Abstract
Accurately characterizing cellular subpopulations is essential for elucidating the mechanisms underlying normal and pathological biology. Isolation of specific cell types can be accomplished by labeling unique, associated proteins with fluorescent antibodies. Cell fixation is commonly used to prepare these samples and allow for long-term storage, but this poses challenges for subsequent protein analysis. We previously established the FITSAR (Formaldehyde-fixed Intracellular Target-Sorted Antigen Retrieval) method in which protein can be isolated and characterized from fixed, enriched cell subpopulations. The current study improves on this method by allowing compatibility with highly sensitive, multiplex protein arrays and demonstrating applicability to long-term stored samples. Feasibility experiments demonstrated parallel detection of cell adhesion molecules using an enzyme-linked immunosorbent assay (ELISA) panel with human, adipose-derived stem cells stored for up to one month.
Keywords: FITSAR, protein characterization, paraformaldehyde, multiplex ELISA
Cell type-specific protein characterization is essential for accurate understanding of normal and pathological biology. Ability to extract protein from fixed cells increases flexibility in sample preparation and potential for intracellular target-based enrichment. We previously established the FITSAR (Formaldehyde-fixed Intracellular Target-Sorted Antigen Retrieval) method, in which protein was isolated and characterized from fixed, enriched cell populations (1). Using adipose-derived stem cells (ASCs) as our cell type of interest, this study expands and improves upon FITSAR via two objectives: 1) validate protein isolation from fixed and stored single cell suspensions and 2) demonstrate detection of isolated proteins on enzyme-linked immunosorbent assay (ELISA) arrays.
A multi-donor, human ASC superlot (Zen-Bio Inc., Research Triangle Park, NC; #36, passage 5) was used for all experiments. ASCs were cultured in medium containing DMEM/F12, 1% antibiotic/antimycotic (HyClone, Logan, UT), 10% fetal bovine serum (Zen-Bio Inc.), 1 ng/mL fibroblast growth factor, 5 ng/mL epidermal growth factor, and 0.25 ng/mL transforming growth factor-β1 (R&D Systems, Minneapolis, MN) (2) and were maintained at 37°C, 5% CO2.
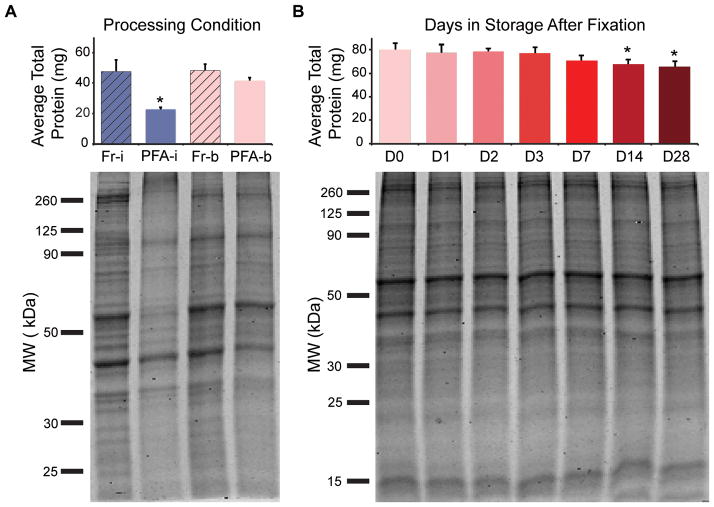
To ensure reproducible protein isolations from cells chemically fixed in 4% paraformaldehyde (PFA) as compared to fresh samples, protein extraction protocols were assessed by total protein amount (BCA Assay, Pierce, ThermoFisher Scientific, Waltham, MA) and quality (Coomassie Blue-stained polyacrylamide gel electrophoresis (PAGE) gel, MP Biomedicals LLC, Santa Ana, CA) (N=3). Fresh and PFA-fixed ASCs were either lysed on ice or by boiling (N=3). Lysis buffer was composed of Tris-HCl, sodium dodecyl sulfate (SDS; Fisher Scientific, Hampton, NH), and protease/phosphatase inhibitors (Pierce) (details in Table 1, Supplementary Protocol) (3). No significant difference in protein yield or banding was observed between the gold standard approach (fresh ASCs lysed on ice, “Fr-i”) (4) and the fixed/boiled method (“PFA-b”) (P > 0.05) (Figure 1A, Supplementary Figure 1S). Additionally, findings were similar when using formalin (4% PFA + 1.5% methanol) or pure 4% PFA fixatives (Supplementary Figure 2S). As corroborated in formalin-fixed paraffin-embedded (FFPE) literature, successful protein extraction from fixed samples requires exposure to formaldehyde scavenging and denaturing reagents (high concentrations of both Tris-HCl and SDS) in addition to high heat to promote protein untangling and methylene bridge thermal hydrolysis (3,5,6).
