Abstract
The biomedical sciences have experienced an explosion of data which promises to overwhelm many current practitioners. Without easy access to data science training resources, biomedical researchers may find themselves unable to wrangle their own datasets. In 2014, to address the challenges posed such a data onslaught, the National Institutes of Health (NIH) launched the Big Data to Knowledge (BD2K) initiative. To this end, the BD2K Training Coordinating Center (TCC; bigdatau.org) was funded to facilitate both in-person and online learning, and open up the concepts of data science to the widest possible audience. Here, we describe the activities of the BD2K TCC and its focus on the construction of the Educational Resource Discovery Index (ERuDIte), which identifies, collects, describes, and organizes online data science materials from BD2K awardees, open online courses, and videos from scientific lectures and tutorials. ERuDIte now indexes over 9,500 resources. Given the richness of online training materials and the constant evolution of biomedical data science, computational methods applying information retrieval, natural language processing, and machine learning techniques are required - in effect, using data science to inform training in data science. In so doing, the TCC seeks to democratize novel insights and discoveries brought forth via large-scale data science training.
Keywords: Education, Metadata, Data Collection, Information Storage and Retrieval, Pattern Recognition, Automated, Classification
INTRODUCTION
Biomedical research has rapidly become a principal focal point for innovation and creativity in modern data management and analysis techniques - often spanning computational, statistical and mathematical disciplines being applied in the biological sciences to extract maximal utility from large-scale data (1, 2). However, the rapidity with which data acquisition is occurring across biomedical research (3), often in lock-step with advances in technology, means that even what might have once been considered having a small-science focus finding themselves facing “big data” challenges (4, 5).
To meet the essential data and computing demands of today’s research biomedical ecosystem, the NIH has made an unprecedented investment in data science research and training through its Big Data to Knowledge (BD2K; https://datascience.nih.gov) program (6). Through a series of career development awards (K01 awards), institutional training awards (T32/T15), a variety of data science training research awards (R25), and the training components of a dozen Centers of Excellence (U54), the NIH has placed a premium on the development of a new generation of biomedical data science professionals (7). These efforts systematically produce unique training materials seeking to introduce researchers to everything from the basics of databasing to data mining, machine learning, and the in-depth examinations of applied biomedical analytics in the investigation of health as well as disease. To catalyze and support all these efforts, the NIH established the BD2K Training Coordinating Center (TCC; http://www.bigdatau.org) with the stated aim to provide biomedical researchers with the tools to negotiate the complex landscape of data science education for biomedical researchers.
1.1 The BD2K Training Coordinating Center
Given the constantly developing analytical needs in biomedical science, varied training modalities are ideal for catering to the differing learning styles of busy researchers. Online training programs like massive open online courses (MOOCs) tend to focus on complete, end-to-end curricula in much the same manner as a traditional university course. Indeed, in some instances universities have adopted the MOOC model to develop entire degree programs. In contrast, scientific seminar and conference presentations tend to provide a narrower scope of content in highly specific domains. Hands-on training programs, done in-person or via the internet, on the other hand, often offer opportunities to directly learn the steps involved with some process, software tool, or analytical approach and then the chance to apply these concepts directly to an example data. While each of these models has their relative advantages and disadvantages, it is often the combination of these which provides the maximal utility for learning. That is, providing the foundational basis of data techniques applied to biomedical research challenges, the step-by-step understanding of computational processes, as well as the focused research rationale for why such measurements are being made. Spanning these levels of understanding help to reinforce a deeper appreciation for what data are telling one and what they represent about underlying biological systems. Consequently, the TCC provides multiple levels of training content, materials, and opportunities for online, as well as, in-person learning.
