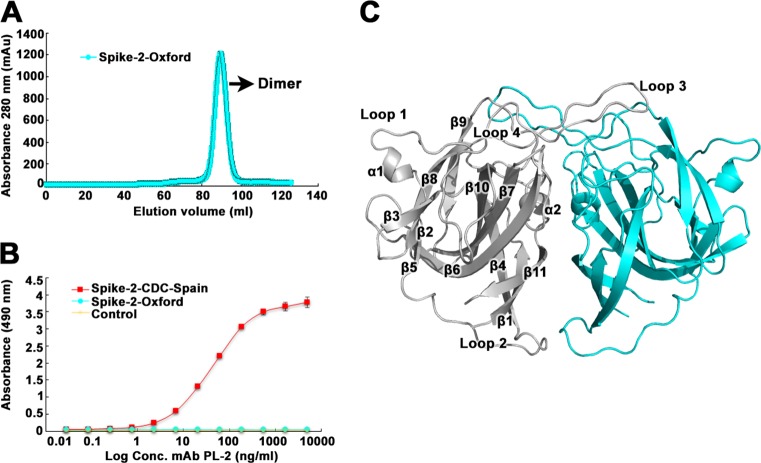
To understand the molecular basis for resistance to MAb PL-2 neutralization, we expressed and purified recombinant HAstV-2-Oxford capsid spike domain (Spike-2-Oxford) and observed that it elutes as a dimer by size exclusion chromatography, consistent with previous observations for recombinant HAstV-2 capsid spike domain (Spike-2-CDC-Spain) (Fig. 2A) (23). We used an enzyme-linked immunosorbent assay (ELISA) to test if MAb PL-2 binds Spike-2-Oxford. Consistent with the lack of MAb PL-2 neutralization of HAstV-2-Oxford infectivity, we observed no binding of MAb PL-2 to Spike-2-Oxford, whereas dose-dependent binding was observed for Spike-2-CDC-Spain (Fig. 2B). We then crystallized Spike-2-Oxford and solved its structure to a 1.35-Å resolution (Fig. 2C; Table 1). The overall structure of Spike-2-Oxford is very similar to that of Spike-2-CDC-Spain, with a root mean square deviation (RMSD) of 0.315 Å.
FIG 2.
Purification, antibody binding, and structure of Spike-2-Oxford. (A) Superdex 200 16/600 size exclusion chromatography trace of Spike-2-CDC-Spain. (B) ELISA results showing specific binding of MAb PL-2 to Spike-2-CDC-Spain and no binding to Spike-2-Oxford. The yellow “Control” sample refers to ELISA wells coated with PBS buffer (no Spike protein). (C) Crystal structure of Spike-2-Oxford, with one half of the dimer in gray and the other half in light blue. The gray half has labeled β-sheets, α-helices, and loops.
TABLE 1.
Data collection and refinement statistics for Spike-2-Oxforda
| Characteristic | Value or code |
|---|---|
| PDB code | 5W1N |
| Data collection | |
| Space group | P 1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 68.63, 71.94, 92.81 |
| α, β, c (°) | 90.00, 111.22, 90.00 |
| Resolution (Å) | 43.97–1.35 (1.42–1.35) |
| Rsym or Rmerge | 0.098 (0.301) |
| I/σI | 10.4 (4.8) |
| Completeness (%) | 96.3 (93.6) |
| Redundancy | 4.6 (4.4) |
| CC1/2 | 0.988 (0.920) |
| Refinement | |
| Resolution (Å) | 43.97–1.35 |
| No. of reflections | 177,796 |
| Rwork/Rfree | 0.156/0.183 |
| No. of atoms | |
| Protein | 6,981 |
| Ligands | 0 |
| Water | 464 |
| B factors | |
| Protein | 14.92 |
| Ligands | 0 |
| Water | 21.90 |
| RMSD | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.962 |
| Ramachandran statistics | |
| Favored (%) | 97.7 |
| Allowed (%) | 2.3 |
| Outliers (%) | 0 |
Data from one crystal were used for structure determination. Values in parentheses are for the highest-resolution shell.