Table 1. Processing parameters for cell lysis.
Check marks indicate process used in preparing samples. Fr-i, fresh/ice; PFA-i, PFA-fixed/ice; Fr-b, fresh/boiled; PFA-b, PFA-fixed/boiled.
| Processing Parameters | Fr-i | PFA-i | Fr-b | PFA-b |
|---|---|---|---|---|
| While in suspension, ASCs were left in phosphate buffered saline (PBS). | ✓ | ✓ | ||
| While in suspension, ASCs were fixed in 4% PFA for 10 minutes at room temperature. | ✓ | ✓ | ||
| ASCs were lysed in a 4°C ice bath for 30 minutes. | ✓ | ✓ | ||
| ASCs were lysed in a boiling water bath at 100°C for 30 minutes followed by 60°C for 2 hours. | ✓ | ✓ | ||
| Lysis buffer was composed of 300 mM Tris-HCl (pH 8) + 2% SDS + 2X protease/phosphatase inhibitor. | ✓ | ✓ | ✓ | ✓ |
Figure 1. Protein extraction amount and quality are similar between fresh and formaldehyde-fixed/stored samples when using appropriate cell lysis buffer and exposure to high heat.
(A) Total protein amount isolated from fresh (Fr) and PFA-fixed (PFA) ASCs was compared under two lysis conditions (i, on ice; b, by boiling). Data represented as mean ± standard deviation (s.d.) from three, independent experiments. A Kruskal-Wallis test with post-hoc Tukey test was performed to determine significance (* significantly different from unmarked groups, P < 0.05). Coomassie Blue-stained PAGE gels indicated protein quality through comparison of banding profiles. Low protein yields and increased aggregation of proteins (>260 KDa) in the “PFA-i” condition underscore importance of lysis process used to isolate proteins from fixed samples. (B) Total isolated protein was compared across PFA-fixed ASCs stored for 0–28 days. Data represented as mean ± s.d. from one independent experiment run in biological triplicate. A one-way ANOVA with Dunnett’s test (D0 as the point of comparison) was performed to determine significance (* significantly different from D0, P < 0.02). Protein quality assessed by Coomassie Blue-stained PAGE gels.
Being able to evaluate stored, fixed samples would increase user flexibility with the FITSAR process. PFA-fixed ASCs were suspended in phosphate buffered saline and stored at 4°C for 0–28 days. After storage, samples were lysed by boiling and assessed for total extracted protein yield and quality (N=3). Fixed cells could be stored for up to 7 days before undergoing a minor, but statistically significant, decrease in total protein yield (P < 0.02) (Figure 1B). The ~15–20% decrease between days 0–14 and 0–28 did not produce visible changes in protein banding profiles among samples isolated across all storage conditions (Figure 1B). This suggested that collected protein is generally representative of the sample’s proteome, and any loss is homogeneously distributed across protein types.
