Abstract
B‐cell differentiation is one of the most recognized examples of the progressive lineage commitment that is distinctive for stem cell systems. However, the characteristics of the stage just before a cell becomes restricted to the B‐cell lineage are less understood. Using single‐cell RNA sequencing technology, Rolink and colleagues are able to define the cellular heterogeneity at this step and challenge our understanding of developmental trajectories in early B‐lymphoid development (Alberti‐Servera et al, 2017).
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Development & Differentiation; Immunology
The ability to prospectively purify and functionally analyze distinct hematopoietic cell fractions has been instrumental for our understanding of how multipotent cells commit to develop into a defined mature blood cell lineage. As a result, hematopoiesis has evolved to an intellectual paradigm that describes stem cell systems as tree‐like structures with the stem cells at the top followed by a hierarchical branching of increasing commitment steps toward the terminally differentiated cells (Reya et al, 2001). However, the hematopoietic tree is continuously refined, especially due to the identification of cellular heterogeneity within previously defined populations (Inlay et al, 2009; Mansson et al, 2010).
Recent technological advances allowing for high‐throughput gene expression analysis on the single‐cell level offer opportunities to define the cellular content of a heterogeneous population based on molecular signatures as an indicator of function (Kolodziejczyk et al, 2015). Appealing targets for such analysis are hematopoietic progenitors that reside at a branching point of commitment since they may provide insight into the molecular mechanisms that regulate cell fate decisions. In this issue of The EMBO Journal, Rolink and colleagues have used this technology to define the heterogeneity within the CD19−B220+ early lymphoid progenitor population (Alberti‐Servera et al, 2017).
Lymphoid specification in general and B‐cell commitment in particular has been intensively studied and now represent a textbook example of how multipotent progenitors are increasingly restricted into single lineages in a stepwise manner characteristic for stem cell systems (Jensen et al, 2016). The common lymphoid progenitor (CLP) is regarded as the branching point between the lymphoid and myeloid lineages. Within this population, expression of the surface marker Ly6D reflects a loss of Natural Killer (NK) cell potential (Inlay et al, 2009), separating the development of cells composing the adaptive and innate immune system. While lineage commitment events in the CLP compartment are fairly well resolved, the classical CD19‐negative B220‐expressing B‐cell progenitor fraction represents a more complex population of cells, and even though advanced sorting protocols to resolve the developmental hierarchies have been established, they do not allow for the isolation of a functionally homogenous population (Rumfelt et al, 2006). The Rolink laboratory has previously characterized a B220+ progenitor denoted early progenitor with lymphoid and myeloid potential (EPLM) with apparent combined lymphoid and macrophage potential (Balciunaite et al, 2005). The existence of such a population downstream of the CLP could not be contained within the current model for B‐cell development, where separation of lymphoid and myeloid lineages occurs before commitment into a defined lymphoid lineage. Even though combined T and myeloid potential has been reported from studies of early thymic progenitors (Bell & Bhandoola, 2008), the results from the functional analysis of the EPLM did not determine if a single cell was truly bi‐potent, or if the population was heterogeneous (Balciunaite et al, 2005). To conclusively resolve this issue, the authors have now used next‐generation sequencing to study the transcriptome of single EPLM (Alberti‐Servera et al, 2017) (Fig 1).
Figure 1. Single cell RNA sequencing resolves heterogeneity within early lymphoid progenitors.
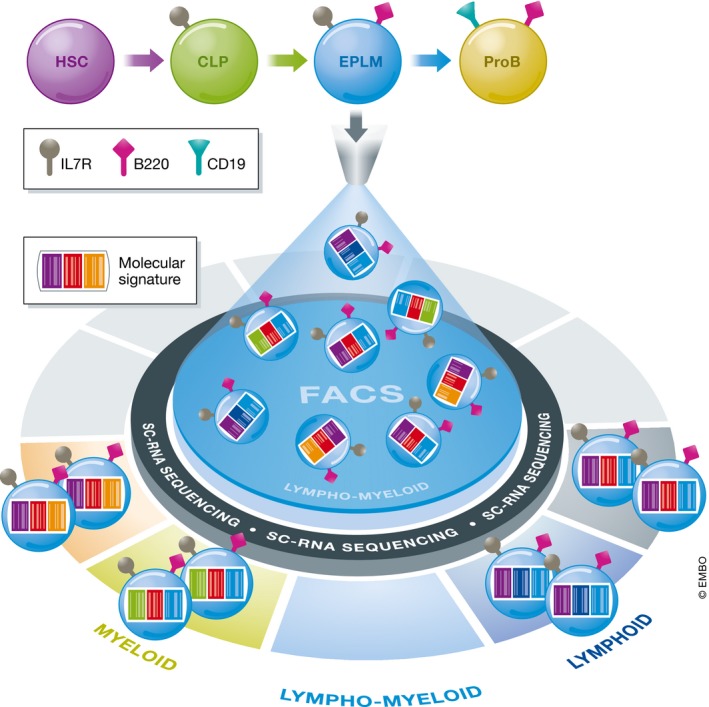
Schematically, B cells are generated in a stepwise manner from hematopoietic stem cells (HSC), and early progenitor with lymphoid and myeloid potential (EPLM) is presumed to be located between common lymphoid progenitors (CLP) and proB cells in the hierarchy. A few selected surface markers are shown for simplicity. The EPLM appears to have lymphoid and myeloid potential when purified by FACS only. However, using single‐cell RNA sequencing Alberti‐Servera et al (2017) reveal heterogeneity in cells’ molecular signatures that divides the EPLM into clusters with distinct lineage programs.
