Abstract
DNA is subjected to major cellular events, such as transcription, replication and DNA repair. To control these processes, the architecture of the DNA is tightly regulated. Recent work, including two studies in this issue of The EMBO Journal, provides compelling evidence that cohesin structures chromosomes through the processive enlargement of loops. While cohesin promotes chromosomal looping, it rather counteracts nuclear compartmentalization.
Subject Categories: Cell Cycle; Chromatin, Epigenetics, Genomics & Functional Genomics; DNA Replication, Repair & Recombination
The genome of a diploid human cell is almost 4 metres long, yet the diameter of a nucleus is only a few micrometres. Within this confined setting, the DNA needs to be folded in such a way that it allows for dynamic processes such as transcription and the DNA damage response. High‐resolution chromatin conformation capture experiments have revealed that the genome is organized into loops and compartments (Rao et al, 2014). Loops are cis contacts ranging up to a few megabases in size. The genome can roughly be divided into two compartments that reflect actively transcribed regions (A) and more repressed regions (B). How the loops are formed has long remained a mystery, and whether loops and compartments share any functional relationship was unknown.
Cohesin is a ring‐shaped protein complex that can entrap DNA inside its lumen. It holds together the sister chromatids from S phase until mitosis to ensure proper chromosome segregation. However, cohesin can also hold together DNA elements in cis to bring together two distant loci. It was already proposed 16 years ago that long‐range loops may be formed by the processive enlargement of tiny loops by the extrusion of DNA through cohesin rings (Nasmyth, 2001). More recently, in silico polymer simulations have revealed that loop extrusion by cohesin within defined genomic regions could in principle fully explain genome organization at the megabase scale (Sanborn et al, 2015; Fudenberg et al, 2016). Based upon the loop extrusion model, one can make a number of predictions. First, loops should simply be cohesin dependent. But more importantly, the duration with which cohesin embraces DNA should determine the length of the loops.
Genome organization by loop extrusion
The past year has seen a number of key publications that together provide overwhelming experimental evidence in support of the loop extrusion model. The cohesin dependency was tested directly through the use of different elegant systems that all allowed the efficient depletion of cohesin's SCC1 subunit (also known as RAD21). SCC1‐deficient cells lacked virtually all loops genomewide (Fig 1A). Cohesin therefore is essential for the formation and/or maintenance of loops (Gassler et al, 2017; Rao et al, 2017; Wutz et al, 2017). Re‐introduction of SCC1 in depleted cells led to the re‐appearance of loops within an hour (Rao et al, 2017). Cohesin‐dependent loop formation apparently is a very rapid process.
