ABSTRACT
Most segmented negative-sense RNA viruses employ a process termed cap snatching, during which they snatch capped RNA leaders from host cellular mRNAs and use the snatched leaders as primers for transcription, leading to the synthesis of viral mRNAs with 5′ heterogeneous sequences (HSs). With traditional methods, only a few HSs can be determined, and identification of their donors is difficult. Here, the mRNA 5′ ends of Rice stripe tenuivirus (RSV) and Rice grassy stunt tenuivirus (RGSV) and those of their host rice were determined by high-throughput sequencing. Millions of tenuiviral HSs were obtained, and a large number of them mapped to the 5′ ends of corresponding host cellular mRNAs. Repeats of the dinucleotide AC, which are complementary to the U1G2 of the tenuiviral template 3′-U1G2U3G4UUUCG, were found to be prevalent at the 3′ termini of tenuiviral HSs. Most of these ACs did not match host cellular mRNAs, supporting the idea that tenuiviruses use the prime-and-realign mechanism during cap snatching. We previously reported a greater tendency of RSV than RGSV to use the prime-and-realign mechanism in transcription with leaders cap snatched from a coinfecting reovirus. Besides confirming this observation in natural tenuiviral infections, the data here additionally reveal that RSV has a greater tendency to use this mechanism in transcribing genomic than in transcribing antigenomic templates. The data also suggest that tenuiviruses cap snatch host cellular mRNAs from translation- and photosynthesis-related genes, and capped RNA leaders snatched by tenuiviruses base pair with U1/U3 or G2/G4 of viral templates. These results provide unprecedented insights into the cap-snatching process of tenuiviruses.
IMPORTANCE Many segmented negative-sense RNA viruses (segmented NSVs) are medically or agriculturally important pathogens. The cap-snatching process is a promising target for the development of antiviral strategies against this group of viruses. However, many details of this process remain poorly characterized. Tenuiviruses constitute a genus of agriculturally important segmented NSVs, several members of which are major viral pathogens of rice. Here, we for the first time adopted a high-throughput sequencing strategy to determine the 5′ heterogeneous sequences (HSs) of tenuiviruses and mapped them to host cellular mRNAs. Besides providing deep insights into the cap snatching of tenuiviruses, the data obtained provide clear evidence to support several previously proposed models regarding cap snatching. Curiously and importantly, the data here reveal that not only different tenuiviruses but also the same tenuivirus synthesizing different mRNAs use the prime-and-realign mechanism with different tendencies during their cap snatching.
KEYWORDS: cap snatching, tenuivirus, base pairing, prime-and-realign mechanism
INTRODUCTION
Eukaryotic organisms cotranscriptionally attach a 7-methylguanosine (m7G) cap structure to the 5′ termini of their mRNAs (1). The cap structure plays important roles in the stability, localization, and translation of mRNAs (2–4). With some exceptions in which cap-independent translation strategies have been developed, viruses also cap their mRNAs (5). Whereas most viruses do so by encoding their own capping systems or by transcribing their genomes with host polymerases, segmented negative-sense RNA viruses (segmented NSVs) of the order Bunyavirales and the families Orthomyxoviridae and Arenaviridae use a unique mechanism termed cap snatching to obtain a cap for their mRNAs (5, 6). During the process of cap snatching, segmented NSVs snatch capped RNA leaders from host mRNAs and use the snatched leaders as primers to transcribe their template RNAs, leading to the synthesis of viral mRNAs with host-derived 5′ heterogeneous sequences (HSs) (5).
The cap-snatching machinery has been studied in detail for the orthomyxovirus influenza virus, a nucleus-replicating segmented NSV. Influenza virus has a transcriptase composed of three subunits, polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2), and polymerase acidic protein (PA). During the process of cap snatching, PB2 binds to the cap structure of a host cellular mRNA, PA cleaves the mRNA at a position 10 to 15 nucleotides (nt) downstream of the cap, and PB1 uses the resulting capped RNA leader as a primer to transcribe viral genome template RNAs (7, 8). Recently, crystal structures of the heterotrimeric polymerases of influenza A and B viruses have been determined, allowing the proposition of an integrated structural model for the concerted action of PB2, PA, and PB1 (9, 10). Segmented NSVs of other families have a monomeric transcriptase and replicate in the cytoplasm of host cells. Although the position of the cap-binding domains remains elusive, an endonuclease activity similar to that of PA has been found from an N-terminal fragment of their transcriptases (11–14).
Despite the above achievements, how a snatched capped RNA leader primes transcription initiation is not clear. One model states that the capped RNA leader base pairs with at least one of the first several nucleotides of the viral template by using its 3′-terminal residues. To date, this base-pairing model has been supported by studies on viruses belonging to the genera Orthomyxovirus, Tospovirus, and Hantavirus (15–19). Importantly, competition experiments have shown a preferential recruitment of capped RNA leaders harboring a multiple-base complementarity to the 3′ ultimate residues of the viral template for influenza virus (17, 18). Another model concerning how viruses use snatched capped RNA leaders is the prime-and-realign mechanism. This mechanism proposes that capped RNA leaders often shift back to realign opposite the 3′ ultimate residues of the viral template after being extended for one to several nucleotides (20). This mechanism is supported by the observation that the 3′ termini of nonviral HSs often have nucleotides that are identical to the first several nucleotides of the template sequences. In a few cases, it has been shown that these nucleotides could not have been donated by the host (20).
Both the base-pairing model and the prime-and-realign mechanism suggest that the so-called 5′ nonviral HSs on viral mRNAs are not necessarily the capped RNA leaders that have been recruited by a segmented NSV. The base-pairing model predicts that capped RNA leaders have at least one 3′-terminal residue that extends into the so-called templated sequences, while the prime-and-realign mechanism suggests that the nonviral HSs may have 3′-terminal additional residues that are not derived from host cellular mRNAs. Apparently, knowledge on the source of the capped RNA leaders is needed to test these ideas. Besides the base-pairing model and the prime-and-realign mechanism, knowledge is also needed to test another idea, i.e., segmented NSVs may preferentially target some host cellular mRNAs. This idea was proposed as early as the 1980s for influenza virus (21, 22). In a later study, it was suggested that influenza virus might predominantly use host cellular mRNAs bearing the dinucleotide CA at approximate positions downstream of the cap (23). More recently, several studies on viruses of the order Bunyavirales suggested that viruses preferentially target host cellular mRNAs encoding cell-cycle-related proteins or mRNAs containing premature translation termination signals (19, 24–26).
Unfortunately, the identity of the host cellular mRNAs that donate capped RNA leaders for segmented NSVs has been a mystery for decades. Generally, the nonviral HSs on a viral mRNA are ≤15 nt in size, and as a whole they have a very high sequence diversity. Using traditional methods, only a few HSs can be obtained, and identification of the host cellular mRNAs involved is difficult. The recent developments in high-throughput sequencing provide unprecedented opportunities to solve this problem. Several recent studies involving high-throughput sequencing have been reported for influenza virus (27–30). However, similar studies are unavailable for other segmented NSVs.