Single‐cell RNA sequencing is evolving as a powerful technology to investigate heterogeneity and trajectories within cell populations. Several different methods have been developed, and Alberti‐Servera et al (2017) chose to utilize the Fluidigm C1 platform and SMARTer chemistry for efficient single‐cell capture and library preparation. In combination with high sequencing depth, this protocol supports the detection of low expressed genes and transcription factors at a high enough coverage to allow for the separation of functionally very similar populations (Ziegenhain et al, 2017). A disadvantage is the large number of cells needed for each run, which makes the method difficult to use for rare cell populations. As a consequence, the authors could not perform the analysis on wild‐type cells but instead took advantage of the Flt3Ltg mouse model, where the EPLM is enriched.
Ly6D marks cells biased toward the B‐cell lineage (Inlay et al, 2009) and is expressed on a subpopulation of the EPLM. Global gene expression analyses of single Ly6D+ and Ly6D− EPLM revealed a cellular heterogeneity where several different lineage programs could be identified. Not surprisingly, the Ly6D‐positive fraction expressed genes associated with B lineage commitment and ought to represent the stage just before restriction to the B‐cell lineage. While Ly6D definitely enrich for cells representing committed B‐cell progenitors, the population is still heterogeneous and an upcoming challenge will be to find surface markers to improve prospective isolation protocols for these cells. Future attempts will benefit from index sorting where single‐cell gene expression signatures can be directly correlated to immunophenotype.
Another major objective of the study was to investigate whether EPLM cells were truly bi‐potent, an important question as it would not fit with the classical hierarchy. The authors looked for co‐expression of key myeloid and lymphoid genes and also performed functional assays, but found no support for a bi‐potent, lymph‐myeloid progenitor. Thus, the EPLM consists of multiple different subgroups of cells with different gene lineage signatures, where the Ly6D‐positive fraction harbors a B‐cell progenitor.
Understanding of B‐cell commitment and how it is regulated during normal development is crucial if we are to understand how the process is dysregulated in leukemia. The stage just before restriction to the B‐cell lineage has been less defined, and with this work, Rolink and colleagues add important knowledge to this part of early lymphoid commitment. No doubt, single‐cell RNA sequencing will be an important tool for resolving heterogeneity and lineage trajectories within both normal and malignant cell populations and will definitely challenge our understanding of lymphoid commitment and lineage specification in general.
Prof. Rolink has for many years contributed with his expertise to the field of hematopoiesis and B‐cell development in particular. It is with regret we received the news that he passed away. He will always be remembered for his innovative research and for being an excellent role model for other scientists in the field.
See also: L Alberti-Servera et al (December 2017)
References
- Alberti‐Servera L, von Muenchow L, Tsapogas P, Capoferri G, Eschbach K, Beisel C, Ceredig R, Ivanek R, Rolink A (2017) Single‐cell RNA sequencing reveals developmental heterogeneity among early lymphoid progenitors. EMBO J 36: 3619–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunaite G, Ceredig R, Massa S, Rolink AG (2005) A B220+ CD117+ CD19− hematopoietic progenitor with potent lymphoid and myeloid developmental potential. Eur J Immunol 35: 2019–2030 [DOI] [PubMed] [Google Scholar]
- Bell JJ, Bhandoola A (2008) The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 452: 764–767 [DOI] [PubMed] [Google Scholar]
- Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL (2009) Ly6d marks the earliest stage of B‐cell specification and identifies the branchpoint between B‐cell and T‐cell development. Genes Dev 23: 2376–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen CT, Strid T, Sigvardsson M (2016) Exploring the multifaceted nature of the common lymphoid progenitor compartment. Curr Opin Immunol 39: 121–126 [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA (2015) The technology and biology of single‐cell RNA sequencing. Mol Cell 58: 610–620 [DOI] [PubMed] [Google Scholar]
- Mansson R, Zandi S, Welinder E, Tsapogas P, Sakaguchi N, Bryder D, Sigvardsson M (2010) Single‐cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood 115: 2601–2609 [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL (2001) Stem cells, cancer, and cancer stem cells. Nature 414: 105–111 [DOI] [PubMed] [Google Scholar]
- Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR (2006) Lineage specification and plasticity in CD19− early B cell precursors. J Exp Med 203: 675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet‐Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W (2017) Comparative analysis of single‐cell RNA sequencing methods. Mol Cell 65: 631–643.e4 [DOI] [PubMed] [Google Scholar]


