Abstract
Macroautophagy is an intracellular degradation system that involves the de novo formation of membrane structures called autophagosomes, although the detailed process by which membrane lipids are supplied during autophagosome formation is yet to be elucidated. Macroautophagy is thought to be associated with canonical membrane trafficking, but several mechanistic details are still missing. In this review, the current understanding and potential mechanisms by which membrane trafficking participates in macroautophagy are described, with a focus on the enigma of the membrane protein Atg9, for which the proximal mechanisms determining its movement are disputable, despite its key role in autophagosome formation.
Keywords: autophagy, endoplasmic reticulum, trafficking
Introduction
Autophagy is an intracellular degradation process that contributes to cellular homeostasis. Because autophagy involves highly dynamic membrane organization, it has occasionally been classified in the broad category of membrane trafficking. For example, microautophagy involves canonical membrane-trafficking events, as it is simply an invagination of the lysosomal membrane, and the mechanism whereby membrane lipids are mobilized is clearly understood [1]. However, in macroautophagy, the double-membrane structure, the autophagosome, engulfs some of the cellular contents and delivers them to the lysosome for degradation. Thus, macroautophagy involves the de novo formation of membrane structures, i.e. autophagosomes, and the detailed process by which membrane lipids are supplied during autophagosome formation is unclear. The basic dogma of membrane biogenesis is that an organelle is generated from a pre-existing organelle. Therefore, various hypotheses have been proposed to explain the formation of de novo membrane structures, such as the involvement of unknown membrane vesicles and the free transfer of lipid molecules [2]. Here, the current understanding and hypotheses regarding the role of membrane trafficking in macroautophagy are summarized, with a focus on the enigma of Atg9, i.e. the mechanism whereby it arrives at the autophagosome-formation site.
Platform for autophagosome formation: omegasome and pre-autophagosomal structure
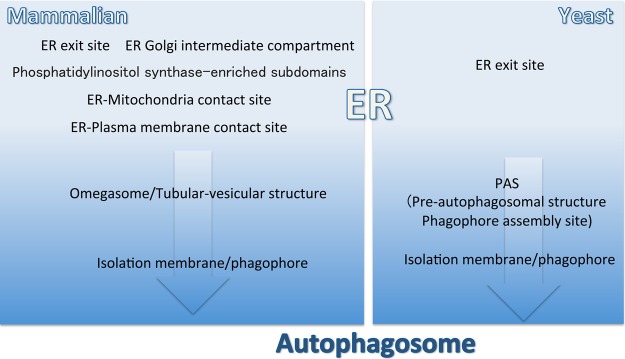
The discovery of the omegasome was a major breakthrough in autophagy research with respect to the membrane-trafficking system. The double FYVE domain-containing protein (DFCP-1) binds to phosphatidylinositol 3-phosphate (PI3P) [3]. Specific types of phosphoinositides are exclusively localized to specific organelles and the endoplasmic reticulum (ER) and Golgi are regarded as being devoid of devoid of PI3P. These findings seem to contradict the fact that DFCP-1 is localized to the ER and Golgi apparatus. During starvation, DFCP-1 accumulates in or near the ER, forming a structure resembling the Greek character omegasome [4] (Figure 1). This accumulation is entirely dependent on the PI3P-binding capacity of the FYVE domain, indicating that PI3P exists in the omegasome. Strikingly, autophagosomes are generated inside, or in close proximity to, the omegasome, suggesting that autophagosomes form in specialized sites within the cell [4]. PI3 kinase, which specifically functions in autophagy and contains the Atg14L subunit, is recruited to the ER, and PI3P generated by the PI3 kinase complex is required for omegasome formation [5]. Detailed electron microscopy utilizing 3D tomography revealed a physical membranous connection between the ER and the forming autophagosome, i.e. the isolation membrane or phagophore [6,7] (Figure 1). More detailed serial and tomographic electron microscopy has shown that a green fluorescence protein–DFCP-1 fusion is localized to the tubular vesicular structure adjacent to the isolation membrane, representing at least part of the omegasome [8] (Figure 1). The tubular vesicular structure was observed as a potentially more nascent seed structure labeled with Atg13 before double-membrane autophagosome formation was initiated [9]. The ER–Golgi-intermediate compartment (ERGIC) is a tubular vesicular organelle that functions in transport from the ER to the Golgi apparatus. Data from an in vitro-reconstitution study suggested that the ERGIC plays a pivotal role in autophagosome formation [10]. ER-exit sites (ERESs) are specialized in the ER involved in the biogenesis of COPII-coated transport vesicles that traffic to the Golgi. In vivo observations by super-resolution microscopy suggest that autophagosome formation takes place, in part, in the vicinity of the ERES and ERGIC, but does not necessarily always occur in this area [9] (Figure 1). FIP200 and a SEC12-binding protein, CTAGE5, participate in remodeling ERESs and may contribute to autophagosome formation [11]. It has recently been reported that specialized regions of the ER are crucial for autophagosome formation. The ER functions in the biosynthesis of membrane lipids, including phosphatidylinositol (PI). The enzyme responsible for PI synthesis (PI synthase) is a transmembrane protein that is mostly localized to the ER, but is enriched in a less well-characterized, highly mobile compartment with an ER origin [12]. Interestingly, the autophagy-related proteins FIP200 and Ulk1, which are now thought to determine the initiation of autophagosome formation, are transiently colocalized with PI synthase in membranes, suggesting a crucial role of this organelle in autophagosome formation [13] (Figure 1). Other reports have shown that autophagosome formation occurs in specialized regions of the ER, such as ER–mitochondria- and ER–plasma membrane-contact sites [14,15] (Figure 1). It is necessary to determine how these specialized regions of the ER are functionally connected to tubular vesicular structure-containing omegasomes.
