Abstract
Triple negative breast cancer (TNBC) is a particular subtype of breast malignant tumor with poorer prognosis than other molecular subtypes. Currently, there is increasing focus on long non-coding RNAs (lncRNAs), which can act as competing endogenous RNAs (ceR-NAs) and suppress miRNA functions involved in post-transcriptional regulatory networks in the tumor. Therefore, to investigate specific mechanisms of TNBC carcinogenesis and improve treatment efficiency, we comprehensively integrated expression profiles, including data on mRNAs, lncRNAs and miRNAs obtained from 116 TNBC tissues and 11 normal tissues from The Cancer Genome Atlas. As a result, we selected the threshold with |log2FC|>2.0 and an adjusted p-value >0.05 to obtain the differentially expressed mRNAs, miRNAs and lncRNAs. Hereafter, weighted gene co-expression network analysis was performed to identify the expression characteristics of dysregulated genes. We obtained five co-expression modules and related clinical feature. By means of correlating gene modules with protein–protein interaction network analysis that had identified 22 hub mRNAs which could as hub target genes. Eleven key dysregulated differentially expressed micro RNAs (DEmiRNAs) were identified that were significantly associated with the 22 hub potential target genes. Moreover, we found that 14 key differentially expressed lncRNAs could interact with the key DEmiRNAs. Then, the ceRNA crosstalk network of TNBC was constructed by utilizing key lncRNAs, key miRNAs, and hub mRNAs in Cytoscape software. We analyzed and described the potential characteristics of biological function and pathological roles of the TNBC ceRNA co-regulatory network; also, the survival analysis was performed for each molecule. These findings revealed that ceRNA crosstalk network could play an important role in the development and progression for TNBC. In addition, we also identified that some molecules in the ceRNA network possess clinical correlation and prognosis.
Keywords: triple negative breast cancer, The Cancer Genome Atlas, weighted gene co-expression network analysis, ceRNA crosstalk network, survival prognosis
Introduction
Breast cancer is a heterogeneous disease with varying histological abnormalities, cytogenetic aberrations, responses to therapy, and prognoses, and accounts for 16% of all female cancers and 22.9% of invasive cancers in women.1 Triple negative breast cancer (TNBC), a particular subtype of BC, is tested with negative for estrogen receptors (ER−), progesterone receptors (PR−), and Erb-B2 receptor tyrosine kinase 2 (HER2−).2 This kind of breast cancer generally has overall poor outcomes, such as more aggressive, higher distant recurrence and poorer survival in metastatic disease.3 Moreover, current targeted therapy strategy is available for most breast cancers that are ER or PR positive, but there are no specific targeted therapy options available for TNBC.4 Therefore, it is urgent and necessary that new treatments are found based on the specific mechanisms of TNBC to improve treatment efficiency and avoid the adverse effects of conventional methods.
Advances in next-generation RNA sequencing technology have brought to light the complexity of our genome. Non-coding RNAs (ncRNAs), such as micro RNAs (miRNAs) and the long non-coding RNAs (lncRNAs), accounting for the majority (98%) of the transcriptome, are defined as gene transcripts with important biological functions.5 Investigating the world of RNA is one of the most important challenges facing biologists today, and lncRNAs can represent potential new biomarkers and drug targets. Salmena et al’s competing endogenous RNA (ceRNA) hypothesis about the multiple molecular involved in post-transcriptional regulatory networks has received much attention in recent years; their study indicated that lncRNAs, cirRNAs, and pseudogenes can act as ceRNAs to suppress miRNA functions and communicate with mRNAs by harboring one or more miRNA response elements (MREs).6 An increasing number of experimental reports are supporting this advanced hypothesis. One such example is lnc-MD1, a muscle-specific lncRNA that has been shown to control muscle differentiation by acting as miR-135 and miR-133 “sponge,” and thus regulating the expression of MAML1 and MEF2C.7 In addition, Wang et al has identified Linc-ROR, which could serve as a ceRNA and acts as miR-145 “sponge”, thereby regulating SOX2, OCT4 and NANOG expression to control self-renewal and maintain pluripotency of human embryonic stem cells.8 However, to date, there is still little research on ceRNA-related lncRNAs’ potential roles in relevant regulatory mechanisms of TNBC.
The Cancer Genome Atlas (TCGA) project was launched in 2005 and has accelerated the comprehensive understanding of cancer genomic profiles that has improved diagnostic methods, therapy standards, and preventive strategies. By the end of 2015, TCGA had expanded to multiple types, reaching 30 different tumor types with genome expression profiles and clinical data through the large-scale public “atlas”.9 Also, the latest progress in the integrated multi-dimensional analyses development of technology bioinformatics has shed new insight on the cancer profiling. Weighted gene co-expression network analysis (WGCNA) is a systems biology algorithm for constructing the correlation patterns among genes across samples.10 Furthermore, WGCNA can be used for uncovering and clustering highly correlated molecules into separate modules that provide information on hub nodes and external sample traits with related modules. The varied studies of WGCNA are widely used in cancer research, genetics of species and other complex diseases to investigate and identify related genes, biological processes, and candidate biomarkers. However, few studies have utilized the open access TCGA database for cancer research with WGCNA method. This research has mined TNBC data in the TCGA database that analyzed the integrated RNA expression profiles from 116 TNBC samples and 11 normal samples. We used the WGCNA and protein–protein interaction (PPI) network to identify hub mRNAs from the differentially expressed mRNAs (DEmRNAs) profile. The databases, miRTarBase and miRcode, were used to screen the key differentially expressed lncRNAs (DElncRNAs) and differentially expressed micro RNAs (DEmiRNAs), as well as DEmiRNAs-DEmRNAs interactions. Hereafter, we constructed a ceRNA network to elaborate on the interactions and potential crosstalk between the hub DElncRNAs, DEmiRNAs and DEmRNAs. In addition, relevant survival analyses were performed to determine the prognostic genes that possess clinical traits.
Materials and methods
Data acquisition and processing
All RNA sequencing data sets were included in this study that met the following criteria: 1) detailed clinical data consisted of age, gender, race, TNM stage, and histological subtype and grade; 2) patients possessed a certain follow-up time. Moreover, details of set grouping conditions were as follows: the tumor group included the breast cancer samples of triple negative; and the control group included normal physical tissue. Overall, publicly available breast cancer level 3 RNAseq and miRNASeq data derived from 127 samples, including 116 TNBC tumor tissues and 11 adjacent normal tissues, were obtained from TCGA. These data were classified into two cohorts. According to the TCGA ethics committee, the data and analysis of this present study did not require further approval.
Screening for DEmRNAs, DEmiRNAs, and DElncRNAs
We merged tumor sample and normal sample data and deleted expressed data that closed to zero. The DEmRNAs and DEmiRNAs of raw count data were processed through the limma package, which assessed the data using linear models and empirical Bayesian methods.11 The threshold of differential expression was defined with an absolute log 2-fold change (FC) >2 and an adjusted p-value <0.05.
In addition, the GENCODE lncRNA annotation (V22) was used to discover and define the lncRNAs from DERNAs.12 In our study, lncRNAs not included in the annotation were discarded.
Weighted gene co-expression network construction
The differential expression genes were applied to find the scale-free gene modules of co-expression and highly correlated genes constructed by WGCNA package.10 The outlier samples were identified and removed to ensure the reliability of co-expression network construction. To build the scale-free co-expression relationship between gene pairs, an appropriate soft threshold power parameter was set to 4, which could penalize weaker connections and strengthen stronger connections. Topological overlap matrix (TOM) was performed by adjacency transformation. Then, we calculated 1-TOM as the distance measured and through the dynamic tree cut method to identify hierarchical clustering genes and modules. The minimum size cut-off of 100 for the resulting dendrogram of each module, and below which separate modules would be merged, was set to 0.25.
Module clinical feature associations
In addition, for investigating the correlation between gene expression modules and clinical features, we plotted the heatmap of all modules with the relation cut-off and p-value at eight different conditions, which included pathology diagnosis age, laterality, histological type, positive lymph nodes, pathology stage, pathology T stage, pathology M stage, pathology N stage. The previously mentioned data were analyzed by Pearson’s correlation tests.
Identifying hub genes though PPI network analysis and functional annotation
In order to further explore the characteristics of each module, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of differentially expressed genes (DEGs) were identified by KEGG Orthology Based Annotation System (KOBAS) web server, which provides abundant annotation information of gene sets from multiple databases covering pathways (KEGG PATHWAY, Reactome, Biocyc, Panther) and supports gene and protein functional annotation.13,14 In addition, the significant enrichment KEGG pathway should have contained at least three DEGs and had a false discovery rate (FDR) of <0.05. We all know that protein interaction data analysis serves as an important tool to unravel the molecular basis of a disease. Thus, it is feasible to discover TNBC-related DEGs of different modules based on protein network. This study constructed interaction networks and retrieved data from STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, http://string.embl.de/), a well-known online database integrating known and predicted protein interactions.15 Additionally, we set the threshold for interaction score at >0.9, which indicates the highest confidence of two protein interaction occurrences.
