ABSTRACT
Patient-derived HIV-1 subtype B Nef clones downregulate HLA-A more efficiently than HLA-B. However, it remains unknown whether this property is common to Nef proteins across primate lentiviruses and how antiviral immune responses may be affected. We examined 263 Nef clones from diverse primate lentiviruses including different pandemic HIV-1 group M subtypes for their ability to downregulate major histocompatibility complex class A (MHC-A) and MHC-B from the cell surface. Though lentiviral Nef proteins differed markedly in their absolute MHC-A and MHC-B downregulation abilities, all lentiviral Nef lineages downregulated MHC-A, on average, 11 to 32% more efficiently than MHC-B. Nef genotype/phenotype analyses in a cohort of HIV-1 subtype C-infected patients (n = 168), together with site-directed mutagenesis, revealed Nef position 9 as a subtype-specific determinant of differential HLA-A versus HLA-B downregulation activity. Nef clones harboring nonconsensus variants at codon 9 downregulated HLA-B (though not HLA-A) significantly better than those harboring the consensus sequence at this site, resulting in reduced recognition of infected target cells by HIV-1-specific CD8+ effector cells in vitro. Among persons expressing protective HLA class I alleles, carriage of Nef codon 9 variants was also associated with reduced ex vivo HIV-specific T cell responses. Our results demonstrate that Nef's inferior ability to downregulate MHC-B compared to that of MHC-A is conserved across primate lentiviruses and suggest that this property influences antiviral cellular immune responses.
IMPORTANCE Primate lentiviruses encode the Nef protein that plays an essential role in establishing persistent infection in their respective host species. Nef interacts with the cytoplasmic region of MHC-A and MHC-B molecules and downregulates them from the infected cell surface to escape recognition by host cellular immunity. Using a panel of Nef alleles isolated from diverse primate lentiviruses including pandemic HIV-1 group M subtypes, we demonstrate that Nef proteins across all lentiviral lineages downregulate MHC-A approximately 20% more effectively than MHC-B. We further identify a naturally polymorphic site at Nef position 9 that contributes to the MHC-B downregulation function in HIV-1 subtype C and show that carriage of Nef variants with enhanced MHC-B downregulation ability is associated with reduced breadth and magnitude of MHC-B-restricted cellular immune responses in HIV-infected individuals. Our study underscores an evolutionarily conserved interaction between lentiviruses and primate immune systems that may contribute to pathogenesis.
KEYWORDS: HLA, Nef, human immunodeficiency virus, immune evasion, lentiviruses
INTRODUCTION
The mammalian major histocompatibility complex class A (MHC-A) and MHC-B loci (also called the human leukocyte antigen [HLA] A and B loci in humans) encode cell surface glycoproteins, expressed in all nucleated cells, that present intracellular pathogen-derived peptide antigens for recognition by CD8+ cytotoxic T lymphocytes (CTL). The MHC class I (MHC-I) loci, particularly MHC-B, rank among the most polymorphic regions in mammalian genomes. In humans, for instance, 3,913 HLA-A and 4,765 HLA-B alleles have been identified to date (Immuno Polymorphism Database-ImMunoGeneTics [IPD-IMGT]/HLA database [https://www.ebi.ac.uk/ipd/imgt/hla/index.html]); similarly, 345 MHC-A and 587 MHC-B alleles have been identified in rhesus macaques (1, 2). Though MHC-I polymorphisms are largely concentrated within exons 2 and 3 (which form the molecule's antigenic peptide-binding cleft and thus play a crucial role in restricting epitope and CTL specificity [3]), polymorphisms also occur outside these regions. For example, HLA-A and HLA-B alleles can be classified into five and two polymorphic types, respectively, based on sequence variations within their cytoplasmic domains (encoded by exons 5 through 7 for HLA-B or by exons 5 through 8 for HLA-A). However, the implications of HLA cytoplasmic polymorphisms on modulating antiviral immunity remain poorly understood.
HLA-I-restricted CD8+ CTL responses modulate immune control of a wide range of human viral infections, including human immunodeficiency virus type 1 (HIV-1) (4, 5), human T-cell leukemia virus type 1 (6), cytomegalovirus (7), and herpes simplex virus (8) as well as simian immunodeficiency virus (SIV) infections in rhesus macaques (9–12) and chimpanzees (13). HLA-B polymorphism plays a particularly key role in immunodeficiency virus control. HIV-1 disease outcomes are disproportionally influenced by the HLA-B alleles expressed (14–16); in particular, CTL responses restricted by certain “protective” HLA-I molecules, notably HLA-B*57, HLA-B*58:01, and HLA-B*81:01, exhibit superior ability to control HIV-1 replication (17–19), likely as a result of specific amino acids located within their peptide-binding grooves (5). In rhesus macaques, the protective MHC-I allele Mamu-B*008 is similarly associated with sustained control of SIVmac infection (20). The mechanisms underlying the dominant role of HLA-B alleles in immunodeficiency virus control, however, remain incompletely understood.
Viruses have evolved various mechanisms to evade HLA-I-restricted antiviral immunity, including inhibition of intracellular antigen processing (e.g., the human cytomegalovirus US3 protein inhibits tapasin, thus promoting peptide retention within the endoplasmic reticulum [21, 22]) and downregulation of HLA-I molecules from the infected cell surface (e.g., the K3 and K5 proteins of Kaposi's sarcoma-associated herpesvirus promote HLA endocytosis and lysosomal degradation [23, 24]). Lentiviruses also downregulate MHC from the infected cell surface. In particular, the lentiviral accessory protein Nef downregulates MHC-A and MHC-B molecules (25–27) by binding to their cytoplasmic domains in conjunction with the μ1 subunit of host adaptor protein 1 (AP1) (28) while the HIV-1 accessory protein Vpu downregulates HLA-C (29). Moreover, it was recently demonstrated that HIV-1 Nef downregulates HLA-A more efficiently than HLA-B in vitro (30, 31), which in turn influenced infected cell recognition by HLA-restricted T cell receptors in a reporter assay system (30) and provides a mechanistic explanation for the dominant influence of HLA-B on antiviral CTL (31). Notably, HIV-1 Nef clones isolated from patients at different infection stages with different disease phenotypes and infected with different HIV-1 group M subtypes display wide-ranging HLA-I downregulation capacities (32–39); extensive variation in Nef-mediated downregulation of HLA-A versus HLA-B has also been reported for HIV-1 subtype B isolates (30). It is not known, however, to what extent Nef's differential downregulation of HLA-A and HLA-B is conserved across lentiviruses and between HIV-1 group M subtypes other than B; furthermore, the in vivo implications of this functional heterogeneity in Nef's key immune evasion activities remain incompletely known.
In this study, we assessed 35 Nef clones from various lentiviruses and 228 patient-derived Nef clones from HIV-1 pandemic group M subtypes A, B, C, and D for their ability to downregulate various HLA-A and HLA-B allotypes. Substantial Nef functional heterogeneity was observed both within and between lentiviral species and HIV-1 group M subtypes; nevertheless, all Nef clones downregulated HLA-A more efficiently than HLA-B such that the average HLA-A/HLA-B downregulation ratio was consistently ∼1.2 across all isolates tested. Differential, Nef-mediated downregulation of HLA-A versus HLA-B molecules in turn modulated the ability of HIV-specific effector T cells to recognize infected target cells in vitro, as well as the breadth and magnitude of HIV-specific CD8 T cell responses ex vivo. Finally, by combining statistical analysis and site-specific mutagenesis, we identified a natural polymorphism within subtype C that modulates Nef's differential ability to downregulate HLA-A and HLA-B molecules.
RESULTS
Differential downregulation of MHC-A and MHC-B by Nef clones from primate lentiviruses.
The cytoplasmic tail regions of MHC-A and MHC-B alleles, where the Nef protein binds with the μ1 subunit of host AP1 for downregulation of MHC (28), are highly conserved among primate species. For example, various nonhuman primate MHC-A and MHC-B alleles share identical cytoplasmic tail sequences to HLA-A*02:01 and HLA-B*35:01, respectively; moreover, these sequences account for 43%, 75%, and 80% of all MHC-A alleles and 61%, 100%, and 36% of all MHC-B alleles in humans, chimpanzees, and rhesus monkeys, respectively (Table 1). MHC-A and MHC-B cytoplasmic tails differ in two key respects: first, MHC-B molecules are three amino acids shorter than MHC-A; second, MHC-A molecules harbor aspartic acid and arginine at codons 314 and 315, respectively (DR314,315), whereas MHC-B molecules harbor dual glycines (GG314,315) at these sites (Table 1).
TABLE 1.
Amino acid sequences of MHC-I cytoplasmic tails in primate species
| MHC type and primate source | Species | Allele name | Amino acid sequencea | No. of alleles with the same cytoplasmic tailb | Total no. of alleles | No. of cytoplasmic tail typesb |
|---|---|---|---|---|---|---|
| MHC-A | ||||||
| Human | Homo sapiens | HLA-A*02:01 | RKSSDRKGGSYSQAASSDSAQGSDVSLTACKV | 68 | 160 | 5 |
| Chimpanzee | Pan troglodytes | Patr-A*04:01 | ................................ | 27 | 36 | 2 |
| Rhesus monkey | Macaca mulatta | Mamu-A*010:01 | ................................ | 151 | 181 | 8 |
| Sooty mangabey | Cercocebus atys | Ceat-A*02:01 | ................................ | 13 | 29 | 8 |
| Blue monkey | Cercopithecus mitis | Cemi-A*01:01 | ................................ | 2 | 3 | 3 |
| MHC-B | ||||||
| Human | Homo sapiens | HLA-B*35:01 | ....GG.......................--- | 162 | 265 | 2 |
| Chimpanzee | Pan troglodytes | Patr-B*11:02 | ....GG.......................--- | 64 | 64 | 1 |
| Rhesus monkey | Macaca mulatta | Mamu-B*039:01 | ....GG.......................--- | 90 | 243 | 21 |
| Sooty mangabey | Cercocebus atys | Ceat-B*03:01 | ....GG.......................--- | 17 | 82 | 16 |
| Blue monkey | Cercopithecus mitis | Cemi-B*01:01 | ....GG.......................--- | 4 | 4 | 1 |
Amino acid sequence of the cytoplasmic tail at residues 310 to 341 in HLA-A*02:01 or 310 to 338 in HLA-B*35:01. Dots denote amino acid residues identical to the sequence of HLA-A*02:01, and dashes denote the absence of amino acids at the indicated positions.