Figure 1.
Relationships between the endoplasmic reticulum and the autophagosome-formation site in mammalian and yeast cells.
Unlike mammalian autophagy, autophagosome formation in the yeast Saccharomyces cerevisiae is thought to occur primarily in the pre-autophagosomal structure (PAS; alternatively known as the phagophore-assembly site). The PAS is characterized as the site where Atg proteins gather [16,17] (Figure 1). The organization of the PAS has gradually become apparent; the dephosphorylation of Atg13 in response to TORC1 suppression during starvation is a key trigger of PAS organization [18]. The PAS is always associated with vacuoles, corresponding to lysosomes in mammals, although only one or a few vacuoles are present in each cell [16,17]. Autophagosome formation and vacuole fusion are interconnected, at least in some types of autophagy involving the Cvt pathway [19]. More importantly, a physical connection appears to exist between the ER and PAS (Figure 1). Markers of ERESs are frequently observed in the vicinity of the PAS [20,21] (Figure 1). Additionally, in COPII mutants, severe autophagy defects have been observed [22–24]. An ER-resident SNARE protein, Ufe1, is translocated to the autophagosome-formation site during autophagy [25]. Therefore, it is highly possible that autophagosome formation may be linked with the ER, as in mammalian cells.
Atg9 vesicles in yeast: a membrane source of autophagosomes or just a reservoir?
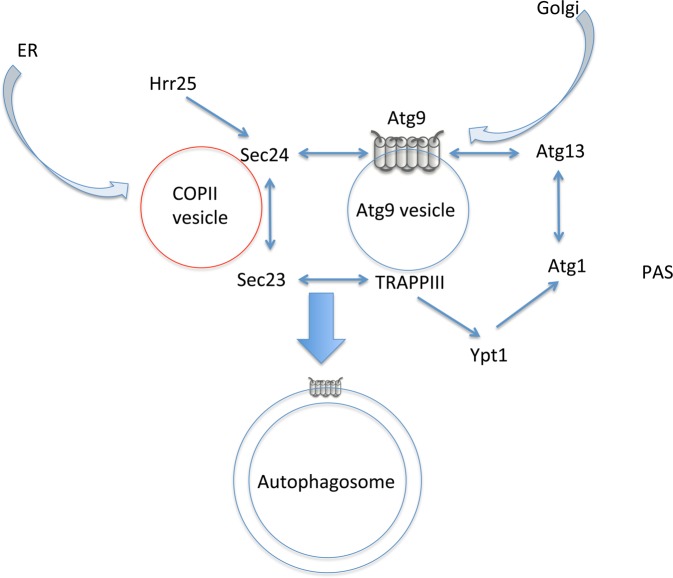
Regarding the relationship between autophagy and membrane trafficking, a key question is related to Atg9, which is a six-transmembrane protein essential for autophagosome formation that was first identified in yeast [26,27] (Figure 2). Atg9 is localized to the outer membrane of double autophagosomal membranes in yeast [28] (Figure 2). In addition to autophagosomes, Atg9 is localized to PASs and numerous cytoplasmic vesicular structures with a 30–60 nm diameter, which move quite rapidly throughout the cytoplasm [28] (Figure 2). Atg9 is thought to cycle between the PAS/autophagosome and the peripheral vesicular pools [29]. Without Atg1 kinase, Atg9 accumulates at the PAS, suggesting that proper autophagosome formation requires this cycling [29]. Atg9 associates with the HORMA domain of Atg13 at the PAS [30] (Figure 2) and then associates with Atg2 and Atg18 [31]. Therefore, Atg9 plays a direct, crucial role in autophagosome formation at the PAS, but its actual function remains unclear.
Figure 2.
Model whereby Atg9 vesicles serve as a direct source of autophagosome membrane in yeast cells.
A key question is how Atg9 arrives at the PAS. Based on the canonical concept that a transmembrane protein does not leave the membrane during membrane trafficking, these cytoplasmic Atg9 proteins in small vesicles and PAS/autophagosomes are expected to be interconnected by canonical membrane-trafficking processes, such as budding and fusion. Two different models have been used to explain the movement of Atg9. One model holds that Atg9-containing vesicles directly arrive at the PAS and the Atg9-vesicular membrane constitutes, at least part of, the autophagosome membrane. An alternative hypothesis is that Atg9 arrives by another route, such as the ER, and this possibility should be carefully considered. Hereafter, these two views will be described in detail.
