ABSTRACT
In prion diseases, an abnormal isoform of prion protein (PrPSc) accumulates in neurons, astrocytes, and microglia in the brains of animals affected by prions. Detailed analyses of PrPSc-positive neurons and glial cells are required to clarify their pathophysiological roles in the disease. Here, we report a novel method for the detection of PrPSc in neurons and glial cells from the brains of prion-infected mice by flow cytometry using PrPSc-specific staining with monoclonal antibody (MAb) 132. The combination of PrPSc staining and immunolabeling of neural cell markers clearly distinguished neurons, astrocytes, and microglia that were positive for PrPSc from those that were PrPSc negative. The flow cytometric analysis of PrPSc revealed the appearance of PrPSc-positive neurons, astrocytes, and microglia at 60 days after intracerebral prion inoculation, suggesting the presence of PrPSc in the glial cells, as well as in neurons, from an early stage of infection. Moreover, the kinetic analysis of PrPSc revealed a continuous increase in the proportion of PrPSc-positive cells for all cell types with disease progression. Finally, we applied this method to isolate neurons, astrocytes, and microglia positive for PrPSc from a prion-infected mouse brain by florescence-activated cell sorting. The method described here enables comprehensive analyses specific to PrPSc-positive neurons, astrocytes, and microglia that will contribute to the understanding of the pathophysiological roles of neurons and glial cells in PrPSc-associated pathogenesis.
IMPORTANCE Although formation of PrPSc in neurons is associated closely with neurodegeneration in prion diseases, the mechanism of neurodegeneration is not understood completely. On the other hand, recent studies proposed the important roles of glial cells in PrPSc-associated pathogenesis, such as the intracerebral spread of PrPSc and clearance of PrPSc from the brain. Despite the great need for detailed analyses of PrPSc-positive neurons and glial cells, methods available for cell type-specific analysis of PrPSc have been limited thus far to microscopic observations. Here, we have established a novel high-throughput method for flow cytometric detection of PrPSc in cells with more accurate quantitative performance. By applying this method, we succeeded in isolating PrPSc-positive cells from the prion-infected mouse brains via fluorescence-activated cell sorting. This allows us to perform further detailed analysis specific to PrPSc-positive neurons and glial cells for the clarification of pathological changes in neurons and pathophysiological roles of glial cells.
KEYWORDS: cell sorting, flow cytometry, prions
INTRODUCTION
Prion diseases are neurodegenerative disorders that are pathologically characterized by the activation of astrocytes and microglia, vacuolation of neurons and neuropils, and the accumulation of an abnormal isoform of prion protein (PrPSc) in the central nervous system (CNS) of humans and animals affected by prions. PrPSc is a major component of prions, the causative agent of prion diseases, and is generated from a cellular isoform of prion protein (PrPC) that is encoded by the Prnp gene of the host. Accumulation of PrPSc is found as a diffused or plaque pattern in neuropils, neurons, and astrocytes in the brains of rodent models for prion diseases or found as a pattern associated with neurons, astrocytes, microglia, and blood vessels in the brains of cattle, deer, and sheep affected with prions (1). Although the formation of PrPSc is considered to be associated closely with neurodegeneration (2–4), the mechanisms of neurodegeneration have not been elucidated fully at this time.
Previous studies have investigated the relationship between the formation of PrPSc and neurodegeneration (5–9). PrP-deficient mice were resistant to prion infection and did not develop neuropathological changes after prion inoculation (5). The transgenic mice expressing PrPC specifically in neurons were susceptible to prion infection and reproduced the neurodegeneration (6). Grafting the prion-infected brain tissues in the brain of PrP-deficient mice did not induce any degeneration in neurons of PrP-deficient mice, even though PrPSc in the grafts neighbored the neurons (7, 8). Furthermore, neuron-specific depletion of the Prnp gene by conditional targeting largely prevented neurodegeneration, even though PrPSc existed in glial cells and extracellular spaces in those mice (9). These reports indicate that neurodegeneration in prion diseases is associated closely with PrPSc formation in neurons.
Considering the findings that astrocytes and oligodendrocytes, as well as neurons, express PrPC (10), the formation of PrPSc in glial cells may contribute to neurodegeneration. The accumulation of PrPSc was found in astrocytes at an early stage of infection after intracerebral inoculation of prions (11), and neurodegeneration was reproduced in the transgenic mice expressing PrPC specifically in astrocytes (12). However, ultrastructural pathologies specific to prion diseases were not found in astrocytes but were in neurons adjacent to PrPSc on astrocytes or to extracellular PrPSc released from astrocytes, although PrPSc is generated from PrPC only in astrocytes of the transgenic mice (13). Oligodendrocytes have been reported as resistant to prion infection (14). Although Schwann cells have been reported as susceptible to prion infection (15), Schwann cells do not appear to be involved in the neurodegenerative process (16). It was reported that prions propagate in microglia isolated from PrPC-overexpressing mice (17) and that microglia isolated from CJD model mice possessed prion infectivity (18). However, the formation or the presence of PrPSc in microglia does not appear to be required for neurodegeneration (19). Taken together, these studies have shown the critical role of neuron-associated PrPSc in neurodegeneration rather than glial cell-associated PrPSc. In contrast, recent studies have proposed important roles for glial cells in PrPSc-associated pathogenesis. Glial cells could be involved in the intracerebral spread of prions (20, 21) and/or in the clearance of PrPSc from the brain (22). Therefore, detailed analyses of glial cells and neurons that contain PrPSc are required to clarify the pathophysiological roles of glial cells in prion diseases.
Until recently, methods available for the cell-type-specific analysis of PrPSc in the brain were largely limited to immunohistochemistry or immunoelectron microscopy using anti-PrP antibodies (1, 23, 24). Although these methods have contributed to the understanding of the cellular distribution and intracellular localization of PrPSc, assessments were made using semiquantitative measurements of the immunoreactivity of PrPSc and were based on observations by individual investigators, both of which are limiting factors for throughput and quantitative performance. Recently, methods for flow cytometric detection of misfolded proteins in cells were reported (25, 26). On the other hand, immunolabeling of markers for neural cells enabled us to analyze neurons (27) and glial cells (28) from brain tissues separately using flow cytometry. The combination of immunolabeling of misfolded proteins and neural cell markers is thought to be applicable to the cell-type-specific analysis of misfolded proteins by flow cytometry with high throughput and better quantitative performance. Previously, we reported the specific detection of PrPSc in an immunofluorescence assay (IFA) using anti-PrP monoclonal antibody (MAb) 132 (29–31). In the present study, we established a novel method for the detection of PrPSc in neurons and glial cells by flow cytometry using a combination of PrPSc-specific staining and immunolabeling of neural cell markers. We also showed the utility of this method for isolating PrPSc-positive cells in the brain.
RESULTS
Flow cytometric detection of PrPSc in prion-infected cells.
We previously reported that specific labeling of PrPSc with MAb 132 is applicable for immunocytochemistry on cells persistently infected with prions (31) and prion-infected primary neuronal cultures (30) after the fixed cells have been treated with guanidine thiocyanate (GdnSCN). We applied this method to detect PrPSc in a neuroblastoma cell line, Neuro2a (N2a-3), infected with prions by flow cytometry.
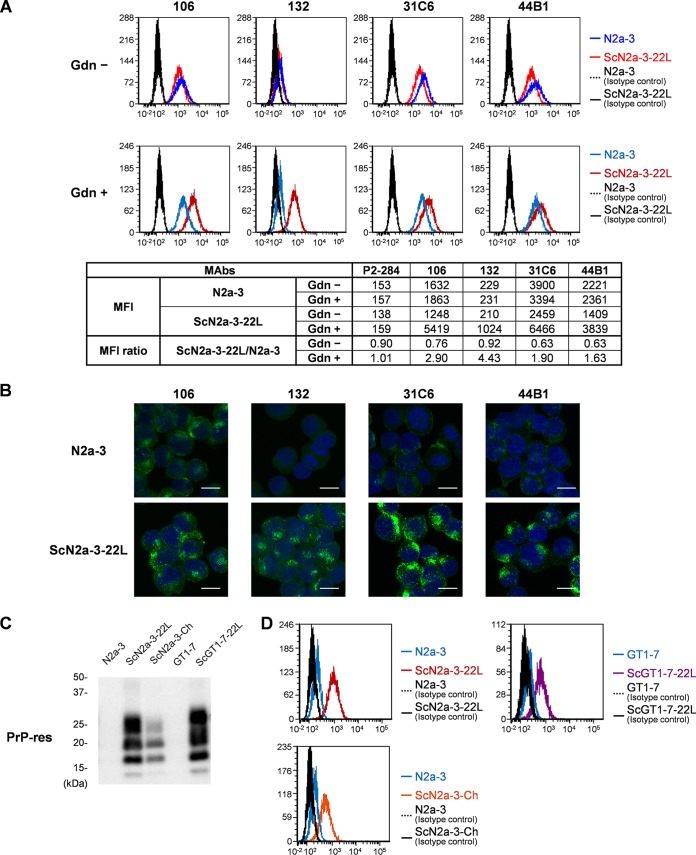
First, we compared the PrP signals between uninfected N2a-3 cells and N2a-3 cells persistently infected with a 22L prion strain (ScN2a-3-22L) using four anti-PrP antibodies with or without cell pretreatment with GdnSCN (Fig. 1A). Without GdnSCN pretreatment, PrP signals were detected by MAbs 106, 31C6, and 44B1 in both N2a-3 and ScN2a-3-22L cells, while PrP signals were hardly detected by MAb 132 (Fig. 1A, Gdn −). The GdnSCN pretreatment enhanced the signals of PrP in ScN2a-3-22L cells that were detected with MAb 132, and the mean fluorescence intensity (MFI) of PrP signals in ScN2a-3-22L cells was 4.4 times higher than those in N2a-3 cells (Fig. 1A, MFI ratio, Gdn +). The GdnSCN pretreatment enhanced PrP signals in ScN2a-3-22L cells by MAbs 106, 31C6, and 44B1 (Fig. 1A, Gdn +). The MFIs of PrP signals by these MAbs were much higher than that by MAb 132 (Fig. 1A). However, PrPC was clearly detected in uninfected N2a-3 cells under the same conditions, thus the MFI ratios of PrP signals for these MAbs were smaller than that obtained by MAb 132 (Fig. 1A, MFI ratio, Gdn +). Consistent with flow cytometric detection, microscopic observation with MAb 132 showed granular PrP signals in ScN2a-3-22L cells but faint or no signals in N2a-3 cells (Fig. 1B). The MAbs 106, 31C6, and 44B1 stained intense PrP signals in ScN2a-3-22L cells; however, PrP signals were also detected at the plasma membranes and perinuclear regions in uninfected N2a-3 cells. Therefore, not the intense PrP signals but a trace or lack of PrPC signals in uninfected cells by MAb 132 enabled us to distinguish PrPSc-positive cells from PrPSc-negative cells by flow cytometry.