Tenuiviruses, which were recently classified into the family Phenuiviridae under the order Bunyavirales, are segmented NSVs that infect plants of the Gramineae family (31). Several tenuiviruses, including Rice stripe tenuivirus (RSV), which has a quadripartite genome, and Rice grassy stunt tenuivirus (RGSV), which has a hexapartite genome, are major viral pathogens of rice in Southeast Asia (31–33). The genomic RNA segments of tenuiviruses share conserved and complementary 5′ and 3′ termini with the sequences 5′-ACACAAAC and GUUUGUGU-3′, respectively (31). Depending on the species, most or all of the genomic segments of a tenuivirus are ambisense, i.e., contain two open reading frames, with one each on the 5′ end of the virion and complementary-sense RNA, respectively. The use of cap snatching of tenuiviruses was reported decades ago (34–36). However, the details of the cap-snatching process of tenuiviruses have remained poorly characterized. Recently, we reported a curious finding that RSV shows a much greater tendency to use the prime-and-realign mechanism than RGSV during the process of cap snatching (37). However, because that study used a heterologous coinfecting plant virus, Rice ragged stunt virus (RRSV), as the donor of the capped RNA leaders, whether this is true in transcription of the two viruses with host-donated capped RNA leaders remains unknown.
Here, besides confirming the species-specific use of the prime-and-realign mechanism in natural infections of tenuiviruses, we found that RSV has a different tendency to use the prime-and-realign mechanism in transcribing its genomic and antigenomic RNA segments. In addition, we obtained clear evidence for a base-pairing requirement in cap snatching of tenuiviruses and found that tenuiviruses cap snatch host cellular mRNAs for translation- and photosynthesis-related proteins.
RESULTS
High-throughput sequencing of the 5′ ends of NP and NCP mRNAs revealed differences in the occurrence of prime-and-realign events between RSV and RGSV.
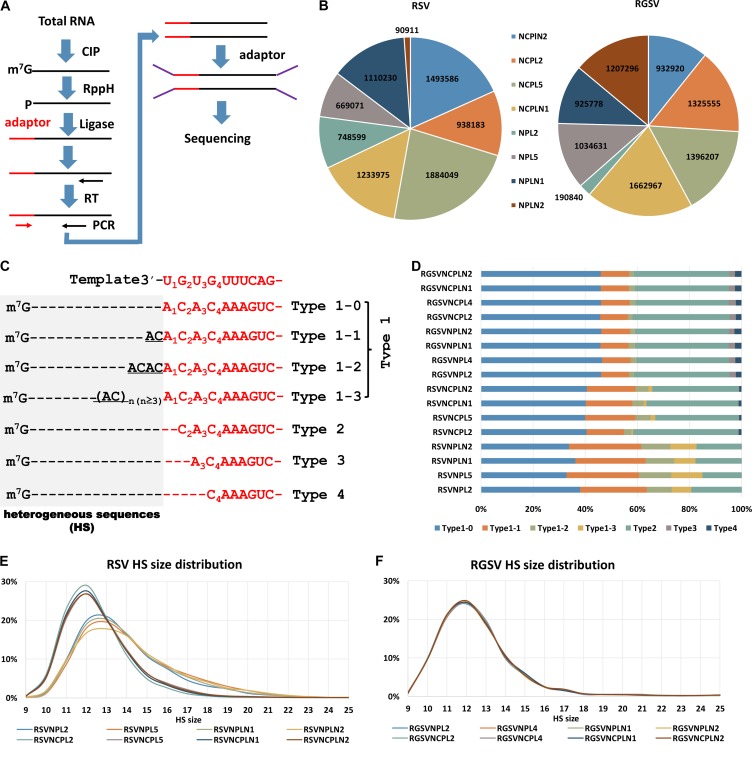
As a first attempt to confirm the greater tendency of RSV than RGSV to use the prime-and-realign mechanism in transcription primed by capped RNA leaders snatched from host cellular mRNAs, a procedure similar to that reported by Sikora et al. (29, 30) (Fig. 1A) was adopted to sequence the 5′ ends of NP and NCP mRNAs of the two tenuiviruses (38–41). For both RSV and RGSV, four different samples were used. NP and NCP mRNAs from different samples were sequenced independently, resulting in 16 data sets, named RSV/RGSVNPL2, RSV/RGSVNPL5, RSV/RGSVNPLN1, RSV/RGSVNPLN2, RSV/RGSVNCPL2, RSV/RGSVNCPL5, RSV/RGSVNCPLN1, and RSV/RGSVNCPLN2. The number of sequences in each data set varied greatly, particularly for NP. However, with the exception of RGSVNPL2 and RSVNPLN2, at least 0.6 million sequences were obtained for each data set, and in total 2.62/3.36 and 5.55/5.32 million sequences were obtained for NP and NCP mRNAs of RSV/RGSV, respectively (Fig. 1B).
FIG 1.
High-throughput sequencing revealed differences in the occurrence of prime-and-realign events between RSV and RGSV. (A) Diagram showing the experimental procedure for sequencing the 5′ ends of tenuiviral mRNAs. Total RNA extracted from virus-infected rice was treated with alkaline phosphatase (CIP) to remove the 5′-phosphate groups of some RNA species and RppH (NEB) to decap mRNAs and leave a monophosphate at their 5′ ends. An RNA oligonucleotide (red) was added to the 5′-monophosphate-bearing mRNAs, and the oligonucleotide-tagged mRNAs were reverse transcribed and PCR amplified. The PCR products were used for library construction and high-throughput sequencing. (B) The number of reads obtained for NP and NCP mRNAs of RSV and RGSV. For each virus, four samples, named NPL2, NPL5, NPLN1, and NPLN2 were used. NP/NCP mRNAs from the same sample were sequenced independently, resulting in 16 data sets in total, with 8 for each virus. (C) Diagram showing the classification of the viral mRNA sequences. The top row shows the conserved 3′ termini of tenuiviral template RNAs (red). NP or NCP mRNAs whose templated sequences (red) lacked residues corresponding to U1, U1G2, and U1G2U3 of the viral template were called type 2, 3, and 4 sequences, respectively, and the remaining sequences (type 1) were classified into type 1-1, 1-2, 1-3, and 1-4 sequences based on the number of AC repeats at the 3′ end of the nonviral HSs (gray). Dotted lines were introduced to align the sequences. (D) Stacked graph showing the percentages of each type of HS in each data set. (E and F) Size distributions of the HSs extracted from each data set for RSV (E) and RGSV (F). The y axis shows the percentage of HSs with a particular size between 9 and 25 nt.
The sequences in each data set were aligned with their template RNAs to delineate the border between the nonviral HSs and the templated sequences. An average of 29%/39% of RSV/RGSV sequences lacked the residues A1, A1C2, or A1C2A3 at positions corresponding to the U1, U1G2, and U1G2U3 of the conserved 3′-U1G2U3G4UUUCAG sequence at the 3′ termini of tenuiviral template RNAs (31). We named these sequences type 2, 3, and 4 sequences, respectively, and called those having an intact 5′-A1C2A3C4AAAGTC sequence the type 1 sequences (Fig. 1C). The HSs extracted from these sequences were called type 1 to type 4 HSs, accordingly. Because the number of the dinucleotide ACs at the 3′ termini of type 1 HSs is of particular interest to us, we further divided type 1 HSs into four subtypes: type 1-0, type 1-1, type 1-2, and type 1-3, which have 0, 1, 2, and at least 3 ACs, respectively, at their 3′ termini (Fig. 1C). According to the model we proposed previously, these four subtypes of type 1 HSs are suggestive of 0 or 1, 1 or 2, 2 or 3, and 3 or more cycles of priming and realignment events, respectively (37).