Numbers were obtained from MHC-I alleles whose cytoplasmic tail sequences are available in the MHC database (https://www.ebi.ac.uk/ipd/index.html).
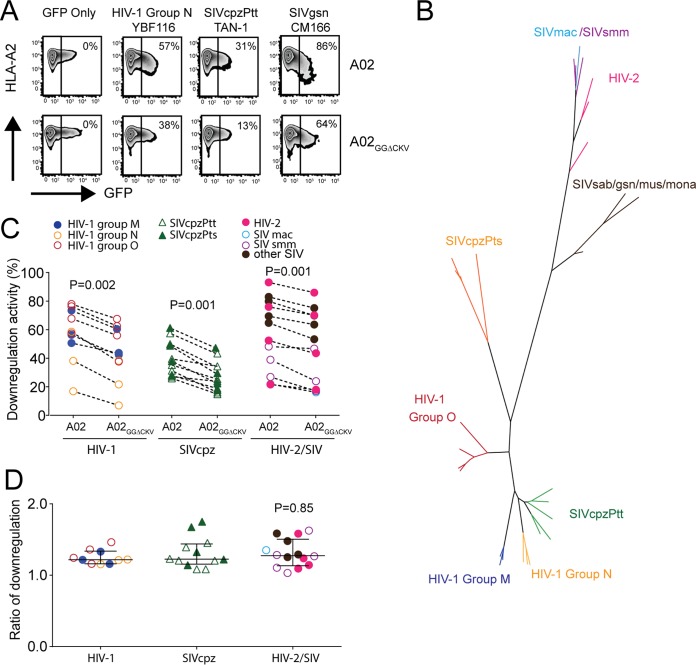
Based on the highly conserved nature of primate MHC-A and MHC-B cytoplasmic tails, we began by examining whether the ability for differential downregulation of MHC-A versus MHC-B is shared across primate lentivirus lineages. We modeled Nef interaction with MHC-A and MHC-B cytoplasmic tails by establishing Jurkat cells stably expressing either HLA-A*02:01 (A02) or an A02 chimera where its cytoplasmic tail was replaced with that of HLA-B*35:01 (B35) (A02GGΔCKV, with the mutations DR314,315 to GG314,315 and deletion of CKV339–341) (30). After confirming that cell surface expression levels of these molecules were comparable (Fig. 1A), we transfected these cells with various green fluorescent protein (GFP)- and Nef-containing plasmids and quantified A02 or A02GGΔCKV downregulation activity on a scale from 0% (denoting no downregulation in the GFP-positive [GFP+] subset) to 100% (denoting complete downregulation). The greater the downregulation activity is, the lower the residual cell surface expression of A02 or A02GGΔCKV is. As expected, no change in cell surface A02 or A02GGΔCKV expression was observed when cells were transfected with a control plasmid expressing GFP alone (Fig. 1A). In contrast, cells transfected with a plasmid encoding GFP and Nef from the YBF116 strain of HIV-1 group N exhibited 57% and 38% reductions in A02 and A02GGΔCKV cell surface expression levels, respectively, in the GFP+ (Nef-expressing) compared to levels in the GFP-negative (GFP−) subsets (Fig. 1A). Similarly, Jurkat cells expressing Nef clones from the SIVcpzPtt strain TAN-1 and SIVgsn strain CM166 also exhibited reduced A02 and A02GGΔCKV cell surface expression (Fig. 1A). Of note, despite substantial functional heterogeneity across isolates (e.g., the SIVgsn strain CM166 displayed particularly high function), all three Nef clones downregulated A02 more efficiently than A02GGΔCKV.
FIG 1.
Downregulation of MHC-A and MHC-B in primate lentiviruses. (A) Jurkat cells stably expressing A02 and A02GGΔCKV were transfected with plasmid DNAs encoding GFP only or GFP plus the indicated lentiviral Nef clones and stained with HLA-A2 antibody. Values shown on flow cytometry plots represent Nef downregulation activities calculated as described in Materials and Methods. The greater the downregulation activity is, the lower the residual cell surface expression of A02 or A02GGΔCKV is. (B) Phylogenetic analysis of lentivirus Nef clones. A maximum likelihood phylogenetic tree of lentivirus Nef clones used, colored according to lentiviral lineage, is shown. (C) Downregulation activities of Nef clones of various lentiviruses determined in Jurkat cells expressing A02 and A02GGΔCKV. Each point represents the mean of triplicate determinations. Statistical analysis was performed using a Wilcoxon matched-pairs test. (D) Ratios of A02 and A02GGΔCKV downregulation activity of these lentivirus Nef clones. Bars and whiskers denote the median and interquartile range for each group. Statistical analysis was performed using a Kruskal-Wallis test.
We then examined 32 additional Nef alleles (n = 35 in total) from primate lentiviruses belonging to HIV-1 groups M, N, and O and their close relative SIVcpz, HIV-2 strains and their close simian relatives SIVmac and SIVsmm, and SIV strains from other primate species (Fig. 1B). Though substantial functional heterogeneity in Nef-mediated HLA downregulation activity was observed within and between lentivirus groups (in particular, SIVcpz Nef clones exhibited overall poorer HLA downregulation than the other lentivirus groups), all Nef clones nevertheless downregulated A02 more efficiently than A02GGΔCKV (Wilcoxon matched-pairs test, all P ≤ 0.002) (Fig. 1C). Moreover, quantification of each Nef clone's function in terms of its A02/A02GGΔCKV downregulation ratio revealed highly comparable values across lentiviruses, with HIV-1, SIVcpz, and HIV-2/SIV strains exhibiting ratios of 1.21 (interquartile range [IQR], 1.16 to 1.30), 1.22 (IQR, 1.14 to 1.45), and 1.27 (IQR, 1.13 to 1.51), respectively (Kruskal Wallis test, P = 0.85) (Fig. 1D). These results suggest that Nef's ability to differentially downregulate MHC-A and MHC-B, as well as the relative efficiency by which it downregulates these molecules, is highly conserved across primate lentivirus lineages.
Differential HLA-A and HLA-B downregulation by HIV-1 group M Nef clones.
We next investigated differential HLA-A versus HLA-B downregulation activities among HIV-1 pandemic group M sequences by quantifying A02 and A02GGΔCKV downregulation in 80 Nef clones (20 each from unique patients infected with HIV-1 subtype A, B, C, or D) (Fig. 2A). Again, some functional heterogeneity within and between subtypes was observed (e.g., as previously reported [35], subtype C Nef proteins displayed the overall poorest A02 downregulation abilities [Kruskal Wallis test, P = 0.003]) (Fig. 2B). Nevertheless, Nef sequences from all HIV-1 subtypes exhibited superior ability to downregulate A02 versus A02GGΔCKV (Fig. 2B) (Wilcoxon matched pairs test, P < 0.001 for all comparisons), and the median ratios of A02/A02GGΔCKV downregulation activity were again highly comparable at 1.15 (IQR, 1.12 to 1.23), 1.23 (IQR, 1.18 to 1.24), 1.24 (IQR, 1.11 to 1.26), and 1.16 (IQR, 1.11 to 1.20) for Nef clones of subtypes A, B, C, and D, respectively (Kruskal Wallis test, P = 0.81) (Fig. 2C).
FIG 2.
Downregulation of HLA-A and HLA-B by patient-derived Nef clones of group M HIV-1. (A) Maximum likelihood phylogenetic tree of Nef clones of patient-derived Nef clones (n = 20 each) of subtypes A, B, C, and D in group M HIV-1, colored according to subtype. Downregulation (B) and downregulation ratios (C) of A02 and A02GGΔCKV by these Nef clones were determined. Each plot represents the mean values of triplicate assays. Statistical analysis was performed using a Wilcoxon matched-pairs test (B) or Kruskal-Wallis test (C).
Nef codons associated with differential HLA-A and HLA-B downregulation.