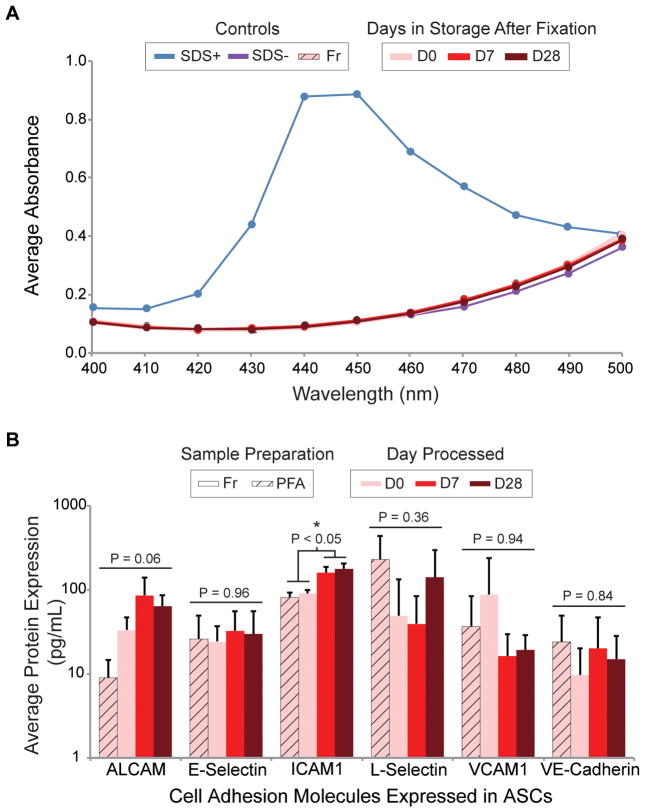
ELISA arrays, a platform in which multiple proteins of interest (POI) may be detected simultaneously at picomolar concentrations, were chosen to demonstrate the expanded compatibility of FITSAR with other protein analysis techniques. However, this technology is incompatible with SDS since detergents interfere with antigen-antibody interactions by forming adducts with protein residues (7). Therefore, sample processing methods were adjusted to include an SDS exclusion step using commercially available, detergent-removing resin spin columns (Pierce). Successful SDS removal from fixed cell lysates was verified using the Stains-All Assay (Acros Organics, Geel, BE) (8,9), and protein yield was quantified. Greater than 99.99% of SDS was removed, as evidenced by the lack of a characteristic SDS absorbance peak at 440–450 nm wavelength for all processed samples (Figure 2A). Protein loss to the columns varied depending on column size. For small columns (10–25 μL sample volume), eluted protein was not significantly different from loaded protein (P = 0.29) (N = 3) (Supplementary Figure S3A). For large columns (25–100 μL sample volume), eluted protein was significantly less than loaded protein (P < 0.001) (N = 3) (Supplementary Figure S3B). Regardless of size, these spin columns function by providing a hydrophobic microenvironment in the resin cavity into which small nonpolar compounds, like SDS, infiltrate and subsequently form an inclusion complex (7). Non-specific adsorption or entrapment of protein is expected to be distributed evenly across protein types, resulting in a detergent-free protein lysate that is representative of the original sample proteome.
Figure 2. Detergent-free protein lysates were successfully detected on CAM ELISA array.
(A) Representative Stains-All Assay absorbance spectra comparing SDS positive (blue) and negative (purple) controls to column-processed ASC storage protein lysates (shades of red). Lack of 440–450 nm peak indicates successful removal of SDS detergent. (B) Quantified protein expression of CAMs expressed in ASC lysates comparing fresh and fixed/stored samples. Data represented as arithmetic mean ± s.d. from one independent experiment run in biological triplicate (technical quadruplicate). Within each POI, one-way ANOVAs were performed to determine significance.
As a proof-of-concept, SDS-purified ASC storage protein lysates (N = 3) were assessed using a multiplexed, human cell adhesion molecule (CAM) ELISA array (QAH-CAM-1, RayBiotech, Norcross, GA). Isolation of cell surface/membrane-bound proteins from fixed samples is notoriously variable compared to proteins isolated from the cytosol or nucleus (10,11), thus providing a challenging test of the reliability/rigorousness of the processing involved in the updated FITSAR method. Each CAM ELISA array consisted of 17 POI (Table 2) printed in technical quadruplet. A 6-point standard curve provided means for quantification. Of the 17 CAM POI available, 8 were potentially expressed in ASCs, given species, origin, and passage number (12–14). Of these 8 POI, 6 were detected, with 5/6 proteins having no significant difference in expression level among fresh, fixed, and fixed/stored samples (P > 0.05) (Figure 2B). Only ICAM1 expression changed significantly, being higher in day-7 and -28 fixed samples compared to day-0 fresh and fixed samples (P < 0.02). This discrepancy is most likely due to the variability associated with these types of microarrays (15). However, another possibility is unforeseen bias favoring ICAM1 isolation during processing (i.e., ICAM1 extraction was more efficient than other proteins, resulting in an artificially inflated value after normalization to total protein levels). For all other proteins assessed, no differences from the gold standard approach was observed.
Table 2. POI available on CAM ELISA arrays.
