Abstract
Epidemiologic studies and studies in rodents point to potential risks from developmental exposure to BPA on cardiometabolic diseases. Furthermore, it is becoming increasingly evident that the manifestation and severity of adverse outcomes is the result of interaction between developmental insults and the prevailing environment. Consistent with this premise, recent studies in sheep found prenatal BPA treatment prevented the adverse effects of postnatal obesity in inducing hypertension. The gene networks underlying these complex interactions are not known. mRNA-seq of myocardium was performed on four groups of four female sheep to assess the effects of prenatal BPA exposure, postnatal overfeeding and their interaction on gene transcription, pathway perturbations and functional effects. The effects of prenatal exposure to BPA, postnatal overfeeding, and prenatal BPA with postnatal overfeeding all resulted in transcriptional changes (85–141 significant differentially expressed genes). Although the effects of prenatal BPA and postnatal overfeeding did not involve dysregulation of many of the same genes, they affected a remarkably similar set of biological pathways. Furthermore, an additive or synergistic effect was not found in the combined treatment group, but rather prenatal BPA treatment led to a partial reversal of the effects of overfeeding alone. Many genes previously known to be affected by BPA and involved in obesity, hypertension, or heart disease were altered following these treatments, and AP-1, EGR1, and EGFR were key hubs affected by BPA and/or overfeeding.
Keywords: Endocrine disruptors, cardiometabolic diseases, obesity, DOHAD, myocardium
INTRODUCTION
The World Health Organization estimates that by the year 2030, on a global basis, more than 23 million people will die annually from cardiovascular diseases (CVD) (WHO 2016 http://www.who.int/cardiovascular_diseases/about_cvd/en). In the United States, CVD is already responsible for a substantial portion of national health care expenditures, with CVD and stroke accounting for more than $315.4 billion in 2010 (Go et al. 2014). In addition, despite the perception that men tend to be susceptible to CVD and women tend to be protected, CVD-related death is one of the leading causes of mortality for both women and men in developed countries. In fact, CVD is among the top 4 causes of death among women aged 25–54 years (Leading cause of death in women, CDC, http://www.cdc.gov/women/lcod/index.htm) and in the past 2–3 decades, the incidence of myocardial infarction has increased significantly in young women (Ablewska et al. 2011). In the INTERHEART study on patients from 52 countries, nine modifiable risk factors were found to account for 94% of attributable risk for a woman having her first myocardial infarction including: dyslipidemia, hypertension, diabetes, and abdominal obesity (Yusuf et al. 2004).
Intrauterine and early environmental influences may be major contributors to the development of adult CVD (Alexander et al. 2015; Blackmore and Ozanne 2015) and its risk factors, and environmental endocrine disrupting chemicals (EDCs) are receiving considerable attention in this context (Ranciere et al. 2015). Common sources of these environmental exposures include the use of anabolic steroids (Hartgens and Kuipers 2004), contraceptives (Waller et al. 2010), and unintended exposure to environmental steroid mimics through food sources or industrial pollutants (Hotchkiss et al. 2008; O’Toole et al. 2008; Cosselman et al. 2015). One of the most controversial EDCs is bisphenol A (BPA), a component of polycarbonate plastic and epoxy resins used in the manufacture of many consumer products (Vandenberg et al. 2013). Humans are exposed ubiquitously to BPA via diet, dust, air, and water, as well as exposures as routine as handling cash receipts (Vandenberg et al. 2010). BPA has been found to act as an estrogen, anti-androgen, and has been shown to affect thyroid function and induce hyperinsulinemia (Acconcia et al. 2015). Questions about potential health risks from developmental exposure to BPA are raised from studies reporting the presence of BPA in human maternal circulation (Padmanabhan et al. 2008; Vandenberg et al. 2010), human tissues (Othman et al. 2016), amniotic and placental fluids (Ikezuki et al. 2002), breast milk (Sun et al. 2004), and the urine of neonates (Calafat et al. 2009). All of these sources represent exposures during critical early developmental windows, which could have very significant long-term health implications.
Epidemiologic and animal studies point to potential risks from early BPA exposures on development of obesity, diabetes, and cardiometabolic diseases (Rezg et al. 2014; Ranciere et al. 2015; Song et al. 2015). Because epidemiologic studies can point to potential associations but cannot address causality, the impact of developmental exposure to BPA has been tested in animal models – primarily in rodents. Considering the marked differences in cardiac physiology between rodents and larger precocial species such as human and sheep (Hew and Keller 2003) and the recent concerns expressed over use of altricial models such as rats for translating findings to humans (Habert et al. 2014), it is necessary to undertake cross-species comparisons for human translation (Neier et al. 2015). An example is the cross-species comparison performed in our recent study (Veiga-Lopez et al. 2015b), which also included a precocial species. Our group and others have extensively used sheep as a model to address the impact of developmental insults stemming from under and over-nutrition (Symonds et al. 2001; Metges 2009; Nathanielsz et al. 2013) as well as abnormal exposure to excess steroids (Padmanabhan and Veiga-Lopez 2013) on metabolic outcomes. Our studies in sheep found that prenatal exposure to excess testosterone, a hormone that is also a precursor for estrogen, from days 30 to 90 of gestation leads to low birth weight offspring, reproductive defects, insulin resistance (Padmanabhan and Veiga-Lopez 2013) and hypertension (King et al. 2007) thereby establishing the critical window for programming adult reproductive and metabolic defects. Using the same critical window of exposure, our studies also found prenatal BPA treatment at levels found in humans (Padmanabhan et al. 2008) induced insulin resistance and adipocyte hypertrophy (Veiga-Lopez et al. 2016). Identification of a critical window for programming cardiometabolic dysfunctions in sheep provides a suitable model for testing the developmental impact of BPA exposure.
Increasingly, evidence indicates that manifestation of final phenotypic outcomes is not only the result of early developmental insults but also a function of the prevailing postnatal environment (Padmanabhan et al. 2016). As such, the prevalence and rise of childhood obesity (Hruby and Hu 2015) creates the metabolic platform for amplifying metabolic disruptions programmed by developmental BPA exposures, or alternatively, prenatal BPA treatment can modulate the responsiveness to postnatal insults. Recently we found that while prenatal BPA increases the gene expression of atrial natriuretic peptide (ANP) in the ventricles of female sheep, it blocked the postnatal obesity-induced increase in diastolic blood pressure and left ventricular surface area (MohanKumar et al. 2016). Because the sheep heart is one of the most commonly used large animal models for studying CVD (Thornburg et al. 2008; Papamatheakis et al. 2013; Albertine 2015; Bensley et al. 2016), the aim of this study was to follow up on the paradoxical interaction between prenatal BPA and postnatal obesity and to gain a comprehensive understanding of both the impact of gestational exposure to BPA on the myocardial transcriptome and the interaction of postnatal obesity in modulating these effects.
MATERIALS AND METHODS
Breeding, Prenatal Treatment, and Maintenance
All procedures used in this experiment were approved by the University Committee on Use and Care of Animals (UCUCA), at the University of Michigan. The study was carried out at the University of Michigan Sheep Research Facility (Ann Arbor, MI; 42° 18′N). Breeder ewes were purchased from a local farm, blocked by weight and body score and randomized to control or BPA treatment groups. Ewes were bred by fertility-proven rams and paint markings left on the rumps of ewes by the raddled rams allowed us to determine the day of mating. BPA treatment spanned days 30 through 90 of gestation (term: ~147 days) and consisted of daily subcutaneous injections of 0.5 mg/kg/day of BPA (purity ≥ 99%, cat# 239658; Aldrich Chemical Co., Milwaukee, WI), with corn oil as the vehicle. Concentrations of unconjugated BPA achieved in umbilical arterial samples using the 0.5 mg/kg/day dose averaged 2.6 ng/mL and has been previously reported (Veiga-Lopez et al. 2013). These levels approach both the median level of BPA measured in maternal circulation of US women (Padmanabhan et al. 2008) and the median level of BPA measured in urine of US women (Calafat et al. 2005). Pregnant ewes in the control group were administered vehicle alone.
Common Conditions
To avoid confounding effects from differences in feeds, all feed was purchased in bulk. Beginning 6 weeks before the expected date of delivery and continuing until the time of delivery, pregnant ewes were group-fed 0.5 kg shelled corn, 2 kg alfalfa hay and 250 mg aureomycin crumbles (chlortetracycline)/ewe/day. Lactating ewes were fed 1 kg shelled corn and 2–2.5 kg of alfalfa hay/ewe/day. All lambs and ewes were provided with water and minerals ad libitum and were treated regularly with anthelmintic to minimize parasitic infection. After weaning at ~2 months, all lambs were subsequently maintained outdoors at the Sheep Research Facility (Ann Arbor, MI; 42°, 18′N). From weaning to 14 weeks of age, all lambs had ad libitum access to commercial feed pellets (Shur-Gain, Elma, NY; contains 18% crude protein) and alfalfa hay. Animals from this cohort have been used in studies addressing the impact of prenatal BPA and postnatal adiposity on insulin sensitivity and adipocyte morphology (Veiga-Lopez et al. 2016) as well as cardiovascular function (MohanKumar et al. 2016).
Treatment Groups
The study consisted of four treatment groups: controls (vehicle-treated and maintenance-fed: n = 11), prenatal BPA-treated and maintenance-fed (BPA: n = 8), control-overfed (no prenatal BPA-treatment but overfed (C + OF: n = 11)) or prenatal BPA-treated and overfed (BPA + OF: n = 11). When twin birth was involved, they were distributed between the maintenance-fed and overfed groups such that only one lamb from each dam was included in a given group.
The daily diet of the maintenance-fed animals (C, BPA), on a per lamb basis, consisted of 1.4 lbs corn, 0.03 lbs of supplement (36% crude protein), and 1.4 lbs hay. The daily diet of overfed animals (C + OF, BPA + OF) consisted of 1.7 lbs corn, 0.03 lbs of supplement, and 1.6 lbs hay initially and then ad libitum. The diet for the maintenance-fed animals was designed to achieve optimal growth without excess fat deposition, while the diet for the overfed animals was designed to make them obese – a body weight ~25% above that of normal weight adult female sheep. To compensate for increased energy demands as the lambs grew, and to meet additional energy demands during inclement weather, the ration was increased in control-fed and overfed females by the same percentages.
Harvesting of heart tissue was conducted during the second breeding season, when the selected animals were 21 months old. All animals were given two intramuscular injections of Prostaglandin F2α (PGF2α 20 mg, Lutalyse, Pfizer Animal Health, Florham Park, NJ), 11 days apart, to induce luteolysis and synchronize the initiation of the follicular phase in cycling females. All animals were weighed prior to euthanasia. Ewes were euthanized 27 h after the second PGF2α injection, during the presumptive follicular phase, by administration of a barbiturate overdose (Fatal Plus, Vortech Pharmaceuticals, Dearborn, MI). Hearts were quickly dissected out and weighed, then chunks of left ventricular free wall myocardium were harvested from a randomly selected subset of four animals per group (n = 4 for each of the control, BPA, C + OF, and BPA + OF groups) and snap frozen in the 2-Methylbutane/isopentane (Sigma-Aldrich, St. Louis, MO)-dry ice bath and stored in −80 freezer until processed.