My interpretation is that the former view stems from the idea that the identities of Atg9-containing vesicles are uniform; these are termed ‘Atg9 vesicles’ [28,32]. It is clear that Atg9 vesicles originate from the Golgi apparatus [28,33] (Figure 2). These vesicles contain factors required for fusion to the nascent autophagosome, such as Trs85 (the sole specific subunit of TRAPPIII) [34], which functions as a guanine nucleotide-exchange factors with Ypt1 [35] (Figure 2). Ypt1 is the yeast homolog of the Rab1 small GTPase; it was originally described as functioning in ER-to-Golgi trafficking [35]. Ypt1 recruits Atg1 to the PAS [32] (Figure 2). TRAPPIII also interacts with Sec23, a subunit of the COPII coat, during vesicle budding from the ER [36] (Figure 2). Another subunit of COPII, Sec24, is phosphorylated by the Hrr25 protein kinase during autophagy, inducing starvation; thereby, it becomes associated with Atg9 [37] (Figure 2). Thus, it has been proposed that Atg9 vesicles and COPII vesicles fuse and form the autophagosome membranes [37,38] (Figure 2). Other membrane sources, such as COPII vesicles, may well be involved in autophagosome formation, because the estimated number of membrane lipids supplied from Atg9 vesicles (calculated from the number of Atg9 molecules on autophagosomes) is not sufficient for the autophagosome membrane [28].
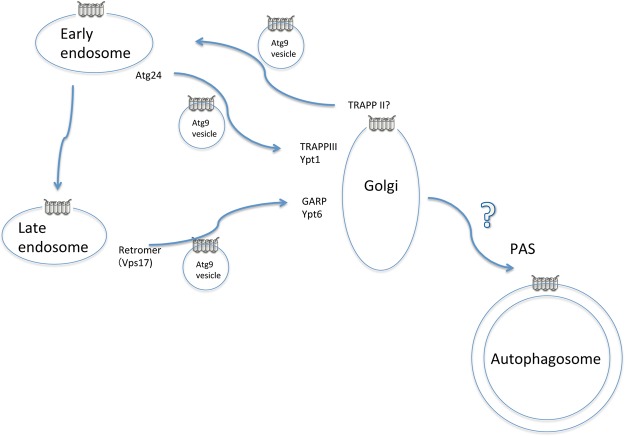
Another view is based on the idea that the so-called Atg9 vesicles are not homogeneous entities, but consist of a mixture of transport vesicles transiting between the Golgi apparatus and endosomes instead (Figure 3). Historically, the involvement of endosomal trafficking has not received substantial attention in yeast autophagy, because endosomal trafficking mutants exhibited no or highly limited defects in autophagy. However, Atg9 is partially distributed to the early and late endosomes [33]. Ohashi and Munro [39] noted that some double mutants for factors involved in endosomal trafficking show severe defects in autophagy and Atg9 trafficking.
Figure 3.
Model whereby Atg9 vesicles function as a reservoir of Atg9 molecules in yeast cells.
This discovery has since been verified using a simpler model [40]. Specifically, Trs85 mutants showed severe defects in the Cvt pathway, a selective autophagy type that occurs even under nutrient-rich conditions, but showed only partial defects during starvation-induced macroautophagy [41]. GARP is a tethering factor complex that receives vesicles from the endosome at the Golgi [42]. GARP mutants show severe defects in the Cvt pathway, but not in starvation-induced macroautophagy [43]. Two small G proteins (Ypt6 and Arl1) appear to be involved in autophagy, at least in the GARP-dependent step [44] (Figure 3). Importantly, the combination of mutations in the GARP subunits vps51 and trs85 results in almost completely defective starvation-induced macroautophagy, and Atg9 trafficking to PAS is abrogated [40] (Figure 3). Likewise, trs85 mutations result in severe defects in autophagy when combined with mutations in the retromer subunit Vps17, which generates vesicles for late endosome-to-Golgi retrograde trafficking [41] (Figure 3).