FIG 1.
Detection of PrPSc in prion-infected cells by flow cytometry. (A) Comparison of PrPC detection in uninfected cells and PrP detection in infected cells with anti-PrP MAbs using flow cytometry. N2a-3 or ScN2a-3-22L cells were harvested with collagenase. The cells were fixed with 4% PFA in PBS and treated with 5 M GdnSCN (Gdn +) or remained untreated (Gdn −). The cells were stained with anti-PrP MAbs 106, 132, 31C6, and 44B1 and then subjected to flow cytometric analysis. In the histograms, red lines show the signals of ScN2a-3-22L cells, blue lines show signals of N2a-3 cells, and the black line and black dashed line show the signals of the infected and uninfected cells, respectively, stained with isotype control antibodies (P2-284). The table shows mean fluorescence intensity (MFI) of PrP signals detected by each MAb. MFI ratios of ScN2a3-22L to N2a-3 cells are also indicated. (B) Detection of PrP by fluorescence microscopy. N2a-3 and ScN2a-3-22L cells stained with the anti-PrP MAbs for flow cytometry were pelleted, and cell nuclei were counterstained with DAPI. The cells were mounted on a glass slide and analyzed by fluorescence microscopy. The PrP signals are shown as green and the cell nuclei are shown as blue (scale, 10 μm). (C) Detection of PrP-res by immunoblotting. N2a-3, ScN2a-3-22L, ScN2a-3-Ch, GT1-7, and ScGT1-7-22L cells were harvested with collagenase. The cell lysates were subjected to immunoblotting after PK treatment. (D) Flow cytometric detection of PrPSc. The cells harvested with collagenase were subjected to PrPSc-specific staining with MAb 132. The histograms show signals of PrP in N2a-3 (blue line) and ScN2a3-22L (red line) cells, PrP signals in N2a-3 (blue line) and ScN2a-3-Ch (orange line) cells, and PrP signals in GT1-7 (blue line) and ScGT1-7-22L (purple line) cells. The black lines and black dashed lines in the histograms show the signals of the isotype control antibody.
To confirm the utility of MAb 132 for the flow cytometric detection of PrPSc in different prion strains or cell lines, we analyzed PrP signals in N2a-3 cells persistently infected with the Chandler prion strain (ScN2a-3-Ch) and GT1-7 cells persistently infected with the 22L prion strain (ScGT1-7-22L). Consistent with the detection of proteinase K (PK)-resistant PrPSc (PrP-res) by immunoblotting (Fig. 1C), ScN2a-3-Ch and ScGT1-7-22L cells were obviously positive for MAb 132, but PrPC signals were hardly detected in the corresponding uninfected cells (Fig. 1D). These results indicated that PrPSc detection with MAb 132 using flow cytometry is applicable for different prion strains or cell lines.
Flow cytometric detection of PrPSc in neurons and glial cells from prion-infected mouse brain.
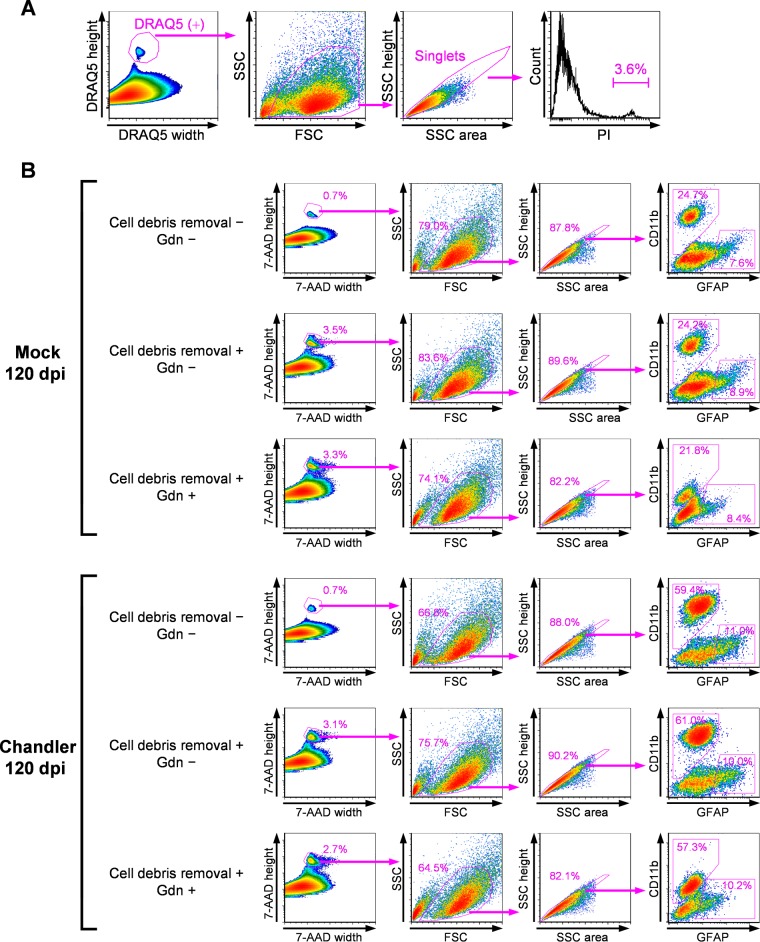
Using PrPSc-specific staining with MAb 132, we attempted to detect PrPSc in neurons and glial cells isolated from mouse brains using flow cytometry. Isolation of adult CNS neurons is problematic because of the tight adhesion of cell bodies and thousands of synapses (32), and mature neurons are damaged easily by enzymatic dissociation and mechanical trituration during the isolation step. These factors increase the chance for cell death and thus hamper subsequent analysis (33). We then evaluated the viability of cells prepared from mouse brain with papain-based enzymes and mechanical trituration. Cell suspensions contained a considerable amount of cell debris. Therefore, the cell nuclei were stained with DRAQ5, a membrane-permeable DNA probe, to distinguish cell bodies from cell debris. In flow cytometric analysis, we selected events corresponding to cell bodies by gating events positive for DRAQ5 (Fig. 2A, DRAQ5 width and DRAQ5 height). Single events were gated based on forward scatter (FSC) and side scatter (SSC) profiles (Fig. 2A, FSC and SSC as well as SSC area and SSC height). Finally, to screen the dead cell population, we analyzed signals of propidium iodide (PI), a DNA probe impermeable to live cell membranes. The proportion of dead cells was less than 3.6% in the mock-infected mouse brain, indicating that cell damage by dissociation could be minimized with our method.
FIG 2.
Method for flow cytometric analysis for neural cells from mouse brains. (A) Cell viability. Mock-infected mouse brain at 120 dpi was dissociated with a papain-based enzyme and mechanical trituration. The cell nuclei were counterstained with DRAQ5, and the dead cells were labeled with PI. Cell bodies positive for DRAQ5 were gated to remove cell debris (far left), and then the single events (singlets) were gated based on the profiles of FSC and SSC (second from the left) and the profiles of SSC area and SSC height (third from the left). The rightmost histogram shows PI signals. The percentage of PI-positive cells is indicated in the histogram. (B) Effects of cell debris removal by density gradient centrifugation and GdnSCN treatment on cell marker signals. Cell suspensions prepared from Chandler strain-infected mouse brains and mock-infected mouse brains at 120 dpi were fixed with 4% PFA. Cell debris was removed by density gradient centrifugation (cell debris removal +), or this step was omitted (cell debris removal −). The cells were subjected to GFAP and CD11b labeling and then fixed again with 4% PFA. The cells were treated with 2.5 M GdnSCN (Gdn +) or left untreated (Gdn −). Immediately before flow cytometric analysis, cell nuclei were counterstained with 7-AAD. In the analysis, 7-AAD-positive cell bodies were gated (far left), and single events were gated based on the profiles of FSC and SSC (second from the left) and the profiles of SSC area and SSC height (third from the left). The rightmost cytograms show GFAP and CD11b signals. Percentages of the gated cell populations against the parent populations are indicated.