RNA Extraction
Total RNA was extracted from the frozen myocardium using TRIzol Reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. The concentration of RNA from each sample was measured by NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Scientific, Waltham, MA). OD 260/280 values were 1.87–1.96 for the RNA samples. The integrity of each RNA sample was monitored through Agilent BioA RNA Integrity Number (RIN) (Agilent Technologies, Santa Clara, CA).
RNA-Seq Library Construction
RNA-Seq was performed by the Genomics, Epigenomics and Sequencing Core (GESC) at the University of Cincinnati. Using the TruSeq RNA sample preparation kit (Illumina, San Diego, CA), one μg of total RNA with RIN (Agilent 2100 Bioanalyzer) ≥ 7.0 was used to purify poly-A containing mRNA using poly-T oligo-attached magnetic beads. The purified mRNA was enzymatically fragmented and random hexamers-primed for the first- and second-strand cDNA synthesis, followed by purification using Agencourt AMPure XP beads (Beckman Coulter, Southfield, MI). The double-strand cDNA with overhangs was converted into blunt ends in the end repair procedure, and adenylated to add a single “A” nucleotide at 3′ ends to prevent them from self-ligation in the following ligation step. The AMPure XP beads-purified fragments were then ligated to sample-specific indexing adapters, and enriched by 10 cycles of PCR using adapter-specific primers.
One μL of purified PCR product (sequencing library, total 30 μL) was analyzed by Bioanalyzer (Agilent) using the DNA 1000 chip to check the DNA fragment size (~260 bp) and yield. To accurately quantify the library concentration for the clustering, the library was 1:104–106 diluted in dilution buffer (10 mM Tris-HCl, pH 8.0 with 0.05% Tween 20), and qPCR analyzed by the Kapa Library Quantification kit (Kapa Biosystems, Wilmington, MA) using ABI’s 9700HT real-time PCR system (Thermo Fisher Scientific, Grand Island, NY).
Cluster Generation and HiSeq Sequencing
The sixteen individually indexed cDNA libraries were pooled for clustering in the cBot system (Illumina). Libraries were clustered at a concentration of 8 pM using Illumina’s TruSeq SR Cluster Kit v3, and sequenced for 50 cycles using TruSeq SBS kit on Illumina HiSeq system. The number of reads generated per sample ranged from 21 – 69 million. The raw fastq files have been submitted to NCBI’s Short Read Archive (SRA) with accession GSE77418.
RNA-Seq Pre-Processing and Differential Expression Analysis
Raw fastq files were checked for quality using FastQC (Babraham Bioinformatics, Babraham Institute, Cambridge, UK, Version 0.10.0) to identify features of the data that may indicate quality problems (e.g., low quality scores, over-represented sequences, and inappropriate GC content). The Tuxedo Suite software package was used for alignment, differential expression analysis, and post-analysis diagnostics (Langmead et al. 2009; Trapnell et al. 2013, 2009). Briefly, reads were aligned to the reference transcriptome (Ensembl Oar_v3.1) (Cunningham et al. 2015) using TopHat (version 2.0.9) and Bowtie (version 2.1.0). Default parameter settings were used for alignment, with the exceptions of: “–b2-very-sensitive,” which allows extra time to search for valid alignments, and “–no-coverage-search” and “–no-novel-juncs” which restricts the mapping to known transcripts. The percent uniquely aligned ranged from 65% to 68%. FastQC was used for a second round of quality control (post-alignment), to ensure that only high quality data would be input to expression quantitation and differential expression analysis. Cufflinks/CuffDiff (version 2.1.1) was used for expression quantitation and differential expression analysis, using Ensembl Oar_v3.1.fa as the reference genome sequence and Ensembl Oar_v3.1.gtf as the reference transcriptome annotation. Parameter settings used were: “–multi-read-correct” to adjust expression calculations for reads that map in more than one locus, “–compatible-hits-norm” and “–upper-quartile –norm” for normalization of expression values. This analysis resulted in FPKM (fragments per kilobase of transcript per million fragments mapped) values, fold changes, P-values, and q-values using the Benjamini-Hochberg False Discovery Rate (FDR) approach for multiple testing adjustment. Differential expression was tested for BPA vs. control (the effect of BPA), overfeeding (C + OF) vs. control (the effect of overfeeding), and for BPA + OF vs. C + OF (the effect of BPA in the presence of overfeeding). Diagnostic plots were generated using the CummeRbund package.
Genes and transcripts satisfying three criteria (test status = “OK,’ FDR < 0.10 and absolute value of fold change |FC | ≥ 1.5) were considered differentially expressed. For the set of differentially expressed genes (DEGs), hierarchical clustering with average linkage and correlation distance measures was performed to identify the main expression profiles observed across the four treatment conditions. Log-transformed expression levels were averaged over the four ewes for each condition, and then data were normalized by subtracting the overall average expression of each gene from each expression value.
Functional Enrichment Testing
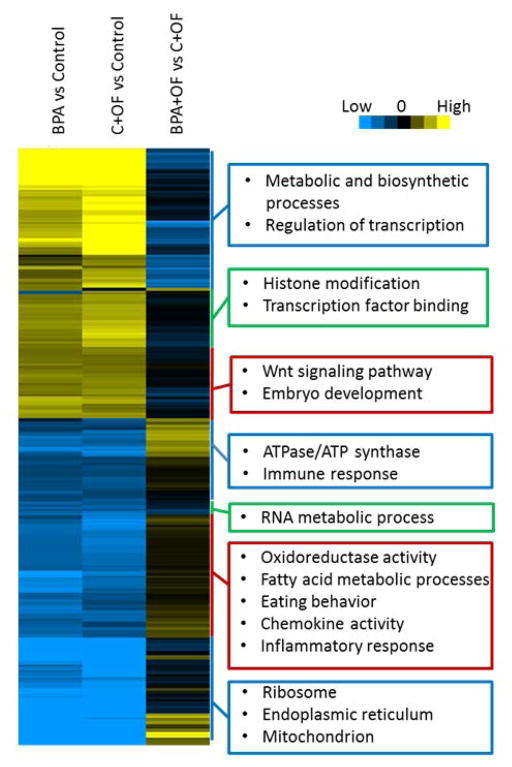
Enriched Gene Ontology (GO) terms, pathways and transcription factors, for each of the tested comparisons were tested using RNA-Enrich (Lee et al. 2015) (http://lrpath.ncibi.org/). RNA-Enrich tests for gene sets that have higher significance values (e.g., for differential expression) than expected at random, and takes into account any relationship between gene read count and significance level. By not requiring a cutoff for significance, RNA-Enrich is able to detect both pathways with a few very significant genes and pathways with many only moderate differentially expressed genes. Because the sheep genome annotation lags far behind that of the human, the sheep Gene IDs were first converted to their orthologous human GeneIDs using Ensembl BioMart (http://www.ensembl.org/biomart/martview). A directional RNA-Enrich test (which tests for significantly up- vs. down- regulated gene sets) was then run for each comparison with the human orthologs and using default settings, with the following databases: Biocarta Pathway, EHMN metabolic pathways, Gene Ontology, KEGG Pathway, Panther Pathway, and transcription factors. Only concepts with less than 1000 genes were considered for this analysis. The LRpath clustering algorithm was then run to assess trends across the RNA-Enrich results using gene sets with FDR < 0.01 in at least two of the three comparisons. RNA-Enrich results are shown in Figure 4 and in Supporting Information, including Table SII. The heatmap files (.ctd, .jtv, gtr, and atr) can be explored in an interactive format using Java TreeView (http://jtreeview.sourceforge.net/).
Figure 4.
Heatmap from hierarchical clustering of significant gene sets from functional enrichment testing. Gene sets with FDR < 0.01 in at least two of the three comparisons were used in clustering. Boxes highlight the main types of gene sets enriched in each cluster.
Gene Network Building
To help interpret the biological relevance of the concepts significantly enriched, MetaCore (Nikolsky et al. 2005) software by Thomson Reuters was used to model interactions among the differentially expressed genes and genes in the top scoring concepts in each of the comparisons. In developing each of the networks, two sets of MetaCore parameters were used to develop parsimonious models of the interactions: (1) the shortest path algorithm with manually curated high-confidence interactions, and (2) parameter settings as previously described (McEachin et al. 2010) where functional, binding, and low-trust interactions were added (Supporting Information, Tables SIIIa–c network statistics for each model).
Comparative Toxicogenomic Database (CTD) Analysis
To understand the results in the context of previous BPA and cardiovascular disease-related literature, the results from this study were compared with relevant gene lists in the CTD database (Mattingly et al. 2003) (http://ctdbase.org/). CTD contains information relevant to understanding how environmental exposures affect human health at the molecular level; this includes interactions and relationships among chemicals, genes/proteins, and diseases. For each of the comparisons, we found the intersection of the differentially expressed genes (FDR < 0.10) and the genes that are documented as being BPA responsive and related to cardiovascular diseases, hypertension, or obesity in the CTD database.
RESULTS
Effect of Prenatal BPA Treatment and Postnatal Overfeeding on Birth Weight and Body Weight
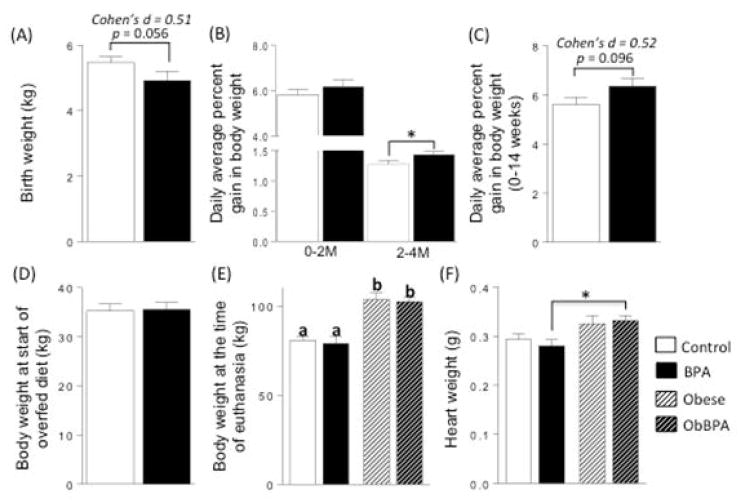
Consistent with previous findings (Savabieasfahani et al. 2006) prenatal BPA treatment tended to reduce the birth weight of female offspring (Fig. 1A; Control vs. BPA, p = 0.056). The average daily percent gain in body weight did not differ between control and prenatal BPA-treated animals during the first 2 months. A significant increase in average daily percent gain in body weight was evident from 2 to 4 months of age in prenatal BPA treated animals relative to controls (Fig. 1B). There was a trend for an increase in daily average percent gain in body weight in prenatal BPA-treated females relative to controls, when the first 14 weeks of life was considered (Fig. 1C). At 14 weeks of age, just prior to the start of overfeeding, there was no significant difference in body weight between control and prenatal BPA treated groups (Fig. 1D). In the subset of females that were used for assessing cardiac tissue gene expression (n = 4/group), overfeeding also increased body weight in both C + OF and BPA + OF groups to ~30% more than their respective maintenance-fed animals (Fig. 1E). The body weight was not different between the two maintenance-fed groups (control and BPA) or between the two overfed groups (C + OF, BPA + OF). Overfeeding significantly increased heart weight, when the entire group was considered (MohanKumar et al. 2016). When analysis was restricted to the four animals used in the present study, this increase was statistically significant only between BPA and BPA + OF groups (Fig. 1F). Cohen’s effect size analysis revealed a large effect size in heart weights between Control and C + OF (d = 1.0) and BPA and BPA + OF (d = 1.2).