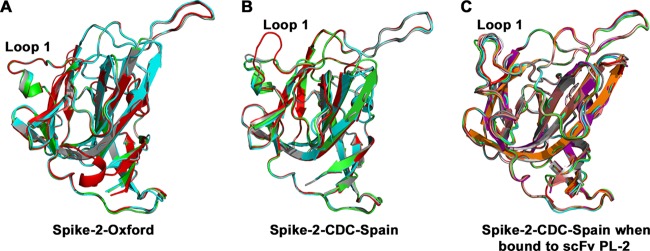
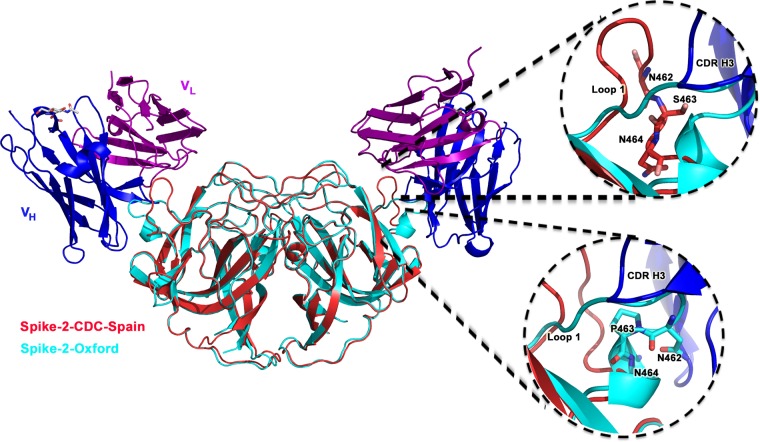
The major structural difference between Spike-2-Oxford and Spike-2-CDC-Spain occurs in loop 1, which interacts with antibody PL-2 (Fig. 3). We observe that the Ser463Pro mutation in loop 1 leads to the formation of a short alpha helix, locking loop 1 into a distinct conformation. Loop 1 adopts this same “down” conformation in all four molecules of Spike-2-Oxford in the crystallographic asymmetric unit (Fig. 3A). In contrast, loop 1 in Spike-2-CDC-Spain appears more flexible and adopts different conformations in each of the four molecules in the crystallographic asymmetric unit, with two molecules in a “down” conformation, one molecule in an intermediate conformation, and one molecule in an “up” conformation (Fig. 3B). Thus, it appears that the flexibility of loop 1 in Spike-2-CDC-Spain may be required for binding of antibody PL-2. Indeed, we observed that loop 1 adopts a single “up” conformation in all eight molecules of the Spike-2-CDC-Spain/scFv PL-2 complex in the crystallographic asymmetric unit (Fig. 3C). Superposition of the Spike-2-Oxford structure onto the structure of the Spike-2-CDC-Spain/scFv PL-2 complex reveals how the “down” conformation of loop 1 in Spike-2-Oxford would sterically clash with antibody heavy-chain complementarity-determining region 3 (CDR 3) (Fig. 4). Overall, our structural studies suggest that the Ser463Pro mutation locks loop 1 into a rigid conformation that clashes with and completely prevents antibody PL-2 binding.
FIG 3.
Structural rigidity of loop 1 in Spike-2-Oxford. (A and B) Structural alignment of all four molecules of Spike-2-Oxford (A) and of Spike2-CDC-Spain (B) in the crystallographic asymmetric unit. (C) Structural alignment of all eight molecules of Spike-2-CDC-Spain in the crystallographic asymmetric unit from the Spike-2-CDC-Spain/scFv PL-2 complex structure.
FIG 4.
Spike-2-Oxford loop 1 sterically clashes with scFv PL-2 binding. The structure of Spike-2-Oxford (cyan) was superimposed onto the structure of Spike-2-CDC-Spain (red) bound to the scFv PL-2 (with the heavy chain colored blue and the light chain colored purple). (Top right) Zoom-in showing Spike-2-CDC-Spain Ser463 and “up” conformation of loop 1; (bottom right) zoom-in showing Spike-2-Oxford Pro463 inducing a helix and “down” conformation in loop 1, which clashes with antibody heavy chain (blue) CDR H3.
Spike-2-Oxford mutant Pro463Ser restores binding to MAb PL-2.