For flow cytometric analysis of cells prepared from the brain, removal of cell debris from samples was required using density gradient centrifugation, because excess debris prolongs measurement duration for this method. In addition to this step, pretreatment with GdnSCN after fixation of the cells was also necessary for the specific staining of PrPSc with MAb 132. These procedures potentially could influence the efficiency of cell labeling to distinguish the cell population; therefore, we assessed the effects of cell debris removal and GdnSCN pretreatment on a proportion of CD11b-positive microglia and glial fibrillary acidic protein (GFAP)-positive astrocytes in the brains of mock-infected or Chandler strain-infected mice in the early clinical stage of prion infection at 120 days postinoculation (dpi) (Fig. 2B). In the flow cytometric analysis, single events corresponding to cell bodies were gated using profiles of nuclear staining with 7-aminoactinomycin D (7-AAD), FSC, and SSC, as described for Fig. 2A. CD11b labeling showed a clear population of CD11b-positive cells and also showed an increase in the proportion of CD11b-positive cells in the Chandler-infected mouse (59.4%) compared with the mock-infected mouse (24.7%) (Fig. 2B, cell debris removal −, far right column). Signal intensities of GFAP in the CD11b-negative cell population in both mock- and Chandler-infected mice ranged over five orders of magnitude (Fig. 2B, cell debris removal −, far right column). Here, we identified GFAP-positive cell populations as only the cells that had strong GFAP signals, as described in previous reports (28, 34). The removal of cell debris increased the proportion of cell bodies labeled with 7-AAD from 0.7% to 3.5% and from 0.7% to 3.1% in mock-infected and Chandler strain-infected mice, respectively. This procedure did not influence the labeling efficiencies of CD11b and GFAP or the proportion of CD11b- and GFAP-positive cells (Fig. 2B, cell debris removal − and +). However, the GdnSCN treatment evidently decreased the intensities of the CD11b and GFAP signals in both the mock- and Chandler-infected mice (Fig. 2B, Gdn − and +). Despite the decreased signal intensities, CD11b- and GFAP-positive cells could be distinguished from cells that were negative for those markers. These results suggest that the procedures did not physically affect the cell populations, even though cell marker signals were influenced.
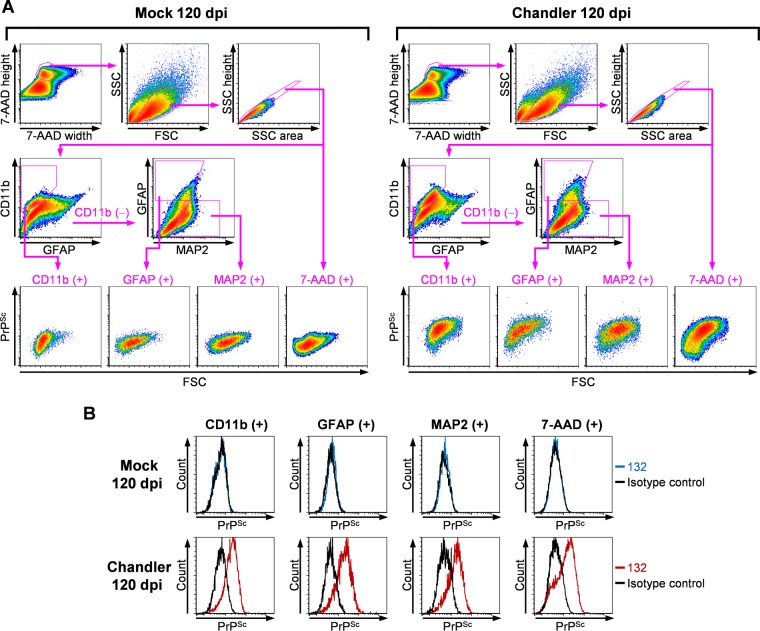
Since the PrPSc detection method using flow cytometry did not appear to affect the viability and composition of cell populations, we next performed a multiple staining procedure with neural cell markers, microtubule-associated protein 2 (MAP2) for neurons, GFAP for astrocytes, and CD11b for microglia, to analyze the PrPSc signals in each cell type using flow cytometry. In this analysis, cell bodies positive for 7-AAD and cells positive for CD11b, GFAP, and MAP2 were separated by the cell marker profiles, and the PrPSc signals from each cell population were analyzed (Fig. 3A). Clear PrP signals were detected in CD11b-positive, GFAP-positive, and MAP2-positive cells, as well as in 7-AAD-positive cell bodies in Chandler-infected samples, and faint or no PrP signals were detected in mock-infected samples (Fig. 3B). This demonstrated the specific detection of PrPSc in neurons, astrocytes, and microglia from the prion-infected mouse brains by flow cytometry.
FIG 3.
Flow cytometric detection of PrPSc in neurons, astrocytes, and microglia from prion-infected mouse brains. (A) Gating strategies for flow cytometric analysis. Cell suspensions were prepared from the mock-infected mouse brains (left) and Chandler strain-infected mouse brains (right) at 120 dpi. The cell suspensions were subjected to the staining of CD11b, GFAP, MAP2, and PrPSc; cell nuclei were stained with 7-AAD. For flow cytometric analysis, single-event gating was performed as shown in Fig. 2B. The single events were separated into CD11b-positive and CD11b-negative cell populations based on the profile of the CD11b signals. The GFAP-positive and MAP2-positive cell populations were gated based on the profiles of the MAP2 and GFAP signals of the CD11b-negative cell populations. PrPSc signals were analyzed in CD11b-positive, GFAP-positive, MAP2-positive, and 7-AAD-positive cell populations. The sequential gating strategy is shown by the magenta-colored polygonal boxes and arrows. (B) PrPSc signals in each cell population. The histograms show the PrPSc signals of CD11b-positive, GFAP-positive, MAP2-positive, and 7-AAD-positive cell populations. The blue and red lines show PrPSc signals in mock-infected samples (top column) and Chandler strain-infected samples (bottom column), respectively. The black lines show the signals labeled with isotype control antibody.
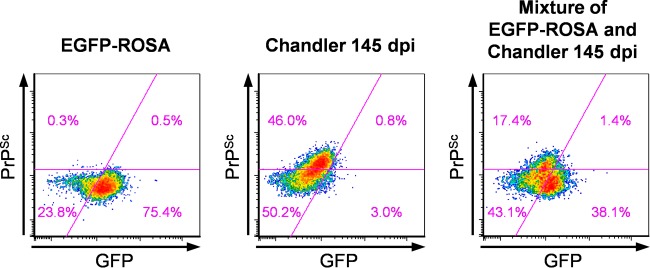
PrPSc is hydrophobic and, thus, more likely to bind to biomolecules nonspecifically. This property could result in false positives in flow cytometric analysis. We needed to exclude the possibility that PrPSc accumulating in the brain extracellular space was binding to cells during sample preparation, particularly during the formalin fixation step, thereby resulting in PrPSc-positive signals. Therefore, we mixed the brain of the Chandler-infected mouse at 145 dpi (terminal stage of the infection) with the brain of an uninfected ROSA26-enhanced green fluorescent protein (EGFP) mouse that ubiquitously expressed EGFP in many cell types (35), and we prepared cell suspensions from this mixture. We performed the entire procedure for the PrPSc-specific staining, including the fixation step (Fig. 4). If PrPSc binding to any cell types was occurring during sample preparation and staining, the PrPSc could be detected in the EGFP-positive cells. However, PrPSc signals were hardly detected in EGFP-positive cells but were clearly detected in EGFP-negative cells from the mixture of the infected and uninfected mouse brains. This result indicates that the detection of PrPSc signals was not attributed to the artifactual binding of PrPSc to cells but instead reflects the presence of PrPSc in cells in the brain.
FIG 4.
Extracellular PrPSc does not affect PrPSc detection. Cell suspensions were prepared at 145 dpi from the left brain hemisphere from an uninfected ROSA26-EGFP reporter mouse (left), from the left brain hemisphere from a Chandler strain-infected mouse (center), and from a mixture of the right brain hemispheres from ROSA26-EGFP and Chandler-infected mice (right). The cell suspensions were subjected to GFP staining with anti-GFP antibodies and BV421-conjugated secondary antibodies. After fixation, the cells subsequently were subjected to PrPSc staining with MAb 132-Af647. The cytograms show GFP and PrPSc signals. The magenta lines represent the threshold levels for GFP and PrPSc. Percentages of each population are indicated in quadrants in the cytograms.