Atg24 is a sorting nexin involved in early endosome-to-Golgi trafficking [45]. Atg24 mutant shows severe defects in the Cvt pathway, similar to the trs85 mutant, but does not show defects in starvation-induced autophagy [46]. In contrast with the vps51 mutant, double mutations in atg24 and trs85 do not have synergistic effects on autophagy and Atg9 trafficking [40] (Figure 3). Therefore, Trs85 is thought to function in the same pathway as Atg24, i.e. early endosome-to-Golgi trafficking [40] (Figure 3). Consistent with these findings, Trs85 is localized to the Golgi [40]. TRAPP III is a guanine nucleotide-exchange factor for Ypt1, and Ypt1 and its GTPase activator gyp1 mutants are also defective in early endosome-to-Golgi trafficking and autophagy [35,47–49]. Therefore, I propose that the defects in autophagy associated with trs85 mutants are due to impaired early endosome-to-Golgi trafficking of Atg9 (Figure 3). These trs85-derived, autophagy-defective phenotypes (under nutrient-rich conditions) are bypassed during starvation by rerouting Atg9 to the Golgi via late endosomes; however, when additional blockages between the late endosome and Golgi occur, Atg9 cannot reach the Golgi and autophagy is defective (Figure 3). A mutant of TRAPPII, a guanine nucleotide-exchange factor for Ypt31/32, exhibits defective membrane trafficking, particularly exiting the Golgi toward the endosome and plasma membrane [50]. The TRAPPII mutant is also defected in Atg9 trafficking [51]; therefore, it is highly possible that TRAPPII-dependent Atg9 trafficking from the Golgi is important (Figure 3). Taking these findings together, I propose that ‘Atg9 vesicles’ are not homogeneous, but form a heterogeneous population representing vesicles found between the Golgi and the endosome (Figure 3) instead. One interpretation of this physiological result is that reserving a membrane protein as cargo of general membrane trafficking may result in less load balancing, compared with reserving in some specialized vesicles.
Atg9 trafficking is supported by other, potentially specific proteins, i.e. Atg27 and Atg23; without these proteins, Atg9 vesicle formation and delivery to the PAS are severely impaired [28,52,53]. Atg23 is a peripheral membrane protein and its localization overlaps substantially with Atg9 peripheral pools, suggesting that it facilitates Atg9 trafficking [33,54]. Atg27 is a single transmembrane protein and localizes to the Golgi, endosomes, and vacuoles, and travels among these sites [52,55]. Atg9 is phosphorylated by Atg1 at PAS, which affects the association efficiency with Atg23, Atg27, and Atg18 [56,57]. In addition to its role in facilitating Atg9 trafficking in the reservoir pool, Atg27 may be involved in the retrieval of Atg9 from the vacuole [55]. The return of Atg27 from the early endosome to the Golgi largely depends on the Snx41–Snx4 (Atg24)-sorting nexin complex, which is different from the Atg9-related Atg20–Snx4 (Atg24) complex [58]. This distinction may be important in the regulation of Atg9 trafficking.
Atg9 trafficking in mammalian systems
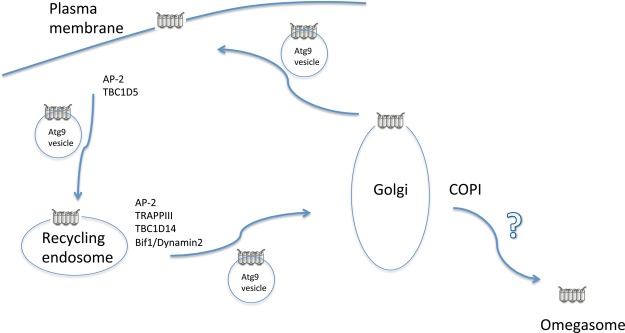
The latter model that Atg9 vesicles are heterogeneous is rather consistent with observations of mammalian ATG9 trafficking (Figure 4). Even in mammals, ATG9 clearly plays a crucial role in autophagosome formation [59–62]. Early studies showed that ATG9 is localized to both the Golgi and endosomes [59]. Live cell imaging has revealed that multiple ATG9-containing vesicles move around in the cytoplasm and that the associations between ATG9-positive areas and areas positive for other Atg markers, such as LC3 and DFCP1, are transient [63,64] (Figure 4). It later became clear that at least some ATG9 molecules reside in recycling endosomes in addition to the Golgi [60,63,65] (Figure 4). ATG9 also travels to the plasma membrane and returns via clathrin-dependent endocytosis using the clathrin coat protein complex AP-2, together with the Rab GAP protein TBC1D5 [61] (Figure 4). It is unclear whether Atg9 is also transported to the plasma membrane in yeast, although the involvement of late secretory pathway components in the plasma membrane has been reported in autophagy [66,67]. Mammalian AP-2 recognizes tyrosine-based and di-leucine-based sorting motifs residing in the N-terminal flanking stretch of ATG9 exposed to the cytosolic face; a sorting motif-deficient ATG9 mutant exhibits accumulation in the recycling endosome [62] (Figure 4). Wild-type ATG9 accumulated in the Golgi, although an ATG9 mutant lacking the tyrosine- and di-leucine-based sorting motifs did not, suggesting that ATG9 returned to the Golgi from the recycling endosome [62]. Sorting nexin 18 functions in the tubulation of the recycling endosome to promote autophagy [68]. The requirement for an endosomal coatomer in autophagy may also relate to this process [69]. The ATG9-containing vesicle detaches from the recycling endosome via the membrane fission machinery containing Bif-1 and Dynamin 2 [70] (Figure 4).
Figure 4.
Model of Atg9 dynamics in mammalian cells.