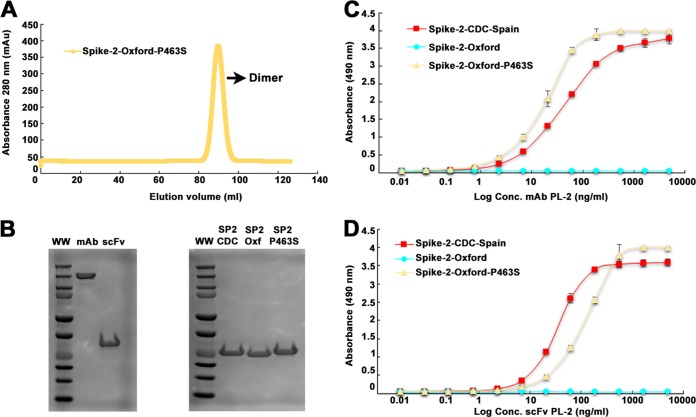
Our structural studies lead to the hypothesis that HAstV-2-Oxford capsid Pro463 is responsible for the lack of binding and neutralization by MAb PL-2. To test our hypothesis, we expressed and purified recombinant Spike-2-Oxford Pro463Ser mutant and observed that it is pure and elutes as a dimer by size exclusion chromatography (Fig. 5A and B). We then performed an ELISA using purified MAb PL-2 and scFv PL-2 (Fig. 5B to D). We observed that both antibody PL-2 samples now bind to the Spike-2-Oxford Pro463Ser mutant at levels similar to those of binding to Spike-2-CDC-Spain. These results confirm that a single amino acid in the capsid protein of HAstV-2-Oxford is responsible for resistance to binding and neutralization by MAb PL-2.
FIG 5.
Purification and antibody binding of Spike-2-Oxford Pro463Ser mutant. (A) Superdex 200 16/600 size exclusion chromatography trace of Spike-2-Oxford Pro463Ser mutant. (B) Coomassie blue-stained nonreducing SDS-PAGE of MAb PL-2 and scFv PL-2 (left). Coomassie blue-stained reducing SDS-PAGE of Spike-2-CDC-Spain, Spike-2-Oxford, and Spike-2-Oxford P463S mutant (right). (C) ELISA results showing specific binding of MAb PL-2 to Spike-2-Oxford Pro463Ser mutant. (D) ELISA results showing specific binding of scFv PL-2 Spike-2-Oxford Pro463Ser mutant.
Spike-2-CDC-Spain elicits HAstV-2 cross-neutralizing antibodies.
We hypothesized that Spike-2-CDC-Spain, which contains Ser463 in loop 1, would elicit antibodies against diverse conformations of loop 1 that can neutralize the infectivity of all four HAstV-2 strains. To test this, we immunized rabbits with purified, recombinant Spike-2-CDC-Spain and found that the anti-Spike-2-CDC-Spain polyclonal antibodies effectively neutralized all four strains of HAstV-2 in a concentration-dependent manner (Fig. 1D). Although the spike most likely contains more than one neutralizing epitope, this study does show that Spike-2-CDC-Spain, which contains Ser463, is able to elicit antibodies that neutralize HAstV-2 strains containing Pro463. Moreover, this is the first demonstration that recombinant HAstV capsid spike can elicit HAstV-neutralizing antibodies and may be an effective immunogen in a HAstV subunit vaccine.
DISCUSSION
Here, we investigated the molecular mechanism by which four strains of HAstV-2 (HAstV-2-Oxford, -RIVMa, -RIVMb, and -RIVMc) are resistant to the HAstV-2-CDC-Spain-neutralizing monoclonal antibody PL-2. Serotype HAstV-2 has been reported to have especially high genetic heterogeneity between strains (13). We describe the crystal structure of Spike-2-Oxford and find that Pro463 induces a rigid helix in loop 1, locking it in a “down” conformation that sterically clashes and prevents binding by MAb PL-2. We further show that the point mutant Pro463Ser recovers binding by MAb PL-2. Finally, we show that recombinant Spike-2-CDC-Spain is immunogenic and elicits antibodies that neutralize all four HAstV-2 strains.
