ABSTRACT
Tight regulation of gene expression is achieved by a variety of protein complexes that selectively bind chromatin, modify it and change its transcription competency. Histone acetylases (HATs) and deacetylases (HDACs) play an important role in this process. They can generate transcriptionally active or inactive chromatin through the addition (HATs) or removal (HDACs) of acetyl groups on histones, respectively.
Repo-Man is a Protein Phosphatase 1 targeting subunit that accumulates on chromosomes during mitotic exit and mediates the removal of mitotic histone H3 phosphorylations. It was shown recently that Repo-Man also regulates heterochromatin formation in interphase and that its depletion favours the switch between transcriptionally inactive and active chromatin, demonstrating that its role goes well beyond mitosis.
Here, we provide the first link between a phosphatase and HDAC complexes. We show that genome-wide Repo-Man binding sites overlap with chromatin regions bound by members of the three HDAC complexes (Sin3a, NuRD and CoREST). We establish that members of the NuRD and Sin3a HDAC complexes interact with Repo-Man by mass spectrometry and that Repo-Man is in close proximity to SAP18 (Sin3a) in interphase as observed by the Proximity Ligation Assay. Altogether, these data suggest a mechanism by which Repo-Man/PP1 complex, via interactions with HDACs, could stabilise gene repression.
Keywords: PP1, Repo-Man, HDAC, epigenetic, heterochromatin
Introduction
Control of gene expression is achieved through the concerted action of regulators that target DNA or histones. Histones can be post-translationally modified providing cues for activation or repression of genes and cell cycle progression signals. For example, at the beginning of mitosis histone H3 is heavily phosphorylated at Thr3, Ser10 and Ser28 by the mitotic kinases Haspin and Aurora B. These phosphorylations have several functions including the release of the chromatin reader heterochromatin protein 1 (HP1)1,2 and the RNA polymerase II (Pol II) basal transcription factor TFIID (TFIID) from their binding sites.3 At the end of mitosis, Protein Phosphatase 1 (PP1), via its targeting subunit Repo-Man, accumulates on the chromosomes to restore the interphase status of histone H3 through the removal of its mitotic phosphorylations.4,5
Apart from being phosphorylated, histones can also be acetylated (by histone acetylases (HATs) and deacetylated by histone deacetylases (HDACs) leading to gene transcription activation and repression, respectively.
HDACs are part of three major complexes: Sin3a, NuRD and CoRest (Fig. 1A).6 These are multi subunit complexes that contain HDACs1 and 2, but differ in the composition of the other subunits. HDACs 1 and 2 are functionally redundant and double knockouts most often trigger abnormal differentiation phenotypes and apoptosis, thus resulting in lethality.6 Similarly, knockouts of Sin3a (Sin3a complex), Lsd1 (CoREST) and Mbd3 (NuRD), all result in embryonic lethality in mice6 proving that they are vital for cellular functions at least during development.
Figure 1.