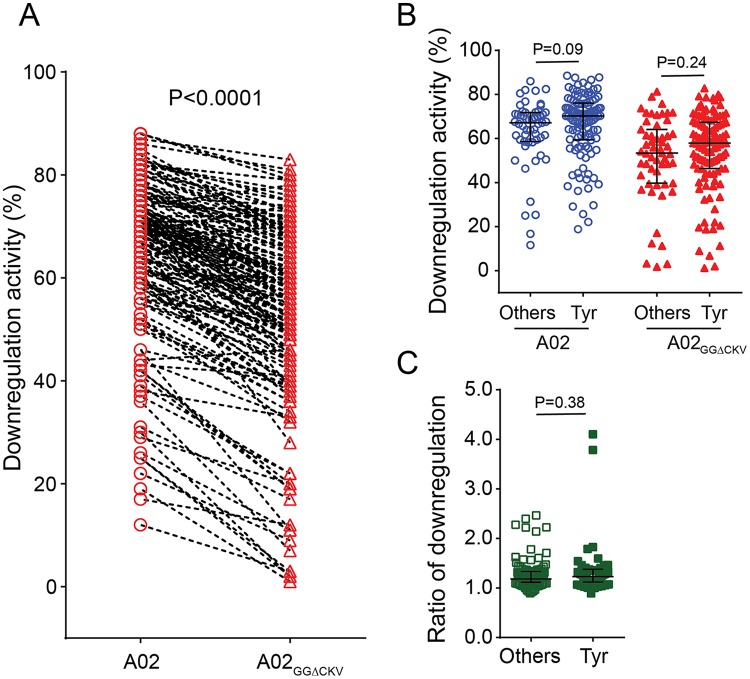
The rare, naturally occurring non-Tyr polymorphism at codon 202 in Nef was previously shown to differentially influence HLA-A versus HLA-B downregulation activity in contrast to the consensus Tyr-202 in HIV-1 subtype B (30). Tyr-202, however, is significantly less prevalent in subtypes A (23.3% frequency in the Los Alamos database, n = 421), C (59.6%, n = 1,640), and D (57.1%, n = 217) than in subtype B (78.3%, n = 3,165) (χ2 test, P < 0.0001). We thus postulated that additional residues may modulate Nef's HLA-A versus HLA-B downregulation ability in non-B subtypes. Because subtype C accounts for >50% of HIV-1 infections globally (40), we isolated 148 additional subtype C Nef clones (n = 168 in total; one clone per patient) and tested their activities. As expected, subtype C Nef clones downregulated A02 more efficiently than A02GGΔCKV (median, 69.7% [IQR, 59.2 to 74.9%] and 56.3% [IQR, 45.3 to 66.9%], respectively) (Fig. 3A) (Wilcoxon matched-pairs test, P < 0.001), yielding a median A02/A02GGΔCKV downregulation ratio of 1.19 [IQR, 1.11 to 1.34]. Stratification of subtype C Nef clones based on codon 202 variation, however, revealed no difference in their abilities to downregulate A02 (Mann-Whitney, P = 0.09) or A02GGΔCKV (P = 0.24) (Fig. 3B) or in the ratio of these values (P = 0.38) (Fig. 3C), suggesting that other Nef residues contribute to differential HLA-A and HLA-B downregulation activity in subtype C. We then performed exploratory codon-function analyses to identify subtype C Nef amino acids associated with these activities. At the predefined threshold of a P value of <0.05 and q value of <0.1, Nef clones carrying Ser-9 exhibited a significantly lower A02GGΔCKV downregulation function (P = 0.00014, q = 0.025) and a higher A02/A02GGΔCKV downregulation ratio (P = 0.00022, q = 0.049) than those carrying other amino acids at this site (Table 2); this was the most significant association observed in the data set. No Nef residue was identified as being associated with A02 downregulation function.
FIG 3.
Downregulation of HLA-A and HLA-B by patient-derived subtype C Nef clones. (A) Downregulation of A02 and A02GGΔCKV by patient-derived Nef clones of subtype C (n = 168). Data represent A02 and A02GGΔCKV downregulation activities (B) and their corresponding A02/A02GGΔCKV downregulation ratios (C) when Nef sequences were stratified based on expression of Tyr-202 (consensus) (n = 55) versus that of other residues at this position (n = 113). Each plot represents the mean values of triplicate assays. Bars and whiskers represent median values and interquartile ranges. Statistical analysis was performed using a Wilcoxon matched-pairs test (A) or Mann-Whitney U test (B and C).
TABLE 2.
Nef amino acid residues associated with A02 and A02GGΔCKV downregulation
| Parameter | Nef codon 9 value for:b |
|
|---|---|---|
| A02GGΔCKV | A02/A02GGΔCKV | |
| Downregulation activitya | ||
| With Ser | 55.5% | 1.22 |
| Without Ser | 71.5% | 1.05 |
| P valuec | 0.00014 | 0.00022 |
| q valuec | 0.02 | 0.04 |
| No. of subjects (clones) testedd | ||
| With Ser | 142 | 142 |
| Without Ser | 12 | 12 |
Results were determined from a predefined threshold using residues observed in n > 5 sequences, at a P value of <0.05 and q value of <0.1. Values represent the median downregulation activity of A02GGΔCKV or pairwise ratio of A02/A02GGΔCKV downregulation.
Nef codon numbers are based on NefHXB2.
Significance was determined by comparing values for Nef clones carrying Ser-9 to those of clones with other amino acids at this site.
The total varies due the gap in the aligned sequence.
Subtype-specific effects of Nef-9 variation on HLA-A and HLA-B downregulation.
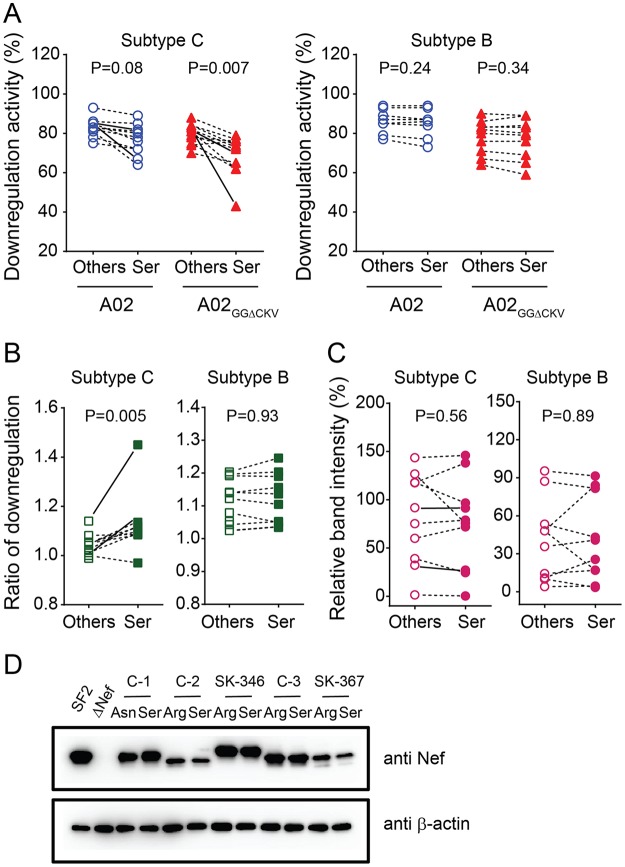
To validate the effects of Nef Ser-9 polymorphism on HLA-A and HLA-B downregulation, we inserted 10 patient-derived subtype C nef alleles encoding arginine (n = 6), lysine (n = 1), leucine (n = 1), cysteine (n = 1), and asparagine (n = 1) residues at position 9 into pNL43 and subsequently reverted these clones to the consensus Ser-9 to generate paired mutant viruses. Reversion to Nef Ser-9 modestly decreased A02 downregulation activity of these recombinant viruses (Wilcoxon matched-pairs test, P = 0.08) but significantly decreased their A02GGΔCKV downregulation activity (P = 0.007) (Fig. 4A, left panel) in HIV-1-infected cells, yielding significant increases in their A02/A02GGΔCKV downregulation ratios from 1.05 (IQR, 1.02 to 1.13) to 1.11 (IQR, 1.08 to 1.15) (P = 0.005) (Fig. 4B, left panel). Even after excluding one Nef clone showing the greatest difference (Fig. 4A and B, SK-367, solid lines) from the analysis, reversion to Ser-9 of the remaining nine clones exhibited significant effects on the A02GGΔCKV downregulation activity and the A02/A02GGΔCKV downregulation ratios (P = 0.01 for both compared to values with non-Ser at the same position). Reversion to Nef Ser-9 yielded no significant changes in Nef steady-state expression level as measured by Western blotting (Fig. 4C, left panel, and D), suggesting that observed results are not attributable to differences in steady-state Nef protein levels.
FIG 4.
Subtype-specific effects of Nef-9 variation on HLA-A and HLA-B downregulation. Subtype C and subtype B Nef clones (n = 10 each) harboring nonconsensus amino acids at position 9 (Others) were cloned into HIV-1 pNL43, and their corresponding Ser-9 consensus reversion mutants were constructed (Ser). Absolute A02 and A02GGΔCKV downregulation (A) and the corresponding ratios of A02 and A02GGΔCKV downregulation (B) by these Nef clones are shown. (C) Nef band intensity by Western blotting (normalized to the control strain NefSF2 as 100% following subtraction of background band intensity observed in the NL43ΔNef negative control) is shown. Solid lines connect the pairwise plots for Nef clones SK-346 and SK-367 that were used for subsequent analyses (Fig. 5 to 7). Each plot represents the mean values of triplicate assays. Statistical analysis was performed by a Wilcoxon matched-pairs test. (D) A representative Western blot is shown. Lysates of HEK-293 T cells transfected with the NL43 strain harboring the indicated subtype C Nef alleles (the control strain NefSF2 and the NL43ΔNef negative control are included) were separately stained by anti-Nef and anti-β-actin antibodies.
Ser-9 is highly conserved in subtypes C (88.1% in the Los Alamos database, n = 1640), A (88.8%, n = 421), and D (86.6%, n = 217) but less so in subtype B (63.3%, n = 3165) (χ2 test, P < 0.0001). To investigate the functional effects of Nef codon 9 variants in HIV-1 subtype B, we inserted 10 patient-derived subtype B nef alleles encoding lysine (n = 4), arginine (n = 1), glycine (n = 1), asparagine (n = 1), leucine (n = 1), isoleucine (n = 1), and methionine (n = 1) at position 9 into pNL43 and constructed their corresponding Ser-9 revertants. In contrast to subtype C, however, Nef Ser-9 revertants exhibited no significant changes in their abilities to downregulate A02 or A02GGΔCKV (Fig. 4A, right panel) or in the ratios thereof (Fig. 4B, right panel). Consistent with subtype C, reversion to Nef Ser-9 yielded no significant changes in the Nef steady-state expression level as measured by Western blotting (Fig. 4C, right panel).