Figure 1.
Birth weight (A), average daily percent gain in body weight from 0 to 2 and 2 to 4 months of age (B), body weight at 14 weeks of age (C), body weight at start of overfed diet (D) and at the time of euthanasia (E), and heart weight (F) comparisons between control and prenatal BPA-treated female sheep that received either maintenance diet or were overfed starting from 14 weeks of age. Data are presented as mean ± SEM. *P < 0.05. Panel E: Different superscripts (a vs. b) indicates significant differences.
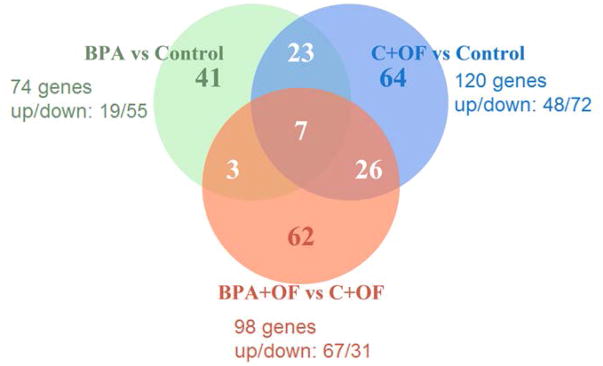
There were 21,586 genes (20265 with human ortholog IDs) that had measurable transcript levels (FPKM > 0) in at least one sample, and 14,289 of these were successfully tested for differential expression as reported by CuffDiff. Of these, 263 genes (229 genes with human ortholog IDs) were significantly differentially expressed (FDR < 0.10 and |FC| ≥ 1.5) in at least one of the comparisons. In particular, BPA treatment altered expression of 85 sheep genes (74 human ortholog IDs) under maintenance-fed conditions. Overfeeding altered expression of 141 genes (120 human orthologs) compared with control while only 112 genes (98 human orthologs) were altered in BPA + OF compared with C + OF (see Supporting Information Table SI with list of all genes with human orthologs and DEGs by the test). The Venn diagram (Fig. 2) shows the overlap between genes (human orthologues) that showed significant differential expression in each of the three comparisons. Seven genes (prostaglandin reductase 1 (PTGR1), carboxypeptidase, vitellogenic like (CPVL), natriuretic peptide receptor 3 (NPR3), family with sequence similarity 155 member B (FAM155B), alcohol dehydrogenase 1A (class I), alpha polypeptide (ADH1A), protease, serine 35 (PRSS35), and major histocompatibility complex, class II, DQ alpha 1 (HLA-DQA1)) were significantly differentially expressed in all three tests. PTGR1 is involved in prostaglandin metabolism, CPVL is a protease expressed in inflammatory cells and endocrine tissues, NPR3 regulates cardiac and circulatory functions, FAM155B is a transmembrane protein involved in calcium ion import, ADH1B is important in a number of metabolic pathways involving drugs and alcohol, PRSS35 is a protease with unknown function mostly highly expressed in female-specific tissues, and HLA-DQA1 is involved in antigen presenting.
Figure 2.
Comparison of differentially expressed genes (FDR < 0.10 and |FC|≥1.5) between three tests: BPA vs. Control, overfeeding (C + OF) vs. Control and BPA + OF vs. C + OF. Presented are the number of human ortholog IDs for sets of differentially expressed genes and number of up- and downregulated genes for each comparison.
Under maintenance fed conditions, several genes important in regulation of growth of the myocardium were upregulated by BPA, including genes involved in the FOXO and GATA1 pathways, epidermal growth factor receptor signaling pathways, fibroblast growth factor receptor binding including the fibroblast growth factor 3 (FGFR3) gene and those of MAP kinase signalling, including transforming growth factor beta 3 (TGFB3) and fibroblast growth factor 18 (FGF-18). BPA treatment also upregulated genes involved in the WNT pathway that are involved in heart development and remodeling (Bergmann 2010), as well as those involved in histone modification. Downregulated genes as a result of BPA exposure included those involved in oxidoreductase activity (e.g., acetyl-CoA acyltransferase 1 (ACAA1), glycerol-3-phosphate dehydrogenase 1 (GPD1), and cytochrome c oxidase subunit 7A1 (COX7A1)), and energy metabolism, which included genes involved in cellular respiration and the electron transport chain. Downregulation of genes invoved in the inflammation process and extracellular space were also evident. With overfeeding, genes upregulated by BPA included those involved in muscle contraction, p38 MAPK pathway, positive regulation of JNK, cellular response to TNF and oxidoreductase activity. However, genes involved in fibroblast growth factor receptor binding were downregulated. Interestingly, the pattern of cardiac relevant upregulated genes with BPA treatment were similar to the overfed animals; these included genes involved in growth mentioned above and also those genes involved in histone modification. Overfeeding alone upregulated genes involved in the apoptosis pathway, and downregulated genes involved in energy metabolism, inflammation pathways and actin mediated contraction genes similar to BPA effects (Table 1).
Table I.
Enriched gene sets for three comparisons: BPA vs. Control, C+OF vs. Control, and BPA+OF vs. C+OF. The first 20 gene sets are the top most significant in at least one comparison. The remaining 15 gene sets are related to the cardiovascular system and significant in at least one comparison. Numbers in parentheses for Gene Set Type represent the number of analyzed genes in the gene set; numbers in parentheses for Direction are the number of significant genes in that category for the corresponding comparison.
| No | Gene Set ID | Gene Set Name | Gene Set Type (Size) | BPA vs. Control | C+OF vs. Control | BPA+OF vs. C+OF | |||
|---|---|---|---|---|---|---|---|---|---|
| Direction (# genes) | q-value | Direction (# genes) | q-value | Direction (#genes) | q-value | ||||
| 1 | GO:0016491 | oxidoreductase activity | GOMF (724) | down (53) | 9.96E-12 | down (102) | 2.05E-13 | up (40) | 2.78E-05 |
| 2 | GO:0045047 | protein targeting to ER | GOBP (84) | down (16) | 3.10E-11 | down (40) | 7.92E-21 | down (3) | 8.01E-02 |
| 3 | GO:0055114 | oxidation-reduction process | GOBP (935) | down (78) | 4.79E-11 | down (127) | 1.24E-12 | up (57) | 1.09E-05 |
| 4 | GO:0006414 | translational elongation | GOBP (87) | down (19) | 2.09E-08 | down (43) | 1.71E-17 | down (3) | 2.93E-01 |
| 5 | GO:0051276 | chromosome organization | GOBP (952) | up (26) | 2.31E-06 | up (53) | 2.97E-08 | down (11) | 1.08E-01 |
| 6 | GO:0016570 | histone modification | GOBP (393) | up (15) | 1.33E-03 | up (34) | 2.82E-05 | down (4) | 3.79E-01 |
| 7 | GO:0006936 | muscle contraction | GOBP (283) | down (25) | 2.80E-01 | down (40) | 5.77E-01 | up (20) | 7.86E-04 |
| 8 | GO:0006954 | inflammatory response | GOBP (452) | down (34) | 1.51E-09 | down (39) | 8.59E-06 | up (22) | 1.24E-02 |
| 9 | GO:0038066 | p38MAPK cascade | GOBP (12) | down (1) | 8.28E-01 | down (3) | 4.71E-01 | up (3) | 1.10E-03 |
| 10 | GO:0046330 | positive regulation of JNK cascade | GOBP (52) | down (6) | 9.92E-01 | down (7) | 3.51E-01 | up (8) | 1.03E-03 |
| 11 | GO:0003735 | structural constituent of ribosome | GOMF (111) | down (22) | 3.77E-18 | down (53) | 2.85E-28 | down (3) | 9.90E-01 |
| 12 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | GOBP (77) | down (16) | 2.93E-11 | down (40) | 7.92E-21 | down (3) | 7.16E-02 |
| 13 | GO:0005615 | extracellular space | GOCC (722) | down (88) | 1.89E-13 | down (108) | 7.88E-16 | up (65) | 4.29E-02 |
| 14 | GO:0005743 | mitochondrial inner membrane | GOCC (417) | down (26) | 2.34E-10 | down (59) | 3.42E-11 | up (11) | 3.56E-03 |
| 15 | GO:0044429 | mitochondrial part | GOCC (938) | down (45) | 1.52E-09 | down (98) | 1.42E-10 | up (32) | 2.99E-04 |
| 16 | GO:0022904 | respiratory electron transport chain | GOBP (127) | down (12) | 5.50E-09 | down (28) | 8.45E-11 | up (4) | 2.21E-01 |
| 17 | GO:0002685 | regulation of leukocyte migration | GOBP (95) | down (8) | 4.44E-02 | down (13) | 1.10E-02 | up (10) | 1.54E-03 |
| 18 | GO:0001071 | nucleic acid binding transcription factor activity | GOMF (883) | up (39) | 8.77E-05 | up (69) | 4.84E-13 | down (21) | 7.94E-02 |
| 19 | GO:0042044 | fluid transport | GOBP (31) | up (5) | 7.44E-02 | up (3) | 1.22E-01 | down (3) | 2.39E-03 |
| 20 | 882901 | 5-Hydroxytryptamine degradation | Panther (14) | down (1) | 3.79E-01 | down (3) | 3.04E-01 | up (4) | 2.65E-06 |
| 21 | GO:0055024 | regulation of cardiac muscle tissue development | GOBP (41) | up (3) | 9.58E-03 | up (6) | 1.03E-02 | down (1) | 9.95E-01 |
| 22 | GO:0055015 | ventricular cardiac muscle cell development | GOBP (11) | up (1) | 2.75E-02 | up (3) | 3.50E-01 | down (1) | 9.40E-01 |
| 23 | GO:0021591 | ventricular system development | GOBP (21) | up (1) | 4.74E-02 | up (2) | 4.19E-02 | down (1) | 3.71E-01 |
| 24 | GO:0033275 | actin-myosin filament sliding | GOBP (40) | down (5) | 5.11E-05 | down (4) | 9.37E-05 | up (1) | 2.53E-01 |
| 25 | hsa04310 | Wnt signaling pathway | KEGG (119) | up (4) | 6.63E-03 | up (12) | 8.84E-03 | down (6) | 3.49E-01 |