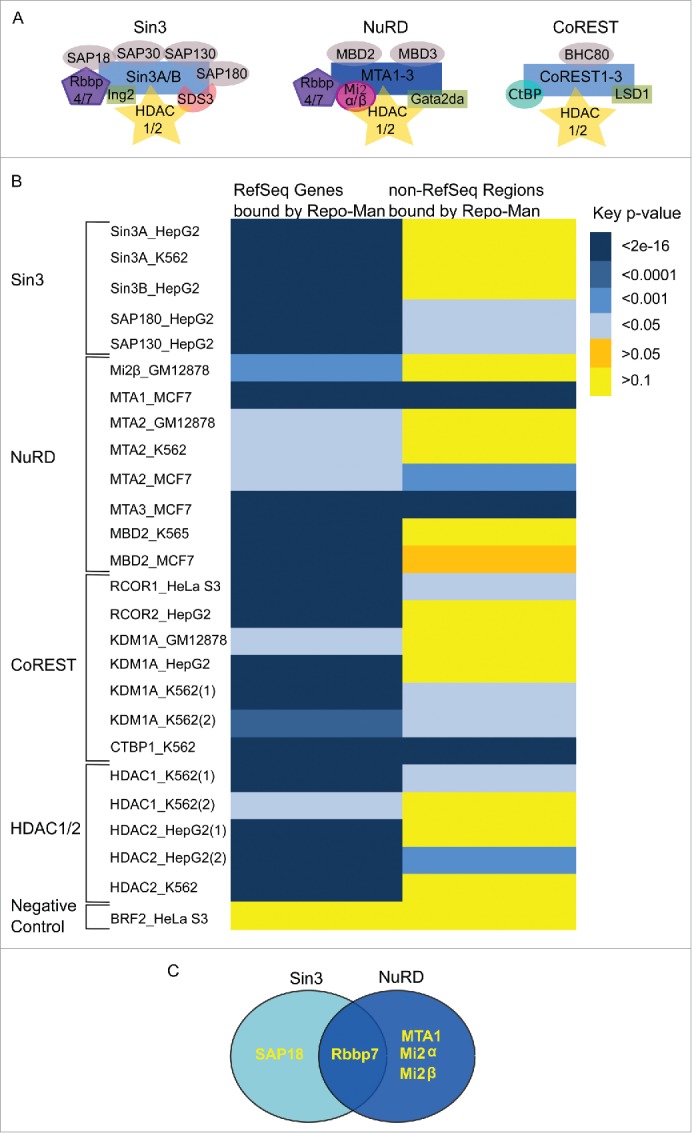
HDAC complexes and interactions with Repo-Man. (A) Three main HDAC complexes members, Sin3A, NuRD and CoREST6. Repo-Man interactors identified by Mass Spectrometry are SAP18, Rbbp7, MTA1 and Mi2α/β. (B) Overlap between Repo-Man bound genes (RefSeq) and the rest of the genome (non-RefSeq) with HDAC available ENCODE data from HeLa and cell lines indicated in the figure; colour key is shown with the correspondent p-value. (C) HDAC complex members interacting with Repo-Man in available Mass Spectrometry datasets.
The HDAC complexes interact with chromatin modifiers and with each other. For example, the coREST complex associates with G9a, the methyltransferase responsible for the methylation of lysine 9 on histone H3, an important marker of repressed chromatin7. REST protein can interact with the coREST complex via its C-terminal domain and the Sin3a complex via its N-terminal domain, thus coREST and Sin3a can be found together in REST regulated sites.7,8 Interestingly, in drosophila embryos, SAP18 (member of the Sin3a complex) and HDAC1 have been found linked to the expression status of the polycomb-regulated gene Fab-7.9 This points to complex cross-talks between different classes of chromatin modifiers that ultimately converge to modulate gene transcription.
Understanding the network of regulators contributing to gene expression has been the focus of many investigations10 that have unveiled the several strategies cells can use in different contexts.
Only recently it was shown that, beside the accepted chromatin writers and erasers, protein phosphatases are also part of this regulatory mechanism.4,11 Repo-Man, a PP1 targeting subunit, associates with repressive chromatin and is both necessary and sufficient to generate a transcriptionally repressed environment characterised by the presence of H3K9me3, H3K27me2/3 and HP1.4 Repo-Man binding sites across the genome were also characterized using a TAG-Proteogenomic approach in HeLa cells.4 The data indicated that Repo-Man has a preference for sub-telomeric genes marked, most often, by repressive modifications, such as the Polycomb deposited mark H3K27me2/3. Depletion of Repo-Man decreased the levels of this modification and increased gene expression. Thus we hypothesize that Repo-Man/PP1 complex could function as a chromatin regulator and therefore possibly interact with other chromatin modifiers, specifically with repressive modulators of gene expression.
Repo-Man appears to bind preferentially to modified H3K27. It has been shown that H3K27 can rapidly transit between an active (K27ac) and inactive (K27me3) state in Embryonic Stem (ES) cells depending on cues such as the availability of the enzymes responsible for the deposition of these markers.12 Importantly, the dynamics between K27ac and K27me has been linked to the phosphorylation status of S28, a substrate of the Repo-Man/PP1 complex. Moreover, during cellular stress, Ser28 is phosphorylated by MSK1 leading to K27ac and gene expression activation. We hypothesise that Repo-Man/PP1 is crucial for reverting this transient signal to its default state and that the removal of Ser28P by Repo-Man/PP1 favours the methylation of K27, thus regulating the acetyl/methyl status of H3K27. This poses the question of the complexity of Repo-Man network of interactions and its implications in gene expression regulation.