TRAPPIII is also involved in mammalian autophagy, and the suppression of its specific subunit TRAPPC8 (Trs85 homolog) or TRAPPC13 results in autophagy defects [62,71–73]. TRAPPIII defects are related to retrograde trafficking of several cargo molecules from the endosome to the Golgi, including Shiga-toxin and ATG9 [62,72–74] (Figure 4). TRAPPC8 interacts with TBC1D14, a negative regulator of autophagy that controls the delivery of membranes from RAB11-positive recycling endosomes to forming autophagosomes [60,72] (Figure 4). Rab1 is also important for autophagy in mammalian cells [75]. TRAPPIII and TBC1D14 affect Rab1, which controls ATG9, similar to the yeast counterpart Ypt1 [72]. In addition to ATG9, another ATG16L-positive vesicle has been identified as a potential autophagosome precursor, but this result may reflect an artifact due to transient overexpression [76,77].
Do we need a new model to explain Atg9 trafficking?
Theoretically, both models (i.e. Atg9 vesicles help form autophagosome membranes, and Atg9 vesicles are reservoirs of Atg9 molecules) are not mutually exclusive, and it is possible that some ‘Atg9 vesicles’ can be directly targeted to the PAS and omegasome. However, the latter model proposes that most ‘Atg9 vesicles’ moving around the cytoplasm represent the Atg9 reservoir pooled in transport vesicles before functioning in the PAS or omegasome. If this is the case, the delivery route from the reservoir to the PAS may be indirect, and several other pathways should be carefully considered. Another model that explains the route from the Golgi to PAS and omegasomes should be established. It is quite possible that before arriving at the PAS and omegasome, Atg9 arrives at the Golgi because the failure to arrive in the Golgi is coupled with autophagy defects [40,62]. For example, Ypt31/32 are involved in vesicle exit from the Golgi, and their mutants show defective autophagy without impaired Atg9 delivery to the PAS [66]. These findings suggest that forward traffic from the Golgi is dispensable, at least for Atg9 delivery to the PAS. Interestingly, the COG complex is also involved in autophagy via Atg9 trafficking [78–80]. COG function in retrograde traffic within the Golgi may be an effector of Ypt1, highlighting the possibility that retrograde traffic inside the Golgi and to the ER may be important [81]. In mammalian cells, COPI-mediated retrograde traffic from the Golgi to the ER is required for autophagy [9]. Calpain is also implicated in playing some role in mobilizing ATG9 from Golgi stacks; therefore, Calpain-dependent processes may be related to Atg9 delivery to autophagosomes [82]. ATG9, localized on tubular appendixes of the ER, subsequently associates with ATG13-positive particles, which grow into autophagosomes [9]. In both yeast and mammals, Atg9-positive tubular vesicular structures have been observed in the vicinity of nascent autophagosomes [33,63]. An interesting possibility is that the lipid composition of such structures plays some roles. Autophagosome formation depends on desaturated fatty acid [83] and at least in yeast, the requisite step is at the Atg9 delivery to PAS [84]. By clarifying the identities of these Atg9-positive signals involved in autophagosome formation, it will be possible to resolve the enigma regarding the movement of Atg9 vesicles.
Abbreviations
- DFCP-1
double FYVE domain-containing protein
- ER
endoplasmic reticulum
- ERESs
ER-exit sites
- ERGIC
ER–Golgi-intermediate compartment
- PAS
pre-autophagosomal structure
- PI
phosphatidylinositol
- PI3P
phosphatidylinositol 3-phosphate
Competing Interests
The Author declares that there are no competing interests associated with this manuscript.
References
- 1.Mijaljica D., Prescott M. and Devenish R.J. (2011) Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 7, 673–682 10.4161/auto.7.7.14733 [DOI] [PubMed] [Google Scholar]
- 2.Noda T., Suzuki K. and Ohsumi Y. (2002) Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 12, 231–235 10.1016/S0962-8924(02)02278-X [DOI] [PubMed] [Google Scholar]
- 3.Cheung P.C., Trinkle-Mulcahy L., Cohen P. and Lucocq J.M. (2001) Characterization of a novel phosphatidylinositol 3-phosphate-binding protein containing two FYVE fingers in tandem that is targeted to the Golgi. Biochem. J. 355, 113–121 10.1042/bj3550113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A. et al. (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsunaga K., Morita E., Saitoh T., Akira S., Ktistakis N.T., Izumi T. et al. (2010) Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 190, 511–521 10.1083/jcb.200911141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ylä-Anttila P., Vihinen H., Jokitalo E. and Eskelinen E.-L. (2009) 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5, 1180–1185 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- 7.Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T. and Yamamoto A. (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- 8.Uemura T., Yamamoto M., Kametaka A., Sou Y.