A greater tendency of RSV to use the prime-and-realign mechanism than RGSV becomes obvious upon such a classification. RSV has a significantly greater tendency to have type 1-1, type 1-2, and type 1-3 HSs than RGSV (chi-square test, P = 0). For RSV, these three types of HSs on average make up proportions of 22.5%, 7.6%, and 5.1%, respectively, of the total HSs, whereas for RGSV, they account for 10.8%, 1.9%, and 0.1%, respectively.
An unexpected finding is that RSV has a greater tendency to use the prime-and-realign mechanism in synthesizing its NP mRNAs than in synthesizing its NCP mRNAs. Although the percentage of each type of HS varied among different data sets for the same mRNAs, type 1 HSs accounted for at least 80% for the mRNAs of NP but at most 66% for those of NCP (chi-square test, P = 0) (Fig. 1D). The mRNAs of NP also had a much greater tendency to contain type 1-2 and type 1-3 HSs (chi-square test, P = 0). These two types of HSs make up proportions of at least 9% and 7%, respectively, for NP mRNAs but account for no more than 6% and 2%, respectively, for NCP mRNAs (Fig. 1D). In addition, the HSs of NP mRNAs have a greater size heterogeneity than those of NCP mRNAs, as would be expected because realignment events may alter the size of an HS (Fig. 1E). In contrast, the NP and NCP mRNAs of RGSV have almost identical frequencies of different HS types and almost overlapping HS size distributions (Fig. 1F).
Mapping HSs to host cellular mRNAs confirmed the virus- and gene-specific usage of the prime-and-realign mechanism of tenuiviruses.
In the preliminary analysis described above, we assumed that all 3′-terminal ACs of type 1 HSs are derived from the prime-and-realign mechanism. However, it is also possible that they are derived from host cellular mRNAs, i.e., the capped RNA leaders snatched by RSV (particularly in synthesizing its NP mRNA) may have a greater tendency to contain the dinucleotide AC at their 3′ termini than those snatched by RGSV. To rule out this possibility and look further into the cap-snatching mechanism of tenuiviruses, we adopted a strategy similar to that reported by Gu et al. (27) to identify host cellular mRNAs for the HSs of RSV and RGSV. Briefly, the 5′ ends of host cellular mRNAs from RSV- or RGSV-infected rice plants were obtained through a high-throughput sequencing strategy. The RSV and RGSV HSs obtained were mapped to the 5′ termini of these host cellular mRNAs.
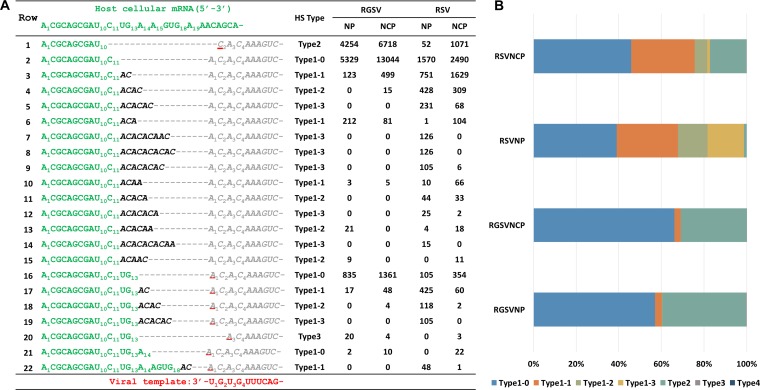
It seems clear that most 3′-terminal ACs of type 1 HSs could not have been derived from the host: of the 940,356, 248,852, and 138,794 type 1-1, type 1-2, and type 1-3 HSs that could be mapped to host cellular mRNAs, 67.8%, 98.8%, and 98.7% had at least one unmatched AC. An example showing the mapping is presented in Fig. 2A. As shown, a single host cellular mRNA can donate dozens of distinct HSs. Whereas some of these HSs are apparently derived from different cleavage events during cap snatching, most of them are generated from different priming and realignment events. Besides the dinucleotide AC, priming and realignment events sometimes generate ACA (see row 7 for an example) or ACAA (see row 10 for an example), but this occurs with an overall frequency of less than 1%. Notably, for the same host cellular mRNA shown here, RSV has a greater HS diversity than RGSV. The HS diversity for the mRNAs of NP and NCP of RSV is comparable. However, NP mRNAs tend to have more type 1 HSs, particularly type 1-2 and type 1-3 HSs, than NCP mRNAs (chi-square test, P = 0) (Fig. 2B).
FIG 2.
Mapping HSs to host cellular mRNAs confirmed the virus- and gene-specific usage of the prime-and-realign mechanism of tenuiviruses. (A) Map of the HSs of NP and NCP mRNAs to a host cellular mRNA (donor; green). The NP and NCP mRNAs were classified into different types according to the criteria shown in Fig. 1C. The reads for each unique mRNA are shown on the right. For each NP/NCP of RSV/RGSV, the reads from the four different samples shown in Fig. 1B were pooled, and the total reads are shown. The templated sequences of the mRNAs are shown in gray. Viral mRNA residues matching the host cellular mRNA are shown in green. Viral mRNA residues derived from priming and realignment are in black and italics. The residues of the templated sequences are numbered according to the template (bottom row), while those of the HSs are numbered according to the host cellular mRNA. (B) Stacked graph showing the percentage of each type of HSs for the NP and NCP mRNAs of RSV/RGSV in panel A.
The frequent occurrence of priming and realignment means that many tenuiviral HSs have additional 3′ ACs compared to the capped RNA leaders. To facilitate investigations into the early steps of tenuiviral cap snatching, we generated new data sets in which the 3′-terminal ACs of type 1 HSs were removed. For simplicity, we refer to all the HSs in the new data sets as tHSs (t for trimmed). tHSs are shown in green in Fig. 2A.
Examination of the nucleotide composition of host cellular mRNAs 3′ to the tHSs provided evidence for a base-pairing requirement in cap snatching of tenuiviruses.
As mentioned in the introduction, a base-pairing model has been proposed for cap snatching. Experimental evidence supporting such a model was first provided by studies on a tospovirus (15), followed by those on a hantavirus (19). More compelling evidence involving competition experiments with different capped RNA leaders was recently reported for the influenza virus (17, 18). The data obtained here allow us to test this model for tenuiviruses.