To uncover the underlying cross talk between Repo-Man and other repressive regulators, we started by exploring the relationship between Repo-Man and repressive complexes. Available datasets for Repo-Man and HDAC members indicate that there is a significant overlap in their binding sites. Moreover, we also identified SAP18 (Sin3 complex) in Repo-Man pull-downs and showed that these proteins are indeed in close proximity in the interphase nucleus.
These findings open a new avenue to investigate the cross-talk between the classic epigenetic modifiers and chromatin-associated phosphatases that ultimately converge to fine tune gene expression regulation.
Results
Repo-Man binding sites genome-wide have been previously characterized4 showing an association with repressive chromatin markers. In order to gain a more detailed picture on the chromatin regulated by Repo-Man/PP1, we have explored the relationship between Repo-Man and repressive HDAC complex members (Fig. 1A). Taking advantage of the Repo-Man genomic binding sites generated through the TAG-Proteogenomics approach in HeLa cells, we have identified overlaps with HDAC ChIP-Seq datasets publically available from HeLa cells and other cell lines when the former was not available (HepG2, MCF7 or K562). This was possible since the same HDAC component shares statistically significant overlaps of binding sites in different cell lines (p <2e-16). A significant overlap was found between the Repo-Man bound Ref-Seq genes and several HDAC complexes binding sites (Fig. 1B). Several members of the three HDAC complexes, Sin3 NuRD and CoREST show significant degrees of overlaps, as do the HDAC enzymes HDAC1 and HDAC2. Overlaps are not observed with a randomly selected dataset (BRF2), or with regions outside genes (non RefSeq). This suggests that Repo-Man and HDAC complexes occupy similar chromatin regions.
Next we have surveyed a number of mass spectrometry datasets previously generated for Repo-Man. The Bollen Laboratory has recently investigated the Repo-Man/PP1 interacting proteome on HEK293T by Split-BioID, a technique established around the BioID technology13. Using TAG-Proteogenomics we have also mapped by Mass Spectrometry the chromatin associated proteins present on the Repo-Man-bound chromatin (enriched for low molecular weight components as only bands across the histones weight range were selected14). Additionally, we have interrogated datasets from Repo-Man pull downs previously generated in chicken (DT40) cells from anaphase cells.15
Members of the HDAC complexes (Fig. 1A) are found across the Repo-Man interactome. SAP18 (Sin3 complex) as well as Rbbp7 (Sin3 and NuRD) were found in two datasets alongside with other three members of the NuRD family (Fig. 1C). This suggests that Repo-Man can interact directly or indirectly with HDAC family members.
Since we have previously shown that Repo-Man has high affinity for H3K27me2/3, we asked if it was particularly enriched in regions containing both H3K27me3 and HDAC: 39.4% of Repo-Man bound Ref-Seq genes have both H3K27me3 and HDAC present (p< 2e−16).
To validate these interactions we have focussed in more detail on the relationship between Repo-Man and Sap18. Transient transfections of HeLa cells with SAP18 fused to GFP show a nuclear localisation with accumulation in foci (Fig. 2A), similar to the pattern observed using an antibody that detects the endogenous protein (Fig. 2B). During mitosis, SAP18 is largely diffused (Fig. 2A and B), a behaviour shared between many chromatin bound proteins.16 To demonstrate the interaction between Repo-Man and SAP18 in cells, we took advantage of the Proximity Ligation Assay, PLA. This assay relies on the availability of antibodies from two different species against the two proteins of interest. Due to the fact that the only available antibodies for both Repo-Man and SAP18 are raised in rabbit, we have used a mouse GFP antibody against the transfected GFP:SAP18 and an antibody targeting the endogenous Repo-Man. This assay shows that Repo-Man and SAP18 do interact in vivo across the nucleus and the presence of PLA signals is almost undetectable in the non-transfected HeLa cells (Fig. 2C and D). This suggests that Repo-Man is in close proximity to SAP18 (and possibly of other HDAC complex members), that indeed they occupy similar nuclear spaces, and possibly chromatin regions, as suggested by the genomic binding sites. Repo-Man and HDAC interactions could perhaps be important to reinforce the repressive state of chromatin regions already bound by Repo-Man.