1.1.1 In-Person Training Activities and Workshops
The TCC has created two specific programs for in-person training and learning which encourage mentorship and collaboration in biomedical data science. First, we developed the Data Science Rotations for Advancing Discovery (RoAD-Trip) program to foster new collaborations among junior biomedical researchers and senior-level data scientists to address the challenge of translating complex data into new knowledge (http://www.bigdatau.org/roadtrip). This program seeks to promote the careers of young biomedical scientists, engage established data scientists, and encourage the development of joint biomedical data science projects which are suitable as new NIH grant proposals.
Second, we have also organized an annual Data Science Innovation Lab, representing another example of fostering new interdisciplinary collaborations among quantitative and biomedical researchers to address data science challenges. This five-day residential workshop is supported by the NIH and the National Science Foundation. With the aid of professional facilitators or mentors, the accepted participants form teams that seek to solve specific data science challenges. In 2016, the Data Science Innovation Lab discussed mobile health and the challenges arising from the use of wearable or ambient sensors. The most recent Data Science Innovation Lab, held in June 2017 with over 30 attendees, put its focus on understanding of the microbiome and the big data derived from microbiota (http://www.bigdatau.org/innovationlab2017).
1.1.2 Online Training Portal and Subject-Specific Training Search Tools
The TCC currently has two initiatives on creating educational video content. In an effort to expand on the general knowledge of Big Data, the TCC alongside the USC School of Cinematic Arts, created the short film: “Big Data: Biomedicine” which focuses on the science of big data and its implications for the future of biomedical research (https://youtu.be/F6CI7jXHGWg). Then, in collaboration with the BD2K Centers-Coordination Center (BD2KCCC) and the NIH Office of Data Science, the TCC has developed a weekly webinar series entitled The BD2K Guide to the Fundamentals of Data Science Series (http://www.bigdatau.org/data-science-seminars). This series consists of lectures from experts across the country covering the basics of data management, representation, computation, statistical inference, data modeling, and other topics relevant to “big data” in biomedicine. These seminar videos are recorded and uploaded to YouTube, while we also include these videos (along with any other archived learning materials from BD2K centers) on the TCC website for discovery and adding to personal educational plans (see below).
1.1.3 The Educational Resource Discovery Index
A major effort of the TCC has been the creation of a sophisticated database of high-quality training materials available from different portals across internet called the Educational Resource Discovery Index (ERuDIte). Through ERuDIte, the TCC provides user-friendly access to a rich catalog of training resources and assembled learning materials. Specific selections of resources permit collections of educational topics tailored to a user’s current and hoped-for knowledge and learning goals. In what follows in Sections 2 and 3, we present the details of our ERuDIte system and a brief summary of how we have applied data science to the organization of data science training materials in this unique platform.
2. The TCC Website and ERuDIte
The TCC’s http://www.bigdatau.org website and ERuDIte have been specifically developed to provide an online platform for fostering and supporting self-directed learning in data science topics as they relate to biomedical research challenges. Users can search ERuDIte using faceted search over several dimensions (cf. Section 3.3), which describe different aspects of the learning resources, to identify those relevant to their training needs. The portal also provides summary visualizations of the ERuDIte catalog contents (http://www.bigdatau.org/statistics). In addition, learners can create individual profiles and then receive access to personalized learning features which allow them to monitor which training resources they have completed and to modify individual learning plans as needed.
Given the breadth and depth of data science training resources, the task of collecting and curating relevant, high-quality learning materials is not a trivial matter and requires a combination of manual and automatic approaches. Data science includes methods from and applications to multiple and diverse fields. As a result, researchers interested in learning about the techniques of data science can be faced with a range of MOOCs about databases, a practical tutorial on a blog which illustrates Matlab processing scripts, an online textbook describing the basics of Bayesian learning using the R programming language, or a video covering the latest advances in deep learning. Such resources provide value for learning, but each may have a different quality, time commitment, and relevance for specific training goals.