Construction and analysis of ceRNA crosstalk network
The hub dysregulated genes and ncRNA associated ceRNA crosstalk network in TNBC was constructed as follows: 1) prediction of target mRNAs of hub mRNAs: we selected the hub DEmRNAs from WGCNA modules as the target genes, and found out the miRNA-mRNA pairs with experimental support by miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/).16 Then, hub DEmRNAs were mapped into the interactions. 2) Prediction of target lncRNAs of miRNAs: the lncRNA-miRNA interactions were predicted through miRcode (http://www.mircode.org/) database.17 The DElncRNAs and DEmiRNAs were mapped into the interactions. 3) Construction of ceRNA network: TNBC-specific lncRNA-miRNA-mRNA interactions were revealed. The ceRNA crosstalk network was constructed with the previously mentioned interactions. The construction of the network was visualized using the Cytoscape 3.4.0 software.
Furthermore, to explore potential biological mechanisms of the predicted ceRNA crosstalk network, the KOBAS web server was used to annotate KEGG pathways of hub genes that could be activated by key lncRNAs and miRNAs. Each item of enrichment had a threshold of FDR <0.05. The network of pathways was visualized using the Cytoscape software.
Survival analysis
To identify prognostic lncRNA and miRNA signatures, combining the clinical data of those patients in TCGA, we established the life curves of those samples with each node in the ceRNA network. Survival analysis was performed using a standard Kaplan–Meier univariate curve through the “survival” package in R 3.3.2.18 Molecule with p-value <0.05 was considered as having statistical significance.
Results
Identification of DEmRNAs, DEmiRNAs and DElncRNAs
The level three expression profiles of TNBC and corresponding clinical data (retrieved up to July 2017) were downloaded by Data Transfer Tool (https://tcga-data.nci.nih.gov/tcga/) in the TCGA database. Data of 127 patients were available. Moreover, to obtain DEmRNAs and DEmiRNAs, we applied the limma algorithm, which can identify the different expressions of TNBC and normal samples. Finally, a total of 2,791 DEmRNAs were identified by the threshold of log2FC >2 and p-value <0.01, of which 1,419 were down-regulated and 1,372 were up-regulated. In the same way, 67 DEmiRNAs were identified by limma, of which 46 were up-regulated in cancer samples and 21 down-regulated.
The GENCODE lncRNA annotation (V22) was used to discover and define the lncRNAs from DERNAs. According to GENECARDS annotation, we identified a total of 723 aberrantly expressed lncRNAs in TNBC tissues compared to the normal tissues. Two hundred and eighty-eight (39.83%) of the DElncRNAs were up-regulated, and 435 (60.17%) were down-regulated. We have shown the top 10 mRNAs, miRNAs and lncRNAs with |log2FC|>2.0 in Table 1.
Table 1.
The top 10 dysregulated mRNAs, miRNAs, lncRNAs
| Gene symbol | |logFC| | False discovery rate | Dysregulated stage |
|---|---|---|---|
| Top 10 DEmRNA | |||
| MYOC | 8.6365 | 1.42E-52 | Down |
| GLYAT | 7.0741 | 1.63E-51 | Down |
| ALB | 6.9597 | 1.74E-43 | Down |
| LEP | 6.7453 | 1.01E-43 | Down |
| DEFB132 | 6.722 | 3.21E-19 | Down |
| MAGEA3 | 11.7565 | 0.001455 | Up |
| MAGEA6 | 10.9583 | 0.002716 | Up |
| CSAG1 | 10.2568 | 0.000183 | Up |
| PRR27 | 9.9301 | 0.007438 | Up |
| NCAN | 8.8403 | 0.000392 | Up |
| Top 10 DEmiRNA | |||
| hsa-mir-486-1 | 5.3091 | 2.90E-46 | Down |
| hsa-mir-486-2 | 5.2976 | 3.71E-46 | Down |
| hsa-mir-4732 | 4.3617 | 3.91E-17 | Down |
| hsa-mir-139 | 3.7614 | 3.71E-46 | Down |
| hsa-mir-451a | 3.7094 | 8.14E-19 | Down |
| hsa-mir-105-1 | 7.8785 | 0.003368 | Up |
| hsa-mir-519a-1 | 7.4736 | 0.003573 | Up |
| hsa-mir-105-2 | 7.247 | 0.005287 | Up |
| hsa-mir-767 | 7.0703 | 0.007391 | Up |
| hsa-mir-516a-1 | 6.8049 | 0.005938 | Up |
| Top 10 DElncRNA | |||
| CH507-513H4.4 | 6.6392 | 2.35E-11 | Down |
| CH17-469D17.1 | 6.0189 | 1.68E-26 | Down |
| LINC01230 | 5.9647 | 1.38E-28 | Down |
| RP11-798K3.2 | 5.9415 | 2.60E-44 | Down |
| RP11-407P15.2 | 5.9326 | 7.73E-31 | Down |
| RP11-54H7.4 | 10.7164 | 0.000396 | Up |
| LINC00392 | 8.5501 | 0.006951 | Up |
| RP11-366F6.2 | 8.5312 | 0.009073 | Up |
| RP11-276H19.4 | 7.9425 | 0.000432 | Up |
| RP13-16H11.9 | 7.8981 | 0.008053 | Up |
Abbreviations: lncRNA, long non-coding RNA; miRNA, micro RNA; DEmiRNA, differentially expressed miRNA; DElncRNA, differentially expressed long non-coding RNA.
Weighted gene co-expression modules construction and analyses
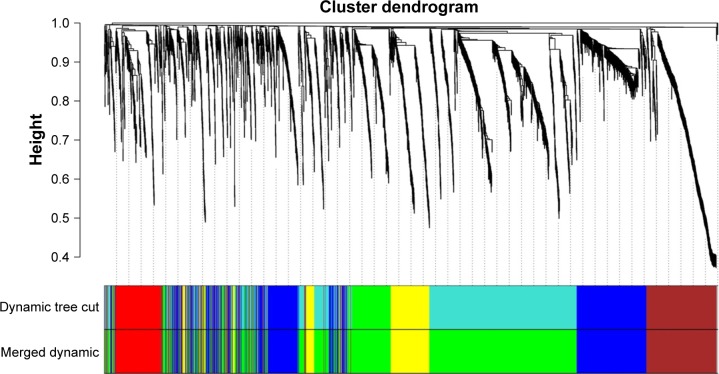
To find the functional clusters in TNBC samples, we took the different regulated mRNAs data to build the correlation network through WGCNA. The hierarchical dendrogram of WGCNA modules, with different colors representing different modules, was constructed with an appropriate soft threshold (Figure 1). Eventually, it gained a total of five color modules, including red, blue, yellow, green, and brown. Additionally, the region of color gray represented those genes that were not divided into modules.
Figure 1.
Hierarchical clustering dendrograms of identified co-expressed genes in modules.
Notes: The dendrogram was generated by unsupervised hierarchical clustering of genes using topological overlap. The colored strips below each dendrogram indicate the module designation that was identified through the clusters of co-expressed genes, and assigned the merged module color to the original module color.
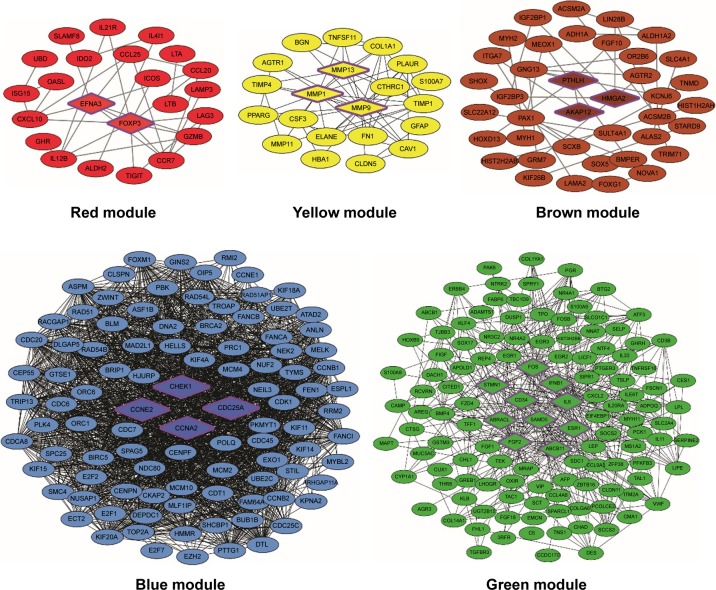
To further understand the characteristics of each module, the annotation of KEGG pathways for each module is listed in Table S1. Furthermore, we constructed the PPI networks to screen out hub DEmRNAs. As is shown in Figure 2, there are 25 nodes and 35 interactions exhibited in the red module. According to the highest confidence of 0.9 as threshold, two hub genes, EFNA3, and FOXP3, were noted in the center in the PPI network. In addition, we found that EFNA3 was mediated by four pathways, including “MicroRNAs in cancer,” “PI3K-Akt signaling pathway,” “Rap1 signaling pathway,” and “Ras signaling pathway.” In the blue module, we found 99 nodes and 869 interactions. And, four hub genes were significantly enriched in the blue PPI network, including CDC25A, CCNE2, STMN1 and CCNA2. The KEGG pathway annotation of blue module revealed the enrichment of genes involved in “cell cycle” (CDC25A, CCNE2, CCNA2), “MicroRNAs in cancer” (CDC25A, CCNE2) and “p53 signaling pathway” (CCNE2).