Figure 1. Cohesin‐dependent loop formation.
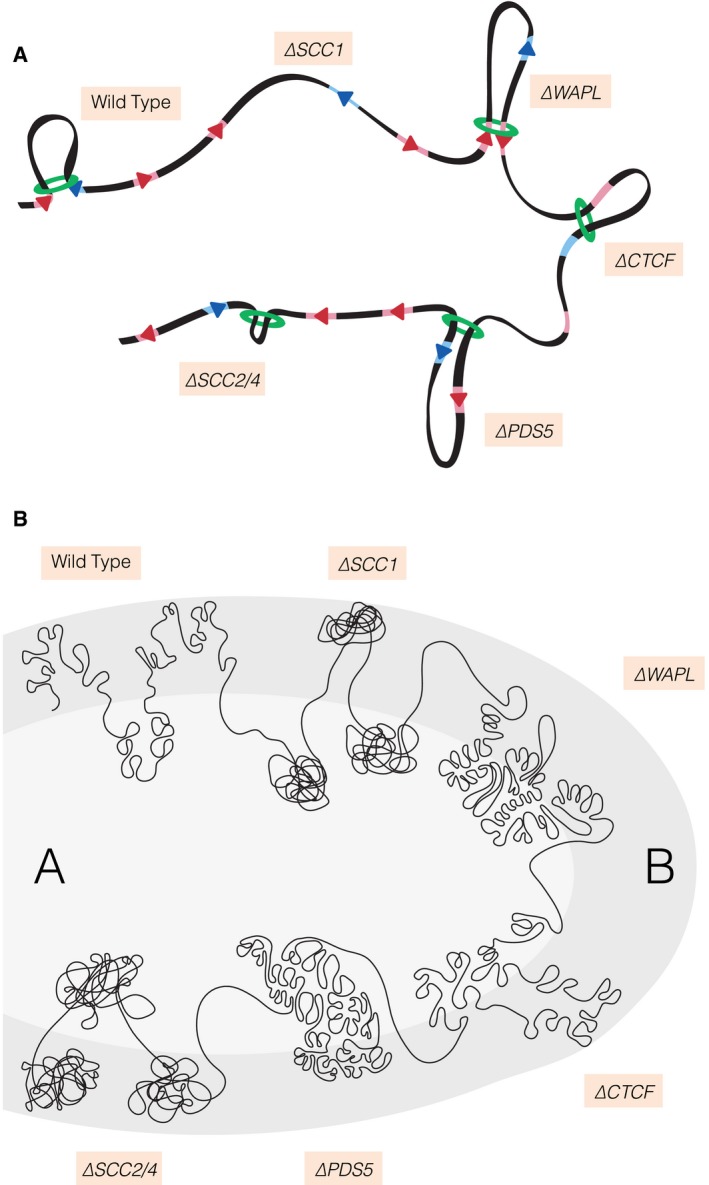
(A) Depletion of the core cohesin subunit SCC1 results in loss of loops. Cohesin is depicted as a green ring. Loss of the SCC2/SCC4 complex results in fewer and shorter loops, while removal of WAPL or PDS5 results in longer loops. CTCF (triangles) binds to CTCF sites (red for forward‐oriented and blue for reverse‐oriented sites) and acts as a boundary element that halts cohesin. (B) Cohesin‐dependent loops negatively affect compartmentalization. Compartments are less well defined if there is an increase in looping. Conversely, a decrease in looping results in a clearer distinction between compartments (light grey is A compartment, and dark grey is B compartment). The A compartment generally contains actively transcribed genes and lies in the inner nucleus, while the B compartment is more repressed and lies in the nuclear periphery.
Important as these findings are, they merely tell us that cohesin is required for loops. It does not provide insight into the mechanism by which the loops are formed. Cohesin binds to chromatin in a dynamic manner that involves a constant cycle of DNA entrapment and release. Cohesin's release factor WAPL is essential for this turnover. WAPL binds to cohesin's PDS5 subunit and opens a DNA exit gate to release cohesin from DNA. If cohesin were to form loops through a processive mechanism, the duration with which it entraps DNA may well determine how far loops can be enlarged. Three studies have now tested this hypothesis in different ways (Gassler et al, 2017; Haarhuis et al, 2017; Wutz et al, 2017). WAPL deficiency led to a far longer residence time of cohesin on the DNA. Importantly, this stabilization of cohesin on DNA led to longer loops genome‐wide (Fig 1A). The duration with which cohesin embraces DNA therefore indeed does determine the length of chromatin loops. This is the strongest evidence yet that cohesin structures the genome by loop extrusion. WAPL‐mediated cohesin turnover ensures that the 3D genome is kept dynamic by the continuous formation, loss and re‐formation of chromatin loops.
CTCF sets the boundaries
Cohesin primarily loops together CTCF sites. CTCF binds to DNA via a consensus motif that has a specific orientation. Intriguingly, loops are almost exclusively found between CTCF sites pointing towards each other (Rao et al, 2014). By using a system that acutely depletes CTCF, two papers have now investigated the genome‐wide role of CTCF. They show that CTCF is required for the formation of loops between CTCF sites (Fig 1A; Nora et al, 2017; Wutz et al, 2017). The formation of loops per se is not affected by CTCF depletion, as this rather is cohesin dependent (Gassler et al, 2017; Rao et al, 2017; Wutz et al, 2017). The available data would support the model that cohesin forms loops until it encounters an inwards‐pointing CTCF site. Here, CTCF then acts as a boundary that stops cohesin.