In the next section, we discuss how we are building ERuDIte using many of the same data science techniques we seek to teach. Specifically, we describe our approaches to 1) identifying high-quality learning resources using both manual and automatic techniques, 2) develop standard schemas and ontologies to describe the resources, 3) automatically assigning rich descriptors to each resource, and 4) provide access to these learning resources.
3. Building the Educational Resource Discovery Index (ERuDIte)
ERuDIte uses techniques from knowledge representation, data modeling, natural language processing, information retrieval, and machine learning to discover, integrate, describe, and organize resources, making ERuDIte a system that uses data science techniques to teach data science. As a result, the multiple components of ERuDIte follow core steps in the data science process: resource identification and extraction (Sect. 3.1), resource description and integration (Sect. 3.2) and automatic modeling (Sect. 3.3).
3.1 Resource Identification and Aggregation Methods
To start our collection process, we first reviewed MOOCs, blogs, e-books, videos, websites, conference presentations and tutorials, and other relevant data science material available on the web (http://www.bigdatau.org/about_erudite). In selecting resources, we consider the reliability of the source provider, the didactic value, and overall quality of the resources. From high-quality sources, such as MOOCs or conference tutorials, we extracted the metadata about the resources using an automatic scraping framework (Sect. 3.1.1). For resources of mixed quality, such as YouTube, we developed automated quality identification techniques (Sect. 3.1.2).
3.1.1 Automatic Website Scraping
Collecting relevant material is essential to the value and success of ERuDIte. As a result, during the initial stages of development, we focused our attention on gathering high quality sources that contained collections of individual resources. This included MOOC sites such as edX (https://www.edx.org/), Coursera (https://www.coursera.org/), and Udacity (https://www.udacity.com/) in addition to the sites of other BD2K centers creating their own training materials (https://commonfund.nih.gov/bd2k).
A few unstructured sources with high-quality resources required completely manual attention, but overall, we focused our early identification efforts on structured sources that would allow us to gather resource data in a semi-automated way. Coursera and Udacity provided rich APIs, but the majority of the other resources required scraping. To streamline the scraping procedure, which demands individual source customization, we created a framework with website-specific modules using the popular Python packages BeautifulSoup and Dryscrape. The framework provides tools to handle dynamic JavaScript pages and to structure, extract, and export the collected data. The scraping framework is then packaged as a Docker image loaded with all the dependencies, and we store the image in a central repository. This allows for parallel development where multiple members of the team can extend the framework as needed without having to manage local software package installation and updates. Consequently, using this framework, we were able to identify and gather large collections quickly. As of September 27, 2017, ERuDIte contains over 9,500 resources. Table 1, above, provides details of the current collection of indexed resources.
Table 1.
Summary of data currently collected in ERuDIte (as of Sept. 2017), by source and by resource type.
| Provider | Types | Total | With Descriptions | With Transcripts | With Additional Text + Slides |
|---|---|---|---|---|---|
| BD2K | Video / Written | 515 | 461 | 264 | 59 |
| edX | Course / Video | 92 | 91 | 70 | 54 |
| Coursera | Course / Video | 77 | 77 | 54 | 56 |
| Udacity | Course / Video | 17 | 17 | 17 | 0 |
| Videolectures.net | Video | 8078 | 5741 | 165 | 4596 |
| YouTube | Video | 410 | 356 | 252 | 0 |
| ELIXIR | Course / Written | 235 | 48 | 0 | 0 |
| Bioconductor | Course / Written | 5 | 2 | 0 | 0 |
| Cornell Virtual Workshop | Course / Written | 38 | 19 | 0 | 0 |
| NIH | Video | 1 | 1 | 0 | 0 |
| OHBM | Video / Written | 78 | 6 | 0 | 51 |
| TOTAL | 9,546 | 6,819 | 822 | 4,816 |
3.1.2 Automated Quality Assessment
To expand our resource collection beyond our manually curated sources, we are developing techniques to identify high-quality learning resources from large open collections, such as YouTube. In this section, we describe how information extraction and machine learning techniques are applied to assess the quality of data science videos in YouTube and include these resources into ERuDIte.