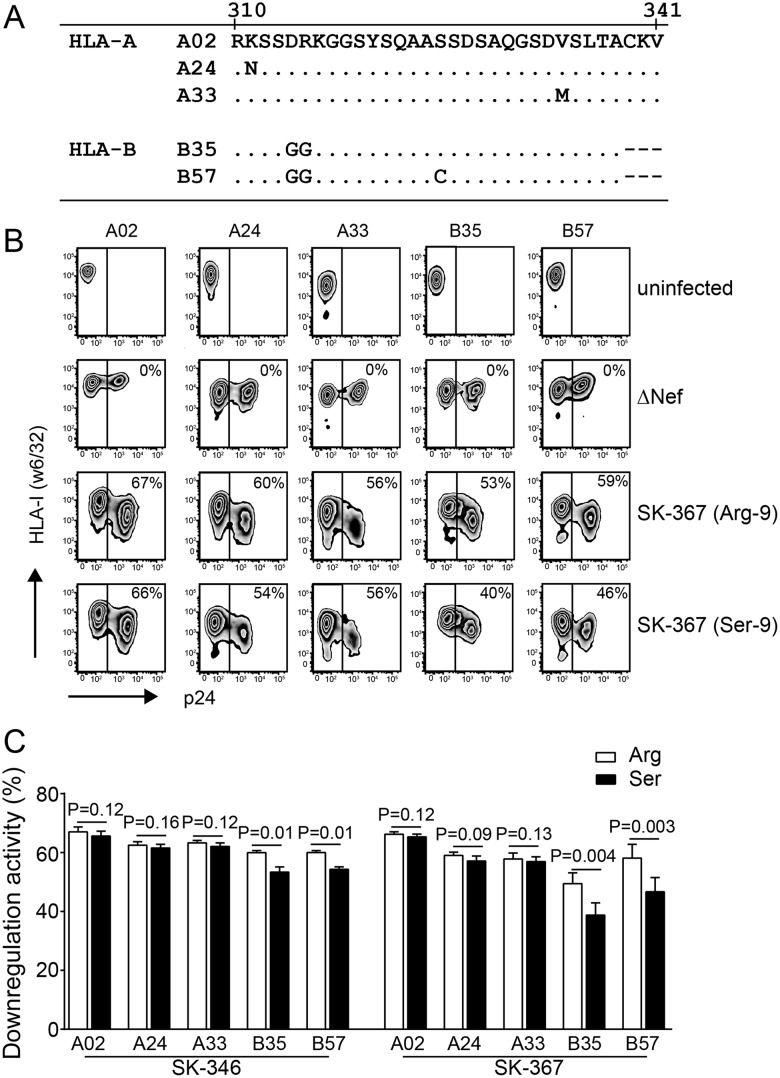
Effect of Nef-9 variations on downregulation of multiple HLA-A and HLA-B allotypes.
Because there are five HLA-A and two HLA-B cytoplasmic tail types in humans, we next tested the ability of two subtype C Nef clones to downregulate various HLA-A and HLA-B molecules using HLA class I-deficient 721.221 cells stably expressing HLA-A*02:01 (A02), HLA-A*24:02 (A24), HLA-A*33:03 (A33), HLA-B*35:01 (B35), or HLA-B*57:01 (B57) (Fig. 5A). We first confirmed comparable surface expression levels of the three HLA-A and two HLA-B allotypes by flow cytometry and confirmed no changes in cell surface HLA-I expression following infection of these cells with vesicular stomatitis virus glycoprotein protein G (VSV-G)-pseudotyped HIV-1NL43 lacking Nef (NL43-ΔNef) (Fig. 5B). Nef sequences from patients SK-346 and SK-367 (Fig. 4A and B, solid lines), both harboring Arg at position 9, were selected for analysis because reversion to Ser-9 substantially increased their A02/A02GGΔCKV downregulation ratios in Jurkat cells (from a mean ± standard deviation [SD] of 1.04 ± 0.02 to 1.14 ± 0.03 for SK-346 and from 1.12 ± 0.04 to 1.45 ± 0.05 for SK-367) (Fig. 4B, solid lines).
FIG 5.
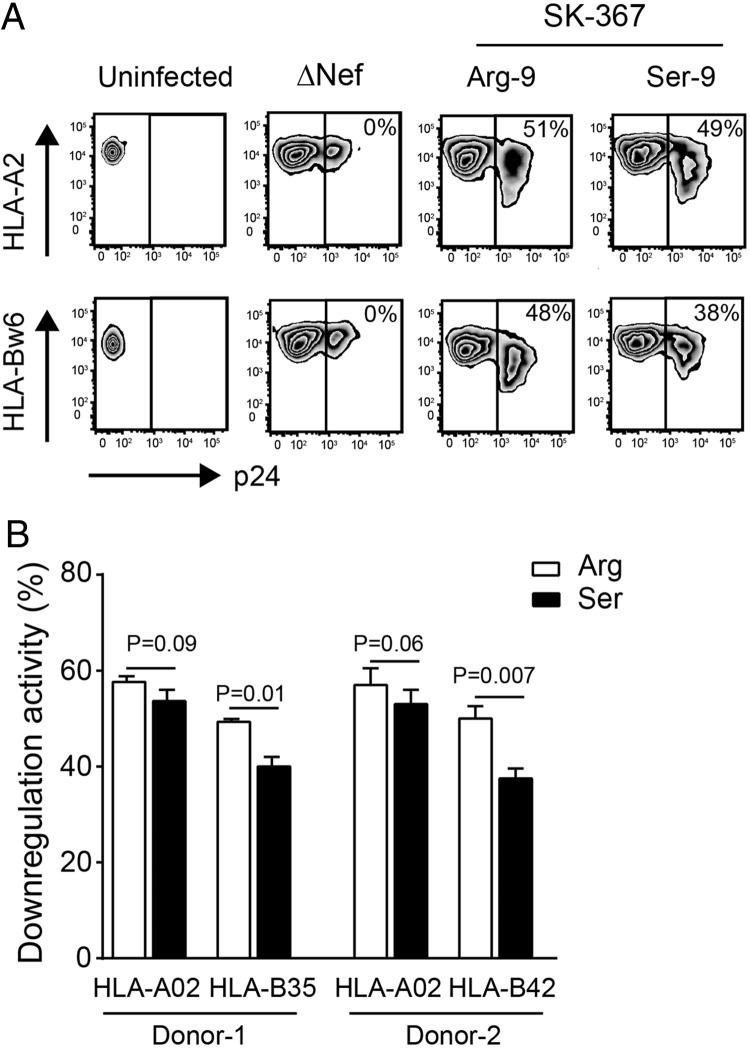
Effect of Nef-9 variations on downregulation of various HLA-A and HLA-B allotypes in 721.221 transfectants. (A) Cytoplasmic tail sequences of HLA-A and HLA-B molecules used. (B) A representative set of flow cytometry plots showing uninfected 721.221 transfectants stably expressing the indicated HLA allotypes, as well as those infected with HIV-1 encoding SK-367 Nef (Arg-9), its revertant (Ser-9), or ΔNef, stained with pan-specific HLA-I MAb w6/32. (C) Quantification of HLA downregulation ability by patient-derived subtype C Nef clones SK-346 and SK-367 and their respective Ser-9 reversions. Data shown are means ± SDs of a minimum of three independent experiments. Statistical analysis was performed using a Wilcoxon matched-pairs test.
VSV-G-pseudotyped HIV-1NL43 expressing SK-346 Nef (containing Arg-9) exhibited mean A02, A24, and A33 downregulation functions of 67.0% (SD, ±1.7%), 62.5% (SD, ±1.2%), and 63.2% (SD, ±1.6%), respectively, and mean B35 and B57 downregulation functions of 59.9% (SD, ±1.5%) and 59.7% (SD, ±1.4%), respectively (Fig. 5C). Reversion of Nef Arg-9 to the consensus Ser-9 did not impact HLA-A downregulation activity (Ser-9 mean activity ± SD of 65.2% ± 1.6%, 61.2% ± 1.4%, and 62.1% ± 1.3% for A02, A24, and A33, respectively; P ≥ 0.12 for all compared to values with Arg-9) but significantly impaired HLA-B downregulation activity (Ser-9 mean activity ± SD of 53.3% ± 1.2% and 54.3% ± 1.1% for B35 and B57, respectively; P = 0.01 for both compared to values with Arg-9) (Fig. 5C). This yielded an increase in the A02/B35 downregulation ratio from 1.11 to 1.23. Similarly, VSV-G-pseudotyped HIV-1NL43 expressing SK-367 Nef (also containing Arg-9) exhibited mean A02, A24, and A33 downregulation functions of 66.2% (SD, ±1.2%), 59.0% (SD, ±1.1%), and 57.8% (SD, ±2.0%), respectively, and mean B35 and B57 downregulation functions of 53.6% (SD, ±3.4%) and 58.1% (SD, ±4.1%), respectively (Fig. 5B and C). Again, reversion of Nef Arg-9 to the consensus Ser-9 in SK-367 did not substantially impact HLA-A downregulation (P ≥ 0.09 for all compared to values with Arg-9) but significantly reduced HLA-B downregulation (Ser-9 mean activity ±SD of 44.3% ± 4.0% and 46.6% ± 4.7% for B35 and B57, respectively; P < 0.005 for both compared values with to Arg-9) (Fig. 5B and C), yielding an increase in the A02/B35 downregulation ratio from 1.22 to 1.45.
We further confirmed the effects of Nef-9 polymorphisms on differential HLA-A and HLA-B downregulation in phytohemagglutinin (PHA)-activated human primary T lymphocytes from two healthy donors: one expressing A02 and B35 (which encodes Ser at codon 324) (Fig. 5A) and the other expressing A02 and B42 (which, like B57, encodes Cys at codon 324) (Fig. 5A). Infection of donor 1's cells with HIV-1NL43 expressing SK-367 Nef (containing Arg-9) yielded mean HLA-A02 and HLA-B35 downregulation values of 57.4% (SD, ±4.0%) and 49.6% (SD, ±5.1%), respectively (Fig. 6A and B). Reversion of Nef Arg-9 to the consensus Ser-9 in SK-367 Nef did not impact HLA-A downregulation but significantly reduced HLA-B downregulation (P = 0.01) (Fig. 6B), yielding an increase in the HLA-A/HLA-B downregulation ratio from 1.16 to 1.29. Consistent results were obtained with cells from donor 2: again, reversion to consensus Ser-9 SK-367 Nef did not significantly impact HLA-A downregulation but significantly reduced HLA-B downregulation (P = 0.007) (Fig. 6B), yielding an increase in the HLA-A/HLA-B downregulation ratio from 1.14 to 1.34.
FIG 6.