Figure 2.
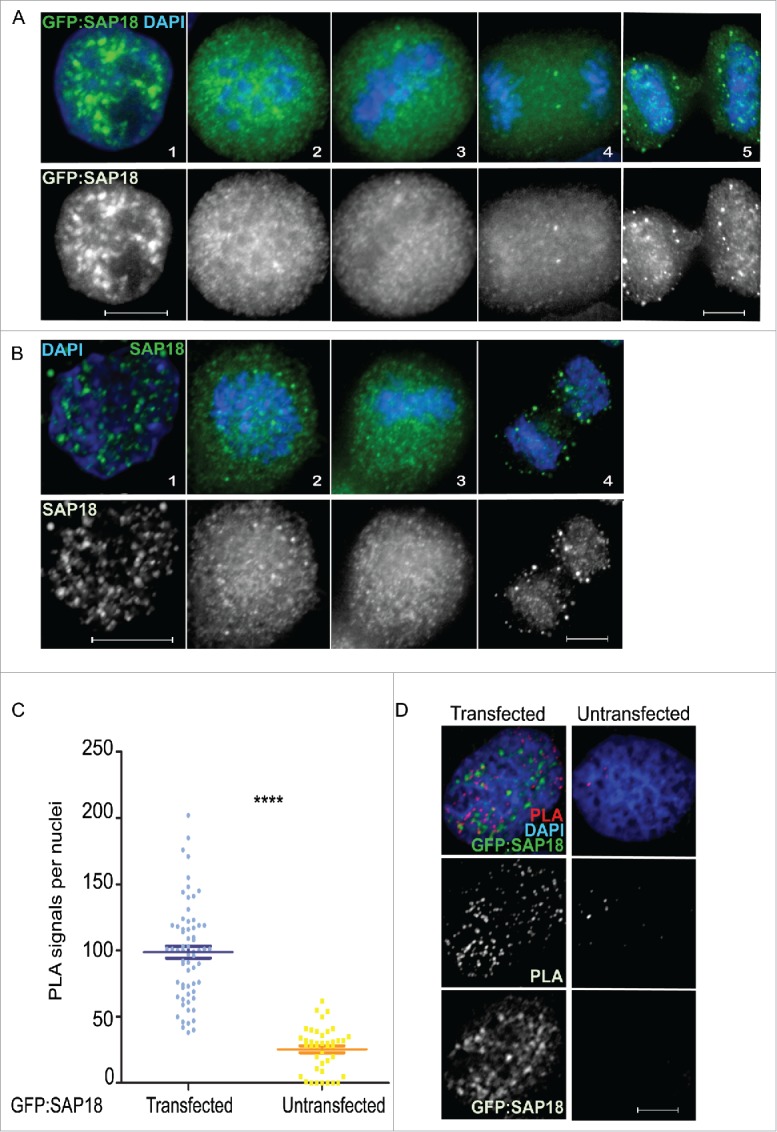
SAP18 (Sin3A complex) is a Repo-Man interactor. (A) Localisation of SAP18 fused to GFP from interphase (1), prometaphase (2), metaphase (3), anaphase (4) and cytokinesis (5), respectively, in HeLa cells. (B) Immunofluorescence of wild-type SAP18 in interphase (1), prometaphase (2), metaphase (3) and anaphase (4) respectively. (C) Counts of experiments in D, amongst ∼65 nuclei, Mann-Whitney test (****p < 0.0001). (D) Proximity ligation assay between Repo-Man (endogenous) and SAP18 (GFP fused) in transfected and non-transfected HeLa cells. Scale bars 10 μm.