Searching for the phrase “data science” on YouTube yields over 190,000 videos (and over 19 million if “data science” is not constrained to be a phrase). However, the amount of relevant and pedagogically valuable videos is a fraction of this number. To filter down the results, we trained a classifier to assess quality using video metadata, such as upload date, views, and “likes”, as well as extracted text, such as title, description, and automatic transcripts.
We use a set of concepts relevant to data science (specifically, from the Data Science Domain from the ontology described in Sect. 3.2.2) to search across YouTube. The search queries include concept names, sometimes with additional clarification terms, for example: “bioinformatics”, (“data science” AND “python”), or ((“data science” OR “machine learning”) AND “regression”). Sixty-two such queries were conducted and the resulting metadata was obtained from those videos and playlists appearing in the first 20 pages of results from YouTube for each query, which yields a dataset of 41,605 videos (35,235 unique). We then manually annotated 986 videos, sampled from across the different pages of results for different queries. These were judged on a scale of 0–4, where 0 is a video that is completely unhelpful as a resource for learning about data science, while 4 is most helpful. These are scored with resources labeled 0–1 considered low-quality and 2+ considered good-quality. This provided us with a roughly balanced data set of 417 low-quality videos and 569 high-quality videos. Finally, using k-fold cross-validation, a logistic regression classifier was trained using a variety of features, including the video text and metadata. The classifier achieves precision of 0.79, recall of 0.85, and a F1 score of 0.82. This performance is sufficient to select promising videos from YouTube for human curation. As training data size increases, we expect that the automatic classification quality will approach human levels of agreement and minimize human effort.
3.2 Resource Description Schema and ERuDIte Integration
With resources originating from different sites and creators, we have developed a metadata standard (Sect 3.2.1) to unify the structure of the ERuDIte index and to provide as much information as possible to learners to select relevant resources. We have also developed an ontology with 6 hierarchical dimensions to further describe the learning resources (Sect 3.2.2). These metadata are organized in the ERuDIte database (Sect 3.2.3), and openly shared with the community using the JSON-LD Linked Data standard (Sect 3.2.4).
3.2.1 ERuDIte’s Learning Resource Metadata Standard
To design the metadata standard for learning resources in ERuDIte, we reviewed previous standards, including Dublin Core, Learning Resource Metadata Initiative (LRMI), IEEE’s Learning Object Metadata (LOM), eXchanging Course Related Information (XCRI), and Metadata for Learning Opportunities (MLO), as well as emerging standards such as Bioschemas.org and the CreativeWork and Course schemas from the Schema.org vocabularies. The key classes of our standard are CreativeWork (used for learning resources), Person (for instructors or material creators), and Organization (for affiliations and resource providers).
We are collaborating with ELIXIR, a large European project organizing life science data, as well as other international organizations (e.g. Goblet, from England, H3Africa, from South Africa, and CSIRO, from Australia), to converge to a common standard for learning resources. In coordination with these groups, we have adopted schema.org vocabularies, defining additional properties when critically needed. Schema.org has the support of major search engines (such as Google, Bing) which facilitates discovery and dissemination of resources indexed in ERuDIte. A white paper on our joint efforts is due in the autumn of 2017.
3.2.2 The Data Science Education Ontology
To further describe the contents of learning resources, we have created the Data Science Education Ontology (DSEO), based on the Python machine learning package scikit-learn (8). The DSEO is specifically organized as a SKOS vocabulary since the flexible “broaderTransitive” property from SKOS best captures the subtle relationships between our concepts. The DSEO is publicly available at http://bioportal.bioontology.org/ontologies/DSEO. Specifically, the DSEO has six hierarchical dimensions, each describing a different facet of a learning resource:
Data Science Process (8 concepts): What stages of the data science process will this resource help me with understanding?