Viruses constantly evolve under the pressure of a host's immune system, and one of the most efficient ways to evade immunity is by acquiring mutations that sterically clash with antibody binding. While serines and prolines are often interchangeable amino acids due to their similarity in size, we find that the mutation of serine to proline in a protein loop can induce a major conformational change. Proline does not follow the typical Ramachandran plot, due to its cyclic side chain that limits motion in its ψ and φ angles, reducing flexibility to the polypeptide backbone. Because loop structures are often associated with antigenic epitopes, it is possible that viruses mutate amino acids in antigenic loops to prolines to evade binding by some antibodies. Indeed, serine codons, as well as threonine, alanine, leucine, histidine, glutamine, and arginine codons, are most susceptible to becoming proline codons, since in these cases the switch requires only a single nucleotide mutation.
One might initially conclude from these studies that vaccine strain antigens with serines in antigenic loops should not be chosen because they are at risk for antibody escape; however, we would actually argue the opposite. Specifically, we propose that antigens with serines in antigenic loops should be chosen for vaccine strains because they would elicit more-diverse polyclonal antibodies against the diverse conformations in the loop, including conformations observed upon mutation to proline. Indeed, our structures reveal that with a serine, the loop will sometimes adopt the conformation that is restrained by a proline mutation. In support of this concept, we show that recombinant Spike-2-CDC-Spain, which has a serine in loop 1, elicits antibodies that neutralize all four HAstV-2 strains with prolines in loop 1.
Notably, we show that recombinant HAstV capsid spike is immunogenic and elicits neutralizing antibodies. These findings demonstrate the high potential to develop a subunit vaccine to prevent HAstV diarrheal disease. In addition, the affordable, simple, and scalable strategy of producing large amounts of recombinant HAstV capsid spike in Escherichia coli hints at the feasibility of global vaccination. Of course, future studies will be required to determine if a vaccine comprised of capsid spikes from multiple serotypes is required to elicit a broadly protective polyclonal antibody response.
MATERIALS AND METHODS
Cells, viruses, and reagents.
C2Bbe1 cells (ATCC), derived from the human colon adenocarcinoma Caco-2 cell line, were propagated in a 10% CO2 atmosphere at 37°C in Dulbecco's modified Eagle's medium with high glucose (DMEM-HG) (Sigma), supplemented with nonessential amino acids (Gibco) and 15% fetal bovine serum (FBS) (Cansera). HAstV-2 strain Oxford (HAstV-2-Oxford) was obtained from J. B. Kurtz (Department of Virology, John Radcliffe Hospital, Oxford, UK). HAstV-2 strains RIVMa, RIVMb, and RIVMc were obtained from S. Guix (Department Microbiologia, Facultat de Biologia, Universitat de Barcelona, Spain). All viral strains were activated and grown as described previously (27), except that 200 μg/ml of trypsin was used to activate the virus infectivity. Anti-Spike-2-CDC-Spain polyclonal sera were generated by immunization of mice with recombinant HAstV-2-CDC-Spain capsid spike (see below). For this, female BALB/c mice (8 weeks old) were immunized intraperitoneally with 50 μg of recombinant HAstV capsid spike at 2-week intervals (a total of four times). The first immunization was done in Freund's complete adjuvant, the second and third with incomplete Freund's adjuvant, and the fourth with no adjuvant. Four days after the fourth immunization, the mice were bled to death. Anti-Core-1 polyclonal sera used to detect HAstV infectivity in C2Bbe1 cells were generated by immunization of New Zealand rabbits with recombinant HAstV-1 capsid core (amino acids 80 to 429) (20). The generation of polyclonal antibodies to Spike-2-CDC-Spain and to Core-1 was approved by the Bioethics Committee of the Institute of Biotechnology UNAM (number 296).
Neutralization assays.
Antibody or scFv at the indicated concentration was preincubated with the indicated HAstV-2 strain at a multiplicity of infection (MOI) of 0.002 for 1 h at room temperature. The virus-antibody mix was then added to confluent monolayers of C2Bbe1 cells grown in 96-multiwell plates, and the plates were incubated for 1 h at 37°C. After this time, the cells were washed three times with phosphate-buffered saline (PBS), and the infection was left to proceed for 18 h at 37°C. Infected cells were detected by an immunoperoxidase focus-forming assay, as described previously (28), with the following modifications. At 18 h postinfection, the cells were fixed for 20 min at room temperature with 2% formaldehyde, washed with PBS, and permeabilized using 0.2% Triton X-100 (in PBS) for 15 min, at room temperature. For detection of infected cells, a polyclonal serum directed to HAstV-1 capsid core was used (see above). Experiments were performed in biological triplicates.