| 26 | 882841 | Endothelin signaling pathway | Panther (64) | up (5) | 1.70E-02 | up (8) | 3.90E-01 | down (2) | 4.66E-01 |
| 27 | GO:0061061 | muscle structure development | GOBP (525) | up (30) | 5.06E-01 | up (60) | 2.17E-02 | down (19) | 9.83E-01 |
| 28 | 882953 | Apoptosis signaling pathway | Panther (95) | up (5) | 8.64E-01 | up (12) | 1.82E-02 | down (7) | 2.13E-01 |
| 29 | GO:0007173 | epidermal growth factor receptor signaling pathway | GOBP (202) | up (13) | 2.91E-02 | up (30) | 8.32E-06 | down (9) | 1.63E-01 |
| 30 | GO:0005104 | fibroblast growth factor receptor binding | GOMF (11) | up (1) | 2.00E-03 | up (3) | 1.92E-04 | down (1) | 1.81E-02 |
| 31 | 884942 | FOXO1_02 | TFs (225) | up (13) | 5.15E-04 | up (28) | 2.40E-04 | down (7) | 1.84E-01 |
| 32 | 885009 | FOXO3_01 | TFs (174) | up (11) | 1.55E-02 | up (16) | 9.45E-03 | down (4) | 1.01E-01 |
| 33 | 884988 | GATA1_05 | TFs (151) | up (13) | 1.55E-02 | up (22) | 2.98E-03 | down (9) | 8.25E-01 |
| 34 | hsa04010 | MAPK signaling pathway | KEGG (220) | up (12) | 1.17E-02 | up (34) | 5.43E-04 | down (18) | 9.96E-01 |
| 35 | GO:0071356 | cellular response to tumor necrosis factor | GOBP (82) | down (6) | 1.94E-01 | down (14) | 5.80E-02 | up (9) | 1.54E-03 |
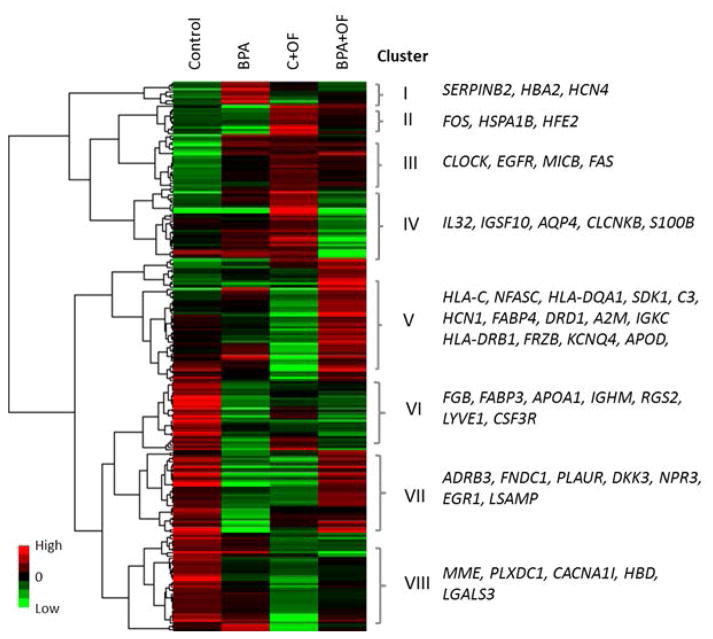
Clustering of the differentially expressed genes across the four treatments revealed a complex relationship between the effects of BPA and overfeeding (Fig. 3). Very few genes exhibited a similar response to BPA under the postnatal maintenance fed and overfed conditions. Instead, BPA in the presence of overfeeding most often had an opposite effect compared with BPA under a maintenance fed diet. A set of genes unchanged by BPA was downregulated in C + OF ewes and upregulated in BPA + OF (cluster V). Since the expected combined effect of BPA and OF – given independently acting individual effects would also be downregulation – this result is indicative of an interaction effect between BPA and overfeeding on gene expression; this group included fatty acid binding protein 4 (FABP4), alpha-2-macroglobulin (A2M) and apolipoprotein D (APOD), and also several genes belonging to the major histocompatibility complex (HLA-C, HLA-DQA1, and HLA-DRB1). FABP4 regulates fatty acid uptake, transport, or metabolism in adipocytes, A2M is a protease inhibitor and cytokine transporter, and APOD is a component of high density lipoprotein. Another group of genes (cluster VII) was downregulated in the C + OF and BPA groups, but upregulated in BPA + OF, also indicative of a strong interaction effect of BPA and overfeeding. This group of genes included several that play a role in cardiovascular function, e.g. adrenoceptor beta 3 (ADRB3), plasminogen activator, urokinase receptor (PLAUR), limbic system-associated membrane protein (LSAMP) and NPR3. ADRB3 is located primarily in the small intestine, adipose tissue and vascular endothelium and involved in the regulation of lipolysis and thermogenesis; PLAUR plays a role in localizing and promoting plasmin formation; LSAMP mediates selective neuronal growth and axon targeting in the developing limbic system. A large group of genes was strongly upregulated in C + OF and BPA groups compared with control, but partially or completely reversed in the BPA + OF group (cluster IV). This group included the genes immunoglobulin superfamily, member 10 (IGSF10), interleukin 32 (IL32), and S100 calcium binding protein B (S100B). IGSF10 may be involved in the maintenance of osteochondroprogenitor cells pool; IL32 induces various cytokines such as TNF-alpha and IL8 and activates typical cytokine signal pathways of NF-kappa-B and p38 MAPK; S100B proteins are involved in the regulation of a number of cellular processes such as cell cycle progression and differentiation.
Figure 3.
Heatmap from hierarchical clustering of significant differentially expressed genes (FDR< 0.10 and |FC|≥1.5) for at least one of the three comparisons performed: BPA vs. Control, C + OF vs. Control and BPA + OF vs. C + OF. The expression values (FPKMs) of the DEGs for the four treatment conditions were log transformed and clustered using criteria indicated in Materials and Methods. Genes listed at right belong to the respective cluster and were chosen based on their association with cardiovascular diseases according to the MetaCore database.
Functional Enrichment Results
Functional enrichment testing using RNA-Enrich found the top scoring gene sets downregulated by overfeeding were related to the extracellular space (FDR= 7.88E-16), oxidation–reduction process (FDR= 1.24E-12), and also to the ribosome, endoplasmic reticulum, and processes related to translation. Conversely, the most significant terms upregulated by overfeeding were related to transcription factor activity (FDR= 4.84E-13) and chromosome organization (FDR= 2.97E-08). Under maintenance-fed conditions, terms related to ribosome, extracellular region, mitochondrion (FDR for these concepts varied from 3.77E-18 to 1.89E-13) and inflammatory response (FDR = 1.51E-09), were downregulated by BPA. The top scoring gene sets upregulated by BPA were chromosome/chromatin organization and embryo development (FDR = 2.31E-06 and 8.09E-05, respectively), which overlapped with the response for overfeeding. The top scoring terms upregulated in BPA + OF groups compared with C + OF group were related to mitochondrion, oxidoreductase activity, and 5-hydroxytryptomine degradation (FDRs ranged from 2.65E-06 to 2.99E-04) (Supporting Information Table SII and Table 1). Visualization of the heatmap of enriched gene sets revealed extensive similarities between the effect of overfeeding and the effect of BPA under maintenance-fed conditions (Fig. 4). Metabolic processes, histone modifications, WNT signaling and other embryo development processes were upregulated by both, while oxidoreductase activity, immune, chemokine, and inflammatory response, and proteins located in the endoplasmic reticulum and mitochondrion were downregulated by both. In contrast to BPA-treatment with maintenance diet, the combination of prenatal BPA-treatment and postnatal overfeeding prevented some effects of postnatal overfeeding alone. Genes related to ATP synthase, immune response and mitochondrion were reduced by either BPA or overfeeding alone, but reversed to higher expression in BPA + OF. Conversely, gene sets related to metabolic and biosynthetic processes or regulation of transcription were increased by either BPA or overfeeding alone, but reversed to lower expression in BPA + OF.
Gene Network Models
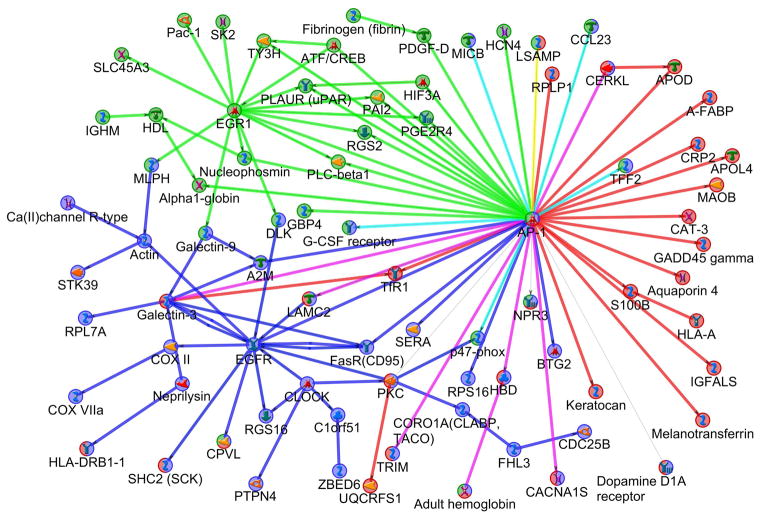
To place the significant genes in biological context, the significant DE genes from each of these tests (BPA vs. control, C + OF vs. control, and BPA + OF vs. C + OF) were analyzed in MetaCore and a protein interaction network was constructed for each. For network construction, two sets of parameters were used: (a) only high-confidence interactions; and (b) high-confidence interactions with binding and low-trust interactions. The three networks derived from the three experiments were combined to present nodes specific and shared for every network (Fig. 5). The BPA vs. control network (see Fig. 5, green nodes and edges) included two main hubs: early growth response 1 (EGR1) and Fos proto-oncogene, AP-1 transcription factor subunit (FOS) (network object AP-1 on Fig. 5). EGR1 is a transcriptional regulator which activates the transcription of target genes whose products are required for mitogenesis and differentiation. FOS participates in forming the transcription factor complex AP-1 and has been implicated as regulators of cell proliferation, differentiation, and transformation. In the BPA vs. control network these two hubs interact both directly – i.e. EGR1 binds to gene c-Fos promoter and activates c-Fos expression (Ishikawa et al. 2007) – and through other genes/proteins, such as regulator of G-protein signaling 2 (RGS2), PLAUR, tyrosine 3-monooxygenase (TY3H), and others. RGS2 plays a role in the regulation of blood pressure in response to signaling via G protein-coupled receptors and GNAQ (G protein subunit alpha q); TY3H plays an important role in the physiology of adrenergic neurons. TY3H and PLAUR were identified as additional new hubs in the BPA vs. control network when functional and low-trust interactions were used for network construction (Supporting Information Table SIIIa).
Figure 5.