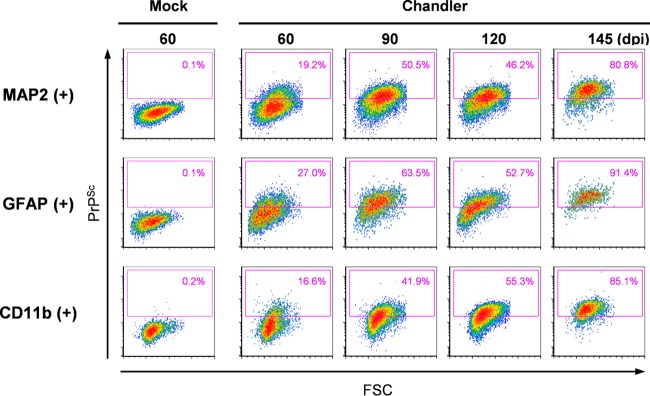
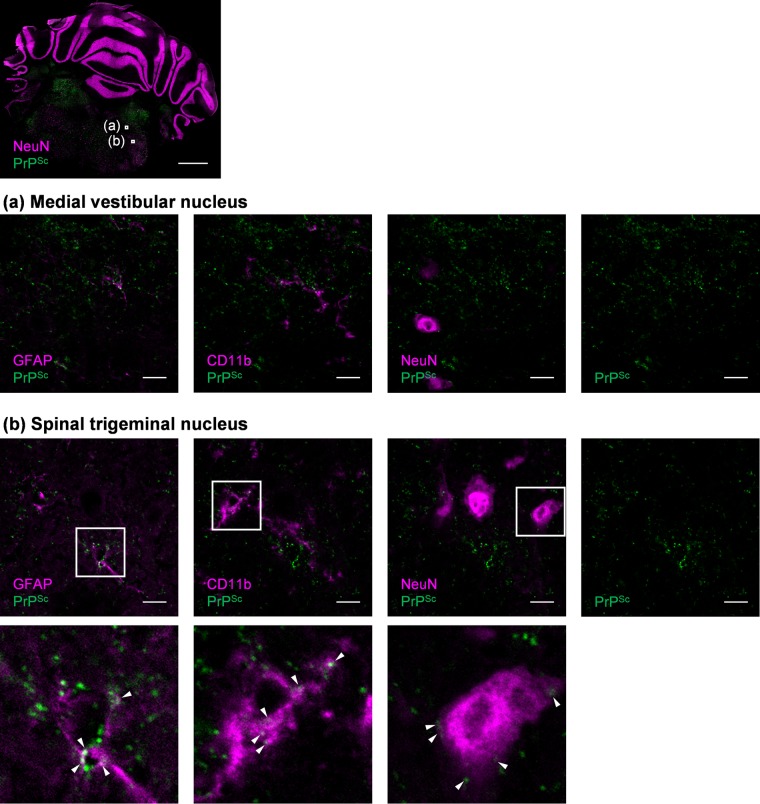
We next analyzed the kinetics of the proportions of PrPSc-positive cells both for neurons and glial cells. Originally, PrPSc was detected at 60 dpi in MAP2-positive, GFAP-positive, and CD11b-positive cells (Fig. 5, 60 dpi). In MAP2-positive neurons, 19.2% of cells were already positive for PrPSc at 60 dpi, and the percentages increased with time; 80.8% of cells were positive for PrPSc at the terminal stage of the disease (at 145 dpi) (Fig. 5, MAP2 +). Surprisingly, percentages of PrPSc-positive astrocytes and microglia were also high (Fig. 5, GFAP + and CD11b +). In GFAP-positive astrocytes, 27.0% of cells were positive for PrPSc at 60 dpi, and more than 90% of cells were positive for PrPSc at 145 dpi. The increase in PrPSc-positive microglia (CD11b-positive cells) was similar to that in MAP2-positive neurons: 16.6% to 85.1% from 60 to 145 dpi. These data suggest that PrPSc is present not only in neurons but also in astrocytes and microglia from the early to terminal stages of infection. To confirm the presence of PrPSc in these cells at the early stages of infection (60 dpi), we used multiple stains for PrPSc, GFAP, CD11b, and a neuronal marker, NeuN, on frozen sections of Chandler-infected mouse brain (Fig. 6). Although most of the PrPSc signals were distributed away from the signals of GFAP, CD11b, and NeuN (Fig. 6, representative image at medial vestibular nucleus), a portion of the PrPSc signals was colocalized either with GFAP, CD11b, or NeuN (Fig. 6, representative image at spinal trigeminal nucleus, arrowheads). This finding is consistent with the detection of PrPSc in astrocytes and microglia, as well as in neurons, using flow cytometry.
FIG 5.
Kinetics of PrPSc-positive neurons, astrocytes, and microglia from Chandler-infected mouse brains. Cell suspensions prepared from the brains of Chandler-infected mice at 60, 90, 120, and 145 dpi and from the brains of mock-infected mice at 60 dpi were subjected to flow cytometric analysis for CD11b, GFAP, MAP2, and PrPSc. Representative data from two independent experiments are shown. Gating strategies are shown in Fig. 3. The panels show the cytograms of FSC and PrPSc signals of MAP2-positive (top), GFAP-positive (middle), and CD11b-positive (bottom) cells. PrPSc-positive cell populations are shown in magenta boxes in the cytograms. The threshold level of PrPSc is the same in all samples. Percentages of PrPSc-positive cells are indicated in the cytograms.
FIG 6.
Detection of PrPSc in neurons, astrocytes, and microglia in a frozen section. The frozen brain section of the Chandler-infected mouse at 60 dpi was subjected to multiple staining of CD11b, GFAP, NeuN, and PrPSc. The images of each cell marker (magenta) and PrPSc (green) are shown. The top image is a merged image of NeuN and PrPSc on the coronal section containing cerebellum and pons (scale, 1 mm). The magnified image of the medial vestibular nucleus (a) and the spinal trigeminal nucleus (b) indicated by white boxes in the top image are shown below (scale, 10 μm). The panel shows merged images of GFAP and PrPSc (far left), CD11b and PrPSc (second from the left), and NeuN and PrPSc (second from the right); a single image of PrPSc is on the far right. The bottom panel shows increased magnifications of the boxed regions in the spinal trigeminal nucleus images. Colocalization of PrPSc and each individual cell marker is shown with arrowheads.
Isolation of PrPSc-positive cells from prion-infected mouse brain by fluorescence-activated cell sorting.
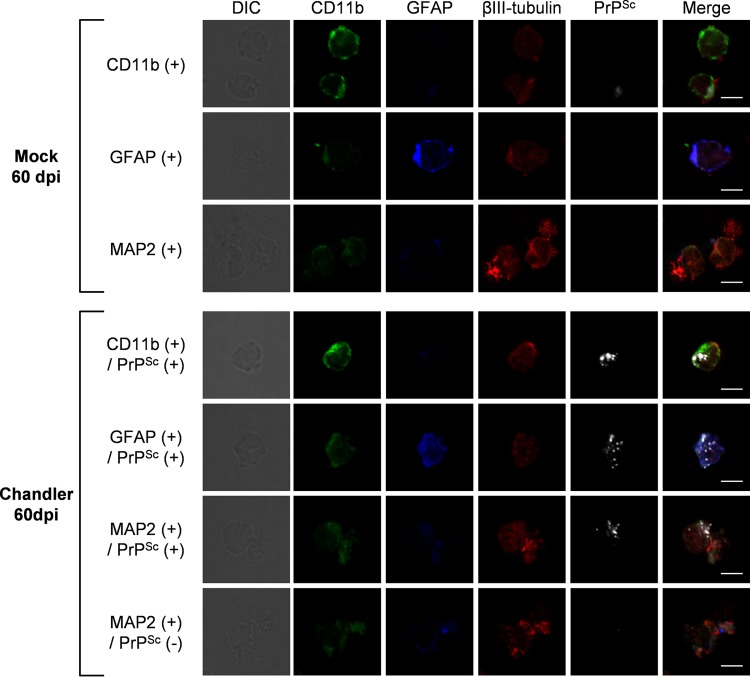
We attempted to apply the flow cytometric analysis of PrPSc to isolation of PrPSc-positive neurons and glial cells by fluorescence-activated cell sorting (FACS). From the brain of mouse infected with the Chandler strain at 60 dpi, cells were isolated that were simultaneously double positive for CD11b and PrPSc, double positive for GFAP and PrPSc, double positive for MAP2 and PrPSc, and positive for MAP2 but negative for PrPSc. As a negative control for the detection of PrPSc, CD11b-, GFAP-, and MAP2-positive cells were isolated from mock-infected mouse brain. Isolated cells were stained for βIII-tubulin to label neurons again and then analyzed by fluorescence microscopy. High-intensity granular stains specific for PrPSc were observed in cells that were sorted as PrPSc-positive and CD11b-, GFAP-, or MAP2-positive fractions. On the contrary, the granular PrPSc stains were hardly observed in cells that were sorted as a PrPSc-negative but MAP2-positive fraction from the brain of Chandler-infected mice or in cells from the brain of mock-infected mice (Fig. 7). These results demonstrate the utility of flow cytometric detection of PrPSc for isolating PrPSc-positive neurons and glial cells by FACS.
FIG 7.
Fluorescence-activated cell sorting of PrPSc-positive cells. Cell suspensions equivalent to one-fifth of a brain of a Chandler-infected mouse at 60 dpi and that of a mock-infected mouse at 60 dpi were subjected to multiple staining of CD11b, GFAP, MAP2, and PrPSc. Approximately 15,000 CD11b-positive cells, 8,500 GFAP-positive cells, and 13,000 MAP2-positive cells were isolated from the mock-infected mouse brain sample. Approximately 500 CD11b- and PrPSc-positive cells, 900 GFAP- and PrPSc-positive cells, 800 MAP2- and PrPSc-positive cells, and 19,000 MAP2-positive and PrPSc-negative cells were isolated from the Chandler-infected mouse brain sample using a cell sorter. Isolated cells subsequently were subjected to βIII-tubulin staining to label neurons. The panels show representative images of isolated cells. The far left column shows differential interference contrast cell images. The far right column shows the merged images of CD11b (green), GFAP (blue), βIII-tubulin (red), and PrPSc (white). The individual images of CD11b, GFAP, βIII-tubulin, and PrPSc are shown on the second to fifth column from the left (scale, 5 μm).
DISCUSSION
Detection of PrPSc by flow cytometry.
Despite a great need for a detailed analysis of PrPSc-positive neurons and glial cells, methods available for cell type-specific PrPSc analyses have been limited to microscopic observation thus far. In the present study, we showed PrPSc detection by flow cytometric analysis in neurons and glial cells using MAb 132 with high throughput and improved quantitative performance.
The immunolabeling of PrPSc using anti-PrP antibodies is the preferred method for the detection of PrPSc-positive cells. However, most of the antibodies against the PrP molecule cannot distinguish PrPSc from PrPC because they have the same primary structure. Some researchers have reported flow cytometric analysis using PrPSc-specific antibodies that can immunoprecipitate PrPSc selectively (36, 37). Paramithiotis et al. showed that PrPSc-specific signals labeled with MAb 9A4 were detected in dendritic cells isolated from lymph node of scrapie-infected sheep but not in those of uninfected sheep (36). Previously, we tried to detect PrPSc in prion-infected cells by flow cytometry using PrPSc-specific antibodies MAb 6H10 (38), 8D5, and 6A12 (39), but our attempts were unsuccessful (data not shown).