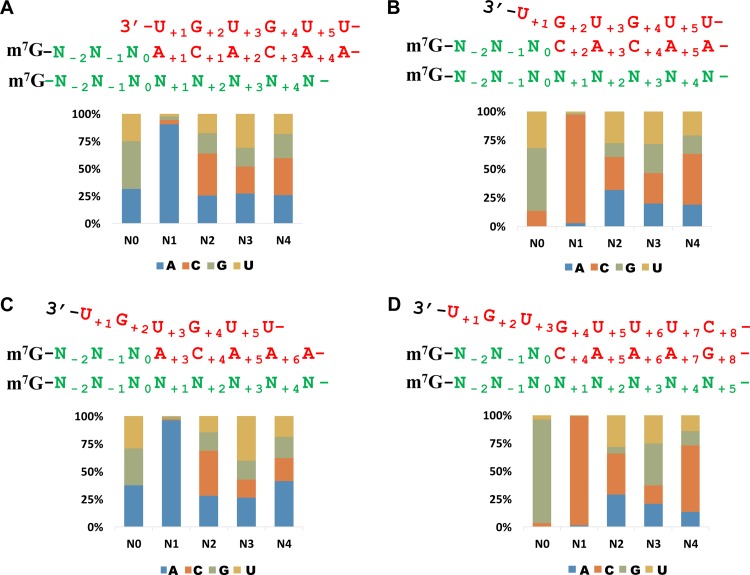
An examination of the sequences presented in Fig. 2A revealed evidence that supports the base-pairing model. For the type 2 sequence in row 1, the first templated residue, C2, corresponding to G2 of the viral template, matches C11 of the host cellular mRNA. Thus, this type 2 sequence can be explained by base pairing of C11 of the capped RNA leader 5′-A1CGCAGCGAU10C11-3′ sequence with the G2 of the template followed by a progressive elongation. Similarly, for the four type 1 and one type 3 sequence in rows 16 to 20, the first templated residue, A1 or A3, corresponding to U1 or U3 of the viral template, coincides with the A14 of the host cellular mRNA, suggesting that these sequences may be derived by a base pairing of the A14 of the capped RNA leader (5′-A1CGCAGCGAU10C11UGA14-3 ′) with U1 or U3 of the template followed by different cycles of priming and realignment or direct elongation. Likewise, the sequences in rows 21 and 22 can be explained by base pairing of A15 and A19 of the capped RNA leaders (5′-A1CGCAGCGAU10C11UGA14A15-3′ and 5′-A1CGCAGCGAU10C11UGA14AGUGA19-3′) with U1 or U3 of the template, respectively. For sequences in rows 2 to 15 (type 1 sequences ending in a C residue), the A1 of the templated sequence does not correspond to U12 of the host cellular mRNA. However, all these sequences can be explained by realignment events after base pairing of C11 of the capped RNA leader (5′-A1CGCAGCGAU10C11) with G2 of the template (37). To test these observations on a larger scale, we designated the last nucleotide matching a tHS on a host cellular mRNA N0 (U10, C11, G13, A14, and G18 of the host mRNA for the tHS in Fig. 2A) and examined the nucleotides at the position N+1 for the 3.9 million tHSs (consisting of 72,133 unique sequences) that have been mapped unambiguously to distinct host cellular mRNAs. As illustrated above, we expected to find an A residue for type 1 tHSs (that do not have a C at their 3′ termini) and type 3 tHSs and a C residue for type 2 and type 4 tHSs at this position. Indeed, the results support this idea, although there were a few exceptions (with a frequency of less than 10%) for each type of tHS (Fig. 3A to D).
FIG 3.
Examination of the nucleotide composition of host cellular mRNAs 3′ to the tHSs provided evidence for a base-pairing requirement in cap snatching of tenuiviruses. The last nucleotide matching a tHS (Fig. 2) on a host cellular mRNA was designated N0. The nucleotide compositions of host cellular mRNAs are shown in stacked graphs for the region N0 to N4 for type 1 tHSs ending in an A, U, or G (A), type 2 tHSs (B), type 3 tHSs (C), and type 4 tHSs (D). Shown above each image are the viral template (red), viral mRNA (green and red), and the host cellular mRNA (green).
A base-pairing requirement for cap snatching means that even tHSs are not always equal to the capped RNA leaders recruited by a tenuivirus. With the exception of type 1 tHSs ending in a C residue, an A (type 1 and type 3 tHSs [see rows 16 to 20 in Fig. 2A for examples]) or C (type 2 and 4 tHSs [see row 1 in Fig. 2A for an example]) residue has been lost during HS identification or trimming. For further analysis, new data sets were generated in which an A or C was added to the 3′ termini of corresponding tHSs. In the text below, we consider these modified tHSs as the capped RNA leaders, although some of them may be one to a few nucleotides smaller than the genuine capped RNA leaders because of a multibase complementarity that is not considered in the tHS modification described here.
Capped RNA leader size influences the frequency of realignment, but differences in capped RNA leader size cannot account for the different tendencies of RSV and RGSV to use the prime-and-realign mechanism.
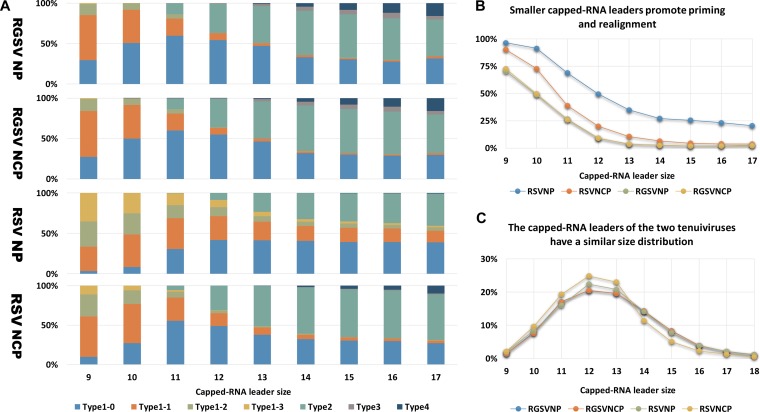
By analyzing RSV mRNAs with capped RNA leaders derived from Cucumber mosaic virus (CMV), Yao et al. (42) earlier proposed that smaller capped RNA leaders might promote the usage of priming and realignment. In light of this, one explanation for the greater tendency of RSV to use the prime-and-realign mechanism is that this virus may prefer smaller capped RNA leaders, particularly in synthesizing the mRNAs of NP. To test this, we first investigated whether capped RNA leader size influenced the realignment frequency of the two tenuiviruses in natural infections. To do this, we examined the HS patterns associated with capped RNA leaders of particular sizes. If smaller capped RNA leaders promoted priming and realignment, we would find that smaller capped RNA leaders have a greater tendency to generate type 1 HSs. Indeed, we observed that for both viruses, the percentage of type1 HSs drops with increasing capped RNA leader size (Fig. 4A and B). Capped RNA leaders of ≤12 nt have a much greater tendency to generate type 1, particularly types 1-1, 1-2, and 1-3 HSs, than those with greater sizes (Fig. 4A and B).
FIG 4.
Capped RNA leader size influences the frequency of realignment, but differences in capped RNA leader size cannot account for the different tendencies of RSV and RGSV to use the prime-and-realign mechanism. (A) Stacked graphs showing the percentages of each type of HS (Fig. 1C) that associates with capped RNA leaders of a particular size, from 9 to 17 nt (x axis) for NP/NCP of RSV/RGSV. (B) The total percentages of type 1-1, 1-2, and 1-3 HSs associated with capped RNA leaders of a particular size (x axis). (C) Size distributions of the capped RNA leaders of the NP and NCP mRNAs of RSV/RGSV. The y axis shows the percentage of capped RNA leaders with a particular size, from 9 to 18 nt. For each mRNA, sequences from the four data sets (Fig. 1B) were pooled for the calculation shown here.