The combined protein interaction network illustrating the response for BPA vs. Control (green nodes and edges), OF vs. Control (blue), and BPA + OF vs. C + OF (red). The network was constructed from the subnetworks of differentially expressed genes with direct, high-confidence interactions from each of the three comparisons. The combination of the node’s colors indicates that the node is involved in more than one subnetwork: for example AP-1 combined green, blue, and red colors meaning that this hub is involved in all three subnetworks. PKC is involved in OF vs. Control and BPA + OF vs. C + OF (combination of blue and red colors). Teal and purple edge colors represent interactions observed in both green and blue, or red and blue, respectively (e.g., the purple edge between AP-1 and LAMC2). A legend for the shapes used for the nodes is provided in MetaCore Quick Refernce Guide https://ftp.genego.com/files/MC_legend.pdf.
The C + OF vs. control network (Fig. 5, blue nodes and edges) includes three main hubs: epidermal growth factor receptor (EGFR), FOS (network object AP-1), and Fas cell surface death receptor (FAS, network object FasR-CD95). EGFR is a cell surface protein that binds to epidermal growth factor and activates several signaling cascades including RAS-RAF-MEK-ERK, PI3 kinase-AKT, PLCgamma-PKC, and STATs modules; FAS is a member of the TNF-receptor superfamily and play a central role in the physiological regulation of programmed cell death. Low-trust interactions additionally identified protein kinase C gamma (gene PRKCG, network object PKC), clock circadian regulator (CLOCK), and A2M as hubs (Supporting Information Table SIIIb). PKC family members phosphorylate a wide variety of protein targets and play roles in modulation of receptors and neuronal functions related to sensitivity to opiates, pain and alcohol, mediation of synaptic function and cell survival after ischemia, and inhibition of gap junction activity after oxidative stress; CLOCK plays a central role in the regulation of circadian rhythms and associated with obesity and metabolic syndrome (Corella et al. 2016).
The network derived from BPA + OF vs. C + OF gene set had less connectivity among nodes compared with the two previous ones (Fig. 5, red nodes and edges). There were no hubs in this network using high-trust interactions, but AP-1 was again identified as a hub when using low-trust interactions (Supporting Information Table SIIIc).
Although the differentially expressed genes in each of these three comparisons were mostly different, the networks that they participate in show important overlaps. Specifically, nuclear AP-1 is a significant interacting transcription factor and a hub in all three networks. PKC is an important regulator of cardiac function and a hub in C + OF vs. control and PKC has also connections in the BPA + OF vs. C + OF network.
Overlaps with Comparative Toxicogenomic Database (CTD)
Given the large numbers of potentially interesting genes, genes were prioritized based on the overlap between the differential expresssion results and CTD annotations. For each of the three comparisons, genes that were both differentially expressed and annotated for cardiovascular diseases and/or BPA response in the CTD were determined (Table 2). Most of the overlapping genes were involved in signal transduction and transcriptional regulation, consistent with a fundamental cellular response to environmental stimulus. An interesting result is that FOS, EGR1, FAS are found to overlap with genes associated with bisphenol A and cardiovascular diseases (from CTD), and they were also hub genes of the BPA vs. control and C + OF vs. control networks. FOS was a primary hub for all three networks.
Table II.
Differentially expressed genes that overlap with genes from the Comparative Toxicogenomic Database (CTD; http://ctdbase.org/) associated with the term “Bisphenol A” and the listed Diseases. The term “Cardiovascular Diseases” (9th row) includes: Hypotension (52 genes), Arrhythmias Cardiac (18), Hypertension (172), Myocarditis (5), Ventricular Premature Complexes (1), Vascular Diseases (16), Atherosclerosis (50), Cardiomegaly (64), Heart Defects Congenital (30), and Cardiomyopathy Hypertrophic (20 genes). Gene names and functions not mentioned in the text are listed in footnote1.
| Genes from CTD: BPA & Disease (# genes) | BPA vs Control (75) | C+OF vs Control (122) | BPA+OF vs C+OF (98) |
|---|---|---|---|
| Bisphenol A & Heart Failure (76) | 1 (ADRB3) | 2(ADRB3, GATM) | 1 (GATM) |
| Bisphenol A & Cardiomyopathies (65) | 0 | 1 (FAS) | 0 |
| Bisphenol A & Myocardial Infarction (82) | 0 | 0 | 1 (S100B) |
| Bisphenol A & Myocardial Ischemia (167) | 0 | 2(GATM, HSPA1A) | 3(GATM, TFRC, UQCRFS1) |
| Bisphenol A & Ventricular Dysfunction, Left (13) | 0 | 1(FAS) | 0 |
| Bisphenol A & Cardiomegaly (64) | 1 (UCP2) | 0 | 0 |
| Bisphenol A & Hypertension (172) | 5 (APOA1, FOS, NCF1, RGS2, UCP2) | 3 (FOS, NCF1, STK39) | 0 |
| Bisphenol A & Obesity (148) | 2 (ADRB3, UCP2) | 1 (ADRB3) | 2 (ALDH1L1, TFRC) |
| Bisphenol A & Cardiovascular Diseases (325) | 5 (APOA1, FOS, NCF1, RGS2, UCP2) | 5 (DRD1, FOS, HSPA1B, NCF1, STK39) | 2 (DRD1, MAOB) |
| Bisphenol A (273) | 5 (APOA1, EGR1, FOS, LPAR1, TH) | 2 (FAS, FOS) | 2 (C3, FABP4) |
ALDH1L1 (aldehyde dehydrogenase 1 family, member L1) loss of function or expression of this gene is associated with decreased apoptosis, increased cell motility, and cancer progression; APOA1 (apolipoprotein A–I) promotes cholesterol efflux from tissues to the liver for excretion, and is a cofactor for lecithin cholesterolacyltransferase (LCAT), an enzyme responsible for the formation of most plasma cholesteryl esters; C3 (complement component 3) C3 peptide modulates inflammation and possesses antimicrobial activity; DRD1 (dopamine receptor D1) this G-protein coupled receptor stimulates adenylyl cyclase and activates cyclic AMP-dependent protein kinases; GATM (glycine amidinotransferase) catalyzes the biosynthesis of guanidinoacetate, the immediate precursor of creatine which plays a vital role in energy metabolism in muscle tissues; HSPA1B (heat shock 70kDa protein 1B) participate in stabilizing of existing proteins against aggregation and mediates the folding of newly translated proteins in the cytosol and in organelles; LPAR1 (lysophosphatidic acid receptor 1) plays a role in the reorganization of the actin cytoskeleton, cell migration, differentiation and proliferation, and thereby contributes to the responses to tissue damage and infectious agents; MAOB (monoamine oxidase B) the protein catalyzes the oxidative deamination of biogenic and xenobiotic amines and plays an important role in the metabolism of neuroactive and vasoactive amines in the central nervous system and peripheral tissues; NCF1 (neutrophil cytosolic factor 1) the protein is subunit of neutrophil NADPH oxidase; STK39 (serine threonine kinase 39) acts as a mediator of stress-activated signals; TFRC (transferrin receptor) participates in cellular iron uptake by the process of receptor-mediated endocytosis; TH (tyrosine hydroxylase) protein is involved in the conversion of tyrosine to dopamine and plays a role in the physiology of adrenergic neurons; UCP2 (uncoupling protein 2) protein separate oxidative phosphorylation from ATP synthesis with energy dissipated as heat - mitochondrial proton leak; UQCRFS1 (ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1) ubiquinol-cytochrome c reductase complex is a respiratory chain that generates an electrochemical potential coupled to ATP synthesis.
DISCUSSION
The findings from this study demonstrate that prenatal BPA treatment, postnatal overfeeding/adiposity and their combination lead to transcriptional changes in the myocardium of female sheep. Although the transcriptional dysregulation induced by prenatal BPA treatment and overfeeding did not involve the same genes for the most part, the biological pathways they affected were remarkably similar. Paradoxically, while the combined treatment failed to produce an additive or synergistic response, prenatal BPA treatment prevented in part the effects of overfeeding. Prenatal BPA treatment altered genes previously known to be involved in obesity, hypertension, or heart disease. The main hubs that were affected by BPA and/or overfeeding include AP-1, EGR1, and EGFR.
Impact of Prenatal BPA Exposure on Cardiovascular System
Recent epidemiogical studies indicate that prenatal BPA treatment is associated with adverse cardiovascular outcomes (Gao and Wang 2014) with a positive correlation existing between BPA exposure and vascular disease, including coronary artery disease and hypertension (Lang et al. 2008; Bae et al. 2012). Prolonged BPA exposure throughout the lifespan has also been found to increase mean arterial pressure in female rats (Patel et al. 2013). To our knowledge data is sparse relative to the effects of prenatal BPA exposure on the cardiovascular system in the offspring. A recent study found prenatal BPA treatment to induce transcriptome changes in the fetal heart suggestive of adverse cardiac programming in rhesus monkeys (Chapalamadugu et al. 2014). Importantly, using the same cohort of animals that included animals used in the present study, we recently found prenatal BPA treatment increased atrial natriuretic peptide (ANP), a marker of left ventricular hypertrophy frequently associated with systemic hypertension, in the left ventricle without having an effect on blood pressure (MohanKumar et al. 2016). This could signify the direct effect of BPA on the left ventricular structure and not secondary to hypertrophic stimuli. In our study BPA downregulated Natriuretic Peptide Receptor 3 (NPR3) gene within the left ventricular myocardium and since NPR system has been shown to play a role in intrinsic growth of the myocardium (Matsukawa et al. 1999; Becker et al. 2014) these findings coupled with our earlier finding of elevated ANP raise the possibility of direct effect of BPA on the NPR system within the left ventricle.
On the one hand, the lack of hypertensive effects in the face of increased left ventricular ANP levels may be a function of the younger age (21 months) at which blood pressure measures were undertaken. Alternatively, since subtle perturbations in blood pressure are better measured with continuous radio telemeters – rather than a single time point measurement such as that use in this study – the impact on blood pressure could have been missed. Further studies detailing structural and cardiovascular functional changes at multiple developmental time points obtained under cardiac stress conditions – an understudied area – would help elucidate the detrimental effects of prenatal BPA exposure on cardiovascular system. The current study, an extension of the previous study at the functional level (MohanKumar et al. 2016), is the first step in this direction and provides a more global perspective of molecular pathways that are altered within the left ventricular myocardium from prenatal exposure to BPA.
Our present findings also provide evidence in support of prenatal BPA treatment having an impact on genes involved in obesity, hypertension, and cardiovascular disease. The key hub, AP1, affected by prenatal BPA treatment, regulates numerous cellular processes including proliferation, cellular differentiation and apoptosis (Ye et al. 2014). The AP-1 pathway is involved in extracellular matrix changes leading to fibrosis, contractile dysfunction and also cardiac hypertrophy (Shaulian and Karin 2002; Hill et al. 2013).