Check marks indicate anticipated or observed expression in passaged ASCs, question marks indicate conflicting reports for expression in passaged ASCs, and X marks indicate lack of observed expression (12–14).
| POI | Proteins expressed in passaged ASCs (P3–6) | |
|---|---|---|
| Expected | Observed | |
| ALCAM | ✓ | ✓ |
| BCAM | ||
| CEACAM1 | ||
| E-Cadherin | ||
| EpCAM | ||
| E-Selectin | ? | ✓ |
| ICAM1 | ✓ | ✓ |
| ICAM2 | ||
| ICAM3 | ||
| L-Selectin | ✓ | ✓ |
| NCAM1 | ||
| NrCAM | ||
| P-Cadherin | ||
| P-Selectin | ? | x |
| PECAM1 | ? | x |
| VCAM1 | ? | ✓ |
| VE-Cadherin | ✓ | ✓ |
Our enhanced technique allows assessment of fixed, stored samples and use of low protein, high throughput ELISA arrays. The improvements to sample processing, specifically detergent removal, also provides potential compatibility with techniques like mass spectrometry and proteomics. By pairing these improvements with our FITSAR method, cell subpopulations of interest can be isolated and their protein expression profiles analyzed with flexibility in timing and use of sensitive, quantitative assays.
Supplementary Material
Replicates (A) 2 and (B) 3 validating protein extraction and quality from fresh and PFA-fixed ASCs as compared by Coomassie Blue-stained PAGE gel. Replicate 1 is presented in Figure 1A.
Total protein amount isolated from fresh (Fr), 10% formalin-fixed (Form) and PFA-fixed (PFA) MG-63 osteosarcoma cells was compared for samples lysed by boiling (“b”). Data represented as arithmetic mean ± s.d. from three biological replicates. One-way ANOVA was performed to determine significance (P < 0.05). A Coomassie Blue-stained PAGE gel indicated protein quality through comparison of banding profiles.
(A) Protein loaded and collected onto small columns, comparing fresh and PFA-fixed protein lysates. Data represented as mean ± s.d. from one independent experiment run in biological triplicate. A two-way ANOVA with Tukey post-hoc test was performed to determine significance. No differences were observed in total protein amount before and after SDS removal (P > 0.09). (B) Protein loaded and collected onto large columns comparing fresh and PFA-fixed/stored protein lysates. Data represented as mean ± s.d. from one independent experiment run in biological triplicate. A two-way ANOVA (processing stage and sample) with post-hoc Tukey test was performed to determine significance (unlike letters are significantly different from each other). Substantial protein loss was experienced when using large SDS removal columns across experimental groups (P < 0.001).
METHOD SUMMARY.
Here, we present a method in which protein from formaldehyde-fixed, stored samples may be assessed on high throughput, multiplex enzyme-linked immunosorbent assay (ELISA) arrays.
Acknowledgments
This work was supported by the National Institutes of Health (R01AR06304 to EMD) and the National Science Foundation (CAREER CBET 1253189 and EAGER CBET 1547819 to EMD, GRFP 2014183678 to JSS). This paper is subject to NIH Public Access Policy.
Footnotes
AUTHOR CONTRIBUTIONS: JSS and EMD designed the study and wrote the manuscript. JSS conducted all experimental work.
COMPETING INTERESTS STATEMENT: The authors declare no competing interests.