Domain (74 concepts): What field of study does this resource focus on?
Datatype (18 concepts): What types of biomedical data are addressed in the resource?
Programming Tool (13 concepts): What programming tools are being used in or taught by this resource?
Resource Format (2 concepts): In what manner is this resource presented?
Resource Depth (2 concepts): How advanced is this resource? At what experience level is it pitched?
3.2.3 Resource Database
To store, integrate, and efficiently query ERuDIte’s resource data and metadata we have adopted the use of a relational database for storing the direct output of our scrapers. We then define a set of database views to integrate data across sources and map source tables to a relational implementation of our metadata standard. This enables us to flexibly extend the metadata schema without modifying collected data. For efficiency, we create a materialized view with appropriate keys and indices which joins standard schema views to form a composite table that powers the resource detail pages on the BD2K TCC website. Additionally, we also generate an Elasticsearch (elastic.co) index over the metadata to power the faceted search of ERuDIte.
3.2.4 Linked Data Representations
To disseminate the resources indexed in ERuDIte as broadly as possible, we embed structured metadata for each resource expressed in JSON-LD (https://json-ld.org/), in addition to our standard schema, on each learning resource webpage. Sharing resource metadata as openly as possible through embedded, concise JSON-LD has several benefits: 1) it complies with the goals of the Semantic Web and Linked Data communities to make the data available on the web. This is, information is not only human readable, but also readable by machines, and to allow for additional content about web-objects to be accrued in a distributed fashion (9); 2) web-based search engines, such as Google, are encouraging the use of the JSON-LD structured data format for webpages, since it aids in their own indexing and the representations of webpage content. This enhanced indexing facilitates discovery of resources by users outside of ERuDIte. We have used our previous work on schema mapping (10) to conveniently translate our relational schema of the resources metadata into JSON-LD format and have successfully applied this approach here, as well. What is more, in order to maximize the reuse of ERuDIte, we have licensed bigdatau.org website content and ERuDIte schema under a Creative Commons Attribution-Non-Commercial-ShareAlike (CC BY-NC-SA) license (https://bigdatau.ini.usc.edu/about_erudite).
3.3 Automated Concept Tagging for ERuDIte
Concept modeling has formed an integral element in the construction of ERuDIte. However, manually tagging the thousands of resources in ERuDIte with concepts from video and other content would be time- and cost-prohibitive. Thus, we have developed automated labeling methods, based on machine learning, natural language processing and information retrieval techniques, to efficiently tag the growing collection of ERuDIte learning resources.
In order to evaluate this concept modeling framework, we created a “gold standard” consisting of 726 manually-curated resources (data science courses from Coursera, Udacity, edX, Cornell’s Virtual Workshop, and videos from Videolectures.net and YouTube) labeled with the appropriate tags from each DSEO dimension. We randomly select 581 resources (~80%) for training and cross validation, and left aside 145 resources for testing. In previous experiments (11) we predicted all concepts across all dimensions; however, upon investigation, classifier performance increased per concept tag if trained on a per dimension basis. We then created fixed fold assignments for the training set of resources, and conducted five-fold cross-validation grid searches over the hyper-parameters defined for each classifier. The hyper-parameter grid included value ranges for parameters specific to the vectorization of each resource and to the classifier method itself. Averaged F1 scores weighted by the support available in each fold were employed to select the best classifier with the best hyper-parameter combination. Predicted tags for the 145 resources in the test set were then obtained.
Table 2 briefly summarizes the performance of the best classifiers along each dimension. Some dimensions are clearly more difficult to resolve than are others. Particularly, our routine struggles with classifying the type of programming tools and datatypes dimensions. We believe that this can be explained by simply having fewer resources are tagged with concepts for these two dimensions. With greater numbers of exemplars along these axes, the better our classification will become. This suspicion has been born out and will be the topic of a subsequent research article from our team. Briefly, but not surprisingly, we observed that classifier performance is markedly improved on tags where there are at least fifteen resources labeled with that tag in the training set. A more comprehensive and detailed article on the validation of ERuDIte and the automated resource tagging approach is in preparation.