There are a few reports on flow cytometric analysis of cells that are positive for misfolded proteins (25, 26, 40, 41). In those reports, cells with misfolded proteins were distinguished from normal cells based on the aggregation or mislocalization of misfolded proteins. For example, monitoring an increase in the intensity of fluorescent reporter proteins that are fused with a target protein is widely used for detection of cells positive for misfolded proteins, such as the mutant superoxide dismutase-1, polyglutamine-expanded huntingtin, and mutant TAR DNA-binding protein 43 (26, 40, 42). However, in the case of prion protein, expression of PrPC fused to fluorescent proteins has been reported to disturb the conversion of PrPC to PrPSc (43); therefore, this method does not appear to be applicable to the detection of PrPSc. Ramdzan et al. reported the use of pulse-shape analysis (PulSA) to monitor the translocation of misfolded proteins into abnormal cellular puncta by flow cytometry (26). The PulSA method utilizes pulse width and height information of fluorescent signals to discriminate differences in the intracellular localization of signals. Therefore, the availability of this method depends on the change in the intracellular localization of signals between misfolded proteins and normal proteins. We previously reported that PrPSc was distributed widely to intracellular compartments, including early, late, and recycling endosomes, lysosomes, and perinuclear Golgi regions, as well as plasma membrane, in prion-infected cultured cells, while much of the PrPC localized at the plasma membrane (30, 31). Despite the differences in intracellular localization between PrPSc and PrPC, we did not succeed in discriminating between PrPSc-positive and PrPSc-negative cells using the PulSA method with anti-PrP antibody labeling (data not shown).
In a previous study, we showed that PrPSc staining with MAb 132 allowed for the detection of PrPSc by fluorescence microscopy without specific manipulations of the threshold settings or digestion of PrPC molecules (31). Here, we showed that discrimination of PrPSc-positive and PrPSc-negative cells by flow cytometry could be achieved with PrPSc-specific staining using MAb 132 after treating fixed cells with GdnSCN (Fig. 1A). Highly intense granular PrPSc signals in infected cells compared with PrPC signals in uninfected cells were clearly detected by microscopy using MAb 106, 31C6, and 44B1, just as with MAb 132 (Fig. 1B). However, antibodies other than MAb 132 could not be used to discriminate prion-infected cells from uninfected cells with the flow cytometry method (Fig. 1A). The difference between MAb 132 and other MAbs is a lack of PrPC signals from uninfected cells (Fig. 1A and B); thus, the lack of PrPC signals rather than the intense PrPSc signals is critical for the clear discrimination between PrPSc-positive cells and PrPSc-negative cells using the flow cytometry method. This conclusion is consistent with our recent study on the detection of PrPSc by cell-based enzyme-linked immunosorbent assay (ELISA); the lack of PrPC detection is also important for the discrimination of PrPSc signals from whole-cell cultures (44). Although the precise reason for the absence of PrPC detection with MAb 132 is not clear currently, that absence is the key factor that enabled the discrimination between PrPSc-positive and PrPSc-negative neurons and glial cells in prion-infected mouse brains.
Cell type-specific detection of PrPSc.
In this study, PrPSc was detected in astrocytes and microglia, as well as in neurons, from prion-infected mouse brains from the early to terminal stages of prion infection using flow cytometry (Fig. 5). For the detection of PrPSc in neurons and glial cells, cell debris was removed largely, and only the cell bodies positive for nuclear staining were used for the analysis (Fig. 3A). This may raise the technical problem that PrPSc in neuronal processes is largely excluded from the flow cytometric analysis. Previous reports suggest that PrPSc is present predominantly in neuropils, which are composed of the processes of neurons and glial cells, unlike neuronal cell bodies (23, 24, 45). Consistent with these reports, a large portion of PrPSc was distributed separately from the neuronal and glial markers, while a relatively small portion of PrPSc was detected by IFA in the cell bodies (Fig. 6). Nevertheless, we could detect PrPSc-positive cells from an early stage of prion infection (Fig. 5), and granular PrPSc signals were detected clearly by fluorescence microscopy in PrPSc-positive cells isolated from the prion-infected mouse brain at 60 dpi by FACS (Fig. 7). These results suggest that this flow cytometric detection of PrPSc is sensitive enough to discriminate between PrPSc-positive and -negative cells, even if only a small portion of the PrPSc in cell bodies is analyzed.
We showed that the proportion of PrPSc-positive neurons, astrocytes, and microglia increased with disease progression (Fig. 5), which may reflect the cell-to-cell spread of PrPSc in the brain. Previous studies have indicated that when prions are inoculated into peripheral tissues, PrPSc is transported from the periphery to the CNS and spreads intracerebrally via neuroanatomical pathways (46–48). Even after intracerebral inoculation of prions, PrPSc appears to spread via neural circuitry within the brain (21). In addition to neuron-to-neuron spread of PrPSc, recent studies also have suggested the important roles of glial cells in the spread of PrPSc between neurons and glial cells. Victoria et al. suggested that astrocytes are involved in the spread of PrPSc between neurons and astrocytes (20), while Zhu et al. suggested that microglia are involved in the clearance of PrPSc from the brain (22). In the current study, mice were intracerebrally inoculated with prions. Thus, it is possible that neurons, microglia, and astrocytes were exposed to inoculum and got infected independently in the early stage after inoculation. After the inoculation of prions via the peripheral route, prions enter CNS through sympathetic and parasympathetic nerves (49, 50); therefore, the entry port of CNS will be neurons. Thus, the kinetic analysis of PrPSc-positive cells in brain after the peripheral infection may address the neuron-to-glial cell spread of prions.
In this study, we succeeded in the isolation of PrPSc-positive neurons and glial cells from prion-infected mouse brain by FACS (Fig. 7). To our knowledge, this is the first study to specifically isolate neurons and glial cells positive for misfolded proteins from the brains of model animals for misfolding diseases, including prion diseases. Our method requires fixation of cells before the PrPSc-specific staining; thus, it is impossible to isolate viable cells. However, recent RNA-sequencing techniques have enabled transcriptome analysis using fixed and FACS-isolated neural cells (34, 51). The comprehensive analyses, including transcriptome analysis, on PrPSc-positive neurons and glial cells isolated by our method will allow a better understanding of the pathological changes that occur in neurons, as well as the roles of glial cells in the PrPSc-associated pathogenesis. Transcriptome analysis experiments specific to PrPSc-positive neurons currently are under way to clarify neuronal cell responses to prion infection.
MATERIALS AND METHODS
Antibodies.
Anti-PrP mouse MAbs 106, 132, 31C6, and 44B1 (52) were used for the detection of PrP by flow cytometry. MAb P2-284, against feline panleukopenia virus, was used as a negative-control antibody (53). The other commercially available primary and secondary antibodies are listed in Table 1. The antibodies were used for flow cytometry and IFA at the dilution rates shown in Table 1. A Zenon R-phycoerythrin (PE) rabbit IgG labeling kit (ThermoFisher Scientific) was used for direct staining of rabbit IgG. Alexa Fluor 647-conjugated MAb 132 (132-Af647) and MAb P2-284 (P2-284-Af647) were prepared by the following procedure. Alexa Fluor 647 succinimidyl ester (ThermoFisher Scientific) was reacted with MAb 132 (1 mg/ml) or MAb P2-284 (1 mg/ml) in phosphate-buffered saline (PBS) at a concentration of 50 μM at room temperature (RT) for 60 min. To inactivate the functional group of the succinimidyl ester, 10 volumes of 100 mM glycine in PBS were added to the reaction solutions. Excess reactive dye and glycine were removed by ultrafiltration using Amicon Ultracel-50k Ultra centrifugal filter units (Millipore). The Alexa Fluor-conjugated antibodies were dialyzed against PBS using Slide-A-Lyzer dialysis cassettes (10,000 molecular weight cutoff; ThermoFisher Scientific). The fluorescent dye/protein (F/P) molar ratios were calculated from absorbance at A280 and A650 for antibodies and Alexa Fluor 647, respectively, according to the manufacturer's instructions. The F/P molar ratios of the antibodies ranged between 1.5 and 2.5. To adsorb antibodies which nonspecifically react to the cells, 400 μg of Alexa Fluor 647-conjugated antibodies was incubated with 2 × 108 N2a-3 cells fixed with 4% paraformaldehyde (PFA) at a concentration of 10 μg/ml in PBS at RT for 2 h. The supernatant was collected after pelleting the cells by centrifugation at 10,000 × g for 10 min. The antibodies were concentrated by ultrafiltration using Amicon Ultracel-50k Ultra centrifugal filter units and stored at 4°C until use.
TABLE 1.
Sources of commercially available antibodies and the dilution rates for staining
| Antibody | Company | Code no. | Dilution |
|---|---|---|---|
| Primary antibodies | |||
| Anti-MAP2 rabbit monoclonal antibody, D5G1 | Cell Signaling Technology | 8707 | 1:400 for flow cytometry |
| Anti-βIII tubulin chicken polyclonal antibodies | Abcam | ab117716 | 1 μg/ml for immunofluorescence staining |
| Anti-GFAP rabbit polyclonal antibodies | Dako | D0344 | 2 μg/ml for flow cytometry |
| Anti-GFAP chicken polyclonal antibodies | Abcam | ab4674 | 5.4 μg/ml for immunofluorescence staining |
| Anti-CD11b rat monoclonal antibody, M1/70 | BioLegend | 101202 | 2.5 μg/ml for flow cytometry, 5 μg/ml for immunofluorescence staining |
| Anti-GFP rabbit polyclonal antibodies | ThermoFisher Scientific | A11122 | 5 μg/ml for flow cytometry |
| Alexa Fluor 488-conjugated anti-NeuN rabbit monoclonal antibody, EPR12763 | Abcam | ab190195 | 0.5 μg/ml for immunofluorescence staining |
| Secondary antibodies | |||
| Horseradish peroxidase-linked sheep F(ab′)2 fragment anti-mouse IgG | GE Healthcare | NA9310 | 1:5,000 for immunoblotting |
| Alexa Fluor 488-conjugated goat F(ab′)2 fragment anti-mouse IgG | ThermoFisher Scientific | A-11017 | 1 μg/ml for flow cytometry |
| Alexa Fluor 488-conjugated goat IgG anti-rat IgG | ThermoFisher Scientific | A-11006 | 2 μg/ml for flow cytometry |
| Alexa Fluor 555-conjugated goat IgG anti-rat IgG | ThermoFisher Scientific | A-21434 | 4 μg/ml for immunofluorescence staining |
| BV421-conjugated donkey IgG anti-rabbit IgG | BioLegend | 406410 | 1:400 for flow cytometry |
| Alexa Fluor 555-conjugated goat IgG anti-chicken IgG | ThermoFisher Scientific | A-21437 | 4 μg/ml for immunofluorescence staining |
| Alexa Fluor 405-conjugated goat IgG anti-chicken IgY | Abcam | ab175675 | 1 μg/ml for immunofluorescence staining |
Cell culture.