If different preferences for small capped RNA leaders were the major reason for the virus- and gene-specific usage of the prime-and-realign mechanism of the two tenuiviruses, we would find that the frequencies of priming and realignment were similar between RSV and RGSV in transcription initiated by capped RNA leaders of the same size. The results presented in Fig. 4A and B contrast with this prediction. As shown, RSV has a greater tendency to have type 1 HSs, particularly type 1-1, 1-2, and 1-3 HSs than RGSV regardless of the size of capped RNA leaders. A similar trend can be found for the NP and NCP mRNAs of RSV. Notably, for capped RNA leaders of ≥13 nt, type 1-1, 1-2, and 1-3 HSs in total account for no more than 3.5% for either mRNA of RGSV and for no more than 10.6% for that of RSV NCP. However, these three types of HSs altogether make up an average proportion of 26% for RSV NP mRNAs (Fig. 4A and B). These observations suggest that although the size of capped RNA leaders has a clear influence on the frequency of priming and realignment, it is unlikely that differences in capped RNA leader size are the major reason for the virus- and gene-specific usage of the prime-and-realign mechanism of the two tenuiviruses. To gain further support for this, we compared the size distribution of the capped RNA leaders of RSV and RGSV. Although RSV and RGSV, as well as the NP and NCP mRNAs of RSV, showed a clear difference in the size distribution of their HSs (Fig. 1E and F), a similar difference was not found between that of their capped RNA leaders. In contrast, the two tenuiviruses (as well as the two mRNAs of them) had nearly identical size distributions for their capped RNA leaders (Fig. 4C).
Sequencing the 5′ ends of other RSV mRNAs revealed that this virus might have a different tendency to use the prime-and-realign mechanism in transcribing genomic and antigenomic RNAs.
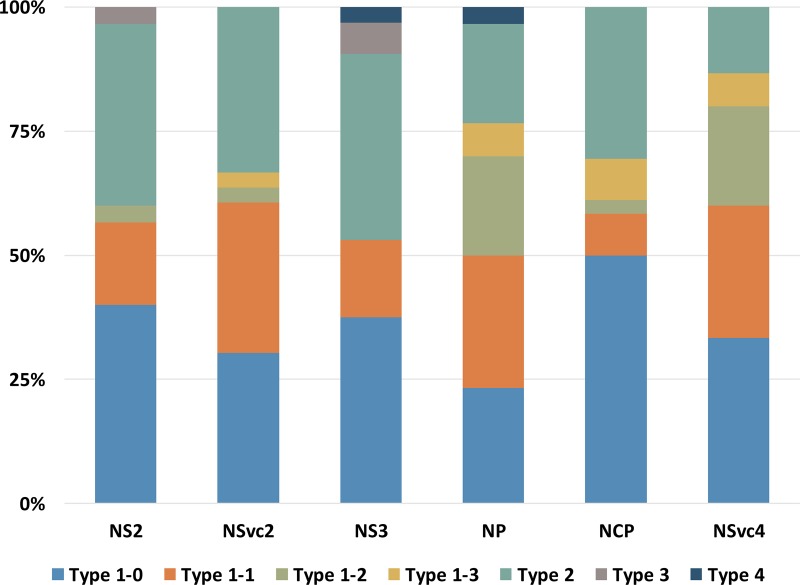
Having observed that RSV has a different tendency to use the prime-and-realign mechanism in synthesizing NP and NCP mRNAs, we were curious whether RSV does so in synthesizing other mRNAs. Initially, we were unable to get the 5′ HSs of other RSV mRNAs. Almost no cDNA clones obtained using the procedure shown in Fig. 1A contained an HS at their 5′ ends. A plausible explanation is that the accumulation levels of these mRNAs are too low relative to their coding RNAs. To circumvent this obstacle, a previously described human eukaryotic initiation factor 4Ea (eIF4Ea) mutant which has a high affinity for capped RNAs was used to enrich RSV mRNAs (43). Using the enriched mRNAs as the substrate, 30 to 32 cDNA clones each for NS2 (encoded on vRNA2 and transcribed from vcRNA2), NSvc2 (encoded on vcRNA2 and transcribed from vRNA2), NS3, and NSvc4 mRNAs were obtained. To facilitate comparison in the same context, a similar number of cDNA clones for the mRNAs of NP (encoded on vcRNA3 and transcribed from vRNA3) and NCP (encoded on vRNA4 and transcribed from vcRNA4) was sequenced with the same method. The HSs for each mRNA obtained in this way were classified into type 1-0, type 1-1, type 1-2, type 1-3, type 2, type 3, and type 4, according to the same criteria as described above. Despite the small number of sequences obtained, a trend similar to that described above was found for NP and NCP mRNAs; the former tends to have more type 1-1, 1-2, and 1-3 HSs (chi-square test, P < 0.005). Interestingly, this trend was found when we compared mRNAs transcribed from vRNAs with those transcribed from vcRNAs (chi-square test P < 2.00E−05) (Fig. 5). On average, type 1-1, 1-2, and 1-3 HSs altogether accounted for 47% of mRNAs transcribed from vRNAs and 18% of mRNAs transcribed from vcRNAs. Thus, RSV has different tendencies to use the prime-and-realign mechanism in transcribing genomic and antigenomic RNAs.
FIG 5.
RSV has a different tendency to use the prime-and-realign mechanism in transcribing genomic and antigenomic RNAs. Thirty to 32 cDNA clones of the mRNAs transcribed from virion-sense (NSVc2, NP, and NSVc4) or complementary-sense template RNAs (NS2, NS3, and NCP) of RSV were obtained. The sequences of the cDNA clones were classified into different types according to the criteria shown in Fig. 1C, and the percentages of each type for each mRNA are shown in the stacked graphs.
Transcripts of several functional groups of genes are frequently targeted by tenuiviruses.
The identity of the host transcripts that donate capped RNA leaders for segmented NSVs has been a mystery for decades. The data obtained here provided a preliminary overview on the donors of the capped RNA leaders of tenuiviruses. As many as 34,917 and 30,565 distinct host transcripts were identified as donors of capped RNA leaders for RSV and RGSV, respectively. Given the fact that we had 51,459 host transcripts for the mapping, this means that most host transcripts are substrates of cap snatching for tenuiviruses. However, these host transcripts are used with very different frequencies. The number of capped RNA leaders that mapped to a host transcript (in a particular data set) ranged from 1 to more than 10,000. As a whole, more than 45% and 49% of the host transcripts matched capped RNA leaders from no more than three of the eight data sets for RSV and RGSV, respectively. Although these may partially due to a sequencing bias and an unequal sequencing depth, respectively, they are also consistent with a scenario in which tenuiviruses cap snatch some host transcripts constantly and frequently but target others occasionally. To focus on host transcripts most intensively used by tenuiviruses, we analyzed the host transcripts that are matched by HSs from at least 7 data sets for each virus. For simplicity, we call these host transcripts frequent donors of capped RNA leaders. The numbers of frequent donors for RSV and RGSV were 6,058 and 6,957, respectively. The number of frequent donors shared by the two viruses was 4,440.