Table 2.
F1 classification scores on ERuDIte test set of resources, by classifier method, for the overall number of tags as well as those with at least a given level of support (from at least 5-to-15 training resources being present). A manuscript providing additional mathematical details on the validation of the ERuDIte autotagging and curation system is in preparation.
| Dimension | Classifier Type | F1 | F1(support >= 5) | F1(support >= 10) | F1(support >= 15) |
|---|---|---|---|---|---|
| Domain | Logistic Regression | 0.762 | 0.778 | 0.793 | 0.807 |
| Resource Depth | SVM | 0.778 | 0.778 | 0.778 | 0.778 |
| Resource Format | SVM | 0.989 | 0.989 | 0.989 | 0.989 |
| Data Science Process | Logistic Regression | 0.705 | 0.704 | 0.704 | 0.704 |
| Programming Tool | Logistic Regression | 0.533 | 0.538 | 0.537 | 0.555 |
| Datatype | Logistic Regression | 0.481 | 0.488 | 0.492 | 0.492 |
3.4 Further Work
3.4.1 Community Validation and Ongoing System Re-Training
The performance of our currently classifiers for ERuDIte resource identification (Sect. 3.1.2) and labeling (Sect. 3.3) has shown promising results (F1~0.8), but are not yet sufficiently accurate for automatically including their results directly into ERuDIte. We plan to leverage our automated classifier system to propose resources and tags to domain expert curators. This community-driven approach will aid in reviewing our classifier predictions thereby ensuring the inclusion of high quality resources and tagging. We have also developed a web-based curation interface for use in accelerating the ERuDIte resource curation process. In addition to validating the predicted tags, reviewers can also propose concepts which do not currently exist in our DSEO vocabulary. As additional curated resources and tags are included, we will systematically retrain our classifiers and expect their performance to improve.
3.4.2 Discovery of Training Pre-requisites
To enable personalized learning plans, automatically inferring which data science concepts are presented in each resource and what other concepts are prerequisites for these is an important step. For example, if a learner is interested in a course on Machine Learning in Matlab but her user profile does not indicate experience in mathematics, ERuDIte might recommend with a resource on Probability. Indeed, there are a number of methods to predict the underlying concepts present in a set of training resources, with topic modeling approaches such as Latent Dirichlet Allocation (LDA) being a commonly employed approach (12). Another approach worth exploring is one that exploits naturally occurring sequential data. Given such a set of sequential data, where each entry is associated with a distribution of concepts, weights are accrued for how likely a concept is to occur in advance of another concept, and a net effect score can be computed by subtracting the weight for the converse of directionality, e.g. a concept occurring after another concept. For instance, the scraping of textbook tables of contents and course syllabi from the Web is one such means of thematic concepts which occur in a regularized order. For example, given the chapter title “Unsupervised Learning: Clustering and Dimensionality Reduction”, an informed algorithm would select the top five (albeit imperfect) results but with diminishing weights: derived via dimensionality reduction, k-means clustering, fuzzy clustering, machine learning, or sparse dictionary learning, to comprise the semantic relevance vector for the topic set (13). By so doing, we envision an evidence-based means of curriculum development.
3.4.3 Personalization of Training Resources
The TCC website is also designed to collect usage data which will allow us to develop personalized and custom learning experiences. First, registered learners can create their own profiles and detail their prior knowledge and their interests, which we can then use to align with our resource concepts to make recommendations and to present search results. Second, registered learners can create educational plans that include resources that they want to review and complete. These can be used as sources of dependency relationships between resources, and also to prevent suggesting resources already known to the user. Third, we have implemented user monitoring, which allows us to understand the sequence of resource browsing activity in order to drive recommendations as users explore ERuDIte. Presently, ERuDIte offers resource-specific recommendations based upon a semantic similarity search on a single resource’s title. However, as TCC website and ERuDIte use grows, personalized recommendations via methods such as collaborative filtering are fully expected.