The following cell lines were used: N2a-3 cells, a subclone of the mouse neuroblastoma cell line Neuro2a (54); the hypothalamic neuronal cell lines GT1 and GT1-7 (55); N2a-3 cells persistently infected with the Chandler or 22L prion strain (ScN2a-3-Ch or ScN2a-3-22L); and GT1-7 cells persistently infected with the 22L prion strain (ScGT1-7-22L) (31).
Detection of PrP in cultured cells by flow cytometry.
Cells cultured in 100-mm dishes for 72 h were harvested by treatment with 0.1% collagenase (Wako) in PBS at RT for 5 min. The cells were pelleted by centrifugation at 300 × g at 4°C for 3 min and washed with cold PBS. For fixation, the cells were resuspended in 2 ml of 4% PFA in PBS and incubated at RT for 10 min. The fixation was terminated by adding 13 ml of 100 mM glycine in PBS containing 0.1% Tween 20 (PBST), and the samples were immediately centrifuged at 2,000 × g at 4°C for 10 min. The pellets were resuspended in 10 ml of 100 mM glycine in PBST and incubated for 10 min. After pelleting, the cells either were treated with 1 ml of 5 M GdnSCN at RT for 15 min or remained untreated. The cells were then washed twice with 10 ml of PBST. For blocking, the cells were incubated with PBST containing 5% fetal bovine serum (FBS) at RT for 30 min. The cells were split to V-bottom 96-well plates (ThermoFisher Scientific) at 1 × 105 cells/well and then pelleted by centrifugation. For the primary antibody reaction, the cells were incubated with 180 μl of anti-PrP MAbs (1 μg/ml) in PBST containing 1% FBS (FBS-PBST) at 4°C overnight. After the primary antibody reaction, the cells were washed three times with 180 μl of PBST. For the secondary antibody reaction, the samples were incubated with 180 μl of Alexa Fluor 488-conjugated anti-mouse IgG F(ab′)2 fragments (1 μg/ml) in FBS-PBST at 4°C for 60 min. After washing the cells with 180 μl of PBST twice and 180 μl of PBS once, the cells were analyzed by a flow cytometer (FACS Verse; equipped with 488-nm and 640-nm lasers; BD Bioscience). Ten thousand events were collected, using a forward scatter threshold of 10,000. Data from Alexa Fluor 488 were collected in pulse height, area, and width parameters. All flow cytometry data were analyzed with FACSuite software (BD Biosciences) and FCS 4 Express (De Novo Software).
Mice and prion strains.
All animal procedures were approved by the Institutional Animal Care and Use Committee of the Graduate School of Veterinary Medicine, Hokkaido University. Four-week-old female Jcl:ICR mice were purchased from CLEA Japan, and all mice were acclimatized for a week prior to use. The mice were intracerebrally inoculated with 20 μl of a 2.5% (wt/vol) brain homogenate from Jcl:ICR mice infected with the mouse-adapted scrapie strain, Chandler. Mice assigned to the mock-infected group were intracerebrally inoculated with 20 μl of a 2.5% (wt/vol) brain homogenate from age-matched uninfected Jcl:ICR mice, as described in the literature (56). ROSA 26-EGFP reporter mice of congenic strain B6.Cg-Tg(Gt(ROSA)26Sor-EGFP)I1Able/J were purchased from Jackson Laboratories and were maintained further by inbreeding.
Preparation of cell suspensions from brains.
The mice were perfused with prechilled modified Hanks' balanced salt solution without Ca2+ and Mg2+ (HBSS) containing KCl (400 mg/liter), KH2PO4 (60 mg/liter), NaHCO3 (350 mg/liter), NaCl (8 g/liter), Na2HPO4 (48 mg/liter), and d-glucose (1 g/liter). The brains were removed surgically from the mice and cut into small pieces in 5 ml of chilled HBSS. The brain pieces were dissociated enzymatically using the neural tissue dissociation kit (P) (Miltenyi Biotec), which contains papain as a protease, according to the manufacturer's instructions, with some modifications. After removal of HBSS by centrifugation at 300 × g at 4°C for 5 min, 1,950 μl of enzyme mix 1 was added to the brain pieces, and the samples were incubated for 15 min at 4°C under slow rotation. Thirty microliters of enzyme mix 2 was added, and the samples were incubated for 15 min at 4°C. The samples were gently triturated 10 times using a Pasteur pipette (Iwaki) and incubated for 15 min at 4°C. The samples again were triturated 10 times, transferred into 5 ml of HBSS in new 15-ml tubes, and then allowed to settle on ice for a few minutes. Five milliliters of the supernatants was filtered through 100-μm nylon mesh (BD Falcon) and collected in new tubes. The remaining sediments again were triturated with a Pasteur pipette 10 times, mixed with 5 ml of HBSS, and allowed to settle on ice for a few minutes. The supernatants then were filtered through 100-μm nylon mesh. This process was repeated and the flowthrough was combined. After centrifugation at 300 × g at 4°C for 10 min, the pellets were resuspended in 10 ml of HBSS. Cell suspensions were used to analyze dead cell populations and PrPSc by flow cytometry.
Flow cytometric analysis for dead cell populations.
Cell suspensions from brains were incubated for 5 min on ice in 200 μl of HBSS containing DRAQ5 (1 μM; Biostatus), a DNA probe that is permeable to living cells, and PI (10 μg/ml), a DNA probe that is not permeable to living cells. The samples were resuspended in 500 μl of chilled HBSS and analyzed by flow cytometer. The PI and DRAQ5 data were collected until the counts of DRAQ5-positive events reached 10,000.
Flow cytometric analysis of PrPSc in neural cells from prion-infected mouse brain.
Cell suspensions were prepared from mouse brains as described above. After pelleting the cell suspensions by centrifugation at 300 × g at 4°C for 10 min, the pellets were resuspended in 2 ml of 4% PFA and then incubated at 4°C for 10 min. This fixation step was terminated by adding 18 ml of 100 mM glycine in PBST, and the samples were immediately centrifuged at 2,000 × g at 4°C for 10 min. The pellets were resuspended in 5 ml of 100 mM glycine in PBST and incubated for 10 min. After pelleting, the cell suspensions were stored in 1 ml of PBST containing 40% FBS and 10% dimethyl sulfoxide at −30°C until use.
Before the immunolabeling of neural cell markers, the cells were thawed rapidly at 37°C, and 100 μl of the samples was transferred to new tubes. The cells were pelleted by centrifugation at 2,000 × g at 4°C for 10 min. For removal of cellular debris by density gradient centrifugation, the cells were resuspended in 300 μl of 15% iodixanol (OptiPrep, Axis-Shield) in PBST, and the cellular debris was floated by centrifugation at 2,000 × g at 4°C for 10 min. After removal of the floating cellular debris, this step was repeated once more. To block nonspecific binding of primary antibodies to cells, the cells were incubated with 180 μl of PBST containing mouse immunoglobulin G1, MAb P2-284 (10 μg/ml), and 5% FBS at 4°C for over 30 min. For staining of astrocytes and microglia, cells were incubated at 4°C overnight in 180 μl of primary antibody solution containing anti-GFAP rabbit polyclonal antibodies and anti-CD11b rat MAb in FBS-PBST. After the primary antibody reaction, cells were washed three times with 200 μl of PBST. The cells then were incubated at 4°C for 60 min in 180 μl of secondary antibody solution containing BV421-conjugated anti-rabbit IgG- and Alexa Fluor 488-conjugated anti-rat IgG. The cells were washed three times with PBST and fixed again with 100 μl of 4% PFA at 4°C for 10 min. The PFA was inactivated by adding 900 μl of 100 mM glycine in PBST following incubation with fresh 100 mM glycine for 10 min.
For PrPSc-specific staining, fixed cells were treated with 100 μl of 2.5 M GdnSCN at RT for 15 min. The GdnSCN was diluted by adding 900 μl of PBST, and the cells were pelleted. After washing with 180 μl of PBST, the cells were incubated at 4°C overnight in 180 μl of antibody solution containing MAb 132-Af647 (2 μg/ml) and Zenon-R-PE-conjugated anti-MAP2 antibodies in FBS-PBST for staining PrPSc and neurons. After two PBST washes, cells were washed once more with PBS. Cells then were resuspended in 100 μl of PBS containing 2.5 ng/ml 7-AAD (Beckman Coulter) for counter staining. The cells were resuspended in 500 μl of PBS and analyzed by a cell sorter (FACS Aria II equipped with 405-nm, 488-nm, and 640-nm lasers; BD Bioscience). The data from BV421, Alexa Fluor 488, R-PE, 7-AAD, and Alexa Fluor 647 were collected in pulse height, area, and width parameters until the counts of 7-AAD-positive events reached 50,000. All flow cytometry data were analyzed with FACS Diva software (BD Biosciences) and FCS 4 Express.