The second hub that was affected by prenatal BPA treatment involved EGR1, a master regulator belonging to the zinc finger transcription factor family. EGR1 regulates cell proliferation, differentiation and apoptosis and is rapidly induced by various stimuli including hypoxia, growth factors, cytokines, and injurous stimuli (Khachigian 2006). EGR1 has been linked to cardiovascular pathologies including cardiac hypertrophy, atherosclerosis (Khachigian 2006). EGR1 silencing has been found to reduce myocardial insult, inflammation and apoptosis (Rayner et al. 2013). The downregulation of the EGR1 gene and the reduction in the inflammtory response pathway evident in prenatal BPA treated animals might therefore represent a compensatory response to overcome adverse changes in the myocardium that are not yet evident at a structural or functional level in these postpubertal animals.
Other pathways relevant to left ventricular function and structure that were altered by prenatal BPA treatment included downregulation of pathways associated with mitochondrial function including oxidoreductase activity, protein targeting to ER, the oxidation–reduction process, those involved with protein biosynthesis including translational and elongation, muscle contraction, and the inflammatory response including p38MAPK cascade and the JNK cascade. The upregulated pathways included those involved with development of cardiac muscle tissue and the WNT pathway, which is involved in cardiac hypertrophy and remodeling (Bergmann 2010).
Our finding of downregulation of pathways involved in mitochondrial function in left ventricular myocardium in prenatal BPA-treted animals is consistent with earlier findings of such dysfunction in neonatal cardiomyocytes of prenatal BPA-treated animals (Jiang et al. 2015). Mitochondrial dysfunction, which includes gene sets involved in regulation of mitochondrial inner membrane, respiratory electron chain, and oxidoreductase activity that are vital for cardiac function and cardiac failure over time (Khan et al. 2016), is a known target for EDCs including BPA (Xia et al. 2014). Additional prenatal BPA-induced disruptions include downregulation of pathways associated with cardiac contractility, such as those associated with actin-myosin filament sliding and myofilament function, which can lead to disruptions in myocardial mechanical properties and contractile functions (Moss et al. 2004).
Pathways associated with epigenetic changes including histone modification (Cheung and Lau 2005) and chromosome organization were also targets of prenatal BPA treatment, suggestive of epigenetic changes underlying such transcriptional changes. An earlier study found early postnatal BPA exposure induced epigenetic changes such as hypermethylation of the mitochondrial gene PGC-1 alpha leading to cardiomyopathy in male rats (Jiang et al. 2015).
Impact of Postnatal Adiposity
It is well known that obesity is detrimental to cardiovascular health. In clinical studies, it has been shown that obesity leads to hypertension and cardiac remodelling including eccentric cardiac hypertrophy and cardiac dysfunction over time (Poirier et al. 2006; Abel et al. 2008). Obesity during early life has also been implicated in the development of cardiovascular disease including cardiac remodeling and left ventricular hypertrophy (Nadeau et al. 2011; Shah et al. 2011). Mechanisms for adverse cardiac remodeling due to obesity are not completely understood, but evidence exists in support of perturbations in cardiac metabolism, mitochondrial dysfunction and oxidative stress (Abel et al. 2008). Findings from the present study are consistent with cardiac remodeling in that overfeeding resulted in increased heart weight in the subset of prenatal BPA-treated animals subjected to transcriptome analyses, along with downregulation of pathways involved in mitochondrial function, such as mitochondrial respiratory chain, and upregulation of pathways associated with cell growth, such as FOXO and GATA pathways that have been linked to cardiac hypertrophy (Kohli et al. 2011).
Strikingly the changes in gene sets evidenced in overfed animals paralleled many of the changes seen in prenatal BPA-treated females including the key hubs affected. The key gene hub affected by overfeeding, AP-1, was also a target of prenatal BPA-treatment. The effects of overfeeding namely an increase in FOS as opposed to a decrease in prenatal BPA-treated females is consistent with finding that long-term high-fat diet upregulates the AP-1 transcription factor c-fos (Foldes et al. 2006). Apart from members of the AP-1 family, pathways relevant to left ventricular function and structure that were affected by prenatal BPA treatment were also affected in the C + OF group. These include downregulation of pathways associated with mitochondrial function, protein biosynthesis, muscle contraction and the inflammatory response and upregulation of pathways involved in cardiac muscle tissue development, cardiac hypertrophy (Bergmann 2010), epigenetic changes, and apoptosis, all supportive of adverse cardiac remodeling. It is well-known that adverse cardiac remodelling leads to poor cardiac efficiency (Abel et al. 2008). EGFR, which is linked to adverse cardiovascular dysfunction including hypertension and cardiac hypertrophy (Smith et al. 2004; Forrester et al. 2016; Peng et al. 2016), was an additional hub that was affected by overfeeding.
Prenatal BPA and Postnatal Diet Interaction
Contrary to the premise that prenatal BPA treatment would accentuate the adverse cardiac effects observed with overfeeding, prenatal BPA treatment prevented the effects of overfeeding, albeit partially; the transcriptome changes seen in BPA + OF group differed from prenatal BPA and C + OF groups. Pathways downregulated in BPA and C + OF groups were either upregulated in BPA + OF group or not significantly altered. While pathways involved in mitochondrial function (oxidoreductase activity, oxidation–reduction process) were upregulated, those involved in epigenetic modification, protein synthesis, muscle contraction, and inflammation that were altered in the BPA and C + OF group were not altered in the BPA + OF group. Our recent findings (MohanKumar et al. 2016) with the cohort of animals that included animals used in the present study found prenatal BPA treatment prevented the effects of postnatal overfeeding in increasing blood pressure and left ventricular area. These findings add functional linkage to the findings at the gene level (present study). Similar to these paradoxcial interactions, a recent study found that a high-fat diet mitigated the adverse effects of BPA on embryo implantation (Martinez et al. 2015). In recent years it has become increasingly clear that the manifestation of a final phenotypic outcome is not merely dictated by the organizational changes that occur during critical periods of differentiation such as those induced by BPA in this study, but also is a function of the prevailing postnatal milieu (Puttabyatappa et al. 2015). Additional studies addressing whether these unexpected findings in the BPA + OF group are adaptive or maladaptive would further our understanding of the interaction between prenatal BPA exposure and the postnatal environment in modulating cardiac function and identifying preventive treatment strategies.
Strength of Bioinformatics Approach
From a bioinformatics perspective, while this would have been a fairly straight-forward study in humans, the use of sheep presented an annotation challenge. However, the use of human orthologs dramatically increased the ability to functionally interpret the results. In addition, the use of RNA-Enrich ensured that we were not merely identifying functions with highly-expressed or long genes, which have more statistical power to detect differential expression. Finally, Metacore network analysis enabled comparison of the interaction networks of the three effects of interest: BPA, BPA in the presence of overfeeding, and overfeeding alone, while CTD provided a disease- and exposure-specific contextual foundation for comparison.
Translational Relevance
Considering the ubiquitous presence of BPA in consumer products and its ability to function through steroid/insulin receptors (Vandenberg et al. 2010), findings from this study are likely to be of both clinical and public health relevance. Importantly, the BPA dose used in this study produced fetal blood levels of BPA well within the range seen in maternal circulation (Padmanabhan et al. 2008) and urine (Calafat et al. 2005) of US women. Because of concerns raised regarding the widespread use of altricial rodent models for risk assessments of EDCs in humans (Habert et al. 2014), use of sheep in this study, a precocial model with similar fetal developmental trajectory as humans (Padmanabhan and Veiga-Lopez 2013) extensively used for understanding developmental origins of metabolic diseases (Green et al. 2010; Nathanielsz et al. 2013; Cardoso et al. 2015) are likely valuable for human translation. Importantly, the chosen window of exposure corresponds with developmental periods of differentiation of metabolic systems in the female sheep (Padmanabhan and Veiga-Lopez 2013), and has been shown to be a critical period for producing metabolic perturbations including insulin resistance (Cardoso et al. 2015) and hypertension (King et al. 2007). Previously we found that this exposure level of BPA induced (in sheep and rats) or was associated with (in human) fetal oxidative stress (Veiga-Lopez et al. 2015b) and low birth weight offspring (Veiga-Lopez et al. 2015a), risk factors for adult onset cardiometabolic diseases (Yajnik 2000; Varvarigou 2010; Perrone et al. 2016). The trend for reduced birth weight in prenatal BPA-treated female offspring in this study, a finding in agreement with our previous studies in sheep (Savabieasfahani et al. 2006) and other studies in rodents (Ranjit et al. 2010), raises the possibility that the effects of prenatal BPA exposure may be the consequence of intrauterine growth restriction of the developing fetus. Future studies are needed to illuminate to what extent the effects of BPA on cardiac function is due to its direct effects on the developing heart as opposed to those programmed via altered fetal growth trajectory. Studies of the interaction of prenatal BPA and postnatal overfeeding are also valuable considering the obesity epidemic facing the nation (Smith and Smith 2016) and the potential for an altered metabolic milieu in modulating the manifestation or severity of prenatally programmed cardiometabolic dysfunctions (Kim et al. 2015; Padmanabhan et al. 2016).
Supplementary Material
Acknowledgments
We thank Douglas Doop for his expert animal care, facility management and help with generation of the experimental animals, Drs. Almudnea Veiga-Lopez, Bachir Abi-Salloum, Ms. Carol Herkimer, Mr. Evan M. Beckett, Mr. Jacob Moeller for assistance with prenatal treatments and or help with tissue harvest, Mr. Xiaoqian Gao for RNA extraction, and Jingyuan Deng and Mario Medvidovic for the RNA sequencing Core services.
Footnotes
Disclosure statement: Authors have nothing to disclose.
AUTHOR CONTRIBUTIONS
LAK and APV participated in the analyses, interpretation of the data, and wrote the manuscript; RCM participated in performing bioinformatics analyses and interpretation of data; MP performed the analyses of data included in Figure 1 and writing of the manuscript; H-SW participated in designing the cardiac transcriptome studies, cardiac tissue harvest, provided oversight of the transcriptome analyses and participated in writing of manuscript; MAS determined and spearheaded the bioinformatic analyses, and participated in the interpretation of data and writing of the manuscript; VP designed the parent study focusing on the interaction between prenatal BPA treatment and overfeeding, provided oversight for integrating the study components, participated in data interpretation and writing of the manuscript. All authors read through the manuscript and provided input in finalizing the manuscript.