Effect of Nef-9 variations on HLA downregulation in human primary T lymphocytes. (A) A representative set of flow cytometry plots of uninfected human primary T lymphocytes obtained from HIV-negative donor 1 and from donors infected with HIV-1 encoding SK-367 Nef (Arg-9), its revertant (Ser-9), or ΔNef, stained with the serotype-specific MAbs for HLA-A02 and HLA-Bw6. Values on flow cytometry plots denote Nef downregulation activities. (B) Quantification of the HLA downregulation ability of SK-367 (Arg) and its corresponding Ser-9 revertant using human primary T lymphocytes from two HIV-negative donors. Data shown are means ± SDs of a minimum of three independent experiments. Statistical analysis was performed using a Wilcoxon matched-pairs test.
Effect on T cell recognition.
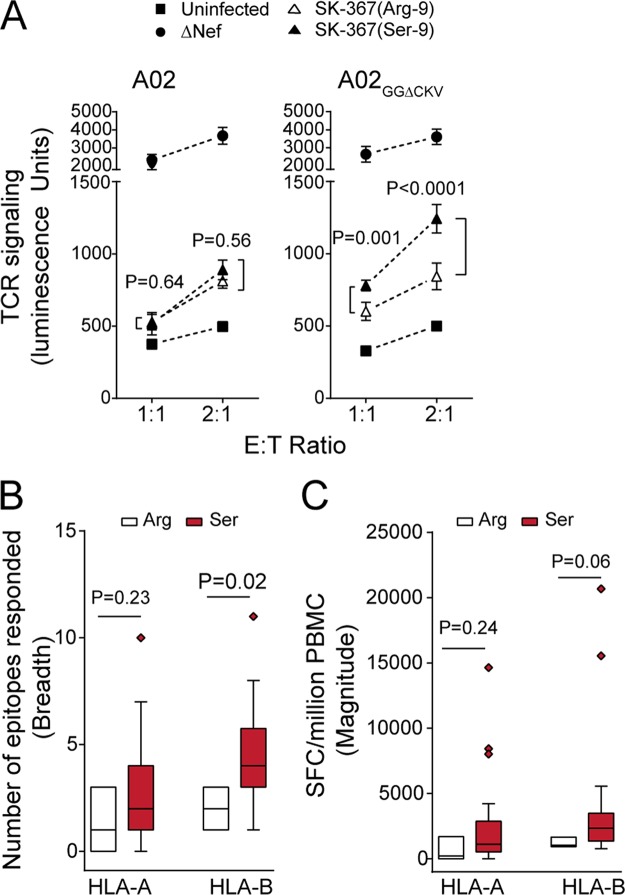
Nef's differential HLA-A versus HLA-B downregulation ability could have consequences for T cell recognition of viral antigens presented on the surface of HIV-infected cells (39, 41, 42). To test this, we used a reporter cell assay featuring HLA-A*02:01-expressing target cells and HIV-1-specific effector reporter cells that transiently express a T cell receptor (TCR) specific for an HLA-A*02:01-restricted HIV-1 epitope in Gag (FK10; Gag433–442, FLGKIWPSYK), human CD8-α chain, and an NFAT-driven luciferase construct (30, 43). Upon coculture of HIV-infected target cells with FK10-specific effector reporter cells, TCR-dependent signaling in the latter was quantified based on luminescence. At effector/target (E/T) ratios of 1:1 and 2:1, effectors cultured with A02-expressing targets infected with HIV-1 NL43-ΔNef produced luminescence more than 5-fold higher than the background level(i.e., when cocultured with uninfected A02 targets) (Fig. 7A, left panel). The same was true when A02GGΔCKV targets were used (Fig. 7A, right panel). In contrast, effectors cocultured with A02-expressing targets infected with HIV-1 expressing parental SK-367 Nef (Arg-9) produced luminescence signals >3-fold lower than those infected with HIV-1 ΔNef (Fig. 7A, left panel). Similar results were obtained when A02GGΔCKV target cells were used (Fig. 7A, right panel). These observations confirm that A02 (and A02GGΔCKV) downregulation by SK-367 Nef reduces antigen recognition by HIV-specific TCR.
FIG 7.
Effect of Nef sensitivity to HLA-A and HLA-B on antigen-specific T cell recognition. (A) Luciferase reporter effector T cells expressing an HLA-A*02-restricted HIV-1 Gag FK10-specific TCR were cocultured with either uninfected target T cells stably expressing A02 or A02GGΔCKV or target T cells infected with HIV-1 encoding SK-367 Nef (Arg-9), its revertant (Ser-9), or ΔNef at the indicated E/T ratios. T cell recognition of target cells by effector cells is quantified as luminescence signal detected at 6 h. Data represent the means ± SDs of a minimum of three independent assays. Statistical analysis was performed by a Wilcoxon matched-pairs test. Over all experiments, background luminescence signal (obtained by incubation of the effector cells alone) was 151.8 ± 17.3, and HIV-1 infection frequency of target cells, determined by intracellular expression of p24Gag protein, was 49.7% ± 11.1%. HLA-A- and HLA-B-restricted epitope response breadth (B) and magnitude (C) were measured using a gamma interferon ELISPOT assay in the subset of patients (n = 30) expressing protective alleles HLA-B*57, HLA-B*58:01, and HLA-B*81:01. Box-and-whisker plots show the median (horizontal line), interquartile range (edges of box), and range (whiskers). Data include outliers. Statistical analysis was performed using a Mann-Whitney U test. SFC, spot-forming cells.
Importantly, however, the reversion of Nef Arg-9 to the consensus Ser-9 in SK-367 differentially affected effector cell recognition of targets expressing A02 versus A02GGΔCKV. Whereas the Ser-9 reversion did not significantly affect effector cell luminescence upon coculture with infected A02 targets (P > 0.5) (Fig. 7A, left panel), it significantly increased effector cell luminescence signals upon coculture with infected A02GGΔCKV targets (P ≤ 0.001) (Fig. 7A, right panel). Together, these observations support the notion that enhanced HLA-B downregulation by subtype C Nef sequences harboring nonconsensus codon 9 residues results in reduced viral antigen presentation upon, and thus reduced T cell recognition of, HIV-infected cells.
We further postulated that enhanced HLA-B downregulation by subtype C Nef sequences harboring nonconsensus residues at codon 9 may have consequences for antiviral immune recognition in vivo. We examined HLA-A- and HLA-B-restricted CD8 T lymphocyte responses to a proteome-wide set of HIV-1 subtype C consensus peptides, measured by gamma interferon enzyme-linked immunosorbent spot (ELISPOT) assay (15). Though no significant associations with HLA-A- and HLA-B-restricted CD8 T lymphocyte responses were observed at the whole-cohort level (Mann Whitney, P > 0.3) (data not shown), when the analysis was restricted to patients expressing protective HLA class I alleles (defined in subtype C as HLA-B*57, HLA-B*58:01, and HLA-B*81:01 [15]) (n = 30), which are anticipated to restrict the most effective anti-HIV CTL responses in vivo, the median number of responses to HLA-B-restricted epitopes (i.e., response breadth) was significantly lower in individuals infected with HIV-1 harboring Nef-9 variants than in those with the consensus Ser-9 (Mann Whitney, P = 0.02) (Fig. 7B). We also observed a trend toward a lower magnitude of HLA-B-restricted epitope responses in individuals infected with HIV-1 harboring Nef codon 9 variants than in those with the consensus Ser-9 (Fig. 7C) (Mann Whitney, P = 0.06). In contrast, Nef codon 9 variants did not significantly associate with HLA-A-restricted response breadth or magnitude (Fig. 7B and C) (P > 0.2). These observations further support the notion that subtype C Nef sequences harboring nonconsensus codon 9 residues possess enhanced ability to evade HLA-B-restricted immune responses.
DISCUSSION
We investigated the ability of Nef clones from different primate lentiviruses, including various subtypes of the pandemic HIV-1 group M, to downregulate MHC-A and MHC-B surface molecules. For all isolates tested, Nef's ability to downregulate HLA-A was consistently 11 to 32% better than its ability to downregulate HLA-B, supporting differential HLA-A versus HLA-B downregulation function as a fundamental property of lentiviral Nef sequences. Furthermore, our analyses of HIV-1 Nef sequences from subtype C, the most predominant subtype globally, revealed Nef codon 9 as a critical mediator of differential HLA-A versus HLA-B downregulation function: target cells infected with recombinant HIV-1 strains expressing subtype C Nef sequences harboring nonconsensus codon 9 residues were poorly recognized by HIV antigen-specific effector cells in vitro, which is consistent with Nef codon 9 variants mediating an enhanced ability to evade HLA-B-restricted immune responses. Moreover, among persons expressing protective HLA-B alleles, carriage of Nef codon 9 variants was associated with poorer ex vivo HIV-specific T cell responses. Our results support in vivo relevance of Nef's differential HLA downregulation functions and provide further mechanistic support for the relative dominance of HLA-B on HIV-1 immune control (14, 15).
Our detailed studies of Nef sequences from HIV-1 subtype B (30) and subtype C in the present study also underscore the existence of subtype-specific HIV-1 polymorphisms that modulate Nef's differential HLA downregulation function. While the rare His-202 mutation substantially diminished Nef's HLA-B downregulation function (but only modestly affected Nef's HLA-A downregulation function) in HIV-1 subtype B (30), Nef codon 9 plays a key role in modulating differential HLA-A versus HLA-B downregulation function in subtype C. Indeed, previous study by our group using a partially overlapping panel of HIV-1 Nef clones identified Ser-9 in subtype C, but not in subtype B, as associated with lower HLA-A02 downregulation (35) (though results of the present study demonstrated an association between Ser-9 and lower sensitivity only to HLA-B cytoplasmic tail sequences at the predefined threshold). The recently reported crystal structure of a ternary complex formed between subtype B Nef, the HLA-A02 cytoplasmic tail, and the cargo-binding μ1 subunit of adaptor protein 1 (AP1) suggests that Nef-202 may be located in close proximity to the CKV339–341 motif of the HLA-A02 cytoplasmic tail (28). In contrast, Nef-9 appears to be distant to the HLA-A02 cytoplasmic tail in this structure. We hypothesize that Nef-9 may indirectly influence Nef-HLA interaction through interacting with other parts of the active ternary complex, possibly the nearby Nef Trp-13 which is critical for interaction with the μ1 subunit of AP1 (44).