The threshold levels of CD11b and MAP2 were set using negative-control samples that were prepared with isotype control antibodies instead of the primary antibodies for CD11b and MAP2. The threshold level of GFAP was set as described in the literature (28, 34). The threshold level of PrPSc was set as the mean MAb 132-Af647 signals from 7-AAD-positive cell populations in mock-infected samples, plus 3-fold to 4-fold of the value of standard deviations.
FACS of PrPSc-positive cells.
Cell sorting was performed using a FACS Aria II equipped with an 85-μm nozzle. The PrPSc-positive cell populations were determined by setting the gate over the threshold levels of PrPSc, as described above. The PrPSc-negative cell populations were determined by setting the gate under the threshold levels of PrPSc so that the bottom 90% of cells in mock-infected samples could be included. For microscopic observations of sorted cells, the collected cells were pelleted and incubated in 50 μl of anti-βIII tubulin chicken antibodies in FBS-PBST at 4°C overnight to label neurons. The cells were washed three times with PBST and incubated with 50 μl of Alexa Fluor 555-conjugated anti-chicken secondary antibodies in FBS-PBST at 4°C for 90 min. After washing three times with PBST, the cells were dried on a 15-well multitest slide (MP Biomedicals) and then mounted with ProLong Gold antifade reagent (ThermoFisher Scientific). The fluorescent images of the samples were acquired using laser scanning confocal microscopy (LSM 700; Zeiss).
Immunoblotting.
Immunoblot detection of PrP-res was performed as previously described (54).
IFA.
The immunofluorescence staining of frozen sections was performed by a method described previously (29), with some modifications. Briefly, the brains were embedded in OCT compound (Sakura Finetek, Japan) and were cut at a thickness of 10 μm. The sections were fixed with 4% PFA for 10 min at RT. The sections were treated with 100 mM glycine in PBS at RT for 10 min and then incubated with blocking solution containing mouse IgG MAb P2-284 (10 μg/ml) and 5% FBS in PBS at RT for over 30 min. The sections then were incubated at 4°C overnight with anti-CD11b rat MAb in PBS containing 1% FBS (FBS-PBS). For the secondary antibody reaction, the sections were incubated at 4°C for 1 h with Alexa Fluor 488-conjugated anti-rat IgG in FBS-PBS. The sections were fixed again with 4% PFA at RT for 15 min and then treated with 2.5 M GdnSCN at RT for 15 min. After incubation with the blocking solution, the sections were incubated at 4°C overnight with MAb 132-Af647 (2 μg/ml), anti-GFAP chicken antibodies, and Alexa Fluor 488-conjugated anti-NeuN rabbit MAb in FBS-PBS. For the secondary antibody reaction, the sections were incubated at 4°C for 1 h with Alexa Fluor 405-conjugated anti-chicken antibodies in FBS-PBS. Finally, the sections were mounted with ProLong Gold antifade reagent. The fluorescent images were acquired using LSM700 and analyzed with ZEN software (Zeiss), as previously described (57, 58).
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Science Research (A) (JSPS KAKENHI grant number JP 15H02475), a Grant-in-Aid for Young Scientists (B) (JSPS KAKENHI grant number JP 17K14954), and a Grant-in-Aid for Challenging Exploratory Research (JSPS KAKENHI grant number JP 16K15033) and by a grant from the Program for Leading Graduate Schools (F01) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan. This work was also supported by grants for TSE research (H29-Shokuhin-Ippan-004) and Research on Measures for Intractable Diseases from the Ministry of Health, Labor and Welfare of Japan.
We thank Zensho Co., Ltd., for the use of the biosafety level 3 facility.
REFERENCES
- 1.Jeffrey M, McGovern G, Sisó S, González L. 2011. Cellular and sub-cellular pathology of animal prion diseases: relationship between morphological changes, accumulation of abnormal prion protein and clinical disease. Acta Neuropathol 121:113–134. doi: 10.1007/s00401-010-0700-3. [DOI] [PubMed] [Google Scholar]
- 2.Senesi M, Lewis V, Kim JH, Adlard PA, Finkelstein DI, Collins SJ. 2017. In vivo prion models and the disconnection between transmissibility and neurotoxicity. Ageing Res Rev 36:156–164. doi: 10.1016/j.arr.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Halliday M, Radford H, Mallucci GR. 2014. Prions: generation and spread versus neurotoxicity. J Biol Chem 289:19862–19868. doi: 10.1074/jbc.R114.568477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soto C, Satani N. 2011. The intricate mechanisms of neurodegeneration in prion diseases. Trends Mol Med 17:14–24. doi: 10.1016/j.molmed.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. 1993. Mice devoid of PrP are resistant to scrapie. Cell 73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 6.Race RE, Priola SA, Bessen RA, Ernst D, Dockter J, Rall GF, Mucke L, Chesebro B, Oldstone MB. 1995. Neuron-specific expression of a hamster prion protein minigene in transgenic mice induces susceptibility to hamster scrapie agent. Neuron 15:1183–1191. doi: 10.1016/0896-6273(95)90105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, Aguzzi A. 1996. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature 379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 8.Brandner S, Isenmann S, Kühne G, Aguzzi A. 1998. Identification of the end stage of scrapie using infected neural grafts. Brain Pathology (Zurich, Switzerland) 8:19–27. doi: 10.1111/j.1750-3639.1998.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallucci G, Dickinson A, Linehan J, Klöhn P-C, Brandner S, Collinge J. 2003. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science (New York, NY) 302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 10.Moser M, Colello RJ, Pott U, Oesch B. 1995. Developmental expression of the prion protein gene in glial cells. Neuron 14:509–517. doi: 10.1016/0896-6273(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 11.Diedrich JF, Bendheim PE, Kim YS, Carp RI, Haase AT. 1991. Scrapie-associated prion protein accumulates in astrocytes during scrapie infection. Proc Natl Acad Sci U S A 88:375–379. doi: 10.1073/pnas.88.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raeber AJ, Race RE, Brandner S, Priola SA, Sailer A, Bessen RA, Mucke L, Manson J, Aguzzi A, Oldstone MB, Weissmann C, Chesebro B. 1997. Astrocyte-specific expression of hamster prion protein (PrP) renders PrP knockout mice susceptible to hamster scrapie. EMBO J 16:6057–6065. doi: 10.1093/emboj/16.20.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffrey M, Goodsir CM, Race RE, Chesebro B. 2004. Scrapie-specific neuronal lesions are independent of neuronal PrP expression. Ann Neurol 55:781–792. doi: 10.1002/ana.20093. [DOI] [PubMed] [Google Scholar]
- 14.Prinz M, Montrasio F, Furukawa H, van der Haar ME, Schwarz P, Rulicke T, Giger OT, Hausler KG, Perez D, Glatzel M, Aguzzi A. 2004. Intrinsic resistance of oligodendrocytes to prion infection. J Neurosci 24:5974–5981. doi: 10.1523/JNEUROSCI.0122-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Follet J, Lemaire-Vieille C, Blanquet-Grossard F, Podevin-Dimster V, Lehmann S, Chauvin JP, Decavel JP, Varea R, Grassi J, Fontes M, Cesbron JY. 2002. PrP expression and replication by Schwann cells: implications in prion spreading. J Virol 76:2434–2439. doi: 10.1128/jvi.76.5.2434-2439.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halliez S, Chesnais N, Mallucci G, Vilotte M, Langevin C, Jaumain E, Laude H, Vilotte JL, Beringue V. 2013. Targeted knock-down of cellular prion protein expression in myelinating Schwann cells does not alter mouse prion pathogenesis. J Gen Virol 94:1435–1440. doi: 10.1099/vir.0.049619-0. [DOI] [PubMed] [Google Scholar]
- 17.Iwamaru Y, Takenouchi T, Ogihara K, Hoshino M, Takata M, Imamura M, Tagawa Y, Hayashi-Kato H, Ushiki-Kaku Y, Shimizu Y, Okada H, Shinagawa M, Kitani H, Yokoyama T. 2007. Microglial cell line established from prion protein-overexpressing mice is susceptible to various murine prion strains. J Virol 81:1524–1527. doi: 10.1128/JVI.01379-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker CA, Martin D, Manuelidis L. 2002. Microglia from Creutzfeldt-Jakob disease-infected brains are infectious and show specific mRNA activation profiles. J Virol 76:10905–10913. doi: 10.1128/JVI.76.21.10905-10913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Priller J, Prinz M, Heikenwalder M, Zeller N, Schwarz P, Heppner FL, Aguzzi A. 2006. Early and rapid engraftment of bone marrow-derived microglia in scrapie. J Neurosci 26:11753–11762. doi: 10.1523/JNEUROSCI.2275-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Victoria GS, Arkhipenko A, Zhu S, Syan S, Zurzolo C. 2016. Astrocyte-to-neuron intercellular prion transfer is mediated by cell-cell contact. Sci Rep 6:20762. doi: 10.1038/srep20762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll JA, Striebel JF, Rangel A, Woods T, Phillips K, Peterson KE, Race B, Chesebro B. 2016. Prion strain differences in accumulation of PrPSc on neurons and glia are associated with similar expression profiles of neuroinflammatory genes: comparison of three prion strains. PLoS Pathog 12:e1005551. doi: 10.1371/journal.ppat.1005551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu C, Herrmann US, Falsig J, Abakumova I, Nuvolone M, Schwarz P, Frauenknecht K, Rushing EJ, Aguzzi A. 2016. A neuroprotective role for microglia in prion diseases. J Exp Med 213:1047–1059. doi: 10.1084/jem.20151000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayers JI, Schutt CR, Shikiya RA, Aguzzi A, Kincaid AE, Bartz JC. 2011. The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Pathog 7:e1001317. doi: 10.1371/journal.ppat.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godsave SF, Wille H, Kujala P, Latawiec D, DeArmond SJ, Serban A, Prusiner SB, Peters PJ. 2008. Cryo-immunogold electron microscopy for prions: toward identification of a conversion site. J Neurosci 28:12489–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen D, Coleman J, Chan E, Nicholson TP, Dai L, Sheppard PW, Patton WF. 2011. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem Biophys 60:173–185. doi: 10.1007/s12013-010-9138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramdzan YM, Polling S, Chia CP, Ng IH, Ormsby AR, Croft NP, Purcell AW, Bogoyevitch MA, Ng DC, Gleeson PA, Hatters DM. 2012. Tracking protein aggregation and mislocalization in cells with flow cytometry. Nat Methods 9:467–470. doi: 10.1038/nmeth.1930. [DOI] [PubMed] [Google Scholar]