Figure 2.
PPI network visualization of modules identifies hub genes.
Notes: Different colors represent the different modules. The hub genes in each module are highlighted with a hexagon shape and purple border.
Abbreviations: MMP, matrix metalloproteinase; PPI, protein–protein interaction.
The yellow module contained 21 nodes and 56 interactions. Also, three hub genes which are members of the peptidase M10 family of matrix metalloproteinases (MMPs), including MMP1, MMP9 and MMP13, were noticed in the PPI network. We identified the KEGG pathways of this module related to critical cellular processes and organismal systems (endocrine system). In the green module, we obtained 125 nodes and 451 interactions. A total of 10 hub genes with significant centrality values were identified in this module, consisting of ABRACL, ABCB11, CHEK1, CD34, FOS, IL6, IFNB1, ESR1, FGF2 and SAMD5. Pathway enrichment analysis of the green module revealed that the enriched terms focused on environment signaling progress and the endocrine system, like “Neuroactive ligand-receptor interaction,” “cAMP signaling pathway,” and “Regulation of lipolysis in adipocytes.” There were 39 nodes and 32 interactions in the brown module. We identified four nodes in center that serve as hub genes in the PPI network, which are as follows: PTHLH, HMGA2 and AKAP12. Moreover, in this module, pathway enriched terms were concentrated in metabolism progress and disease-related pathways; for instance, the hub gene HMGA2 was enriched in “MicroRNAs in cancer”, “Transcriptional misregulation in cancer”.
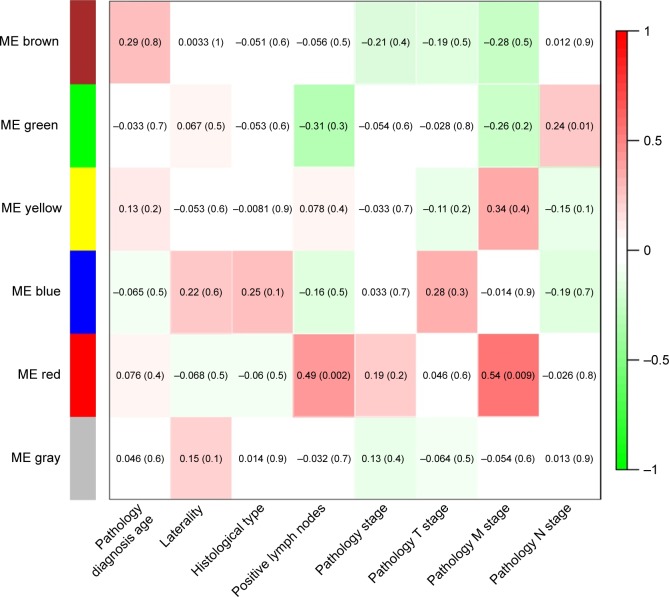
Module-trait relationships
After gaining the summarized clinical and pathological information (Table 2), we constructed the feature heatmap for investigating the relation between modules and clinical characteristics (Figure 3). As a result, we noticed that the red module for two variables, encompassing positive lymph nodes (r=0.49, p=0.002) and pathology M stage (r=0.54, p=0.009), displayed the highest correlation coefficient. Moreover, the yellow module also showed the higher association in pathology M stage (r=0.34), though no statistical significance was revealed (p=0.4). The green module exhibited the highest correlation in pathology N stage (r=0.24, p=0.01). Taken together, the DEmRNAs of the previously mentioned modules may be the key regulators for the development of TNBC.
Table 2.
The clinical features of triple negative breast cancer patients
| Clinical feature | Variable | Patients n (%) |
|---|---|---|
| Pathology diagnosis age (years) | >65 | 19 (14.96) |
| ≤65 | 108 (85.04) | |
| Laterality | Left | 63 (49.61) |
| Right | 64 (50.39) | |
| Histological type | Infiltrating ductal carcinoma | 108 (85.04) |
| Infiltrating lobular carcinoma | 4 (3.15) | |
| Medullary carcinoma | 3 (2.36) | |
| Metaplastic carcinoma | 5 (3.94) | |
| Other | 7 (5.11) | |
| Positive lymph nodes | Negative | 72 (56.69) |
| Positive | 55 (43.31) | |
| Pathology stage | I | 25 (19.69) |
| II | 76 (59.84) | |
| III | 22 (17.32) | |
| IV | 4 (3.15) | |
| Pathology T stage | T1 | 30 (23.62) |
| T2 | 78 (61.42) | |
| T3 | 14 (11.02) | |
| T4 | 5 (3.94) | |
| Pathology M stage | M0 | 108 (85.04) |
| M1 | 2 (1.57) | |
| Mx | 17 (13.39) | |
| Pathology N stage | N0 | 77 (60.63) |
| N1 | 31 (24.41) | |
| N2 | 14 (11.02) | |
| N3 | 5 (3.94) |
Figure 3.
Module–trait relationships.
Notes: Correlation analysis of the identified gene modules with clinical features of triple negative breast cancer. The relationships were analyzed through Pearson’s correlation coefficient (r), accompanied with p-value in brackets.
Abbreviation: ME, module eigengenes.
ceRNA correlation network in TNBC
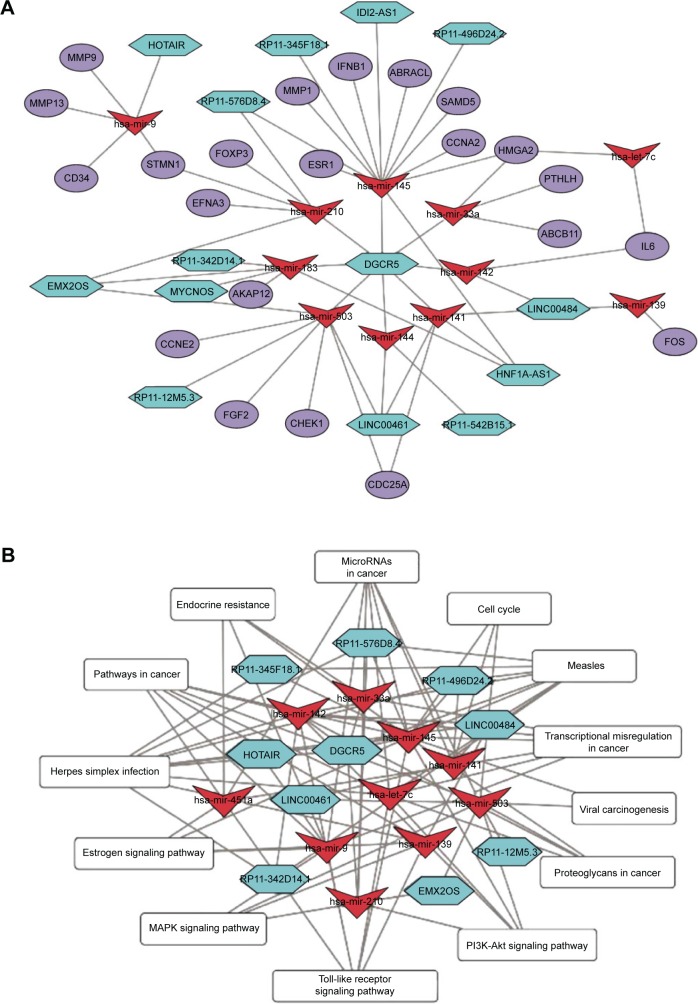
To further investigate the role and relation of lncRNA and miRNA mediation in TNBC, we searched for DEmiRNAs targeted by these 22 hub mRNAs obtained from WGCNA and PPI network analysis. Through the predicted miRNAs and algorithm analyzed dysregulated expressed 67 miRNAs. As a result, 11 intersection miRNAs related to the 22 hub genes were identified by miRTarBase database, and some of them have been reported to be cancer-associated miRNAs such as hsa-mir-145, hsa-mir-9, and hsa-mir-503. The miRNAs target with lncRNA through MREs in the ceRNA network, and thus, we detected whether key miRNAs and the above 723 differently expressed lncRNAs have the potential MREs based on the information retrieved from the miRcode database. The predicted results showed that the key miRNAs interacted with 14 DElncRNAs, which could act as key lncRNAs. Finally, we obtained 22 hub DEmRNAs mediating with 11 DEmiRNAs and 14 DElncRNAs, which could be the key nodes involved in the ceRNA network.
Based on combination of the above data, the lncRNA-miRNA-mRNA ceRNA network was established and visualized using the Cytoscape software (Figure 4A). The detailed information of each node is shown in Table 3. In the ceRNA network, some of them have been reported to be cancer-associated moleculars such as MMP1, ESR1, STMN1 and HOTAIR. We noticed that DGCR5 is the key lncRNA interrelated with seven key miRNAs (hsa-mir-145, hsa-mir-33a, hsa-mir-142, hsa-mir-144, hsa-mir-503, hsa-mir-183, and hsa-mir-210), and may serve as ceRNA to inhibit target miRNAs, and mediated related hub genes translation such as HMGA2, CCNA2, SAMD5, ABRACL, and IFNB1. And, HNF1A-AS1 may act as an endogenous sink by binding hsa-mir-183, hsa-mir-141, hsa-mir-145 and may be associated with targeted genes transcripts such as AKAP12, ESR1, MMP1, CCNA2. In addition, RP11-576D8.4 is a lncRNA, with no current research to find its function. In our study, it may have played a role in TNBC by its interactions with hsa-mir-145, hsa-mir-210, which then regulate the target hub mRNAs FOXP3, STMN1, EFNA3, ESR1, MMP1, and IFNB1. Moreover, EMX2OS, LINC00461, and LINC00484 also serve as the center molecule of several miRNAs. Thus, we suppose that DGCR5, HNF1A-AS1, LINC00484, LINC00461, RP11-576D8.4 and EMX2OS may closely contribute to the pathogenesis of TNBC.