References
- 1.Sadick JS, Boutin ME, Hoffman-Kim D, Darling EM. Protein characterization of intracellular target-sorted, formalin-fixed cell subpopulations. Sci Rep. 2016;6:33999. doi: 10.1038/srep33999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estes BT, Diekman BO, Guilak F. Monolayer cell expansion conditions affect the chondrogenic potential of adipose-derived stem cells. Biotechnol Bioeng. 2008;99:986–995. doi: 10.1002/bit.21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawashima Y, Kodera Y, Singh A, Matsumoto M, Matsumoto H. Efficient extraction of proteins from formalin-fixed paraffin-embedded tissues requires higher concentration of tris(hydroxymethyl)aminomethane. Clinical Proteomics. 2014;11:4. doi: 10.1186/1559-0275-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahmood T, Yang PC. Western blot: technique, theory, and trouble shooting. N Am J Med Sci. 2012;4:429–434. doi: 10.4103/1947-2714.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magdeldin S, Yamamoto T. Toward deciphering proteomes of formalin-fixed paraffin-embedded (FFPE) tissues. PROTEOMICS. 2012;12:1045–1058. doi: 10.1002/pmic.201100550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Addis M, Tanca A, Pagnozzi D, Crobu S, Fanciulli G, Cossu-Rocca P, Uzzau S. Generation of high-quality protein extracts from formalin-fixed, paraffin-embedded tissues. PROTEOMICS. 2009;9:3815–3823. doi: 10.1002/pmic.200800971. [DOI] [PubMed] [Google Scholar]
- 7.Antharavally BS, Mallia KA, Rosenblatt MM, Salunkhe AM, Rogers JC, Haney P, Haghdoost N. Efficient removal of detergents from proteins and peptides in a spin column format. Anal Biochem. 2011;416:39–44. doi: 10.1016/j.ab.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Rupprecht KR, Lang EZ, Gregory SD, Bergsma JM, Rae TD, Fishpaugh JR. A precise spectrophotometric method for measuring sodium dodecyl sulfate concentration. Anal Biochem. 2015;486:78–80. doi: 10.1016/j.ab.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Rusconi F, Valton E, Nguyen R, Dufourc E. Quantification of sodium dodecyl sulfate in microliter-volume biochemical samples by visible light spectroscopy. Anal Biochem. 2001;295:31–37. doi: 10.1006/abio.2001.5164. [DOI] [PubMed] [Google Scholar]
- 10.Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Development and validation of a novel protein extractionmethodology for quantitation of protein expression informalin-fixed paraffin-embedded tissues using westernblotting. J Pathol. 2009;217:497–506. doi: 10.1002/path.2504. [DOI] [PubMed] [Google Scholar]
- 11.Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KJS. Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Laboratory Investigation. 2005;85:1405–1415. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 12.Zuk P, Zhu M, Ashjian P, De Ugarte D, Huang J, Mizuno H, Alfonso Z, Fraser J, et al. Human Adipose Tissue Is a Source of Multipotent Stem Cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz AJ, Tholpady A, Tholpady SS, Shang H, Ogle RC. Cell Surface and Transcriptional Characterization of Human Adipose-Derived Adherent Stromal (hADAS) Cells. Stem Cells. 2005;23:412–423. doi: 10.1634/stemcells.2004-0021. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell JB, McIntosh K, Zvonic S, Garrett S, Floyd ZE, Kloster A, Halvorsen YD, Storms RW, et al. Immunophenotype of Human Adipose-Derived Cells: Temporal Changes in Stromal-Associated and Stem Cell–Associated Markers. Stem Cells. 2006;24:376–385. doi: 10.1634/stemcells.2005-0234. [DOI] [PubMed] [Google Scholar]
- 15.Berg D, Hipp S, Malinowsky K, Böllner C, Becker KF. Molecular profiling of signalling pathways in formalin-fixed and paraffin-embedded cancer tissues. Eur J Cancer. 2010;46:47–55. doi: 10.1016/j.ejca.2009.10.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Replicates (A) 2 and (B) 3 validating protein extraction and quality from fresh and PFA-fixed ASCs as compared by Coomassie Blue-stained PAGE gel. Replicate 1 is presented in Figure 1A.
Total protein amount isolated from fresh (Fr), 10% formalin-fixed (Form) and PFA-fixed (PFA) MG-63 osteosarcoma cells was compared for samples lysed by boiling (“b”). Data represented as arithmetic mean ± s.d. from three biological replicates. One-way ANOVA was performed to determine significance (P < 0.05). A Coomassie Blue-stained PAGE gel indicated protein quality through comparison of banding profiles.
(A) Protein loaded and collected onto small columns, comparing fresh and PFA-fixed protein lysates. Data represented as mean ± s.d. from one independent experiment run in biological triplicate. A two-way ANOVA with Tukey post-hoc test was performed to determine significance. No differences were observed in total protein amount before and after SDS removal (P > 0.09). (B) Protein loaded and collected onto large columns comparing fresh and PFA-fixed/stored protein lysates. Data represented as mean ± s.d. from one independent experiment run in biological triplicate. A two-way ANOVA (processing stage and sample) with post-hoc Tukey test was performed to determine significance (unlike letters are significantly different from each other). Substantial protein loss was experienced when using large SDS removal columns across experimental groups (P < 0.001).