PCR and sequencing.
For HAstV-2 sequencing, RNA was isolated from viral lysates using the PureLink viral RNA/DNA minikit (Invitrogen), and cDNA was synthesized with SuperScript III reverse transcriptase (Thermo Fisher Scientific) using as a primer the sequence 5′-GCGGTCTCCAGAAAGTTTG-3′ (HastV2LW), corresponding to nucleotide positions 2369 to 2387 of the HAstV-2 capsid gene (accession number L06802.1). For PCR amplification, Vent DNA Polymerase (New England BioLabs) and the oligonucleotides HAstV2LW and HAstV2Up (5′-CAGTTCACTCAAATGAACCA-3′), corresponding to nucleotides 1215 to 1234 of the HAstV-2 capsid gene (accession number L06802.1), were used. The PCR product was purified using the DNA clean and Concentrator-5 kit (Zymo Research) and sequenced in the sequencing facility of the Instituto de Biotecnología, UNAM.
Expression and purification of Spike-2.
Synthetic genes codon optimized for Escherichia coli encoding HAstV-2-CDC-Spain capsid spike amino acids 431 to 674 (GenBank accession AAA62427.1) and HAstV-2-Oxford capsid spike (GenBank accession number KY964327) were purchased. To make spike expression plasmids, genes were cloned into pET52b (Addgene) in frame with a C-terminal thrombin cleavage site and a 10-histidine purification tag. To make Spike-2-Oxford Pro463Ser mutant expression plasmid, the Phusion Site-Direct Mutagenesis kit (Thermo Scientific) was used with phosphorylated mutagenesis primers. All plasmids were verified by DNA sequencing. Plasmids were transformed into E. coli strain BL21(DE3), and protein production was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 18°C for 16 h. E. coli cells were lysed by ultrasonication in 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 20 mM imidazole (buffer A) containing 2 μM MgCl2, 1,250 U benzonase (Millipore), and 1× protease inhibitor cocktail Set V EDTA-free (Millipore). Proteins were batch purified from soluble lysates by Talon metal affinity chromatography and eluted with buffer A containing 500 mM imidazole. Proteins were dialyzed into 20 mM Tris-HCl (pH 8.0) and 25 mM NaCl and purified by anion-exchange chromatography on a HiTrap Q FF column with a gradient elution to 20 mM Tris-HCl (pH 8.0) and 1 M NaCl. Proteins were buffer exchanged into PBS and further purified by size exclusion chromatography on a Superdex 200 column in PBS.
ELISA.
Purified spike proteins at a concentration of 5 μg/ml in PBS (150 μl total) were incubated overnight at room temperature in 96-well ELISA microtiter plates. Plates were then washed three times with PBS containing 0.05% Tween 20 (PBST). Wells were blocked by adding 150 μl of 5% bovine serum albumin (BSA) in PBS and incubating the mixture at room temperature for 1 h followed by three PBST washes. Antibody sample MAb PL-2 or scFv PL-2 was diluted to 5 μg/ml with 1% BSA in PBS and serially diluted 1:3 with 1% BSA in PBS. Wells were incubated with 150 μl antibody for 1 h at room temperature, and the plates were washed three times with PBST. For ELISAs in which the primary antibody was MAb PL-2, plates were incubated for 1 h at room temperature with 150 μl horseradish peroxidase (HRP)-conjugated, secondary goat anti-mouse IgG antibody diluted 1:5,000 in 1% BSA in PBS. For ELISAs in which the primary antibody was scFv PL-2, plates were incubated for 1 h at room temperature with 150 μl HRP-conjugated Strep-Tactin protein, diluted 1:5,000 in 1% BSA in PBS. Plates were washed three times with PBST and developed by adding peroxidase substrate o-phenylenediamine dihydrochloride (OPD) in 0.05 M phosphate-citrate buffer (pH 5.0) and 1.5% hydrogen peroxide for 10 min at room temperature. The reactions were stopped by incubation with 2 N sulfuric acid for 10 min at room temperature, and the absorbance was measured at 490 nm.