References
- 1.Abel ED, Litwin SE, Sweeney G. Cardiac remodeling in obesity. Physiol Rev. 2008;88:389–419. doi: 10.1152/physrev.00017.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ablewska U, Jankowski K, Rzewuska E, Liszewska-Pfejfer D, Hryniewiecki T. A levels of endogenous gonadal hormones and their relationship with selected coronary artery disease risk factors among young women post myocardial infarction. Acta Biochim Pol. 2011;58:385–389. [PubMed] [Google Scholar]
- 3.Acconcia F, Pallottini V, Marino M. Molecular mechanisms of action of BPA. Dose Response. 2015;13:1–9. doi: 10.1177/1559325815610582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albertine KH. Utility of large-animal models of BPD: chronically ventilated preterm lambs. Am J Physiol Lung Cell Mol Physiol. 2015;308:L983–L1001. doi: 10.1152/ajplung.00178.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander BT, Dasinger JH, Intapad S. Fetal programming and cardiovascular pathology. Compr Physiol. 2015;5:997–1025. doi: 10.1002/cphy.c140036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bae S, Kim JH, Lim YH, Park HY, Hong YC. Associations of bisphenol A exposure with heart rate variability and blood pressure. Hypertension. 2012;60:786–793. doi: 10.1161/HYPERTENSIONAHA.112.197715. [DOI] [PubMed] [Google Scholar]
- 7.Becker JR, Chatterjee S, Robinson TY, Bennett JS, Panakova D, Galindo CL, Zhong L, Shin JT, Coy SM, Kelly AE, et al. Differential activation of natriuretic peptide receptors modulates cardiomyocyte proliferation during development. Development. 2014;141:335–345. doi: 10.1242/dev.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bensley JG, De Matteo R, Harding R, Black MJ. The effects of preterm birth and its antecedents on the cardiovascular system. Acta Obstet Gynecol Scand. 2016;95:652–663. doi: 10.1111/aogs.12880. [DOI] [PubMed] [Google Scholar]
- 9.Bergmann MW. WNT signaling in adult cardiac hypertrophy and remodeling: lessons learned from cardiac development. Circ Res. 2010;107:1198–1208. doi: 10.1161/CIRCRESAHA.110.223768. [DOI] [PubMed] [Google Scholar]
- 10.Blackmore HL, Ozanne SE. Programming of cardiovascular disease across the life-course. J Mol Cell Cardiol. 2015;83:122–130. doi: 10.1016/j.yjmcc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Calafat AM, Kuklenyik Z, Reidy JA, Caudill SP, Ekong J, Needham LL. Urinary concentrations of bisphenol A and 4-nonylphenol in a human reference population. Environ Health Perspect. 2005;113:391–395. doi: 10.1289/ehp.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calafat AM, Weuve J, Ye X, Jia LT, Hu H, Ringer S, Huttner K, Hauser R. Exposure to bisphenol A and other phenols in neonatal intensive care unit premature infants. Environ Health Perspect. 2009;117:639–644. doi: 10.1289/ehp.0800265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardoso RC, Puttabyatappa M, Padmanabhan V. Steroidogenic versus metabolic programming of reproductive neuroendocrine, ovarian and metabolic dysfunctions. Neuroendocrinology. 2015;102:226–237. doi: 10.1159/000381830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapalamadugu KC, Vandevoort CA, Settles ML, Robison BD, Murdoch GK. Maternal bisphenol a exposure impacts the fetal heart transcriptome. PLoS One. 2014;9:e89096. doi: 10.1371/journal.pone.0089096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Mol Endocrinol. 2005;19:563–573. doi: 10.1210/me.2004-0496. [DOI] [PubMed] [Google Scholar]
- 16.Corella D, Asensio EM, Coltell O, Sorli JV, Estruch R, Martinez-Gonzalez MA, Salas-Salvado J, Castaner O, Aros F, Lapetra J, et al. CLOCK gene variation is associated with incidence of type-2 diabetes and cardiovascular diseases in type-2 diabetic subjects: dietary modulation in the PREDIMED randomized trial. Cardiovasc Diabetol. 2016;15:4. doi: 10.1186/s12933-015-0327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosselman KE, Navas-Acien A, Kaufman JD. Environmental factors in cardiovascular disease. Nat Rev Cardiol. 2015;12:627–642. doi: 10.1038/nrcardio.2015.152. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foldes G, Vajda S, Lako-Futo Z, Sarman B, Skoumal R, Ilves M, deChatel R, Karadi I, Toth M, Ruskoaho H, Lepran I. Distinct modulation of angiotensin II-induced early left ventricular hypertrophic gene programming by dietary fat type. J Lipid Res. 2006;47:1219–1226. doi: 10.1194/jlr.M500550-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Forrester SJ, Kawai T, O’Brien S, Thomas W, Harris RC, Eguchi S. Epidermal growth factor receptor transactivation: mechanisms, pathophysiology, and potential therapies in the cardiovascular system. Annu Rev Pharmacol Toxicol. 2016;56:627–653. doi: 10.1146/annurev-pharmtox-070115-095427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, Wang HS. Impact of bisphenol a on the cardiovascular system – epidemiological and experimental evidence and molecular mechanisms. Int J Environ Res Public Health. 2014;11:8399–8413. doi: 10.3390/ijerph110808399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, et al. Heart disease and stroke statistics – 2014 update: a report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green AS, Rozance PJ, Limesand SW. Consequences of a compromised intrauterine environment on islet function. J Endocrinol. 2010;205:211–224. doi: 10.1677/JOE-09-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habert R, Muczynski V, Grisin T, Moison D, Messiaen S, Frydman R, Benachi A, Delbes G, Lambrot R, Lehraiki A, et al. Concerns about the widespread use of rodent models for human risk assessments of endocrine disruptors. Reproduction. 2014;147:R119–R129. doi: 10.1530/REP-13-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartgens F, Kuipers H. Effects of androgenic-anabolic steroids in athletes. Sports Med. 2004;34:513–554. doi: 10.2165/00007256-200434080-00003. [DOI] [PubMed] [Google Scholar]
- 26.Hew KW, Keller KA. Postnatal anatomical and functional development of the heart: a species comparison. Birth Defects Res B Dev Reprod Toxicol. 2003;68:309–320. doi: 10.1002/bdrb.10034. [DOI] [PubMed] [Google Scholar]
- 27.Hill C, Wurfel A, Heger J, Meyering B, Schluter KD, Weber M, Ferdinandy P, Aronheim A, Schulz R, Euler G. Inhibition of AP-1 signaling by JDP2 overexpression protects cardiomyocytes against hypertrophy and apoptosis induction. Cardiovasc Res. 2013;99:121–128. doi: 10.1093/cvr/cvt094. [DOI] [PubMed] [Google Scholar]
- 28.Hotchkiss CE, Weis C, Blaydes B, Newbold R, Delclos KB. Multigenerational exposure to ethinyl estradiol affects bone geometry, but not bone mineral density in rats. Bone. 2008;43:110–118. doi: 10.1016/j.bone.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Hruby A, Hu FB. The epidemiology of obesity: a big picture. Pharmacoeconomics. 2015;33:673–689. doi: 10.1007/s40273-014-0243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikezuki Y, Tsutsumi O, Takai Y, Kamei Y, Taketani Y. Determination of bisphenol A concentrations in human biological fluids reveals significant early prenatal exposure. Hum Reprod. 2002;17:2839–2841. doi: 10.1093/humrep/17.11.2839. [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa H, Shozu M, Okada M, Inukai M, Zhang B, Kato K, Kasai T, Inoue M. Early growth response gene-1 plays a pivotal role in down-regulation of a cohort of genes in uterine leiomyoma. J Mol Endocrinol. 2007;39:333–341. doi: 10.1677/JME-06-0069. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Xia W, Yang J, Zhu Y, Chang H, Liu J, Huo W, Xu B, Chen X, Li Y, Xu S. BPA-induced DNA hypermethylation of the master mitochondrial gene PGC-1alpha contributes to cardiomyopathy in male rats. Toxicology. 2015;329:21–31. doi: 10.1016/j.tox.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Khachigian LM. Early growth response-1 in cardiovascular pathobiology. Circ Res. 2006;98:186–191. doi: 10.1161/01.RES.0000200177.53882.c3. [DOI] [PubMed] [Google Scholar]
- 34.Khan S, Beigh S, Chaudhari BP, Sharma S, Aliul Hasan Abdi S, Ahmad S, Ahmad F, Parvez S, Raisuddin S. Mitochondrial dysfunction induced by bisphenol A is a factor of its hepatotoxicity in rats. Environ Toxicol. 2016;31:1922–1934. doi: 10.1002/tox.22193. [DOI] [PubMed] [Google Scholar]
- 35.Kim SH, Despres JP, Koh KK. Obesity and cardiovascular disease: friend or foe? Eur Heart J. 2015 doi: 10.1093/eurheartj/ehv509. [DOI] [PubMed] [Google Scholar]
- 36.King AJ, Olivier NB, Mohankumar PS, Lee JS, Padmanabhan V, Fink GD. Hypertension caused by prenatal testosterone excess in female sheep. Am J Physiol Endocrinol Metab. 2007;292:E1837–E1841. doi: 10.1152/ajpendo.00668.2006. [DOI] [PubMed] [Google Scholar]
- 37.Kohli S, Ahuja S, Rani V. Transcription factors in heart: promising therapeutic targets in cardiac hypertrophy. Curr Cardiol Rev. 2011;7:262–271. doi: 10.2174/157340311799960618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lang IA, Galloway TS, Scarlett A, Henley WE, Depledge M, Wallace RB, Melzer D. Association of urinary bisphenol A concentration with medical disorders and laboratory abnormalities in adults. JAMA. 2008;300:1303–1310. doi: 10.1001/jama.300.11.1303. [DOI] [PubMed] [Google Scholar]
- 39.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee C, Patil S, Sartor MA. RNA-Enrich: a cut-off free functional enrichment testing method for RNA-seq with improved detection power. Bioinformatics. 2015;32:1100–1102. doi: 10.1093/bioinformatics/btv694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez AM, Cheong A, Ying J, Xue J, Kannan K, Leung YK, Thomas MA, Ho SM. Effects of high-butterfat diet on embryo implantation in female rats exposed to bisphenol A. Biol Reprod. 2015;93:147. doi: 10.1095/biolreprod.115.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsukawa N, Grzesik WJ, Takahashi N, Pandey KN, Pang S, Yamauchi M, Smithies O. The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc Natl Acad Sci USA. 1999;96:7403–7408. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattingly CJ, Colby GT, Forrest JN, Boyer JL. The comparative toxicogenomics database (CTD) Environ Health Perspect. 2003;111:793–795. doi: 10.1289/ehp.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McEachin RC, Chen H, Sartor MA, Saccone SF, Keller BJ, Prossin AR, Cavalcoli JD, McInnis MG. A genetic network model of cellular responses to lithium treatment and cocaine abuse in bipolar disorder. BMC Syst Biol. 2010;4:158. doi: 10.1186/1752-0509-4-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metges CC. Early nutrition and later obesity: animal models provide insights into mechanisms. Adv Exp Med Biol. 2009;646:105–112. doi: 10.1007/978-1-4020-9173-5_11. [DOI] [PubMed] [Google Scholar]