Figure 4.
Network diagram.
Notes: (A) The ceRNA crosstalk network of TNBC. Key lncRNAs, miRNAs, and hub mRNAs are indicated as turquoise hexagons, red inverted triangles, and slate blue ellipses. (B) Co-expression network of the key lncRNAs and miRNAs mediated in the pathways. Pathways are depicted in rounded rectangle. The number of connections indicate the association between the key miRNAs and lncRNAs with signaling pathways.
Abbreviations: ceRNA, competing endogenous RNA; lncRNA, long non-coding RNA; miRNA, microRNA; TNBC, triple negative breast cancer.
Table 3.
Each node of ceRNA crosstalk network
| Nodes | Dysregulated stage | |logFC| | False discovery rate |
|---|---|---|---|
| Hub mRNAs | |||
| CDC25A | Up | 3.578 | 1.58E-18 |
| CCNE2 | Up | 3.023 | 1.02E-07 |
| CHEK1 | Up | 2.634 | 1.46E-13 |
| CCNA2 | Up | 3.998 | 4.88E-20 |
| AKAP12 | Down | 3.094 | 1.02E-22 |
| HMGA2 | Up | 4.919 | 0.00159 |
| PTHLH | Down | 2.352 | 0.00012 |
| FOS | Down | 3.699 | 1.83E-28 |
| IL6 | Down | 3.275 | 6.98E-17 |
| ABCB11 | Down | 2.266 | 4.80E-05 |
| ESR1 | Down | 3.127 | 4.45E-10 |
| SAMD5 | Down | 2.227 | 1.36E-06 |
| CD34 | Down | 2.398 | 1.21E-26 |
| IFNB1 | Up | 3.746 | 0.00574 |
| STMN1 | Up | 2.891 | 3.46E-14 |
| FGF2 | Down | 2.423 | 1.98E-09 |
| ABRACL | Up | 2.217 | 3.59E-08 |
| EFNA3 | Up | 2.121 | 0.00019 |
| FOXP3 | Up | 2.546 | 1.54E-08 |
| MMP9 | Up | 6.325 | 1.39E-05 |
| MMP13 | Up | 8.159 | 2.19E-07 |
| MMP1 | Up | 7.547 | 1.25E-08 |
| Key miRNAs | |||
| hsa-mir-139 | Down | 3.761 | 3.71E-46 |
| hsa-let-7c | Down | 2.026 | 3.46E-06 |
| hsa-mir-141 | Up | 2.589 | 4.70E-05 |
| hsa-mir-142 | Up | 2.813 | 1.09E-05 |
| hsa-mir-144 | Down | 3.623 | 2.67E-17 |
| hsa-mir-145 | Down | 2.529 | 6.66E-15 |
| hsa-mir-183 | Up | 2.106 | 0.00039 |
| hsa-mir-210 | Up | 4.541 | 2.55E-08 |
| hsa-mir-33a | Up | 3.083 | 8.12E-07 |
| hsa-mir-451a | Down | 3.709 | 8.14E-19 |
| hsa-mir-503 | Up | 2.595 | 7.32E-06 |
| hsa-mir-9 | Up | 2.929 | 0.00813 |
| Key lncRNAs | |||
| DGCR5 | Up | 2.388 | 5.63E-05 |
| EMX2OS | Down | 2.563 | 6.28E-12 |
| HOTAIR | Up | 2.53 | 0.00323 |
| LINC00461 | Up | 3.763 | 0.00076 |
| LINC00466 | Up | 3.45 | 0.00187 |
| LINC00484 | Down | 2.645 | 3.53E-12 |
| LINC00517 | Down | 2.659 | 4.38E-14 |
| MYCNOS | Up | 3.219 | 0.00104 |
| RP11-122M14.1 | Up | 2.556 | 7.40E-05 |
| RP11-12M5.3 | Up | 3.108 | 0.00791 |
| RP11-345F18.1 | Down | 4.065 | 2.15E-11 |
| RP11-496D24.2 | Up | 2.882 | 0.00633 |
| RP11-542B15.1 | Up | 2.996 | 2.54E-05 |
| RP11-576D8.4 | Up | 3.102 | 0.00304 |
Abbreviations: ceRNA, competing endogenous RNA; miRNA, micro RNA; lncRNA, long non-coding RNA.
Co-expression network of the key miRNA and lncRNAs mediated in the pathways
To evaluate whether the ceRNA network involved in specific enrichment biological pathways could be activated by the 19 key DEmiRNAs and DElncRNAs, we annotated by KOBAS web server, a total of significantly enriched 14 KEGG pathways (FDR <0.05) (Table 4). Then, a network involved in key DElncRNAs, DEmiRNAs, and signal pathways interactions was constructed using Cytoscape (Figure 4B). Several critical tumor-related signaling pathways were involved in our ceRNA crosstalk network; for instance, “Pathways in cancer, MicroRNAs in cancer, p53 signaling pathway and PI3K-Akt signaling pathway”. Also, revealed were the enriched terms focused on critical cellular processes and organismal systems, such as “cell cycle, Estrogen signaling pathway, endocrine resistance, EGFR tyrosine kinase inhibitor resistance”.
Table 4.
KEGG annotation of ceRNA crosstalk network
| KEGG pathways | Input number | False discovery rate | Genes |
|---|---|---|---|
| MicroRNAs in cancer | 6 | 5.20E-07 | HMGA2, CDC25A, STMN1, MMP9, CCNE2, EFNA3 |
| Pathways in cancer | 6 | 1.81E-06 | MMP1, CCNE2, MMP9, IL6, FOS, FGF2 |
| Endocrine resistance | 4 | 5.53E-06 | MMP9, FOS, ESR1, ABCB11 |
| Cell cycle | 4 | 1.14E-05 | CCNA2, CHEK1, CDC25A, CCNE2 |
| PI3K-Akt signaling pathway | 5 | 1.52E-05 | FGF2, IFNB1, EFNA3, IL6, CCNE2 |
| Estrogen signaling pathway | 3 | 0.00024 | MMP9, FOS, ESR1 |
| Toll-like receptor signaling pathway | 3 | 0.00024 | FOS, IL6, IFNB1 |
| TNF signaling pathway | 3 | 0.00024 | MMP9, FOS, IL6 |
| Measles | 3 | 0.00041 | IL6, IFNB1, CCNE2 |
| Transcriptional misregulation in cancer | 3 | 0.00085 | MMP9, IL6, HMGA2 |
| Herpes simplex infection | 3 | 0.00087 | FOS, IL6, IFNB1 |
| Proteoglycans in cancer | 3 | 0.00101 | MMP9, FGF2, ESR1 |
| Viral carcinogenesis | 3 | 0.00101 | CCNA2, CHEK1, CCNE2 |
| MAPK signaling pathway | 3 | 0.00167 | FOS, FGF2, STMN1 |
Abbreviations: ceRNA, competing endogenous RNA; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; TNF, tumor necrosis factor.
Among these key DElncRNAs, we noticed that three lncRNAs had more interactions than other regulators, and they may be key regulators related to the progression of TNBC, including DGCR5, RP11-576D8.4, and EMX2OS. It is interesting that DGCR5 has enriched a total of 21 pathogenesis-related signaling pathways by mediating six DEmiRNAs, such as “MAPK signaling pathway”, “EGFR tyrosine kinase inhibitor resistance”, and “Transcriptional misregulation in cancer”. The lncRNA EMX2OS can directly regulate hsa-mir-210 and hsa-mir-503 to activate 12 pivotal pathways, such as “p53 signaling pathway”, “Proteoglycans in cancer”, and “Ras signaling pathway”. In addition, we found that RP11-576D8.4, a novel lncRNA, can mediate hsa-mir-145 and hsa-mir-210 to regulate 19 pathways, including “Endocrine resistance”, “Estrogen signaling pathway”, “MicroRNAs in cancer”.