Structure determination of Spike-2-Oxford.
Purified Spike-2-Oxford in PBS was concentrated to 28.3 mg/ml. Crystals were grown by hanging-drop vapor diffusion at 22°C with a well solution of 20% polyethylene glycol (PEG) 3350, 0.2 M magnesium acetate, and 0.1 HEPES buffer (pH 7.5). Crystals were transferred into a cryoprotectant solution of 25% PEG 3350, 25% glycerol, 0.2 M magnesium acetate, and 0.1 M HEPES (pH 7.5) and flash frozen in liquid nitrogen. Diffraction data were collected at cryogenic temperature at the Advanced Photon Source on beamline 23-ID-B using a wavelength of 1.033 Å. Diffraction data from a single crystal were processed with iMosflm (29) and Scala (30) (Table 1). The Spike-2-Oxford structure was solved by molecular replacement, using the HAstV-2-CDC-Spain capsid spike (PDB ID 5KOU) (22) and the program PHASER (31). The Spike-2-Oxford structure was refined and manually rebuilt using PHENIX (32) and Coot (33), respectively. The final Spike-2-Oxford structure had two dimers in the asymmetric unit of the crystal.
Accession number(s).
Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5W1N. The HAstV-2-Oxford sequence was deposited in GenBank under accession number KY964327.
ACKNOWLEDGMENTS
We thank Rafaela Espinosa for the production of hyperimmune sera to HAstV spike and core domains and Marco A. Espinoza for sequencing the capsid regions of HAstV-2 viruses. We thank Susana Guix for sharing of the HAstV-2-RIVM strains. We thank Phillip Berman for the use of ELISA instruments and Alicia Sanchez-Fauquier for donation of MAb PL-2 in ascites fluid. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases grants AI095369 and AI130073 (to R.M.D.) and by the Hellman Fellows Fund (to R.M.D.). The National Institutes of Health “Initiative for Maximizing Student Development” provided fellowship support (to W.A.B.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Mendez E, Arias CF. 2013. Astroviruses, p 609–628. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Koci MD, Schultz-Cherry S. 2002. Avian astroviruses. Avian Pathol 31:213–227. doi: 10.1080/03079450220136521. [DOI] [PubMed] [Google Scholar]
- 3.Glass RI, Noel J, Mitchell D, Herrmann JE, Blacklow NR, Pickering LK, Dennehy P, Ruiz-Palacios G, de Guerrero ML, Monroe SS. 1996. The changing epidemiology of astrovirus-associated gastroenteritis: a review. Arch Virol Suppl 12:287–300. doi: 10.1007/978-3-7091-6553-9_31. [DOI] [PubMed] [Google Scholar]
- 4.Goodgame RW. 2001. Viral causes of diarrhea. Gastroenterol Clin North Am 30:779–795. doi: 10.1016/S0889-8553(05)70210-7. [DOI] [PubMed] [Google Scholar]
- 5.Walter JE, Mitchell DK. 2003. Astrovirus infection in children. Curr Opin Infect Dis 16:247–253. doi: 10.1097/00001432-200306000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Bosch A, Pinto RM, Guix S. 2014. Human astroviruses. Clin Microbiol Rev 27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, Shah D, Brooks T, Paine SM, Anderson G, Virasami A, Tong CY, Clark DA, Plagnol V, Jacques TS, Qasim W, Hubank M, Breuer J. 2015. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis 60:881–888. doi: 10.1093/cid/ciu940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naccache SN, Peggs KS, Mattes FM, Phadke R, Garson JA, Grant P, Samayoa E, Federman S, Miller S, Lunn MP, Gant V, Chiu CY. 2015. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 60:919–923. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King AMQ, Lefkowitz E, Adams MJ, Carstens EB (ed). 2011. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses, 9th ed Elsevier Inc., Philadelphia, PA. [Google Scholar]
- 11.Kurtz JB, Lee TW. 1984. Human astrovirus serotypes. Lancet ii:1405. [DOI] [PubMed] [Google Scholar]