It is intriguing that Nef consensus Ser-9 occurs in >88% of HIV-1 subtype C sequences despite its association with reduced HLA-B downregulation. We speculate that Ser-9 could enhance another Nef function at the expense of HLA downregulation activity or that Ser-9 may represent an adapted form (i.e., escape variant) specific for one or more HLA alleles (as selective pressure from CTL responses is a major driver of Nef variability [39, 45]). In support of Nef codon 9 as being under HLA-associated immune pressures in HIV-1 subtype C, Lys-9 was recently identified as being significantly overrepresented among HLA-C*07-expressing persons (46); nevertheless, further analyses are necessary to establish why subtype C HIV-1 maintains Nef Ser-9 in the majority of infected hosts.
Some limitations merit mention. Although we investigated 268 Nef clones from various lentiviruses including HIV-1, this panel does not capture the entirety of lentivirus Nef genetic diversity. Nevertheless, despite a substantial dynamic range of MHC-A and MHC-B downregulation functions across lentiviral lineages, the vast majority of Nef clones downregulated MHC-A more efficiently than MHC-B molecules, an observation that was consistent across all immortalized and primary cell models and all HLA allotypes and cytoplasmic variants tested (noting, however, that all cells used in the present study were human-derived; we acknowledge that MHC cell surface expression levels and, by extension, the Nef activity required to downregulate these molecules may vary among primate species). We employed transient-transfection systems and recombinant virus approaches to assess Nef functions; the former are limited by Nef overexpression and potential cytotoxicity during plasmid delivery, and the latter are inherently limited by potential incompatibilities between insert and backbone although a recent study demonstrated that relative functional defects in subtype C patient-derived sequences were detectable in a recombinant virus system regardless of HIV-1 genetic context (47). Nevertheless, both systems yielded consistent results for Nef clones across subtypes. We synthesized overlapping peptides based on the subtype C consensus amino acid sequence for ELISPOT assays. Although this approach has been widely used in analyzing HIV-specific CD8 T cell activity across the HIV-1 proteome, consensus peptides do not capture the full extent of HIV-1 sequence variation within the host. Lastly, it is not possible to disentangle cause and effect using cross-sectional approaches. Therefore, longitudinal studies of Nef function and antiviral T cell responses, beginning in acute/early infection, are warranted. Despite these limitations, our study, the largest of its kind to date, significantly extends the body of evidence supporting differential Nef-mediated evasion of HLA-A- and HLA-B-restricted immune responses as a key modulator of antiviral T cell responses in HIV-infected hosts.
MATERIALS AND METHODS
Nef clones. (i) Nef clones from various lentiviruses.
A total of 35 Nef clones from various lentiviruses were made as part of previous studies and ligated into the pCGCG plasmid that coexpresses Nef and GFP (25, 48). The nef clones used were from the following lentiviruses (GenBank accession number): from HIV-1 group M, NL43 (M19921), NA7 (DQ242535), and JR-CSF (M38429); from HIV-1 group O, MVP8161 (AY536905), MVP13127 (AY536904), HJ162 (GQ925932), and HJ735 (GQ925935); from HIV-1 group N, YBF30 (AJ006022), YBF116 (AY536907), and DJ131 (AY532635); from HIV-2, BEN (M30502), CBL-23 (DQ222472), 60415K (DQ092764), and 310319 (DQ092766); from SIVcpzPtt, US (AY536908), Cam5k2 (AY536911), GAB2 (AF382828), MT145 (DQ373066), MB897 (EF535994), GAB1 (X52154), and GAB2 (AF382828); from SIVcpzPts, Nok5 (AY536915), Nik4 (AY536916), TAN1 (EF394356), TAN2 (DQ374657), and TAN3 (DQ374658); from SIVsmm, FMm1 (DQ092762), FYr1 (DQ092760), and FWr1 (DQ092758); from other SIVs, SIVgsn CM 166 (AF468659), SIVmus CMS1085 (AY340700), SIVtan 1 (AF395566), SIVsab 1 (U04005), CML1 (AY340701), and SIVmac 239 (M33262).
(ii) Nef clones from HIV-1 subtypes A to D.
A total of 228 nef clones isolated from plasma HIV RNA of unique antiretroviral-naive patients chronically infected with HIV-1 subtypes A (n = 20), B (n = 20), C (n = 168), and D (n = 20) (one nef clone per patient) were made as part of previous studies and cloned into the pSELECT plasmid that coexpresses Nef and GFP under separate promoters (35). Subtypes A- and D-infected individuals were recruited from the Uganda AIDS Rural Treatment Outcomes (UARTO) Cohort, Kampala and Mbarara in Uganda (49), subtype B infected individuals were recruited from British Columbia, Canada (50), and subtype C infected individuals were recruited from Durban, South Africa. For the subtype C-infected patients, median plasma viral loads were 4.8 (IQR, 2.0 to 6.4) RNA log10 copies/ml, and median CD4 counts were 333 (IQR, 11 to 943) cells/mm3. All individuals were HLA class I typed and antiretroviral naive. This study was approved by the Institutional Review Boards at all relevant institutions; all participants provided written informed consent, or specimens were anonymized according to IRB-approved procedures. GenBank accession numbers for clonal nef sequences of subtypes A to D used in this study are as follows: KC906734, KC906735, KC906737, KC906739, KC906741, KC906743 to KC906755, KC906756 to KC906776, KC906778 to KC906785, KC906787 to KC906789, KC906791 to KC906795, KC906797 to KC906799, KC906800, KC906805, KC906812, KC906813, KC906817, KC906821, KC906823, KC906827, KC906828, KC906832, KC906839, KC906841, KC906845, KC906853, KC906862 to KC906865, KC906868, KC906880, KC906900, KC906902, KC906905, KC906907, KC906910, KC906913, KC906918, KC906919, KC906921, KC906922, KC906927, KC906933, KC906942, KC906944, KC906948, KC906949, KC906955, KC906966, KC906975, KC906978, KC906982, KC906984 to KC906986, KC906989, KC906993, KC906996, KC906997, KC906999, KC907000, KC907002, KC907004, KC907015, KC907024, KC907026, KC907027, KC907037, KC907048, KC907051, KC907054, KC907058, KC907070, KC907073, KC907075, KC907076, KM263030 to KM263046, KM263049, KM263052 to KM263077, KM263079 to KM263101, KM263103 to KM263107, KM263109 to KM263012, KM263114, and KM263116 to KM263141.
To facilitate a consistent codon numbering scheme, all clonal Nef sequences were pairwise-aligned to the HIV-1 subtype B reference strain HXB2 (HIV-1HXB2; GenBank accession number K03455), and insertions with respect to HIV-1HXB2 were stripped out prior to analysis.
Preparation of recombinant viruses.
Recombinant viruses expressing patient-derived HIV-1 subtype C nef clones in an HIV-1NL43 proviral backbone were generated as previously published (34, 37, 51). Specific mutations (i.e., at Nef position 9) were also introduced into patient-derived nef clones using overlapping PCR (36, 39), after which these mutant nef sequences were inserted into pNL43. The nef region of all recombinant and mutant viruses was verified by DNA sequencing. The resultant plasmid DNA (5 μg) along with a plasmid encoding vesicular stomatitis virus envelope G glycoprotein (VSV-G) (1 μg) were transfected into 106 293T cells, and the virus-containing culture supernatants were harvested 48 h later. Recombinant HIV-1NL43 viruses lacking nef (ΔNef) were used as a negative control as previously described (37, 39).
Analysis of Nef-mediated HLA downregulation.
Nef-mediated downregulation of cell surface HLA-I molecules was assessed in three different cell systems: (a) Jurkat, a human CD4+ T cell line stably transfected with the gene encoding HLA-A*02:01 or its cytoplasmic tail mutant; (b) 721.221, an HLA-I-null human lymphoblastoid cell line stably transfected to express a single HLA-I allele; (c) human primary T lymphocytes isolated from HIV-negative donors expressing autologous HLA-A2 and HLA-Bw6 serotype alleles.
For the Jurkat system, human cDNA encoding HLA-A*02:01 was cloned into the pcDNA3.1 plasmid. Mutations DR314,315 to GG314,315 and deletion of CKV339–341 were then introduced, giving rise to HLA-A02GGΔCKV, whose cytoplasmic tail is identical to that of HLA-B*35:01, as previously described (30). Cell surface expression of HLA-A*02 and its cytoplasmic tail mutant HLA-A02GGΔCKV were confirmed using an HLA-A2 serotype-specific MAb. A total of 6 × 105 HLA-A02- or HLA-A02GGΔCKV-expressing Jurkat cells were electroporated with 4 μg of pGCGC or pSELECT plasmid DNA encoding GFP alone or various Nef alleles at 250 V and 950 μF using a Gene Pulser Xcell electroporation system (Bio-Rad Laboratories, Inc.). Twenty-four hours later, cells were stained with the HLA-A2 serotype-specific antibody (clone BB7.2; BioLegend Co) followed by 7-amino-actinomycin D (7-AAD) (BioLegend Co).
For the second system, 721.221 cell lines lacking HLA-A, HLA-B, and HLA-C were engineered to stably express a single HLA class I allele (HLA-A*02:01, A*24:02, A*33:03, B*35:01, or B*57:01; kindly provided by M. Takiguchi, Kumamoto University, Japan). HLA gene delivery was confirmed by genotyping as described previously (52), and HLA-I cell surface expression was confirmed by staining with a pan-HLA-I-specific antibody (clone w6/32; BioLegend Co.) followed by flow cytometry. Cells were infected with VSV-G-pseudotyped recombinant HIV-1 stocks, harvested 48 h later, and stained with the pan-HLA-I-specific antibody. 7-AAD and intracellular p24Gag staining (anti-p24 Gag-fluorescein isothiocyanate [FITC] monoclonal antibody [MAb] KC57; Beckman-Coulter) was also performed as previously described (37).