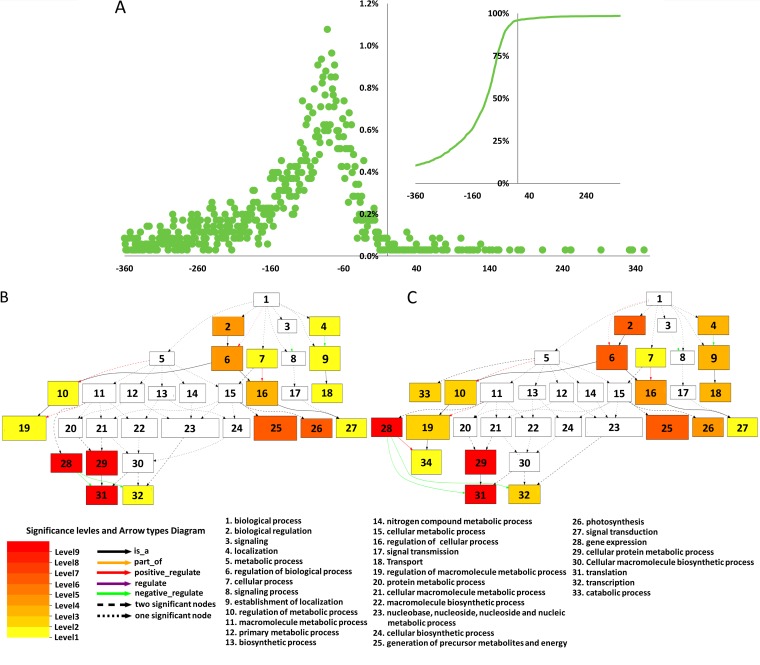
As shown in Fig. 6A, most of the frequent donors that could be mapped to the rice genome (http://rice.plantbiology.msu.edu/) had a transcriptional start site (TSS) that localized to putative promoter regions of protein-encoding rice genes. However, the TSSs are very far from or near to the start codons of corresponding host genes for some donors, suggesting that these donors may be noncoding RNAs or aberrant transcripts.
FIG 6.
Transcripts of several functional groups of genes are frequently targeted by tenuiviruses. (A) Mapping of frequent donors of capped RNA leaders to the genome of rice. The x axis indicates the positions of donor TSSs relative to the start codons (position 0) of corresponding rice genes. The y axis indicates the percentages of donors having TSSs at particular positions. The small image on the right side shows the total percentages (y axis) of donors having TSSs upstream of a particular position (x axis). (B and C) Hierarchical graph showing the results of singular enrichment analysis (SEA) with genes frequently cap snatched by RSV (B) and RGSV (C). The enrichment analysis was carried out using agriGO (45). The nodes in the image were classified into 10 levels, which are shown with the indicated colors. The smaller a term's adjusted P value, the more statistically significant the difference (indicated by the node's color being darker and redder).
To identify relevant rice genes with high confidence, only donors with a TSS located at the region −360 to −20 upstream of the start codon of a corresponding rice gene locus were considered. In this way, we identified 1,134 and 1,430 rice genes for the frequent donors of RSV and RGSV, respectively. Among them, 946 genes were shared by the two viruses. GO enrichment analysis was done to see whether tenuiviruses preferentially use some functional groups of genes. A total of 19 GO biological process terms were found for the two viruses, with 17 of them shared by the two tenuiviruses and another 2 specific to RGSV (Fig. 6B and C). Strikingly, for both viruses, most of the enriched GO slim terms pointed to two groups of genes: 83/97 (RSV/RGSV) were genes related to translation, and 19/19 were genes related to photosynthesis (Fig. 6B and C). A further examination revealed that 71/86 (RSV/RGSV) of the 83/97 translation-related genes encode large or small subunits of the ribosome, whereas 12/11 (RSV/RGSV) of the 19/19 photosynthesis-related genes encode chlorophyll A- or B-binding proteins. The enrichment level of the genes encoding chlorophyll A- or B-binding proteins was impressive, because only 17 similar genes can be found in the annotated gnome of rice.
DISCUSSION
In all, we determined the 5′ ends of the NP and NCP mRNAs of two tenuiviruses and mapped a large number of the HSs of these two mRNAs to host cellular mRNAs by using a high-throughput sequencing procedure. This revealed a frequent presence of single or multiple repetitive dinucleotides that could not have been derived from host cellular mRNAs at the 3′ ends of tenuiviral HSs, which provided clear evidence that tenuiviruses use the prime-and-realign mechanism in natural infections. In addition, this gave us an opportunity to examine the sequences of the host cellular mRNAs on the 3′ side of the tHSs, which revealed that tenuiviral capped RNA leaders have at least one residue that extends into the so-called templated sequences, providing strong evidence for the involvement of base pairing in tenuiviral cap snatching. We also found that tenuiviruses target host cellular mRNAs encoding translation- and photosynthesis-related proteins. This may be an important piece of information for further studies on the involvement of cap snatching in symptom induction of tenuiviruses. Most interestingly, besides confirming a previous finding that RSV has a greater tendency than RGSV to use the prime-and-realign mechanism, we found that RSV uses this mechanism with different tendencies in transcribing its genomic and antigenomic RNAs. To our knowledge, this study represents the first attempt to investigate the cap-snatching mechanism of cytoplasm-replicating segmented NSVs by using a high-throughput sequencing strategy. Besides providing insights into the transcription initiation mechanisms of tenuiviruses, our results may also be useful for studies on a large number of related viruses.
Overall, the model we proposed previously to explain how RSV and RGSV use capped RNA leaders snatched from RRSV is in accordance with the present data, after slight modifications (37). According to this model, a 9- to 20-nt, preferentially 11- to 14-nt (Fig. 4C), capped RNA leader ending in an A or C residue is recruited by the transcriptase of a tenuivirus. This capped RNA leader aligns opposite the U1, G2, U3, or G4, but more typically U1 or G2 of the template to prime mRNA synthesis (Fig. 1C). With a capped RNA leader ≤10 nt in size, tenuiviruses tend to use at least one cycle of priming and realignment before progressive elongation (Fig. 4A and B). Different tenuiviruses or even the same tenuivirus synthesizing different mRNAs may use the prime-and-realign mechanism with different tendencies. As evidenced by the observation that priming and realignment events usually generate repeats of the dinucleotide AC, the realignment normally occurs after adding no more than 3 nucleotides to a capped RNA leader (Fig. 2A). Because a single-base complementarity is enough for the priming activity of a capped RNA leader and most transcripts in a host cell contain at least one A or C residue in a window of 12 nt (9 to 20 nt from the cap), tenuiviruses can cap snatch almost all the transcripts accumulated within a host cell. However, a subset of host transcripts from some functional groups of genes are used more frequently (Fig. 6B and C).
It should be noted that the nonhost ACs at the 3′ termini of type 1 HSs might be explained in several different ways. One explanation is that tenuiviruses may snatch capped RNA leaders from their own mRNAs. This may indeed occur, as demonstrated recently for Tomato spotted wilt virus in an in vitro system (16). However, it is not unreasonable to assume that a virus does not use such a mechanism frequently in natural infections. In addition, it has been demonstrated that the transcriptase of the influenza virus could protect viral mRNAs from cleavage (44). Therefore, before strong in vivo evidence for such a resnatching mechanism is available, we favor the explanation that these ACs are derived from the prime-and-realign mechanism.