- 12.Koopmans MP, Bijen MH, Monroe SS, Vinje J. 1998. Age-stratified seroprevalence of neutralizing antibodies to astrovirus types 1 to 7 in humans in The Netherlands. Clin Diagn Lab Immunol 5:33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Grazia S, Medici MC, Pinto P, Moschidou P, Tummolo F, Calderaro A, Bonura F, Banyai K, Giammanco GM, Martella V. 2012. Genetic heterogeneity and recombination in human type 2 astroviruses. J Clin Microbiol 50:3760–3764. doi: 10.1128/JCM.02102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurtz J, Lee T. 1978. Astrovirus gastroenteritis age distribution of antibody. Med Microbiol Immunol 166:227–230. doi: 10.1007/BF02121154. [DOI] [PubMed] [Google Scholar]
- 15.Kurtz JB, Lee TW, Craig JW, Reed SE. 1979. Astrovirus infection in volunteers. J Med Virol 3:221–230. doi: 10.1002/jmv.1890030308. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell DK. 2002. Astrovirus gastroenteritis. Pediatr Infect Dis J 21:1067–1069. doi: 10.1097/00006454-200211000-00018. [DOI] [PubMed] [Google Scholar]
- 17.Bjorkholm M, Celsing F, Runarsson G, Waldenstrom J. 1995. Successful intravenous immunoglobulin therapy for severe and persistent astrovirus gastroenteritis after fludarabine treatment in a patient with Waldenstrom's macroglobulinemia. Int J Hematol 62:117–120. doi: 10.1016/0925-5710(95)00396-A. [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama CC, Loh J, Zhao G, Stappenbeck TS, Wang D, Huang HV, Virgin HW, Thackray LB. 2012. Adaptive immunity restricts replication of novel murine astroviruses. J Virol 86:12262–12270. doi: 10.1128/JVI.02018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dryden KA, Tihova M, Nowotny N, Matsui SM, Mendez E, Yeager M. 2012. Immature and mature human astrovirus: structure, conformational changes, and similarities to hepatitis E virus. J Mol Biol 422:650–658. doi: 10.1016/j.jmb.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.York RL, Yousefi PA, Bogdanoff W, Haile S, Tripathi S, DuBois RM. 2016. Structural, mechanistic, and antigenic characterization of the human astrovirus capsid. J Virol 90:2254–2263. doi: 10.1128/JVI.02666-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toh Y, Harper J, Dryden KA, Yeager M, Arias CF, Mendez E, Tao YJ. 2016. Crystal structure of the human astrovirus capsid protein. J Virol 90:9008–9017. doi: 10.1128/JVI.00694-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong J, Dong L, Mendez E, Tao Y. 2011. Crystal structure of the human astrovirus capsid spike. Proc Natl Acad Sci U S A 108:12681–12686. doi: 10.1073/pnas.1104834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogdanoff WA, Campos J, Perez EI, Yin L, Alexander DL, DuBois RM. 2017. Structure of a human astrovirus capsid-antibody complex and mechanistic insights into virus neutralization. J Virol 91:e01859-16. doi: 10.1128/JVI.01859-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Fauquier A, Carrascosa AL, Carrascosa JL, Otero A, Glass RI, Lopez JA, San Martin C, Melero JA. 1994. Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization. Virology 201:312–320. doi: 10.1006/viro.1994.1296. [DOI] [PubMed] [Google Scholar]
- 25.Bass DM, Upadhyayula U. 1997. Characterization of human serotype 1 astrovirus-neutralizing epitopes. J Virol 71:8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogdanoff WA, Morgenstern D, Bern M, Ueberheide BM, Sanchez-Fauquier A, DuBois RM. 2016. De novo sequencing and resurrection of a human astrovirus-neutralizing antibody. ACS Infect Dis 2:313–321. doi: 10.1021/acsinfecdis.6b00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendez-Toss M, Romero-Guido P, Munguia ME, Mendez E, Arias CF. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J Gen Virol 81:2891–2897. doi: 10.1099/0022-1317-81-12-2891. [DOI] [PubMed] [Google Scholar]
- 28.Mendez E, Salas-Ocampo E, Arias CF. 2004. Caspases mediate processing of the capsid precursor and cell release of human astroviruses. J Virol 78:8601–8608. doi: 10.1128/JVI.78.16.8601-8608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]