Survival analysis of key nodes in ceRNA network
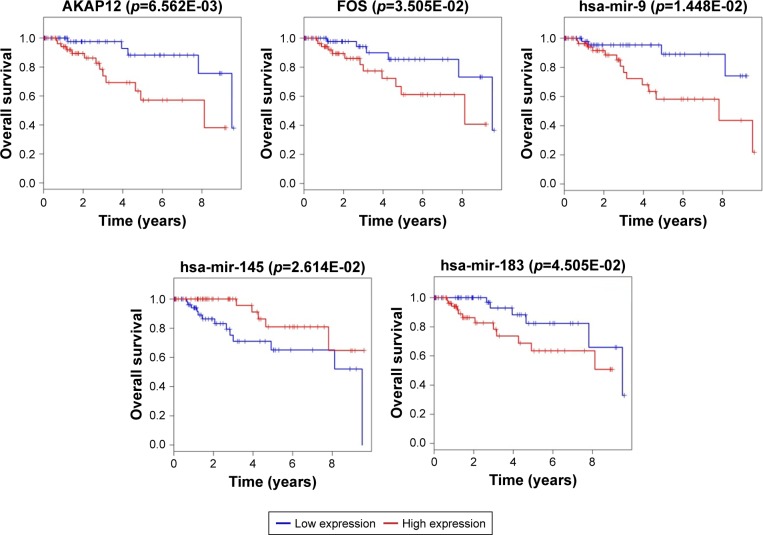
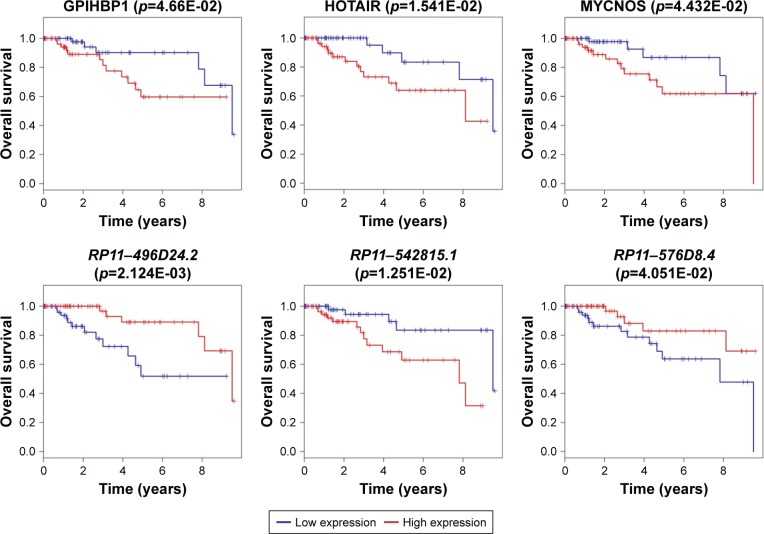
The Kaplan–Meier curves were performed to correlate expression pattern and clinical information of all TNBC samples to identify key nodes significantly associated with patients’ survival (p<0.05). A total of 22 hub genes, 14 key lncRNAs and 12 key miRNAs involved in ceRNA network were subjected to survival analysis. Finally, we noticed that the over-expression levels of seven key molecules (AKAP12, FOS, EMX2OS, MYCNOS, RP11-542B15.1, hsa-mir-9, and hsa-mir-183) predicted patients with poor outcome. On the contrary, the patients with high expression of hsa-mir-145, LINC00461, RP11-576D8.4 and RP11-496D24.2 have shown longer overall survival time than patients with the lower expression. All survival analyses plotted are shown in Figures 5 and 6.
Figure 5.
Survival curve analysis of ceRNA network for overall survival in triple negative breast cancer.
Notes: Two hub DEGs (FOS and AKAP12), and three key miRNAs (hsa-mir-145, hsa-mir-183, hsa-mir-9) are presented (p<0.05). Abscissa axis: overall survival time: days; vertical axis: survival function.
Abbreviations: ceRNA, competing endogenous RNA; DEGs, differentially expressed genes; miRNA, micro RNA.
Figure 6.
Survival curve analysis of ceRNA network for the overall survival in triple negative breast cancers.
Notes: Six key lncRNAs (LINC00461, RP11-496D24.2, RP11-542B15.1, RP11-576D8.4, EMX2OS, MYCNOS) are presented (p<0.05). Abscissa axis: overall survival time: days; vertical axis: survival function.
Abbreviations: ceRNA, competing endogenous RNA; lncRNA, long non-coding RNA.
Discussion
TNBC is a subtype of breast malignant tumor with poorer prognosis than other molecular subtypes. Even the development of chemotherapy has made progress and the patients can achieve pathological complete response after neoadjuvant chemotherapy, TNBC patients still have a higher metastasis rate and poorer prognosis than other breast cancer subtypes.19 With the increasing amounts of evidence indicating that ncRNAs have important roles in various biological processes, the correlated ceRNA networks have also been implicated in many kinds of tumor and regarded as a mechanism potentially driving tumors progression and metastasis,20,21 including in lung cancer,22 gastric cancer,23 and hepatocellular carcinoma.24 In addition, the ceRNA crosstalk could also relate to the exact regulatory mechanism of TNBC. Therefore, to find the proven effective target therapies for TNBC, we comprehensively integrated mRNA and miRNA data from TCGA, adopting WGCNA and other methods to identify the lncRNA-miRNA-mRNA interactions mediated in the ceRNA network for further investigation of the regulatory mechanisms of lncRNAs in this study.
lncRNAs are defined as non-protein coding transcripts longer than 200 nucleotides. Nevertheless, much research in recent years has shown that some lncRNAs can encode proteins to regulate cellular processes. In our work, 11 hub lncRNAs were identified as hub lncRNAs, and six of them (DGCR5, LINC00484, LINC00461, EMX2OS, RP11-576D8.4, and HNF1A-AS1) were distinctly enriched in the construction of the ceRNA network by integrated analysis. After detailed observation of the ceRNA network, we noticed that DGCR5 is the center of the network that may be involved in regulating the target genes by competing for five common shared miRNAs (hsa-mir-145, hsa-mir-141, hsa-mir-144, hsa-mir-183, and hsa-mir-210). In previous studies, DGCR5 was shown to deregulate expression in various tumors that could serve as a potential biomarker for diagnosis and prognosis. Sun et al had identified that DGCR5 was also down-regulated in pancreatic ductal adenocarcinoma cell lines, and its dysregulated expression could interact with miR-320a to inhibit tumor cell proliferation and migration.25 Huang et al had found that down-regulation of lncRNA DGCR5 correlated with poor prognosis in hepatocellular carcinoma.26 In our study, we noticed that DGCR5 with high-expression can compete with miRNAs to regulate the expression of the WGCNA green and red module target genes, such as ESR1, CHEK1, SAMD5, FGF2, EFNA3 and FOXP3 involved in the ceRNA network. Moreover, combined with clinical characteristics, the WGCNA green and red module demonstrated that they had the highest correlation coefficient in pathology N stage, pathology M stage, and positive lymph nodes of TNBC. Thus, it was hypothesized that knocking down the expression of DGCR5 may inhibit cell proliferation and lymphatic metastasis in TNBC. However, the survival analysis for DGCR5 showed no significant p-value, and these findings require further research to annotate its impact on breast cancer.
Besides, LINC00461 has the second-largest interactions in the ceRNA network. Recent study identified that dysfunctional expression of LINC00461 was significantly upregulated in both human and mouse glioma tissues. Deguchi et al demonstrated that LINC00461 could be a potential oncogene to regulate topoisomerase 2 alpha with sponging miR-411-5p in glioma.27 In this study, we identified that LINC00461 with high-expression may compete with three hub DEmiRNAs (miR-503, miR-144, miR-141) to regulate the expression of the target genes. In addition, the survival analysis showed that patients with highly expressed LINC00461 have prolonged overall survival time. In the construction of the ceRNA network, another two key DElncRNAs, RP11-496D24.2 and RP11576D8.4, were also related to the clinical outcome of TNBC patients. Low expression of these two lncRNAs suggested a poor prognosis in patients. It can be hypothesized that inducing the expression of the lncRNAs, including LINC00461, RP11-496D24.2 and RP11576D8.4, can prevent cell proliferation, and they could become the potential diagnosis and treatment targets for TNBC.
Even though miRNAs are defined as small ncRNA molecules, which have been well investigated in various human diseases, only a few lncRNAs have been functionally and mechanically characterized. In this study, we constructed the new ceRNA network to comprehensively illuminate the relations of lncRNAs and miRNAs. In our study, we identified 11 miRNAs, namely, mir-9, mir-33a, mir-139, mir-141, mir-142, mir-144, mir-145, mir-183, mir-210, mir-503 and let-7c, which should be further investigated. The mir-9, mir-183 and mir-145 were particularly important in the core of the ceRNA network. Tang et al verified that exosome mir-9 can suppress the expression of HBGF-5 and be a functional biomarker in hepatocellular carcinoma.28 Our study revealed that over-expression of mir-9 predicted a shorter lifetime compared to patients with lower mir-9 expression in TNBC. As for mir-183, Peng et al showed that mir-183 can be a potential biomarker for lung adenocarcinoma.29 Our survival analysis indicated that high expression of mir-183 was negatively linked to overall survival of TNBC patients. Regarding mir-145, Wang et al proved that the loss of mir-145 can induce Ras signaling pathway to promote aggressive Pten-deficient basal-like breast cancer.30 Our data also confirmed that the low expression of mir-145 predicted a shorter lifetime in TNBC. However, these findings need more research to investigate whether these aberrant miRNAs have a specific role in TNBC tumorigenesis. Many protein coding genes involved in the new ceRNA network were recently reported as key oncogenes participating in breast cancer development and progression, such as CCNE2, ESR1, HMGA2 and STMN1, which have also proven promising therapeutic targets in TNBC.31–33 Moreover, another two DEmRNAs, namely, FOS and AKAP12, demonstrated interactions with hub DEmiRNAs, and their high-expression predicted poor outcome in TNBC.