4. CONCLUSION
The BD2K TCC seeks to promote and support biomedical data science learning with a multi-pronged approach. We routinely organize in-person training events to engage researchers in data science learning through applied projects in biomedicine. These are used to actively create online learning materials with a potential to guide learners through data science concepts used in current biomedical research. Moreover, we use data science techniques to collect and organize learning resources widely available on the internet in order to help self-directed learners easily maneuver through the data science landscape - creating an online space where the knowledge and skills being taught are those being used in the learning ecosystem itself.
As the field of biomedicine increasingly demands multi-disciplinary skills and data science knowledge, resources for easily available constant learning are sorely needed. Data science is now providing support to a broad range of translational scientists (14) who require skills in data management, analytics, and visualization in order to better understand the richness and nuances of the data they are collecting. We believe that the work of the TCC and its emphasis on ERuDIte will contain the materials that not only establish the fundamentals of data science but also track the interest levels associated with new methods and techniques (15). Finally, even though the TCC is primarily focused on biomedical data science, we expect that the materials and approaches we provide will help the scientific community at large – helping to democratize training in data science to the widest possible audience.
Figure 1.
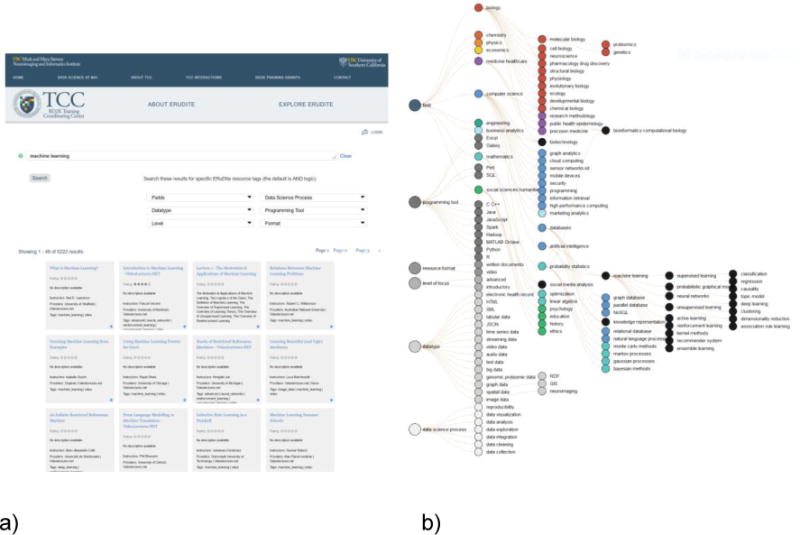
a) TCC ERuDIte faceted search page. b) One representation of the ERuDIte “Knowledge Map”.
Figure 2.
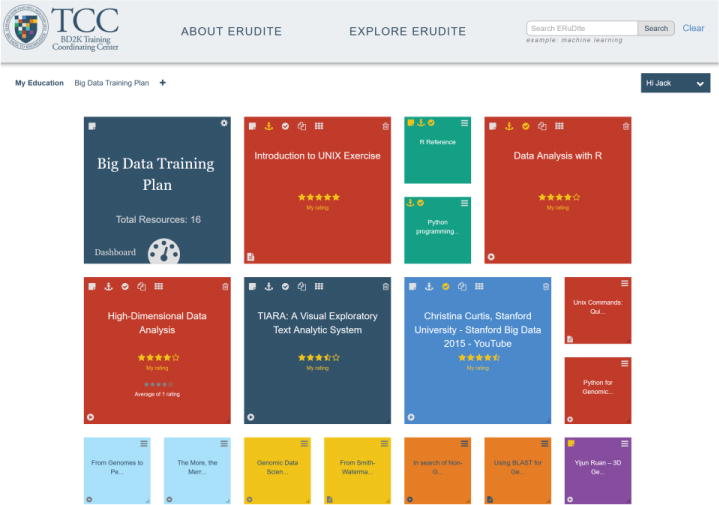
An example of an ERuDIte “training plan” comprised of resources indexed in the ERuDIte database. Users select, gather, arrange, color-code, rate, and then can utilize the educational resources in a prescribed, pre-requisite order or in any order they wish.