For the third system, primary T lymphocytes were isolated from peripheral blood mononuclear cell (PBMCs) from two HIV-negative donors and activated with phytohemagglutinin for 5 days. HLA-I cell surface expression was evaluated by serotype-specific MAbs for HLA-A2 (for HLA-A*02:01) (clone BB7.2; BioLegend) and HLA-Bw6 (for HLA-B*35:01 and B*42:01) (clone SFR8-B6; kindly provided by M. Takiguchi). Primary T cells were infected with recombinant HIV-1 stocks, harvested 48 h later, and stained with the serotype-specific MAbs. 7-AAD and intracellular p24Gag staining was also performed as described above (37).
In all cases, live (7-AAD-negative) cells were gated, and the mean fluorescence intensity (MFI) of HLA-I in Nef-expressing cells (defined as the GFP+ subset in the Jurkat system or the p24Gag+ subset in the 721.221 and primary cell systems, denoted MFIHLA-I Nef+ in the below calculation) and non-Nef-expressing cells (defined as the GFP− or p24Gag− subsets in the Jurkat and 721.221/primary cell systems, respectively, denoted MFIHLA-I Nef−) was analyzed by flow cytometry (FACSVerse; BD Biosciences). The following formula was used to calculate the HLA-I downregulation activity of each Nef clone: (MFIHLA-I Nef− − MFIHLA-I Nef+)/MFIHLA-I Nef−. All Nef functional values are reported as the mean of a minimum of triplicate experiments.
Western blotting.
HEK-293T cells were transfected with proviral DNAs encoding patient-derived nef alleles or their mutants for preparation of cell lysates as described previously (37, 53). Briefly, lysates were prepared in duplicate, subjected to SDS-PAGE, transferred to nitrocellulose membranes, and separately stained with rabbit anti-Nef polyclonal antiserum (NIH AIDS Research and Reference Reagent Program) and mouse anti-β actin monoclonal antibody (Wako Pure Chemical Industries, Ltd., Osaka, Japan) followed by secondary antibodies (37). Band intensities were quantified using an Amersham Imager 600 instrument (GE Healthcare Life Sciences).
T cell recognition assay.
The effect of Nef-mediated HLA-I downregulation on T cell recognition was analyzed using a T cell receptor (TCR)-based reporter cell coculture assay as previously described (30, 43). Briefly, effector cells were prepared by electroporation of Jurkat cells with expression plasmids encoding TCR-α and -β chains specific for the HLA-A*02:01-restricted HIV-1 Gag FK10 epitope (Gag433–442, FLGKIWPSYK), human CD8α chain (Invivogen), and NFAT-driven luciferase reporter (Affymetrix). The resultant effector cells were incubated for 24 h, after which the CD8-expressing fraction was isolated by magnetic bead sorting (Milteyni). Target cells were prepared by infecting Jurkat cells engineered to express A02 or A02GGΔCKV (or negative-control parental Jurkat cells lacking A02) with recombinant HIV-1 encoding nef alleles of interest. A total of 5 × 104 effector cells were cocultured with 5 × 104 target cells (unless otherwise specifically indicated). Coculture results in TCR-mediated calcium-flux and robust luciferase expression in the effector cells that is dependent on endogenously processed HIV-1 FK10Gag peptide antigen presented on HLA-A*02 on the surface of the target cells. Six hours later, NFAT-driven luciferase signal in effector cells was quantified using a Steady-Glo luciferin kit (Promega) and was measured using a CentroXS3 plate reader (Berthhold Technologies).
ELISPOT assays.
HIV-specific CD8 T cell responses were characterized by gamma interferon ELISPOT assay in the HIV-1 subtype C-infected individuals recruited in the Durban area, South Africa, as part of previously published studies (15). Briefly, assays were carried out with a panel of 410 18-mer peptides spanning the entire HIV-1 subtype C proteome (synthesized based on the published 2001 HIV-1 subtype C consensus sequence [15]) that were arranged in a matrix of 11 to 12 peptides per pool (54); responses to individual 18-mer peptides within a given pool were subsequently confirmed in a separate ELISPOT assay with a single peptide.
Statistical analysis.
Statistical analyses were performed using GraphPad Prism, version 6.0. For codon-function analyses, a Mann-Whitney U test was used to identify amino acids associated with differential ability to downregulate A02 and A02GGΔCKV (expressed as A02/A02GGΔCKV downregulation ratios). Multiple comparisons were addressed using q values, the P value analogue of the false discovery rate (FDR), which denotes the expected proportion of false positives among results deemed significant at a given P value threshold (55). For example, at a q value of ≤0.1, we expect 10% of identified associations to represent false positives. Statistical significance was defined as a P value of <0.05 (for univariate analyses) or a P value of <0.05 and q value of <0.1 (for analyses correcting for multiple hypothesis testing).
ACKNOWLEDGMENTS
We thank Xiaofei Jia, Yong Xiong, Masaru Yokoyama, and Hironori Sato for discussions, Richard Harrigan for access to clinical specimens, and Helen Byakwaga, Xiaomei Kuang, and Guinevere Lee for technical support.
This study was supported by a grant from the Japan Agency for Medical Research and Development (AMED; Research Program on HIV/AIDS), a JSPS KAKENHI Grant-in-Aid for Scientific Research and an AIDS International Collaborative Research Grant from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and Kumamoto University (to T.U.). M.T. received funding from a JSPS KAKENHI Grant-in-Aid for Scientific Research. F.M. is supported by a scholarship for The International Priority Graduate Programs, Advanced Graduate Courses for International Students (Doctoral Course), MEXT, Japan. F.K. received funding from the Deutsche Forschungsgemeinschaft and an Advanced ERC investigator grant. T.N. received funding from the South African Research Chairs Initiative of the National Research Foundation and was also supported by the Howard Hughes Medical Institute. J.K.M. received funding for this research from the National Research Foundation of South Africa; J.K.M., T.N., and M.A.B. also received support from the Canada-Sub Saharan Africa (CANSSA) HIV/AIDS Network through funding provided by the Global Health Research Initiative (GHRI) and the Canadian Institutes for Health Research (CIHR). Z.L.B. and M.A.B. received funding from the CIHR (HOP-115700 and PJT-148621). M.A.B. is a Tier II Canada Research Chair in Viral Pathogenesis and Immunity. Z.L.B. is supported by a Scholar Award from the Michael Smith Foundation for Health Research. The UARTO cohort is supported by grants from the National Institutes of Health (P30 AI027763, R01 MH054907, and UM1 CA181255 (to D.B. and J.N.M.)). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Fukami-Kobayashi K, Shiina T, Anzai T, Sano K, Yamazaki M, Inoko H, Tateno Y. 2005. Genomic evolution of MHC class I region in primates. Proc Natl Acad Sci U S A 102:9230–9234. doi: 10.1073/pnas.0500770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. 2004. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res 14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parham P. 1988. Function and polymorphism of human leukocyte antigen-A,B,C molecules. Am J Med 85:2–5. doi: 10.1016/0002-9343(88)90369-5. [DOI] [PubMed] [Google Scholar]
- 4.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol 68:6103–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker CE, Nightingale S, Taylor GP, Weber J, Bangham CR. 1994. Circulating anti-Tax cytotoxic T lymphocytes from human T-cell leukemia virus type I-infected people, with and without tropical spastic paraparesis, recognize multiple epitopes simultaneously. J Virol 68:2860–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddehase MJ, Mutter W, Munch K, Buhring HJ, Koszinowski UH. 1987. CD8-positive T lymphocytes specific for murine cytomegalovirus immediate-early antigens mediate protective immunity. J Virol 61:3102–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen JI. 1998. Infection of cells with varicella-zoster virus down-regulates surface expression of class I major histocompatibility complex antigens. J Infect Dis 177:1390–1393. doi: 10.1086/517821. [DOI] [PubMed] [Google Scholar]
- 9.Heeney JL, van Els C, de Vries P, ten Haaft P, Otting N, Koornstra W, Boes J, Dubbes R, Niphuis H, Dings M, Cranage M, Norley S, Jonker M, Bontrop RE, Osterhaus A. 1994. Major histocompatibility complex class I-associated vaccine protection from simian immunodeficiency virus-infected peripheral blood cells. J Exp Med 180:769–774. doi: 10.1084/jem.180.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 11.Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med 189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukamoto T, Dohki S, Ueno T, Kawada M, Takeda A, Yasunami M, Naruse T, Kimura A, Takiguchi M, Matano T. 2008. Determination of a major histocompatibility complex class I restricting simian immunodeficiency virus Gag241–249 epitope. AIDS 22:993–994. doi: 10.1097/QAD.0b013e3282f88d1b. [DOI] [PubMed] [Google Scholar]
- 13.Balla-Jhagjhoorsingh SS, Koopman G, Mooij P, Haaksma TG, Teeuwsen VJ, Bontrop RE, Heeney JL. 1999. Conserved CTL epitopes shared between HIV-infected human long-term survivors and chimpanzees. J Immunol 162:2308–2314. [PubMed] [Google Scholar]