-s., Yabashi A., Yamada A. et al. (2014) A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol. Cell. Biol. 34, 1695–1706 10.1128/MCB.01327-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karanasios E., Walker S.A., Okkenhaug H., Manifava M., Hummel E., Zimmermann H. et al. (2016) Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 7, 12420 10.1038/ncomms12420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge L., Zhang M. and Schekman R. (2014) Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife 3, e04135 10.7554/eLife.04135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge L., Zhang M., Kenny S.J., Liu D., Maeda M., Saito K. et al. (2017) Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 18, 1586–1603 10.15252/embr.201744559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y.J., Guzman-Hernandez M.L. and Balla T. (2011) A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell 21, 813–824 10.1016/j.devcel.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimura T., Tamura N., Kono N., Shimanaka Y., Arai H., Yamamoto H. et al. (2017) Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 36, 1719–1735 10.15252/embj.201695189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N. et al. (2013) Autophagosomes form at ER–mitochondria contact sites. Nature 495, 389–393 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- 15.Nascimbeni A.C., Giordano F., Dupont N., Grasso D., Vaccaro M.I., Codogno P. et al. (2017) ER–plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J. 36, 2018–2033 10.15252/embj.201797006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T. and Ohsumi Y. (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981 10.1093/emboj/20.21.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J., Huang W.-P., Stromhaug P.E. and Klionsky D.J. (2002) Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277, 763–773 10.1074/jbc.M109134200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noda T. (2017) Regulation of autophagy through TORC1 and mTORC1. Biomolecules 7, 52 10.3390/biom7030052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X., Mao K., Yu A.Y.H., Omairi-Nasser A., Austin J., Glick B.S. et al. (2016) The Atg17-Atg31-Atg29 complex coordinates with Atg11 to recruit the Vam7 SNARE and mediate autophagosome-vacuole fusion. Curr. Biol. 26, 150–160 10.1016/j.cub.2015.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graef M., Friedman J.R., Graham C., Babu M. and Nunnari J. (2013) ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 24, 2918–2931 10.1091/mbc.E13-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki K., Akioka M., Kondo-Kakuta C., Yamamoto H. and Ohsumi Y. (2013) Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 126, 2534–2544 10.1242/jcs.122960 [DOI] [PubMed] [Google Scholar]
- 22.Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A. et al. (2001) Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 12, 3690–3702 10.1091/mbc.12.11.3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamasaki M., Noda T. and Ohsumi Y. (2003) The early secretory pathway contributes to autophagy in yeast. Cell Struct. Funct. 28, 49–54 10.1247/csf.28.49 [DOI] [PubMed] [Google Scholar]
- 24.Reggiori F., Wang C.-W., Nair U., Shintani T., Abeliovich H. and Klionsky D.J. (2004) Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 2189–2204 10.1091/mbc.E03-07-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemus L., Ribas J.L., Sikorska N. and Goder V. (2016) An ER-localized SNARE protein is exported in specific COPII vesicles for autophagosome biogenesis. Cell Rep. 14, 1710–1722 10.1016/j.celrep.2016.01.047 [DOI] [PubMed] [Google Scholar]
- 26.Noda T., Kim J., Huang W.-P., Baba M., Tokunaga C., Ohsumi Y. et al. (2000) Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465–480 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang T., Reiche S., Straub M., Bredschneider M. and Thumm M. (2000) Autophagy and the cvt pathway both depend on AUT9. J. Bacteriol. 182, 2125–2133 10.1128/JB.182.8.2125-2133.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto H., Kakuta S., Watanabe T.M., Kitamura A., Sekito T., Kondo-Kakuta C. et al. (2012) Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 198, 219–233 10.1083/jcb.201202061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reggiori F., Tucker K.A., Stromhaug P.E. and Klionsky D.J. (2004) The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6, 79–90 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- 30.Suzuki S.W., Yamamoto H., Oikawa Y., Kondo-Kakuta C., Kimura Y., Hirano H. et al. (2015) Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc. Natl Acad. Sci. U.S.A. 112, 3350–3355 10.1073/pnas.1421092112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C.-W., Kim J., Huang W.-P., Abeliovich H., Stromhaug P.E., Dunn W.A. Jr et al. (2001) Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J. Biol. Chem. 276, 30442–30451 10.1074/jbc.M102342200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., Menon S., Yamasaki A., Chou H.-T., Walz T., Jiang Y. et al. (2013) Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl Acad. Sci. U.S.A. 110, 9800–9805 10.1073/pnas.1302337110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D.J. and Reggiori F. (2010) An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190, 1005–1022 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kakuta S., Yamamoto H., Negishi L., Kondo-Kakuta C., Hayashi N. and Ohsumi Y. (2012) Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J. Biol. Chem. 287, 44261–44269 10.1074/jbc.M112.411454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch-Day M.A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C.R. et al. (2010) Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl Acad. Sci. U.S.A. 107, 7811–7816 10.1073/pnas.1000063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan D., Cai Y., Wang J., Zhang J., Menon S., Chou H.