Whereas the different tendencies of RSV and RGSV to use the prime-and-realign mechanism are consistent with our previous report and are understandable because the two viruses may have differences in their transcriptases, the different tendency of RSV to use the prime-and-realign mechanism in transcribing genomic and antigenomic template RNAs is unexpected and difficult to explain at present. However, it is interesting that a similar observation was obtained in a recent study with influenza virus (28). Unlike RSV, the influenza virus does not employ an ambisense coding strategy. However, exactly one-half of its eight mRNAs is produced with more frequent priming and realignment events than the other half. Koppstein et al. (28) explained this observation by using a single-nucleotide polymorphism at position +4 of viral templates: influenza virus RNAs with a C residue at this position are transcribed with less priming and realignment events, while those with a U are transcribed with more priming and realignment events. It was postulated that a U would favor dissociation of the newly extended capped RNA leaders from the template relative to a C at position +4. Apparently, such an explanation cannot be applied to RSV, because the template RNAs of RSV have a long stretch of identical 3′ termini (31). Therefore, besides providing deeper insights into the cap snatching of tenuiviruses, our results suggest some new features of priming and realignment, a mechanism whose biological meaning remains unknown but seems to be used commonly by segmented NSVs during their cap snatching.
MATERIALS AND METHODS
Plant material and virus inoculation.
The rice variety Shuhui no. 1 was used for this study. RSV- and RGSV-infected rice plants were obtained by inoculating rice with viruliferous Laodelphax striatellus Fallen and Nilaparvata lugens (Hemiptera, Delphacidae), respectively, at their seedling stage. Samples LN1 and LN2 were collected 1 week after the appearance of symptoms typical of RSV or RGSV infeciton, respectively. Samples L2 and L5 were collected at the jointing and tilling stages, respectively. For each sample, leaves of a single plant were collected and mixed for RNA extraction.
High-throughput sequencing of the 5′ ends viral mRNAs.
Twelve micrograms of total RNA extracted from virus-infected rice plants using an RNeasy plant minikit (Qiagen) was treated with Terminator (Epicentre), a 5′-exonuclease specific for monophosphorylated RNA. The Terminator-digested RNA (with about 2 μg left) was sequentially treated with alkaline phosphatase (NEB), which dephosphorylates uncapped RNAs, and RppH (NEB), which removes the 5′ cap of mRNAs and leaves a monophosphate at the 5′ end. An RNA oligonucleotide (TCTACrArGrUrCrCrGrArCrGrArUrC) was ligated to the 5′-monophosphate-containing mRNA with T4 RNA ligase 1 (NEB). Primers specific to NP and NCP mRNAs of RSV/RGSV were used to reverse transcribe the 5′-oligonucleotide-tagged RNA with the SuperScript III first-strand synthesis system (Invitrogen). The cDNA was purified using DNA clean and concentrator 5 (Zymo), and one-half was used for PCR with a forward primer that annealed to the adaptor (GTTCTACAGTCCGACGATC) and gene-specific reverse primers, which are available from the authors upon request. To reduce PCR bias, the lowest number of PCR cycles allowing observation of corresponding bands for each mRNA was used. The PCR products were sent to Biomarker Technologies for sequencing with a HiSeq 2500 platform.
High-throughput sequencing of the donor RNAs, HS mapping, and GO enrichment analysis.
The 5′ ends of rice mRNAs were determined in a procedure similar to those described above except that a random primer was used in the reverse transcription (CAGCTCTTCCCGAACCAACATCNNNNNN). In addition, the adaptor that tags the 5′ ends of rice mRNAs was biotin labeled. This allowed an enrichment of RNAs that had been tagged with the adaptor before reverse transcription. PCR products of about 150 to 200 nt in size were recovered from the gel and sequenced. To obtain different rice mRNAs as much as possible, three different samples were used and sequenced independently. The sequences obtained from the three sequencing reaction mixtures were pooled to build a nonredundant library for HS mapping. For HS mapping, the 30-nt 5′ fragments of the library sequences obtained were used. To map the donors onto the genome of rice, the 150-nt 5′ fragments of the sequences were used. GO enrichment analysis was done with agriGO (45). GO slims that were enriched at the level of P < 0.01were recorded.
Small-scale sequencing of RSV mRNAs.
One hundred micrograms of total RNA was purified from pooled RSV-infected rice plants. Capped RNA was enriched using a bacterially expressed eIF4E mutant according to a procedure described by Choi and Hagedorn (43). After purification, the 5′ ends of each RSV mRNA were sequenced using a procedure similar to that described above except that the PCR products were cloned using E. coli and Sanger sequencing.
ACKNOWLEDGMENTS
This work was supported by grants from the National Basic Research Program 973 (2014CB138402), Natural Science Foundation of China (31672005 and 31401715), and Natural Science Foundation of Fujian, China (2016J05071), a fund from the State Tobacco Monopoly Administration (110201601024 [LS-04]), and FAFU Funds for Excellent Young Scholars or Innovation and Development (xjq201622, k80nd800101b, KFA17130A, KFA17455A, and CXZX2016132).
REFERENCES
- 1.Shatkin AJ. 1976. Capping of eucaryotic mRNAs. Cell 9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Darnell JE., Jr 1979. Transcription units for mRNA production in eukaryotic cells and their DNA viruses. Prog Nucleic Acid Res Mol Biol 22:327–353. doi: 10.1016/S0079-6603(08)60803-X. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Furuichi Y, Sierra JM, Muthukrishnan S, Shatkin AJ, Ochoa S. 1976. A protein binding the methylated 5′-terminal sequence, m7GpppN, of eukaryotic messenger RNA. Proc Natl Acad Sci U S A 73:1559–1563. doi: 10.1073/pnas.73.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schibler U, Perry RP. 1977. The 5′-termini of heterogeneous nuclear RNA: a comparison among molecules of different sizes and ages. Nucleic Acids Res 4:4133–4149. doi: 10.1093/nar/4.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decroly E, Ferron F, Lescar J, Canard B. 2011. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walia JJ, Falk BW. 2012. Fig mosaic virus mRNAs show generation by cap-snatching. Virology 426:162–166. doi: 10.1016/j.virol.2012.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RWH, Ortin J, Hart DJ, Cusack S. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol 15:500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 8.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 9.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 10.Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crepin T, Hart D, Lunardi T, Nanao M, Ruigrok RWH, Cusack S. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 11.Morin B, Coutard B, Lelke M, Ferron F, Kerber R, Jamal S, Frangeul A, Baronti C, Charrel R, de Lamballerie X, Vonrhein C, Lescar J, Bricogne G, Gunther S, Canard B. 2010. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog 6:e1001038. doi: 10.1371/journal.ppat.1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reguera J, Weber F, Cusack S. 2010. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog 6:e1001101. doi: 10.1371/journal.ppat.