Based on the results of co-expression by KEGG pathways network, we could identify how the lncRNAs from ceRNA network activated the crucial pathways to regulate TNBC development and progression. Two enriched pathways, PI3K-Akt signaling pathway and mitogen-activated protein kinase (MAPK) signaling pathway, were reported as critical pathways in TNBC. Previous studies have identified that the crosstalk between the PI3K/Akt and MAPK pathways can overly activate to cause cell proliferation/metabolism, which impacts the prognosis of TNBC patients.34 Among the KEGG pathways network, five DElncRNAs, including DGCR5, EMX2OS, LINC00461, LINC00484, and RP11-576D8.4, may play specific roles for tumor-suppressing effects in TNBC, via down-regulation of two signal pathways.
Currently, although lncRNA-miRNA-mRNA network is receiving increasingly more attention, investigations focusing on that the way remarkable ncRNA mediate human diseases. Chen et al constructed a lncRNA-miRNA-mRNA ceRNA network to provide new insight into the common and specific subtype ceRNA interactions, which investigate the different categories of breast tumor (luminal A, luminal B, HER-2 enriched and TNBC subtype).35 However, this study only focused on different ncRNA data, and there were no combinations with clinical data to predict the prognosis in TNBC. Additionally, a similar study reported by Zhang et al also used the other kind of network-based method to analyse four different TCGA datasets for TNBC.36 The research used different molecular features, including expression profiles, gene copy number variants datasets and DNA methylation datasets, to identify some molecules that may serve as potential biomarkers.37 Nonetheless, the previous reports still could not provide an integrated WGCNA method to construct the ceRNA network for TNBC. And thus, the lncRNAs, miR-NAs, or other types of biological molecules have not been mechanically and functionally characterized.
Our research has some limitations. After a comprehensive search of Gene Expression Omnibus database, an international public repository that archives and freely distributes microarray, next-generation sequencing, and other forms of high-throughput functional genomic datasets, we found that there is no such independent data set that contains both coding and non-coding expression profiles for TNBC. Therefore, we lacked the validation step using independent datasets to demonstrate our findings. Moreover, since our research was only based on limited samples and information from TCGA databases, there is no more detailed information in TCGA for considering the subtypes analysis. Thus, further research is needed for verifying our study and unraveling the mechanism of ceRNA in TNBC.
Taken together, our study used an integrative biology approach to analyze non-coding and coding RNA expression profiles between normal and tumor samples from the TCGA database. Moreover, we performed WGCNA algorithm to select hub genes and construct the new lncRNA-miRNA-mRNA ceRNA crosstalk network to display a novel regulatory mechanism for further investigation of TNBC.
Supplementary material
Table S1.
The enriched KEGG pathways for each module
| Module | KEGG pathways | Input number | False discovery rate | Top genes |
|---|---|---|---|---|
| Red module | Cytokine–cytokine receptor interaction | 21 | 1.50E-19 | IL12B, CXCL9, CCR8, CCR7, IFNB1 |
| Chemokine signaling pathway | 11 | 1.03E-08 | CXCL9, CCR7, CCR8, CCL7, CXCL10 | |
| Type I diabetes mellitus | 5 | 4.33E-05 | IL12B, IFNG, GZMB, RBPJL, CD80 | |
| Jak-STAT signaling pathway | 7 | 8.04E-05 | IL12B, IL21, IFNB1, IFNG, GHR | |
| Toll-like receptor signaling pathway | 6 | 9.21E-05 | IL12B, CXCL9, IFNB1, CXCL10, CD80 | |
| Allograft rejection | 4 | 0.00031 | IL12B, IFNG, GZMB, CD80 | |
| Tryptophan metabolism | 4 | 0.00031 | IDO2, IDO1, IL4I1, ALDH2 | |
| RIG-I-like receptor signaling pathway | 4 | 0.00215 | IL12B, IFNB1, ISG15, CXCL10 | |
| PI3K-Akt signaling pathway | 7 | 0.00347 | TCL1A, IFNB1, EFNA2, FGF21, CD19, GHR | |
| Primary immunodeficiency | 3 | 0.00455 | TAP1, ICOS, CD19 | |
| Transcriptional misregulation in cancer | 5 | 0.00525 | ETV7, PAX5, CCR7, BCL2A1, GZMB | |
| TNF signaling pathway | 4 | 0.00622 | CREB3L3, RBPJL, CXCL10, CCL20 | |
| Intestinal immune network for IgA production | 3 | 0.00715 | CCL25, ICOS, CD80 | |
| Cytosolic DNA-sensing pathway | 3 | 0.01276 | ZBP1, IFNB1, CXCL10 | |
| CAMs | 4 | 0.01403 | ICOS, TIGIT, PDCD1, CD80 | |
| Blue module | Cell cycle | 29 | 1.26E-23 | CCNA2, MAD2L1, BUB1B, E2F1, ORC6 |
| Oocyte meiosis | 18 | 1.74E-11 | CDC25C, CCNB1, PLK1, CDC20, MAD2L1 | |
| Progesterone-mediated oocyte maturation | 13 | 7.85E-08 | CDC25C, CCNB1, PLK1, MAD2L1, CDC25A | |
| Fanconi anemia pathway | 10 | 3.27E-07 | RMI2, BRCA2, FANCI, BLM, UBE2T | |
| Homologous recombination | 7 | 9.22E-06 | RAD51, RAD54L, BLM, XRCC2, BRCA2 | |
| p53 signaling pathway | 9 | 1.59E-05 | CCNB2, CCNB1, CCNE2, CCNE1, CDKN2A | |
| HTLV-I infection | 13 | 0.00101 | CCNB2, MYBL2, CDC20, MAD2L1, CDKN2A | |
| DNA replication | 5 | 0.00309 | RNASEH2A, DNA2, MCM2, FEN1, MCM4 | |
| MicroRNAs in cancer | 13 | 0.00309 | CDC25C, DNMT3B, CDC25A, CCNE2, CCNE1 | |
| Protein digestion and absorption | 7 | 0.00371 | COL10A1, COL11A1, COL11A2, SLC15A1, SLC6A19 | |
| Pancreatic cancer | 6 | 0.00441 | BRCA2, CDKN2A, E2F2, MAPK10, E2F1 | |
| Neuroactive ligand-receptor interaction | 11 | 0.01537 | CHRNB4, CHRNA5, GABRA5, CHRM3, HTR6 | |
| Glycine, serine and threonine metabolism | 4 | 0.0293 | AGXT, PSAT1, CBS, CBSL | |
| Hepatitis B | 7 | 0.03896 | CCNA2, CCNE2, CCNE1, BIRC5, E2F2 | |
| Cholinergic synapse | 6 | 0.04077 | CHRNB4, CACNA1B, GNG4, KCNQ3, KCNJ4 | |
| Yellow module | Neuroactive ligand-receptor interaction | 9 | 0.00038 | NTSR1, ADRB3, GABRD, GRIA1, ADRA1A |
| Focal adhesion | 7 | 0.00166 | ITGA8, CAV1, FN1, FLNC, COL1A1 | |
| Calcium signaling pathway | 6 | 0.003759 | NTSR1, ADRB3, ADRA1A, AGTR1, ITPKA | |
| CAMs | 5 | 0.008612 | ITGA8, CLDN6, CLDN14, CLDN5, CDH15 | |
| ECM-receptor interaction | 4 | 0.008612 | ITGA8, TNN, FN1, COL1A1 | |
| Proteoglycans in cancer | 5 | 0.021599 | MMP9, CAV1, FN1, FLNC, PLAUR | |
| Renin secretion | 3 | 0.029087 | ADCYAP1R1, ADRB3, AGTR1 | |
| PI3K-Akt signaling pathway | 6 | 0.030187 | ITGA8, FN1, COL1A1, TNN, CSF3, SGK2 | |
| Drug metabolism – cytochrome P450 | 3 | 0.030187 | GSTM2, ADH7, UGT2B4 | |
| PPAR signaling pathway | 3 | 0.030471 | PPARG, FABP4, MMP1 | |
| Metabolism of xenobiotics by cytochrome P450 | 3 | 0.030471 | GSTM2, ADH7, UGT2B4 | |
| Chemical carcinogenesis | 3 | 0.038852 | GSTM2, ADH7, UGT2B4 | |
| cGMP-PKG signaling pathway | 4 | 0.038852 | ADRB3, MYLK2, ADRA1A, AGTR1 | |
| Pathways in cancer | 6 | 0.043731 | MMP1, FN1, MMP9, KLK3, AGTR1, PPARG | |
| Green module | Neuroactive ligand-receptor interaction | 28 | 4.01E-10 | ADRA2A, CCKBR, ADRB2, AVPR2, GRPR, P2RY6 |
| cAMP signaling pathway | 20 | 2.07E-07 | CALML5, GRIN2D, RAPGEF3, PLN, OXTR, NPR1 | |
| Regulation of lipolysis in adipocytes | 11 | 1.46E-06 | PLIN1, PLA2G16, ADRB2, NPR1, PNPLA2 | |
| Metabolic pathways | 52 | 2.04E-06 | MGLL, ALDH1A1, CKMT2, AK5, KL, AOX1 | |
| Tyrosine metabolism | 9 | 2.51E-06 | MAOA, TPO, AOC3, AOX1, PNMT | |
| Adipocytokine signaling pathway | 11 | 6.3E-06 | RXRG, CD36, LEPR, LEP, SLC2A1 | |
| Retinol metabolism | 10 | 2.37E-05 | ALDH1A1, CYP1A1, RDH5, AOX1, CYP2A6 | |
| AMPK signaling pathway | 13 | 2.92E-05 | PFKFB1, CD36E, F4EBP1, LEPR, IGF1 | |
| Drug metabolism – cytochrome P450 | 10 | 3.09E-05 | MAOA, AOX1, CYP2A6, UGT2B15, ADH1B | |
| Focal adhesion | 16 | 4.09E-05 | VEGFD, TNXB, COL2A1, VWF, CHAD | |
| Metabolism of xenobiotics by cytochrome P450 | 10 | 4.09E-05 | AKR1C1, CYP1A1, CYP2A6, ADH1B, GSTM5 | |
| PI3K-Akt signaling pathway | 21 | 4.51E-05 | VEGFD, FGF2, TNXB, COL2A1, IL6, IGF1 | |
| ECM-receptor interaction | 10 | 9.01E-05 | CHAD, TNXB, COL2A1, CD36, SDC1 | |
| Calcium signaling pathway | 14 | 0.00016 | CALML5, CCKBR, GRIN2D, PLN, OXTR, GRPR | |
| Protein digestion and absorption | 10 | 0.000167 | COL2A1, COL14A1, CPA1, COL17A1, COL4A6 | |
| Brown module | Transcriptional misregulation in cancer | 3 | 9.86E-12 | HIST1H3J, HMGA2, IBSP |
| PI3K-Akt signaling pathway | 6 | 0.040831 | IBSP, IGF2BP1, LAMA2, GNG13, ITGA7, FGF10 | |
| ECM-receptor interaction | 3 | 0.042262 | ITGA7, LAMA2, IBSP | |
| GABAergic synapse | 3 | 0.044141 | GNG13, KCNJ6, GABRA2 | |
| Morphine addiction | 3 | 0.045198 | GNG13, KCNJ6, GABRA2 |
Abbreviations: AMPK, AMP-activated protein kinase; CAM, cell adhesion molecule; ECM, extracellular matrix; GABA, gamma-aminobutyric acid; HTLV, human T-lymphotropic virus; IgA, immunoglobulin A; JAK, Janus kinase; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPAR, peroxisome proliferator-activated receptors; STAT, signal transducers and activators of transcription; TNF, tumor necrosis factor.