-T. et al. (2013) The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl Acad. Sci. U.S.A. 110, 19432–19437 10.1073/pnas.1316356110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis S., Wang J., Zhu M., Stahmer K., Lakshminarayan R., Ghassemian M. et al. (2016) Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. eLife 5, e21167 10.7554/eLife.21167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farhan H., Kundu M. and Ferro-Novick S. (2017) The link between autophagy and secretion: a story of multitasking proteins. Mol. Biol. Cell 28, 1161–1164 10.1091/mbc.E16-11-0762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohashi Y. and Munro S. (2010) Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol. Biol. Cell 21, 3998–4008 10.1091/mbc.E10-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirahama-Noda K., Kira S., Yoshimori T. and Noda T. (2013) TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J. Cell Sci. 126, 4963–4973 10.1242/jcs.131318 [DOI] [PubMed] [Google Scholar]
- 41.Meiling-Wesse K., Epple U.D., Krick R., Barth H., Appelles A., Voss C. et al. (2005) Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 280, 33669–33678 10.1074/jbc.M501701200 [DOI] [PubMed] [Google Scholar]
- 42.Bonifacino J.S. and Hierro A. (2011) Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol. 21, 159–167 10.1016/j.tcb.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reggiori F., Wang C.-W., Stromhaug P.E., Shintani T. and Klionsky D.J. (2003) Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 278, 5009–5020 10.1074/jbc.M210436200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang S. and Rosenwald A.G. (2016) Autophagy in Saccharomyces cerevisiae requires the monomeric GTP-binding proteins, Arl1 and Ypt6. Autophagy 12, 1721–1737 10.1080/15548627.2016.1196316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hettema E.H., Lewis M.J., Black M.W. and Pelham H.R. (2003) Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22, 548–557 10.1093/emboj/cdg062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nice D.C., Sato T.K., Stromhaug P.E., Emr S.D. and Klionsky D.J. (2002) Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277, 30198–30207 10.1074/jbc.M204736200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sclafani A., Chen S., Rivera-Molina F., Reinisch K., Novick P.J. and Ferro-Novick S. (2010) Establishing a role for the GTPase Ypt1p at the late Golgi. Traffic 11, 520–532 10.1111/j.1600-0854.2010.01031.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lafourcade C., Galan J.-M., Gloor Y., Haguenauer-Tsapis R. and Peter M. (2004) The GTPase-activating enzyme Gyp1p is required for recycling of internalized membrane material by inactivation of the Rab/Ypt GTPase Ypt1p. Mol. Cell. Biol. 24, 3815–3826 10.1128/MCB.24.9.3815-3826.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kramer M.H., Farré J.-C., Mitra K., Yu M.K., Ono K., Demchak B. et al. (2017) Active interaction mapping reveals the hierarchical organization of autophagy. Mol. Cell 65, 761–774.e5 10.1016/j.molcel.2016.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sciorra V.A., Audhya A., Parsons A.B., Segev N., Boone C. and Emr S.D. (2005) Synthetic genetic array analysis of the PtdIns 4-kinase Pik1p identifies components in a Golgi-specific Ypt31/rab-GTPase signaling pathway. Mol. Biol. Cell 16, 776–793 10.1091/mbc.E04-08-0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou S., Chen Y., Liu Y., Segev N., Yu S., Liu Y. et al. (2013) Trs130 participates in autophagy through GTPases Ypt31/32 in Saccharomyces cerevisiae. Traffic 14, 233–246 10.1111/tra.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yen W.-L., Legakis J.E., Nair U. and Klionsky D.J. (2007) Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell 18, 581–593 10.1091/mbc.E06-07-0612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Backues S.K., Orban D.P., Bernard A., Singh K., Cao Y. and Klionsky D.J. (2015) Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae. Traffic 16, 172–190 10.1111/tra.12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tucker K.A., Reggiori F., Dunn W.A. Jr and Klionsky D.J. (2003) Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J. Biol. Chem. 278, 48445–48452 10.1074/jbc.M309238200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segarra V.A., Boettner D.R. and Lemmon S.K. (2015) Atg27 tyrosine sorting motif is important for its trafficking and Atg9 localization. Traffic 16, 365–378 10.1111/tra.12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng Y., Backues S.K., Baba M., Heo J.-M., Harper J.W. and Klionsky D.J. (2016) Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy 12, 648–658 10.1080/15548627.2016.1157237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papinski D., Schuschnig M., Reiter W., Wilhelm L., Barnes C.A., Maiolica A. et al. (2014) Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol. Cell 53, 471–483 10.1016/j.molcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma M., Burd C.G. and Chi R.J. (2017) Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic 18, 134–144 10.1111/tra.12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young A.R.J., Chan E.Y.W., Hu X.W., Köchl R., Crawshaw S.G., High S. et al. (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- 60.Longatti A., Lamb C.A., Razi M., Yoshimura S.-i., Barr F.A. and Tooze S.A. (2012) TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J. Cell Biol. 197, 659–675 10.1083/jcb.201111079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Popovic D. and Dikic I. (2014) TBC1D5 and the AP2 complex regulate ATG9 trafficking and initiation of autophagy. EMBO Rep. 15, 392–401 10.1002/embr.201337995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imai K., Hao F., Fujita N., Tsuji Y., Oe Y., Araki Y. et al. (2016) Atg9a trafficking through the recycling endosomes is required for autophagosome formation. J. Cell Sci. 129, 3781–3791 10.1242/jcs.196196 [DOI] [PubMed] [Google Scholar]