Acknowledgments
This work is supported by an NIH Big Data to Knowledge (BD2K) award to the BD2K Training Coordinating Center (U24 ES026465; http://www.bigdatau.org). Further information on the NIH BD2K program, its contributing NIH institutes, and all of its supported projects may be found via the NIH Data Science Office website (https://datascience.nih.gov/). Any opinions, findings, and conclusions expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health. The authors would also like to express their gratitude to the staff of the USC Mark and Mary Stevens Neuroimaging and Informatics Institute at the USC Keck School of Medicine.
Footnotes
The authors declare no competing financial interests.
References
- 1.Margolis R, et al. The National Institutes of Health’s Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. J Am Med Inform Assoc. 2014;21:957–958. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam NR, Wieder R, Ghosh D. Data science, learning, and applications to biomedical and health sciences. Ann N Y Acad Sci. 2017;1387:5–11. doi: 10.1111/nyas.13309. [DOI] [PubMed] [Google Scholar]
- 3.Van Horn JD, Toga AW. Human neuroimaging as a “Big Data” science. Brain Imaging Behav. 2014;8:323–331. doi: 10.1007/s11682-013-9255-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinkus GJ. Sparsey: event recognition via deep hierarchical sparse distributed codes. Front Comput Neurosci. 2014;8:160. doi: 10.3389/fncom.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Althoff T, et al. Large-scale physical activity data reveal worldwide activity inequality. Nature. 2017;547:336–339. doi: 10.1038/nature23018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourne PE, et al. The NIH Big Data to Knowledge (BD2K) initiative. J Am Med Inform Assoc. 2015;22:1114. doi: 10.1093/jamia/ocv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garmire LX, et al. THE TRAINING OF NEXT GENERATION DATA SCIENTISTS IN BIOMEDICINE. Pac Symp Biocomput. 2016;22:640–645. doi: 10.1142/9789813207813_0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 9.Taheriyan M, Knoblock CA, Szekely P, Ambite JL. Proceedings of the 2012 ESWC Confernce on Linked APIs for the Semantic Web Workshop (LAPIS); 2012. [Google Scholar]
- 10.Ambite JL, et al. paper presented at the Proceedings of the 26th International Conference on World Wide Web Companion; Perth, Australia. 2017. [Google Scholar]
- 11.Ambite JL, et al. Procs 26th Intl Conf on World Wide Web; ACM, New York, NY, USA. 2017. Vol. WWW’17 Companion. [Google Scholar]
- 12.Gabrilovich E, Markovitch S. Proceedings of International Joint Conference on Artificial Intelligence; Hyderabad, India. 2007. [Google Scholar]
- 13.Gordon J, Zhu L, Galstyan A, Natarajan P, Burns GAPC. Proceedings of the Association for Computational Linguistics (ACL) 2016 [Google Scholar]
- 14.Jackson RD, Gabriel S, Pariser A, Feig P. Training the Translational Scientist. Science Translational Medicine. 2010;2:63mr62. doi: 10.1126/scitranslmed.3001632. [DOI] [PubMed] [Google Scholar]
- 15.Dunn MC, Bourne PE. Building the biomedical data science workforce. PLOS Biology. 2017;15:e2003082. doi: 10.1371/journal.pbio.2003082. [DOI] [PMC free article] [PubMed] [Google Scholar]