Acknowledgments
We gratefully acknowledge The Cancer Genome Atlas pilot project for providing all data in this research. Additionally, we are very grateful to Dr Chuiguo Huang for constructive and insightful suggestions. Financially supported by National Natural Science Foundation of China (nos 81673979, 81473688, 81373314, 81173265); Education Program of China for New Century Excellent Talents (no NCET-13-0827); Traditional Chinese Medicine Administration Project of Guangdong Province, China (no 20141070); Science and Technology Program of Guangzhou, China (no 2014J4100104); Science and Technology Program of Guangdong, China (nos 2014A020212672, and 2014A020210015); Natural Science Foundation of Guangdong Province, China (nos 2016A030313114; 2015A030313333); National Undergraduate Training Programs for Innovation and Entrepreneurship in 2015 (no 201510559046); Guangzhou Municipal Enterprise Research and Development Institutions Construction Project (no 201503010064); The Fundamental Research Funds for the Central Universities\Research and Cultivation and Innovation Fund of Jinan University, Guangzhou, China (nos 21615412, 21615464, and 21617467); Guangzhou Municipal Science and Technology Program in 2018 (project leader: Min Ma).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2012;62(1):10. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. Breast Dis Year Book Q. 2010;22(3):302–304. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 3.Rakha EA, Elrehim DA, Paish C, et al. Basal phenotype identifies a poor prognostic subgroup of breast cancer of clinical importance. Eur J Cancer. 2006;42(18):3149. doi: 10.1016/j.ejca.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Zaky SS, Lund MJ, May KA, et al. The negative effect of triple-negative breast cancer on outcome after breast-conserving therapy. Ann Surg Oncol. 2011;18(10):2858. doi: 10.1245/s10434-011-1669-4. [DOI] [PubMed] [Google Scholar]
- 5.Baltimore D. Our genome unveiled. Nature. 2001;409(6822):814–816. doi: 10.1038/35057267. [DOI] [PubMed] [Google Scholar]
- 6.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Xu Z, Jiang J, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25(1):69. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Tomczak K, Czerwińska P, Wiznerowicz M. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19(1A):A68. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1):559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7(1):252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stelzer G, Dalah I, Stein TI, et al. In-silico human genomics with GeneCards. Hum Genomics. 2011;5(6):709–717. doi: 10.1186/1479-7364-5-6-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34(Web Server issue):W720. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(Database issue):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu SD, Tseng YT, Shrestha S, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42(Database issue):D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goeman JJ, Oosting J, Cleton-Jansen AM, Anninga JK, van Houwelingen HC. Testing association of a pathway with survival using gene expression data. Bioinformatics. 2005;21(9):1950–1957. doi: 10.1093/bioinformatics/bti267. [DOI] [PubMed] [Google Scholar]
- 19.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sui J, Li YH, Zhang YQ, et al. Integrated analysis of long non-coding RNA-associated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol. 2016;49(5):2023. doi: 10.3892/ijo.2016.3716. [DOI] [PubMed] [Google Scholar]
- 23.Li CY, Liang GY, Yao WZ, et al. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol. 2016;48(5):1965. doi: 10.3892/ijo.2016.3407. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Fan D, Jian Z, Chen GG, Lai PBS. Cancer specific long noncoding RNAs show differential expression patterns and competing endogenous rna potential in hepatocellular carcinoma. PLoS One. 2015;10(10):e0141042. doi: 10.1371/journal.pone.0141042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yong S, Yabin Y, Bing Z, et al. Reciprocal regulation of DGCR5 and miR-320a affects the cellular malignant phenotype and 5-FU response in pancreatic ductal adenocarcinoma. Oncotarget. 2017;8(53):90868–90878. doi: 10.18632/oncotarget.18377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang R, Wang X, Zhang W, et al. Down-regulation of LncRNA DGCR5 correlates with poor prognosis in hepatocellular carcinoma. Cell Physiol Biochem. 2016;40(3–4):707. doi: 10.1159/000452582. [DOI] [PubMed] [Google Scholar]
- 27.Deguchi S, Katsushima K, Hatanaka A, et al. Oncogenic effects of evolutionarily conserved noncoding RNA ECONEXIN on gliomagenesis. Oncogene. 2017;6(32):4629–4640. doi: 10.1038/onc.2017.88. [DOI] [PubMed] [Google Scholar]
- 28.Tang J, Li Y, Liu K, et al. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Medica. 2017 doi: 10.23736/S0026-4806.17.05167-9. [DOI] [PubMed] [Google Scholar]
- 29.Peng Z, Pan L, Niu Z, et al. Identification of microRNAs as potential biomarkers for lung adenocarcinoma using integrating genomics analysis. Oncotarget. 2017;8(38):64143–64156. doi: 10.18632/oncotarget.19358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Liu JC, Ju Y, et al. MicroRNA-143/145 loss induces Ras signaling to promote aggressive Pten-deficient basal-like breast cancer. JCI Insight. 2017;2(15):93313. doi: 10.1172/jci.insight.93313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tormo E, Adamartigues A, Ballester S, et al. The role of miR-26a and miR-30b in HER2+ breast cancer trastuzumab resistance and regulation of the CCNE2 gene. Sci Rep. 2017;7:41309. doi: 10.1038/srep41309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Comparison of ESR1 mutations in tumor tissue and matched plasma samples from metastatic breast cancer patients. Transl Oncol. 2017;10(5):766–771. doi: 10.1016/j.tranon.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obayashi S, Horiguchi J, Higuchi T, et al. Stathmin1 expression is associated with aggressive phenotypes and cancer stem cell marker expression in breast cancer patients. Int J Oncol. 2017;51(3):781–790. doi: 10.3892/ijo.2017.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto K, Tsuda H, Koizumi F, et al. Activated PI3K/AKT and MAPK pathways are potential good prognostic markers in node-positive, triple-negative breast cancer. Ann Oncol. 2014;25(10):1973. doi: 10.1093/annonc/mdu247. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Xu J, Li Y, et al. Competing endogenous RNA network analysis identifies critical genes among the different breast cancer subtypes. Oncotarget. 2016;8(6):10171–10184. doi: 10.18632/oncotarget.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Ren C, Lau KK, et al. A network medicine approach to build a comprehensive atlas for the prognosis of human cancer. Briefings Bioinf. 2016;17(6):1044. doi: 10.1093/bib/bbw076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan Z, Ren C, Zhao H, et al. Identification of novel prognostic indicators for triple-negative breast cancer patients through integrative analysis of cancer genomics data and protein interactome data. Oncotarget. 2016;7(44):71620. doi: 10.18632/oncotarget.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