- 46.MohanKumar SM, Rajendran TD, Vyas AK, Hoang V, Asirvatham-Jeyaraj N, Veiga-Lopez A, Olivier NB, Padmanabhan V, MohanKumar PS. Effects of prenatal bisphenol-A exposure and postnatal overfeeding on cardiovascular function in female sheep. J Dev Orig Health Dis. 2016 doi: 10.1017/S204017441600057X. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moss RL, Razumova M, Fitzsimons DP. Myosin crossbridge activation of cardiac thin filaments: implications for myocardial function in health and disease. Circ Res. 2004;94:1290–1300. doi: 10.1161/01.RES.0000127125.61647.4F. [DOI] [PubMed] [Google Scholar]
- 48.Nadeau KJ, Maahs DM, Daniels SR, Eckel RH. Childhood obesity and cardiovascular disease: links and prevention strategies. Nat Rev Cardiol. 2011;8:513–525. doi: 10.1038/nrcardio.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nathanielsz PW, Ford SP, Long NM, Vega CC, Reyes-Castro LA, Zambrano E. Interventions to prevent adverse fetal programming due to maternal obesity during pregnancy. Nutr Rev. 2013;71(Suppl 1):S78–S87. doi: 10.1111/nure.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neier K, Marchlewicz EH, Dolinoy DC, Padmanabhan V. Assessing human health risk to endocrine disrupting chemicals: a focus on prenatal exposures and oxidative stress. Endocr. Disruptors (Austin) 2015:3. doi: 10.1080/23273747.2015.1069916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikolsky Y, Ekins S, Nikolskaya T, Bugrim A. A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol Lett. 2005;158:20–29. doi: 10.1016/j.toxlet.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Othman ER, Al-Adly DM, Elgamal DA, Ghandour N, El-Sharkawy S. Bisphenol A concentrates preferentially in human uterine leiomyoma and induces proliferation in rat myometrium. Reprod Sci. 2016;23:508–514. doi: 10.1177/1933719115608001. [DOI] [PubMed] [Google Scholar]
- 53.O’Toole TE, Conklin DJ, Bhatnagar A. Environmental risk factors for heart disease. Rev Environ Health. 2008;23:167–202. doi: 10.1515/reveh.2008.23.3.167. [DOI] [PubMed] [Google Scholar]
- 54.Padmanabhan V, Cardoso RC, Puttabyatappa M. Developmental programming, a pathway to disease. Endocrinology. 2016;157:1328–1340. doi: 10.1210/en.2016-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Padmanabhan V, Siefert K, Ransom S, Johnson T, Pinkerton J, Anderson L, Tao L, Kannan K. Maternal bisphenol-A levels at delivery: a looming problem? J Perinatol. 2008;28:258–263. doi: 10.1038/sj.jp.7211913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Padmanabhan V, Veiga-Lopez A. Animal models of the polycystic ovary syndrome phenotype. Steroids. 2013;78:734–740. doi: 10.1016/j.steroids.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papamatheakis DG, Chundu M, Blood AB, Wilson SM. Prenatal programming of pulmonary hypertension induced by chronic hypoxia or ductal ligation in sheep. Pulm Circ. 2013;3:757–780. doi: 10.1086/674767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel BB, Raad M, Sebag IA, Chalifour LE. Lifelong exposure to bisphenol a alters cardiac structure/function, protein expression, and DNA methylation in adult mice. Toxicol Sci. 2013;133:174–185. doi: 10.1093/toxsci/kft026. [DOI] [PubMed] [Google Scholar]
- 59.Peng K, Tian X, Qian Y, Skibba M, Zou C, Liu Z, Wang J, Xu Z, Li X, Liang G. Novel EGFR inhibitors attenuate cardiac hypertrophy induced by angiotensin II. J Cell Mol Med. 2016;20:482–494. doi: 10.1111/jcmm.12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perrone S, Santacroce A, Picardi A, Buonocore G. Fetal programming and early identification of newborns at high risk of free radical-mediated diseases. World J Clin Pediatr. 2016;5:172–181. doi: 10.5409/wjcp.v5.i2.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poirier P, Giles TD, Bray GA, Hong Y, Stern JS, Pi-Sunyer FX, Eckel RH American Heart Association Obesity Committee of the Council on Nutrition PA Metabolism. Obesity and cardiovascular disease: pathophysiology, evaluation, and effect of weight loss: an update of the 1997 American Heart Association Scientific Statement on Obesity and Heart Disease from the Obesity Committee of the Council on Nutrition, Physical Activity, and Metabolism. Circulation. 2006;113:898–918. doi: 10.1161/CIRCULATIONAHA.106.171016. [DOI] [PubMed] [Google Scholar]
- 62.Puttabyatappa M, Cardoso RC, Padmanabhan V. Effect of maternal PCOS and PCOS-like phenotype on the offspring’s health. Mol Cell Endocrinol. 2015;435:29–39. doi: 10.1016/j.mce.2015.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ranciere F, Lyons JG, Loh VH, Botton J, Galloway T, Wang T, Shaw JE, Magliano DJ. Bisphenol A and the risk of cardiometabolic disorders: a systematic review with meta-analysis of the epidemiological evidence. Environ Health. 2015;14:46. doi: 10.1186/s12940-015-0036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ranjit N, Siefert K, Padmanabhan V. Bisphenol-A and disparities in birth outcomes: a review and directions for future research. J Perinatol. 2010;30:2–9. doi: 10.1038/jp.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rayner BS, Figtree GA, Sabaretnam T, Shang P, Mazhar J, Weaver JC, Lay WN, Witting PK, Hunyor SN, Grieve SM, et al. Selective inhibition of the master regulator transcription factor Egr-1 with catalytic oligonucleotides reduces myocardial injury and improves left ventricular systolic function in a preclinical model of myocardial infarction. J Am Heart Assoc. 2013;2:e000023. doi: 10.1161/JAHA.113.000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rezg R, El-Fazaa S, Gharbi N, Mornagui B. Bisphenol A and human chronic diseases: current evidences, possible mechanisms, and future perspectives. Environ Int. 2014;64:83–90. doi: 10.1016/j.envint.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 67.Savabieasfahani M, Kannan K, Astapova O, Evans NP, Padmanabhan V. Developmental programming: differential effects of prenatal exposure to bisphenol-A or methoxychlor on reproductive function. Endocrinology. 2006;147:5956–5966. doi: 10.1210/en.2006-0805. [DOI] [PubMed] [Google Scholar]
- 68.Shah AS, Khoury PR, Dolan LM, Ippisch HM, Urbina EM, Daniels SR, Kimball TR. The effects of obesity and type 2 diabetes mellitus on cardiac structure and function in adolescents and young adults. Diabetologia. 2011;54:722–730. doi: 10.1007/s00125-010-1974-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 70.Smith KB, Smith MS. Obesity statistics. Prim Care. 2016;43:121–135. doi: 10.1016/j.pop.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 71.Smith NJ, Chan HW, Osborne JE, Thomas WG, Hannan RD. Hijacking epidermal growth factor receptors by angiotensin II: new possibilities for understanding and treating cardiac hypertrophy. Cell Mol Life Sci. 2004;61:2695–2703. doi: 10.1007/s00018-004-4244-3. [DOI] [PubMed] [Google Scholar]
- 72.Song Y, Chou EL, Baecker A, You NY, Song Y, Sun Q, Liu S. Endocrine-disrupting chemicals, risk of type 2 diabetes, and diabetes-related metabolic traits: a systematic review and meta-analysis. J Diabetes. 2015;8:516–32. doi: 10.1111/1753-0407.12325. [DOI] [PubMed] [Google Scholar]
- 73.Sun Y, Irie M, Kishikawa N, Wada M, Kuroda N, Nakashima K. Determination of bisphenol A in human breast milk by HPLC with column-switching and fluorescence detection. Biomed Chromatogr. 2004;18:501–507. doi: 10.1002/bmc.345. [DOI] [PubMed] [Google Scholar]
- 74.Symonds ME, Budge H, Stephenson T, McMillen IC. Fetal endocrinology and development – manipulation and adaptation to long-term nutritional and environmental challenges. Reproduction. 2001;121:853–862. doi: 10.1530/rep.0.1210853. [DOI] [PubMed] [Google Scholar]
- 75.Thornburg KL, Louey S, Giraud GD. The role of growth in heart development. Nestle Nutr Workshop Ser Pediatr Program. 2008;61:39–51. doi: 10.1159/000113169. [DOI] [PubMed] [Google Scholar]
- 76.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vandenberg LN, Chahoud I, Heindel JJ, Padmanabhan V, Paumgartten FJ, Schoenfelder G. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ Health Perspect. 2010;118:1055–1070. doi: 10.1289/ehp.0901716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vandenberg LN, Hunt PA, Myers JP, Vom Saal FS. Human exposures to bisphenol A: mismatches between data and assumptions. Rev Environ Health. 2013;28:37–58. doi: 10.1515/reveh-2012-0034. [DOI] [PubMed] [Google Scholar]
- 80.Varvarigou AA. Intrauterine growth restriction as a potential risk factor for disease onset in adulthood. J Pediatr Endocrinol Metab. 2010;23:215–224. doi: 10.1515/jpem.2010.23.3.215. [DOI] [PubMed] [Google Scholar]
- 81.Veiga-Lopez A, Kannan K, Liao C, Ye W, Domino SE, Padmanabhan V. Gender-specific effects on gestational length and birth weight by early pregnancy BPA exposure. J Clin Endocrinol Metab. 2015a;100:E1394–E1403. doi: 10.1210/jc.2015-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Veiga-Lopez A, Luense LJ, Christenson LK, Padmanabhan V. Developmental programming: gestational bisphenol-A treatment alters trajectory of fetal ovarian gene expression. Endocrinology. 2013;154:1873–1884. doi: 10.1210/en.2012-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Veiga-Lopez A, Moeller J, Sreedharan R, Singer K, Lumeng C, Ye W, Pease A, Padmanabhan V. Developmental programming: interaction between prenatal BPA exposure and postnatal adiposity on metabolic variables in female sheep. Am J Physiol Endocrinol Metab. 2016;310:E238–E247. doi: 10.1152/ajpendo.00425.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Veiga-Lopez A, Pennathur S, Kannan K, Patisaul HB, Dolinoy DC, Zeng L, Padmanabhan V. Impact of gestational bisphenol A on oxidative stress and free fatty acids: human association and interspecies animal testing studies. Endocrinology. 2015b;156:911–922. doi: 10.1210/en.2014-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waller DK, Gallaway MS, Taylor LG, Ramadhani TA, Canfield MA, Scheuerle A, Hernandez-Diaz S, Louik C, Correa A National Birth Defects Prevention S. Use of oral contraceptives in pregnancy and major structural birth defects in offspring. Epidemiology. 2010;21:232–239. doi: 10.1097/EDE.0b013e3181c9fbb3. [DOI] [PubMed] [Google Scholar]
- 86.Xia W, Jiang Y, Li Y, Wan Y, Liu J, Ma Y, Mao Z, Chang H, Li G, Xu B, et al. Early-life exposure to bisphenol a induces liver injury in rats involvement of mitochondria-mediated apoptosis. PLoS One. 2014;9:e90443. doi: 10.1371/journal.pone.0090443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yajnik C. Interactions of perturbations in intrauterine growth and growth during childhood on the risk of adult-onset disease. Proc Nutr Soc. 2000;59:257–265. doi: 10.1017/s0029665100000288. [DOI] [PubMed] [Google Scholar]
- 88.Ye N, Ding Y, Wild C, Shen Q, Zhou J. Small molecule inhibitors targeting activator protein 1 (AP-1) J Med Chem. 2014;57:6930–6948. doi: 10.1021/jm5004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–952. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.