- 14.Bihl F, Frahm N, Di Giammarino L, Sidney J, John M, Yusim K, Woodberry T, Sango K, Hewitt HS, Henry L, Linde CH, Chisholm JV III, Zaman TM, Pae E, Mallal S, Walker BD, Sette A, Korber BT, Heckerman D, Brander C. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol 176:4094–4101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- 15.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 16.Leslie A, Matthews PC, Listgarten J, Carlson JM, Kadie C, Ndung'u T, Brander C, Coovadia H, Walker BD, Heckerman D, Goulder PJ. 2010. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol 84:9879–9888. doi: 10.1128/JVI.00320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frater AJ, Brown H, Oxenius A, Gunthard HF, Hirschel B, Robinson N, Leslie AJ, Payne R, Crawford H, Prendergast A, Brander C, Kiepiela P, Walker BD, Goulder PJ, McLean A, Phillips RE. 2007. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J Virol 81:6742–6751. doi: 10.1128/JVI.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford H, Prado JG, Leslie A, Hué S, Honeyborne I, Reddy S, van der Stok M, Mncube Z, Brander C, Rousseau C, Mullins JI, Kaslow R, Goepfert P, Allen S, Hunter E, Mulenga J, Kiepiela P, Walker BD, Goulder PJR. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol 81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol 80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol 81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones TR, Wiertz EJ, Sun L, Fish KN, Nelson JA, Ploegh HL. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci U S A 93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park B, Kim Y, Shin J, Lee S, Cho K, Fruh K, Lee S, Ahn K. 2004. Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity 20:71–85. doi: 10.1016/S1074-7613(03)00355-8. [DOI] [PubMed] [Google Scholar]
- 23.Coscoy L, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci U S A 97:8051–8056. doi: 10.1073/pnas.140129797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hewitt EW, Duncan L, Mufti D, Baker J, Stevenson PG, Lehner PJ. 2002. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J 21:2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Specht A, DeGottardi MQ, Schindler M, Hahn B, Evans DT, Kirchhoff F. 2008. Selective downmodulation of HLA-A and -B by Nef alleles from different groups of primate lentiviruses. Virology 373:229–237. doi: 10.1016/j.virol.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 26.DeGottardi MQ, Specht A, Metcalf B, Kaur A, Kirchhoff F, Evans DT. 2008. Selective downregulation of rhesus macaque and sooty mangabey major histocompatibility complex class I molecules by Nef alleles of simian immunodeficiency virus and human immunodeficiency virus type 2. J Virol 82:3139–3146. doi: 10.1128/JVI.02102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams M, Roeth JF, Kasper MR, Fleis RI, Przybycin CG, Collins KL. 2002. Direct binding of human immunodeficiency virus type 1 Nef to the major histocompatibility complex class I (MHC-I) cytoplasmic tail disrupts MHC-I trafficking. J Virol 76:12173–12184. doi: 10.1128/JVI.76.23.12173-12184.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia X, Singh R, Homann S, Yang H, Guatelli J, Xiong Y. 2012. Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat Struct Mol Biol 19:701–706. doi: 10.1038/nsmb.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apps R, Del Prete GQ, Chatterjee P, Lara A, Brumme ZL, Brockman MA, Neil S, Pickering S, Schneider DK, Piechocka-Trocha A, Walker BD, Thomas R, Shaw GM, Hahn BH, Keele BF, Lifson JD, Carrington M. 2016. HIV-1 Vpu mediates HLA-C downregulation. Cell Host Microbe 19:686–695. doi: 10.1016/j.chom.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahiti M, Toyoda M, Jia X, Kuang XT, Mwimanzi F, Mwimanzi P, Walker BD, Xiong Y, Brumme ZL, Brockman MA, Ueno T. 2016. Relative resistance of HLA-B to downregulation by naturally occurring HIV-1 Nef sequences. mBio 7:e01516-16. doi: 10.1128/mBio.01516-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajapaksa US, Li D, Peng YC, McMichael AJ, Dong T, Xu XN. 2012. HLA-B may be more protective against HIV-1 than HLA-A because it resists negative regulatory factor (Nef) mediated down-regulation. Proc Natl Acad Sci U S A 109:13353–13358. doi: 10.1073/pnas.1204199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchhoff F, Easterbrook PJ, Douglas N, Troop M, Greenough TC, Weber J, Carl S, Sullivan JL, Daniels RS. 1999. Sequence variations in human immunodeficiency virus type 1 Nef are associated with different stages of disease. J Virol 73:5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis MJ, Balamurugan A, Ohno A, Kilpatrick S, Ng HL, Yang OO. 2008. Functional adaptation of Nef to the immune milieu of HIV-1 infection in vivo. J Immunol 180:4075–4081. doi: 10.4049/jimmunol.180.6.4075. [DOI] [PubMed] [Google Scholar]
- 34.Mahiti M, Brumme ZL, Jessen H, Brockman MA, Ueno T. 2015. Dynamic range of Nef-mediated evasion of HLA class II-restricted immune responses in early HIV-1 infection. Biochem Biophys Res Commun 463:248–254. doi: 10.1016/j.bbrc.2015.05.038. [DOI] [PubMed] [Google Scholar]
- 35.Mann JK, Byakwaga H, Kuang XT, Le AQ, Brumme CJ, Mwimanzi P, Omarjee S, Martin E, Lee GQ, Baraki B, Danroth R, McCloskey R, Muzoora C, Bangsberg DR, Hunt PW, Goulder PJ, Walker BD, Harrigan PR, Martin JN, Ndung'u T, Brockman MA, Brumme ZL. 2013. Ability of HIV-1 Nef to downregulate CD4 and HLA class I differs among viral subtypes. Retrovirology 10:100. doi: 10.1186/1742-4690-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mwimanzi P, Hasan Z, Hassan R, Suzu S, Takiguchi M, Ueno T. 2011. Effects of naturally arising HIV Nef mutations on cytotoxic T lymphocyte recognition and Nef's functionality in primary macrophages. Retrovirology 8:50. doi: 10.1186/1742-4690-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mwimanzi P, Markle TJ, Martin E, Ogata Y, Kuang XT, Tokunaga M, Mahiti M, Pereyra F, Miura T, Walker BD, Brumme ZL, Brockman MA, Ueno T. 2013. Attenuation of multiple Nef functions in HIV-1 elite controllers. Retrovirology 10:1. doi: 10.1186/1742-4690-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noviello CM, Pond SL, Lewis MJ, Richman DD, Pillai SK, Yang OO, Little SJ, Smith DM, Guatelli JC. 2007. Maintenance of Nef-mediated modulation of major histocompatibility complex class I and CD4 after sexual transmission of human immunodeficiency virus type 1. J Virol 81:4776–4786. doi: 10.1128/JVI.01793-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueno T, Motozono C, Dohki S, Mwimanzi P, Rauch S, Fackler OT, Oka S, Takiguchi M. 2008. CTL-mediated selective pressure influences dynamic evolution and pathogenic functions of HIV-1 Nef. J Immunol 180:1107–1116. doi: 10.4049/jimmunol.180.2.1107. [DOI] [PubMed] [Google Scholar]
- 40.Hemelaar J. 2013. Implications of HIV diversity for the HIV-1 pandemic. J Infect 66:391–400. doi: 10.1016/j.jinf.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 41.Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 42.Tomiyama H, Akari H, Adachi A, Takiguchi M. 2002. Different effects of Nef-mediated HLA class I down-regulation on human immunodeficiency virus type 1-specific CD8+ T-cell cytolytic activity and cytokine production. J Virol 76:7535–7543. doi: 10.1128/JVI.76.15.7535-7543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anmole G, Kuang XT, Toyoda M, Martin E, Shahid A, Le AQ, Markle T, Baraki B, Jones RB, Ostrowski MA, Ueno T, Brumme ZL, Brockman MA. 2015. A robust and scalable TCR-based reporter cell assay to measure HIV-1 Nef-mediated T cell immune evasion. J Immunol Methods 426:104–113. doi: 10.1016/j.jim.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Iijima S, Lee YJ, Ode H, Arold ST, Kimura N, Yokoyama M, Sato H, Tanaka Y, Strebel K, Akari H. 2012. A noncanonical mu-1A-binding motif in the N terminus of HIV-1 Nef determines its ability to downregulate major histocompatibility complex class I in T lymphocytes. J Virol 86:3944–3951. doi: 10.1128/JVI.06257-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, Kadie C, Bhattacharya T, Chui C, Szinger J, Mo T, Hogg RS, Montaner JS, Frahm N, Brander C, Walker BD, Harrigan PR. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog 3:e94. doi: 10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin TH, Peng J, Seese AM, Shapiro R, Frater J, Ndung'u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilembe W, Heckerman D, Goulder PJ, Allen TM, Allen S, Hunter E. 2014. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345:1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiguoya MW, Mann JK, Chopera D, Gounder K, Lee GQ, Hunt PW, Martin JN, Ball TB, Kimani J, Brumme ZL, Brockman MA, Ndung'u T. 2017. Subtype-specific differences in Gag-protease-driven replication capacity are consistent with intersubtype differences in HIV-1 disease progression. J Virol 91:e00253-17. doi: 10.1128/JVI.00253-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heigele A, Schindler M, Gnanadurai CW, Leonard JA, Collins KL, Kirchhoff F. 2012. Down-modulation of CD8αβ is a fundamental activity of primate lentiviral Nef proteins. J Virol 86:36–48. doi: 10.1128/JVI.00717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunt PW, Cao HL, Muzoora C, Ssewanyana I, Bennett J, Emenyonu N, Kembabazi A, Neilands TB, Bangsberg DR, Deeks SG, Martin JN. 2011. Impact of CD8+ T-cell activation on CD4+ T-cell recovery and mortality in HIV-infected Ugandans initiating antiretroviral therapy. AIDS 25:2123–2131. doi: 10.1097/QAD.0b013e32834c4ac1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, Swenson LC, Tao I, Szeto S, Rosato P, Sela J, Kadie CM, Frahm N, Brander C, Haas DW, Riddler SA, Haubrich R, Walker BD, Harrigan PR, Heckerman D, Mallal S. 2009. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One 4:e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mwimanzi P, Markle TJ, Ogata Y, Martin E, Tokunaga M, Mahiti M, Kuang XT, Walker BD, Brockman MA, Brumme ZL, Ueno T. 2013. Dynamic range of Nef functions in chronic HIV-1 infection. Virology 439:74–80. doi: 10.1016/j.virol.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 52.Itoh Y, Mizuki N, Shimada T, Azuma F, Itakura M, Kashiwase K, Kikkawa E, Kulski JK, Satake M, Inoko H. 2005. High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Immunogenetics 57:717–729. doi: 10.1007/s00251-005-0048-3. [DOI] [PubMed] [Google Scholar]
- 53.Toyoda M, Ogata Y, Mahiti M, Maeda Y, Kuang XT, Miura T, Jessen H, Walker BD, Brockman MA, Brumme ZL, Ueno T. 2015. Differential ability of primary HIV-1 Nef isolates to downregulate HIV-1 entry receptors. J Virol 89:9639–9652. doi: 10.1128/JVI.01548-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, Feeney ME, Rodriguez WR, Basgoz N, Draenert R, Stone DR, Brander C, Goulder PJ, Rosenberg ES, Altfeld M, Walker BD. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol 77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]