The enriched KEGG pathways for each module
| Module | KEGG pathways | Input number | False discovery rate | Top genes |
|---|---|---|---|---|
| Red module | Cytokine–cytokine receptor interaction | 21 | 1.50E-19 | IL12B, CXCL9, CCR8, CCR7, IFNB1 |
| Chemokine signaling pathway | 11 | 1.03E-08 | CXCL9, CCR7, CCR8, CCL7, CXCL10 | |
| Type I diabetes mellitus | 5 | 4.33E-05 | IL12B, IFNG, GZMB, RBPJL, CD80 | |
| Jak-STAT signaling pathway | 7 | 8.04E-05 | IL12B, IL21, IFNB1, IFNG, GHR | |
| Toll-like receptor signaling pathway | 6 | 9.21E-05 | IL12B, CXCL9, IFNB1, CXCL10, CD80 | |
| Allograft rejection | 4 | 0.00031 | IL12B, IFNG, GZMB, CD80 | |
| Tryptophan metabolism | 4 | 0.00031 | IDO2, IDO1, IL4I1, ALDH2 | |
| RIG-I-like receptor signaling pathway | 4 | 0.00215 | IL12B, IFNB1, ISG15, CXCL10 | |
| PI3K-Akt signaling pathway | 7 | 0.00347 | TCL1A, IFNB1, EFNA2, FGF21, CD19, GHR | |
| Primary immunodeficiency | 3 | 0.00455 | TAP1, ICOS, CD19 | |
| Transcriptional misregulation in cancer | 5 | 0.00525 | ETV7, PAX5, CCR7, BCL2A1, GZMB | |
| TNF signaling pathway | 4 | 0.00622 | CREB3L3, RBPJL, CXCL10, CCL20 | |
| Intestinal immune network for IgA production | 3 | 0.00715 | CCL25, ICOS, CD80 | |
| Cytosolic DNA-sensing pathway | 3 | 0.01276 | ZBP1, IFNB1, CXCL10 | |
| CAMs | 4 | 0.01403 | ICOS, TIGIT, PDCD1, CD80 | |
| Blue module | Cell cycle | 29 | 1.26E-23 | CCNA2, MAD2L1, BUB1B, E2F1, ORC6 |
| Oocyte meiosis | 18 | 1.74E-11 | CDC25C, CCNB1, PLK1, CDC20, MAD2L1 | |
| Progesterone-mediated oocyte maturation | 13 | 7.85E-08 | CDC25C, CCNB1, PLK1, MAD2L1, CDC25A | |
| Fanconi anemia pathway | 10 | 3.27E-07 | RMI2, BRCA2, FANCI, BLM, UBE2T | |
| Homologous recombination | 7 | 9.22E-06 | RAD51, RAD54L, BLM, XRCC2, BRCA2 | |
| p53 signaling pathway | 9 | 1.59E-05 | CCNB2, CCNB1, CCNE2, CCNE1, CDKN2A | |
| HTLV-I infection | 13 | 0.00101 | CCNB2, MYBL2, CDC20, MAD2L1, CDKN2A | |
| DNA replication | 5 | 0.00309 | RNASEH2A, DNA2, MCM2, FEN1, MCM4 | |
| MicroRNAs in cancer | 13 | 0.00309 | CDC25C, DNMT3B, CDC25A, CCNE2, CCNE1 | |
| Protein digestion and absorption | 7 | 0.00371 | COL10A1, COL11A1, COL11A2, SLC15A1, SLC6A19 | |
| Pancreatic cancer | 6 | 0.00441 | BRCA2, CDKN2A, E2F2, MAPK10, E2F1 | |
| Neuroactive ligand-receptor interaction | 11 | 0.01537 | CHRNB4, CHRNA5, GABRA5, CHRM3, HTR6 | |
| Glycine, serine and threonine metabolism | 4 | 0.0293 | AGXT, PSAT1, CBS, CBSL | |
| Hepatitis B | 7 | 0.03896 | CCNA2, CCNE2, CCNE1, BIRC5, E2F2 | |
| Cholinergic synapse | 6 | 0.04077 | CHRNB4, CACNA1B, GNG4, KCNQ3, KCNJ4 | |
| Yellow module | Neuroactive ligand-receptor interaction | 9 | 0.00038 | NTSR1, ADRB3, GABRD, GRIA1, ADRA1A |
| Focal adhesion | 7 | 0.00166 | ITGA8, CAV1, FN1, FLNC, COL1A1 | |
| Calcium signaling pathway | 6 | 0.003759 | NTSR1, ADRB3, ADRA1A, AGTR1, ITPKA | |
| CAMs | 5 | 0.008612 | ITGA8, CLDN6, CLDN14, CLDN5, CDH15 | |
| ECM-receptor interaction | 4 | 0.008612 | ITGA8, TNN, FN1, COL1A1 | |
| Proteoglycans in cancer | 5 | 0.021599 | MMP9, CAV1, FN1, FLNC, PLAUR | |
| Renin secretion | 3 | 0.029087 | ADCYAP1R1, ADRB3, AGTR1 | |
| PI3K-Akt signaling pathway | 6 | 0.030187 | ITGA8, FN1, COL1A1, TNN, CSF3, SGK2 | |
| Drug metabolism – cytochrome P450 | 3 | 0.030187 | GSTM2, ADH7, UGT2B4 | |
| PPAR signaling pathway | 3 | 0.030471 | PPARG, FABP4, MMP1 | |
| Metabolism of xenobiotics by cytochrome P450 | 3 | 0.030471 | GSTM2, ADH7, UGT2B4 | |
| Chemical carcinogenesis | 3 | 0.038852 | GSTM2, ADH7, UGT2B4 | |
| cGMP-PKG signaling pathway | 4 | 0.038852 | ADRB3, MYLK2, ADRA1A, AGTR1 | |
| Pathways in cancer | 6 | 0.043731 | MMP1, FN1, MMP9, KLK3, AGTR1, PPARG | |
| Green module | Neuroactive ligand-receptor interaction | 28 | 4.01E-10 | ADRA2A, CCKBR, ADRB2, AVPR2, GRPR, P2RY6 |
| cAMP signaling pathway | 20 | 2.07E-07 | CALML5, GRIN2D, RAPGEF3, PLN, OXTR, NPR1 | |
| Regulation of lipolysis in adipocytes | 11 | 1.46E-06 | PLIN1, PLA2G16, ADRB2, NPR1, PNPLA2 | |
| Metabolic pathways | 52 | 2.04E-06 | MGLL, ALDH1A1, CKMT2, AK5, KL, AOX1 | |
| Tyrosine metabolism | 9 | 2.51E-06 | MAOA, TPO, AOC3, AOX1, PNMT | |
| Adipocytokine signaling pathway | 11 | 6.3E-06 | RXRG, CD36, LEPR, LEP, SLC2A1 | |
| Retinol metabolism | 10 | 2.37E-05 | ALDH1A1, CYP1A1, RDH5, AOX1, CYP2A6 | |
| AMPK signaling pathway | 13 | 2.92E-05 | PFKFB1, CD36E, F4EBP1, LEPR, IGF1 | |
| Drug metabolism – cytochrome P450 | 10 | 3.09E-05 | MAOA, AOX1, CYP2A6, UGT2B15, ADH1B | |
| Focal adhesion | 16 | 4.09E-05 | VEGFD, TNXB, COL2A1, VWF, CHAD | |
| Metabolism of xenobiotics by cytochrome P450 | 10 | 4.09E-05 | AKR1C1, CYP1A1, CYP2A6, ADH1B, GSTM5 | |
| PI3K-Akt signaling pathway | 21 | 4.51E-05 | VEGFD, FGF2, TNXB, COL2A1, IL6, IGF1 | |
| ECM-receptor interaction | 10 | 9.01E-05 | CHAD, TNXB, COL2A1, CD36, SDC1 | |
| Calcium signaling pathway | 14 | 0.00016 | CALML5, CCKBR, GRIN2D, PLN, OXTR, GRPR | |
| Protein digestion and absorption | 10 | 0.000167 | COL2A1, COL14A1, CPA1, COL17A1, COL4A6 | |
| Brown module | Transcriptional misregulation in cancer | 3 | 9.86E-12 | HIST1H3J, HMGA2, IBSP |
| PI3K-Akt signaling pathway | 6 | 0.040831 | IBSP, IGF2BP1, LAMA2, GNG13, ITGA7, FGF10 | |
| ECM-receptor interaction | 3 | 0.042262 | ITGA7, LAMA2, IBSP | |
| GABAergic synapse | 3 | 0.044141 | GNG13, KCNJ6, GABRA2 | |
| Morphine addiction | 3 | 0.045198 | GNG13, KCNJ6, GABRA2 |
Abbreviations: AMPK, AMP-activated protein kinase; CAM, cell adhesion molecule; ECM, extracellular matrix; GABA, gamma-aminobutyric acid; HTLV, human T-lymphotropic virus; IgA, immunoglobulin A; JAK, Janus kinase; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPAR, peroxisome proliferator-activated receptors; STAT, signal transducers and activators of transcription; TNF, tumor necrosis factor.