- 63.Orsi A., Razi M., Dooley H.C., Robinson D., Weston A.E., Collinson L.M. et al. (2012) Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 23, 1860–1873 10.1091/mbc.E11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koyama-Honda I., Itakura E., Fujiwara T.K. and Mizushima N. (2013) Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 9, 1491–1499 10.4161/auto.25529 [DOI] [PubMed] [Google Scholar]
- 65.Puri C., Renna M., Bento C.F., Moreau K. and Rubinsztein D.C. (2013) Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154, 1285–1299 10.1016/j.cell.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geng J., Nair U., Yasumura-Yorimitsu K. and Klionsky D.J. (2010) Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 21, 2257–2269 10.1091/mbc.E09-11-0969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nair U., Jotwani A., Geng J., Gammoh N., Richerson D., Yen W.-L. et al. (2011) SNARE proteins are required for macroautophagy. Cell 146, 290–302 10.1016/j.cell.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knævelsrud H., Søreng K., Raiborg C., Håberg K., Rasmuson F., Brech A. et al. (2013) Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J. Cell Biol. 202, 331–349 10.1083/jcb.201205129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Razi M., Chan E.Y.W. and Tooze S.A. (2009) Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 185, 305–321 10.1083/jcb.200810098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takahashi Y., Tsotakos N., Liu Y., Young M.M., Serfass J., Tang Z. et al. (2016) The Bif-1-Dynamin 2 membrane fission machinery regulates Atg9-containing vesicle generation at the Rab11-positive reservoirs. Oncotarget 7, 20855–20868 10.18632/oncotarget.8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Behrends C., Sowa M.E., Gygi S.P. and Harper J.W. (2010) Network organization of the human autophagy system. Nature 466, 68–76 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lamb C.A., Nühlen S., Judith D., Frith D., Snijders A.P., Behrends C. et al. (2016) TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 35, 281–301 10.15252/embj.201592695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramírez-Peinado S., Ignashkova T.I., van Raam B.J., Baumann J., Sennott E.L., Gendarme M. et al. (2017) TRAPPC13 modulates autophagy and the response to Golgi stress. J. Cell Sci. 130, 2251–2265 10.1242/jcs.199521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bassik M.C., Kampmann M., Lebbink R.J., Wang S., Hein M.Y., Poser I. et al. (2013) A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922 10.1016/j.cell.2013.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang J., Birmingham C.L., Shahnazari S., Shiu J., Zheng Y.T., Smith A.C. et al. (2011) Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy 7, 17–26 10.4161/auto.7.1.13840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moreau K., Ravikumar B., Renna M., Puri C. and Rubinsztein D.C. (2011) Autophagosome precursor maturation requires homotypic fusion. Cell 146, 303–317 10.1016/j.cell.2011.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J., Chen Z., Stang M.T. and Gao W. (2017) Transiently expressed ATG16L1 inhibits autophagosome biogenesis and aberrantly targets RAB11-positive recycling endosomes. Autophagy 13, 345–358 10.1080/15548627.2016.1256521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yen W.-L., Shintani T., Nair U., Cao Y., Richardson B.C., Li Z. et al. (2010) The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 188, 101–114 10.1083/jcb.200904075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller V.J. and Ungar D. (2012) Re‘COG'nition at the Golgi. Traffic 13, 891–897 10.1111/j.1600-0854.2012.01338.x [DOI] [PubMed] [Google Scholar]
- 80.Wang I.-H., Chen Y.-J., Hsu J.-W. and Lee F.-S. (2017) The Arl3 and Arl1 GTPases co-operate with Cog8 to regulate selective autophagy via Atg9 trafficking. Traffic 18, 580–589 10.1111/tra.12498 [DOI] [PubMed] [Google Scholar]
- 81.Suvorova E.S., Duden R. and Lupashin V.V. (2002) The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157, 631–643 10.1083/jcb.200111081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marcassa E., Raimondi M., Anwar T., Eskelinen E.-L., Myers M.P., Triolo G. et al. (2017) Calpain mobilizes Atg9/Bif-1 vesicles from Golgi stacks upon autophagy induction by thapsigargin. Biol. Open 6, 551–562 10.1242/bio.022806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ogasawara Y., Itakura E., Kono N., Mizushima N., Arai H., Nara A. et al. (2014) Stearoyl-CoA desaturase 1 activity is required for autophagosome formation. J. Biol. Chem. 289, 23938–23950 10.1074/jbc.M114.591065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ogasawara Y., Kira S., Mukai Y., Noda T. and Yamamoto A. (2017) Ole1, fatty acid desaturase, is required for Atg9 delivery and isolation membrane expansion during autophagy in Saccharomyces cerevisiae. Biol. Open 6, 35–40 10.1242/bio.022053 [DOI] [PMC free article] [PubMed] [Google Scholar]