Thanks to WAPL‐mediated turnover, cohesin creates dynamic loops, but CTCF then determines the genomic regions within which these loops can be enlarged. How CTCF creates a boundary for cohesin is unclear. Possible scenarios would be that CTCF forms a physical barrier, that cohesin has affinity for CTCF, or that CTCF locally slows down cohesin's “extrusion activity”. Interestingly, Wutz et al (2017) show that PDS5 is required to maintain CTCF boundary integrity. PDS5 deficiency stabilizes cohesin on DNA, which leads to longer loops, but in contrast to WAPL‐deficient cells, cohesin now less frequently loops together CTCF sites (Fig 1A). How PDS5 in fact controls CTCF's boundary function remains an interesting question for the future.
A processivity factor?
Cohesin's loading onto DNA to a large degree is dependent on the cohesin loader complex consisting of SCC2 and SCC4 (also known as NIPBL and MAU2, respectively). In correspondence with the finding that cohesin is required for looping, these loading factors are also important for looping (Haarhuis et al, 2017; Schwarzer et al, 2017). Strikingly though, the SCC2/SCC4 complex is not only important for the amount of loops, but also for the length of the loops (Fig 1A; Haarhuis et al, 2017). What actually drives cohesin‐dependent loop enlargement remains unknown. Cohesin harbours an ABC‐like ATPase machinery whose activity is dependent on SCC2 (Murayama & Uhlmann, 2014). This raises the fascinating possibility that SCC2 controls cohesin's enzymatic activity and thereby its processivity on DNA to drive the enlargement of chromatin loops. Further studies will be needed to decipher how exactly SCC2 controls loop formation.
Cohesin counteracts nuclear compartmentalization
Whereas deficiency for either cohesin or one of its loader subunits impaired loop formation, this did not abrogate the formation of compartments (Gassler et al, 2017; Haarhuis et al, 2017; Rao et al, 2017; Schwarzer et al, 2017; Wutz et al, 2017). In fact, these cells displayed a more pronounced distinction between the A and B compartments. Conversely, compartmentalization was less pronounced in WAPL‐deficient cells (Gassler et al, 2017; Haarhuis et al, 2017; Wutz et al, 2017). Together this would suggest that there are two distinct modes of genome organization. One mode entails cohesin‐dependent loop formation, and the other is nuclear compartmentalization. Loop formation does however affect the latter mode, but in a negative sense (Fig 1B). It may well be that cohesin‐mediated looping limits the macro‐scale mobility of DNA within the nucleus and thereby antagonizes the forces that drive compartmentalization. Cohesin‐mediated loop formation could also be used to relocate DNA from one compartment to another to control gene expression.
All in all, these different papers have provided major insight into different aspects of chromosomal organization. We have learned a great deal about the basic principles behind loop formation, what determines which DNA elements are bridged together, what may drive loop formation, and even how all of this affects nuclear organization. It has been quite a year for chromosome biology.
See also: J Gassler et al (December 2017) and G Wutz et al (December 2017)
References
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA (2016) Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassler J, Brandão HB, Imakaev M, Flyamer IM, Ladstätter S, Bickmore WA, Peters JM, Mirny LA, Tachibana K (2017) A mechanism of cohesin‐dependent loop extrusion organizes zygotic genome architecture. EMBO J 36: 3600–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez‐Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, Brummelkamp TR, de Wit E, Rowland BD (2017) The cohesin release factor WAPL restricts chromatin loop extension. Cell 169: 693–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Uhlmann F (2014) Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505: 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35: 673–745 [DOI] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG (2017) Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer‐Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, Huang X, Shamim MS, Shin J, Turner D, Ye Z, Omer AD, Robinson JT, Schlick T, Bernstein BE, Casellas R et al (2017) Cohesin loss eliminates all loop domains. Cell 171: 305–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, Geeting KP, Gnirke A, Melnikov A, McKenna D, Stamenova EK, Lander ES, Aiden EL (2015) Chromatin extrusion explains key features of loop and domain formation in wild‐type and engineered genomes. Proc Natl Acad Sci USA 112: E6456–E6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe‐Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, Spitz F (2017) Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsitis RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, Walther N, Koch B, Kueblbeck M, Ellenberg J, Zuber J, Fraser P, Peters JM (2017) Topologically Associating Domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36: 3573–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]