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallat GD, Huang QF, Wang WJ, Dong HH, Ly H, Liang YY, Dong CJ. 2014. High-resolution structure of the N-terminal endonuclease domain of the Lassa virus L polymerase in complex with magnesium ions. PLoS One 9:e87577. doi: 10.1371/journal.pone.0087577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reguera J, Gerlach P, Rosenthal M, Gaudon S, Coscia F, Gunther S, Cusack S. 2016. Comparative structural and functional analysis of bunyavirus and arenavirus cap-snatching endonucleases. PLoS Pathog 12:e1005636. doi: 10.1371/journal.ppat.1005636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duijsings D, Kormelink R, Goldbach R. 2001. In vivo analysis of the TSWV cap-snatching mechanism: single base complementarity and primer length requirements. EMBO J 20:2545–2452. doi: 10.1093/emboj/20.10.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Knippenberg I, Lamine M, Goldbach R, Kormelink R. 2005. Tomato spotted wilt virus transcriptase in vitro displays a preference for cap donors with multiple base complementarity to the viral template. Virology 335:122–130. doi: 10.1016/j.virol.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 17.Geerts-Dimitriadou C, Goldbach R, Kormelink R. 2011. Preferential use of RNA leader sequences during influenza A transcription initiation in vivo. Virology 409:27–32. doi: 10.1016/j.virol.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Geerts-Dimitriadou C, Zwart MP, Goldbach R, Kormelink R. 2011. Base-pairing promotes leader selection to prime in vitro influenza genome transcription. Virology 409:17–26. doi: 10.1016/j.virol.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Cheng E, Mir MA. 2012. Signatures of host mRNA 5′ terminus for efficient hantavirus cap snatching. J Virol 86:10173–10185. doi: 10.1128/JVI.05560-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D. 1995. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol 69:5754–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beaton AR, Krug RM. 1981. Selected host cell capped RNA fragments prime influenza viral RNA transcription in vivo. Nucleic Acids Res 9:4423–4436. doi: 10.1093/nar/9.17.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw MW, Lamb RA. 1984. A specific sub-set of host-cell mRNAs prime influenza virus mRNA synthesis. Virus Res 1:455–467. doi: 10.1016/0168-1702(84)90003-0. [DOI] [PubMed] [Google Scholar]
- 23.Rao P, Yuan W, Krug RM. 2003. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J 22:1188–1198. doi: 10.1093/emboj/cdg109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mir MA, Duran WA, Hjelle BL, Ye C, Panganiban AT. 2008. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc Natl Acad Sci U S A 105:19294–19299. doi: 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panganiban AT, Mir MA. 2009. Bunyavirus N: eIF4F surrogate and cap-guardian. Cell Cycle 8:1332–1337. doi: 10.4161/cc.8.9.8315. [DOI] [PubMed] [Google Scholar]
- 26.Hopkins KC, McLane LM, Maqbool T, Panda D, Gordesky-Gold B, Cherry S. 2013. A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of Dcp2-accessible targets for cap-snatching. Genes Dev 27:1511–1525. doi: 10.1101/gad.215384.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu WF, Gallagher GR, Dai WW, Liu P, Li RD, Trombly MI, Gammon DB, Mello CC, Wang JP, Finberg RW. 2015. Influenza A virus preferentially snatches noncoding RNA caps. RNA 21:2067–2075. doi: 10.1261/rna.054221.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koppstein D, Ashour J, Bartel DP. 2015. Sequencing the cap-snatching repertoire of H1N1 influenza provides insight into the mechanism of viral transcription initiation. Nucleic Acids Res 43:5052–5064. doi: 10.1093/nar/gkv333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikora D, Rocheleau L, Brown EG, Pelchat M. 2014. Deep sequencing reveals the eight facets of the influenza A/HongKong/1/1968 (H3N2) virus cap-snatching process. Sci Rep 4:6181. doi: 10.1038/srep06181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sikora D, Rocheleau L, Brown EG, Pelchat M. 2017. Influenza A virus cap-snatches host RNAs based on their abundance early after infection. Virology 509:167–177. doi: 10.1016/j.virol.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 31.Falk BW, Tsai JH. 1998. Biology and molecular biology of viruses in the genus Tenuivirus. Annu Rev Phytopathol 36:139–163. doi: 10.1146/annurev.phyto.36.1.139. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez BC, Haenni AL. 1994. Molecular biology of tenuiviruses, a remarkable group of plant viruses. J Gen Virol 75:467–475. doi: 10.1099/0022-1317-75-3-467. [DOI] [PubMed] [Google Scholar]
- 33.Hibino H. 1996. Biology and epidemiology of rice viruses. Annu Rev Phytopathol 34:249–274. doi: 10.1146/annurev.phyto.34.1.249. [DOI] [PubMed] [Google Scholar]
- 34.Huiet L, Feldstein PA, Tsai JH, Falk BW. 1993. The Maize stripe virus major noncapsid protein messenger RNA transcripts contain heterogeneous leader sequences at their 5′ termini. Virology 197:808–812. doi: 10.1006/viro.1993.1662. [DOI] [PubMed] [Google Scholar]
- 35.Ramirez BC, Garcin D, Calvert LA, Kolakofsky D, Haenni AL. 1995. Capped nonviral sequences at the 5′ end of the mRNAs of Rice hoja blanca virus RNA4. J Virol 69:1951–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu T, Toriyama S, Takahashi M, Akutsu K, Yoneyama K. 1996. Non-viral sequences at the 5′ termini of mRNAs derived from virus-sense and virus-complementary sequences of the ambisense RNA segments of rice stripe tenuivirus. J Gen Virol 77:541–546. doi: 10.1099/0022-1317-77-3-541. [DOI] [PubMed] [Google Scholar]
- 37.Liu XJ, Xiong GH, Qiu P, Du ZG, Kormelink R, Zheng LP, Zhang J, Ding XL, Yang L, Zhang SB, Wu ZJ. 2016. Inherent properties not conserved in other tenuiviruses increase priming and realignment cycles during transcription of Rice stripe virus. Virology 496:287–298. doi: 10.1016/j.virol.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Hayakawa T, Toriyama S. 1992. Complete nucleotide sequence of RNA 4 of rice stripe virus isolate T, and comparison with another isolate and with maize stripe virus. J Gen Virol 73:1309–1312. doi: 10.1099/0022-1317-73-5-1309. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Y, Hayakawa T, Toriyama S, Takahashi M. 1991. Complete nucleotide sequence of RNA 3 of Rice stripe virus: an ambisense coding strategy. J Gen Virol 72:763–767. doi: 10.1099/0022-1317-72-4-763. [DOI] [PubMed] [Google Scholar]
- 40.Toriyama S, Kimishima T, Takahashi M. 1997. The proteins encoded by Rice grassy stunt virus RNA5 and RNA6 are only distantly related to the corresponding proteins of other members of the genus Tenuivirus. J Gen Virol 78:2355–2363. doi: 10.1099/0022-1317-78-9-2355. [DOI] [PubMed] [Google Scholar]
- 41.Toriyama S, Kimishima T, Takahashi M, Shimizu T, Minaka N, Akutsu K. 1998. The complete nucleotide sequence of the rice grassy stunt virus genome and genomic comparisons with viruses of the genus Tenuivirus. J Gen Virol 79:2051–2058. doi: 10.1099/0022-1317-79-8-2051. [DOI] [PubMed] [Google Scholar]
- 42.Yao M, Zhang TQ, Zhou T, Zhou YJ, Zhou XP, Tao XR. 2012. Repetitive prime-and-realign mechanism converts short capped RNA leaders into longer ones that may be more suitable for elongation during rice stripe virus transcription initiation. J Gen Virol 93:194–202. doi: 10.1099/vir.0.033902-0. [DOI] [PubMed] [Google Scholar]
- 43.Choi YH, Hagedorn CH. 2003. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc Natl Acad Sci U S A 100:7033–7038. doi: 10.1073/pnas.1232347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shih SR, Krug RM. 1996. Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology 226:430–435. doi: 10.1006/viro.1996.0673. [DOI] [PubMed] [Google Scholar]
- 45.Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community Nucleic Acids Res 38:W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]