- 27.Guez-Barber D, Fanous S, Harvey BK, Zhang Y, Lehrmann E, Becker KG, Picciotto MR, Hope BT. 2012. FACS purification of immunolabeled cell types from adult rat brain. J Neurosci Methods 203:10–18. doi: 10.1016/j.jneumeth.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith SM, Kimyon RS, Watters JJ. 2014. Cell-type-specific Jumonji histone demethylase gene expression in the healthy rat CNS: detection by a novel flow cytometry method. ASN Neuro 6:193–207. doi: 10.1042/AN20130050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai K, Hasebe R, Takahashi Y, Song CH, Suzuki A, Yamasaki T, Horiuchi M. 2013. Absence of CD14 delays progression of prion diseases accompanied by increased microglial activation. J Virol 87:13433–13445. doi: 10.1128/JVI.02072-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka M, Fujiwara A, Suzuki A, Yamasaki T, Hasebe R, Masujin K, Horiuchi M. 2016. Comparison of abnormal isoform of prion protein in prion-infected cell lines and primary-cultured neurons by PrPSc-specific immunostaining. J Gen Virol 97:2030–2042. doi: 10.1099/jgv.0.000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamasaki T, Suzuki A, Shimizu T, Watarai M, Hasebe R, Horiuchi M. 2012. Characterization of intracellular localization of PrP(Sc) in prion-infected cells using a mAb that recognizes the region consisting of aa 119-127 of mouse PrP. J Gen Virol 93:668–680. doi: 10.1099/vir.0.037101-0. [DOI] [PubMed] [Google Scholar]
- 32.Brewer GJ, Torricelli JR. 2007. Isolation and culture of adult neurons and neurospheres. Nat Protoc 2:1490–1498. doi: 10.1038/nprot.2007.207. [DOI] [PubMed] [Google Scholar]
- 33.Saxena A, Wagatsuma A, Noro Y, Kuji T, Asaka-Oba A, Watahiki A, Gurnot C, Fagiolini M, Hensch TK, Carninci P. 2012. Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. Biotechniques 52:381–385. doi: 10.2144/0000113878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivasan K, Friedman BA, Larson JL, Lauffer BE, Goldstein LD, Appling LL, Borneo J, Poon C, Ho T, Cai F, Steiner P, van der Brug MP, Modrusan Z, Kaminker JS, Hansen DV. 2016. Untangling the brain's neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun 7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giel-Moloney M, Krause DS, Chen G, Van Etten RA, Leiter AB. 2007. Ubiquitous and uniform in vivo fluorescence in ROSA26-EGFP BAC transgenic mice. Genesis 45:83–89. doi: 10.1002/dvg.20269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paramithiotis E, Pinard M, Lawton T, LaBoissiere S, Leathers VL, Zou W-Q, Estey LA, Lamontagne J, Lehto MT, Kondejewski LH, Francoeur GP, Papadopoulos M, Haghighat A, Spatz SJ, Head M, Will R, Ironside J, O'Rourke K, Tonelli Q, Ledebur HC, Chakrabartty A, Cashman NR. 2003. A prion protein epitope selective for the pathologically misfolded conformation. Nat Med 9:893–899. doi: 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- 37.Tayebi M, Jones DR, Taylor WA, Stileman BF, Chapman C, Zhao D, David M. 2011. PrP(Sc)-specific antibodies with the ability to immunodetect prion oligomers. PLoS One 6:e19998. doi: 10.1371/journal.pone.0019998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horiuchi M, Karino A, Furuoka H, Ishiguro N, Kimura K, Shinagawa M. 2009. Generation of monoclonal antibody that distinguishes PrPSc from PrPC and neutralizes prion infectivity. Virology 394:200–207. doi: 10.1016/j.virol.2009.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Masujin K, Kaku-Ushiki Y, Miwa R, Okada H, Shimizu Y, Kasai K, Matsuura Y, Yokoyama T. 2013. The N-terminal sequence of prion protein consists an epitope specific to the abnormal isoform of prion protein (PrP(Sc)). PLoS One 8:e58013. doi: 10.1371/journal.pone.0058013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whiten DR, San Gil R, McAlary L, Yerbury JJ, Ecroyd H, Wilson MR. 2016. Rapid flow cytometric measurement of protein inclusions and nuclear trafficking. Sci Rep 6:31138. doi: 10.1038/srep31138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olzscha H, Fedorov O, Kessler BM, Knapp S, La Thangue NB. 2017. CBP/p300 bromodomains regulate amyloid-like protein aggregation upon aberrant lysine acetylation. Cell Chem Biol 24:9–23. doi: 10.1016/j.chembiol.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herrera F, Tenreiro S, Miller-Fleming L, Outeiro TF. 2011. Visualization of cell-to-cell transmission of mutant huntingtin oligomers. PLoS Curr 3:Rrn1210. doi: 10.1371/currents.RRN1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barmada SJ, Harris DA. 2005. Visualization of prion infection in transgenic mice expressing green fluorescent protein-tagged prion protein. J Neurosci 25:5824–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shan Z, Yamasaki T, Suzuki A, Hasebe R, Horiuchi M. 2016. Establishment of a simple cell-based ELISA for the direct detection of abnormal isoform of prion protein from prion-infected cells without cell lysis and proteinase K treatment. Prion 10:305–318. doi: 10.1080/19336896.2016.1189053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godsave SF, Wille H, Pierson J, Prusiner SB, Peters PJ. 2013. Plasma membrane invaginations containing clusters of full-length PrP(Sc) are an early form of prion-associated neuropathology in vivo. Neurobiol Aging 34:1621–1631. doi: 10.1016/j.neurobiolaging.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayers JI, Kincaid AE, Bartz JC. 2009. Prion strain targeting independent of strain-specific neuronal tropism. J Virol 83:81–87. doi: 10.1128/JVI.01745-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride PA, Schulz-Schaeffer WJ, Donaldson M, Bruce M, Diringer H, Kretzschmar HA, Beekes M. 2001. Early spread of scrapie from the gastrointestinal tract to the central nervous system involves autonomic fibers of the splanchnic and vagus nerves. J Virol 75:9320–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimberlin RH, Walker CA. 1982. Pathogenesis of mouse scrapie: patterns of agent replication in different parts of the CNS following intraperitoneal infection. J R Soc Med 75:618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Keulen LJ, Vromans ME, van Zijderveld FG. 2002. Early and late pathogenesis of natural scrapie infection in sheep. APMIS 110:23–32. [DOI] [PubMed] [Google Scholar]
- 50.Kaatz M, Fast C, Ziegler U, Balkema-Buschmann A, Hammerschmidt B, Keller M, Oelschlegel A, McIntyre L, Groschup MH. 2012. Spread of classic BSE prions from the gut via the peripheral nervous system to the brain. Am J Pathol 181:515–524. doi: 10.1016/j.ajpath.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Molyneaux BJ, Goff LA, Brettler AC, Chen HH, Brown JR, Hrvatin S, Rinn JL, Arlotta P. 2015. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim C-L, Umetani A, Matsui T, Ishiguro N, Shinagawa M, Horiuchi M. 2004. Antigenic characterization of an abnormal isoform of prion protein using a new diverse panel of monoclonal antibodies. Virology 320:40–51. doi: 10.1016/j.virol.2003.10.026. [DOI] [PubMed] [Google Scholar]
- 53.Horiuchi M, Mochizuki M, Ishiguro N, Nagasawa H, Shinagawa M. 1997. Epitope mapping of a monoclonal antibody specific to feline panleukopenia virus and mink enteritis virus. J Vet Med Sci 59:133–136. doi: 10.1292/jvms.59.133. [DOI] [PubMed] [Google Scholar]
- 54.Uryu M, Karino A, Kamihara Y, Horiuchi M. 2007. Characterization of prion susceptibility in Neuro2a mouse neuroblastoma cell subclones. Microbiol Immunol 51:661–669. doi: 10.1111/j.1348-0421.2007.tb03954.x. [DOI] [PubMed] [Google Scholar]
- 55.Schätzl HM, Laszlo L, Holtzman DM, Tatzelt J, DeArmond SJ, Weiner RI, Mobley WC, Prusiner SB. 1997. A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J Virol 71:8821–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song CH, Honmou O, Ohsawa N, Nakamura K, Hamada H, Furuoka H, Hasebe R, Horiuchi M. 2009. Effect of transplantation of bone marrow-derived mesenchymal stem cells on mice infected with prions. J Virol 83:5918–5927. doi: 10.1128/JVI.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamasaki T, Baron GS, Suzuki A, Hasebe R, Horiuchi M. 2014. Characterization of intracellular dynamics of inoculated PrP-res and newly generated PrP(Sc) during early stage prion infection in Neuro2a cells. Virology 450-451: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamasaki T, Suzuki A, Hasebe R, Horiuchi M. 2014. Comparison of the anti-prion mechanism of four different anti-prion compounds, anti-PrP monoclonal antibody 44B1, pentosan polysulfate, chlorpromazine, and U18666A, in prion-infected mouse neuroblastoma cells. PLoS One 9:e106516. doi: 10.1371/journal.pone.0106516. [DOI] [PMC free article] [PubMed] [Google Scholar]