Based on these observations, it would be expected that changes in Repo-Man occupancy at chromatin would result in alterations of the methylation/acetylation status. This is exactly what we have observed. Depletion of Repo-Man causes a global decrease of H2K27me2/3 as well as at Repo-Man bound genes, leading to their activation; moreover, targeting of Repo-Man to a specific locus, causes a local increase of H2K27me2/3 levels.4
Together our data suggests a working model where Repo-Man maintains H3 in a de-phosphorylated status and, by interacting (directly or indirectly) with HDAC complexes, favours the removal of histone acetylation groups ultimately maintaining gene repression (Fig. 3).
Figure 3.
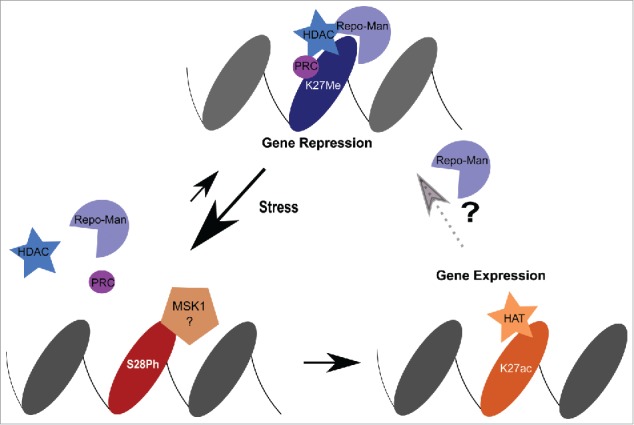
Model. Repo-Man is found together with HDAC complexes to maintain repression at H3K27me2/3. Upon stress induction, MSK1 phosphorylates Ser28 and leads to the dispersal of Repo-Man/HDAC from the chromatin sites. Consequently, HATs mediate the acetylation of Lysine 27 activating gene expression. This can cause either a permanent or transient re-activation. For the latter, the decrease in kinase activity will favour the binding of Repo-Man to the acetylated H3K27 (Repo-Man has affinity for this residues as well in vitro) and promote the recruitment of HDAC to restore a repressed state.
Discussion
Repo-Man/PP1 is an important mitotic exit complex functioning in the reorganisation of the nuclear envelope and re-modelling of mitotic chromatin. The phosphorylation on histone H3 by the mitotic AuroraB and Haspin kinases are reverted at the end of mitosis by Repo-Man/PP1.4,5,15 De-phosphorylation of H3S10 is vital for the re-establishment of a repressive environment in the daughter cells, enabling the re-association of HP1, ejected from its docking site (H3K9me3) upon phosphorylation of S10 at mitosis onset.2 Thereafter the role of Repo-Man was poorly understood.
Recently, we have shown that the formation of heterochromatin is regulated by Repo-Man and its depletion disrupts the nuclear distribution of both HP1 and the repressive marks H3K9me3 and H3K27me2/3.4 Together with reduction of H3K27me2/3 levels, Repo-Man depletion alleviates gene repression of some polycomb repressed genes.4 Post-translational modifications at Lysine 27 have been reported to be highly dynamic, thus priming genes for rapid transitions between activation and repression.17 For example, Ser28 phosphorylation by the stress-activated kinase MSK1, has been linked to the transition of K27me3 into K27ac in a panel of genes, resulting in their activation.18,19 We propose that Repo-Man/PP1 counteracts this kinase and, by removing the Ser28P, allows the reinstatement of K27me3. In this scenario, it would be therefore expected that the enzymes responsible for the transition between K27ac and K27me3 have to be present, transiently or more stably, at these regions. The identification of this epigenetic network will be important to understand the regulatory mechanisms underlying this transition.
HDAC enzymes remove the acetylation of lysines and are part of three main complexes, CoRest, NuRD and Sin3A. These complexes can 1) recognize a modification20 or be recruited by transcription factors to catalyse a reaction;21 2) catalyse other modifications, for example through LSD1, a H3K4 demethylase present within a complex;22 3) act in concerted action with other histone modifiers to propagate an epigenetic landscape. The latter is a clear example of cross talk between different chromatin modifiers. For example, the methyltransferase G9a (K9me) and HDAC are both recruited by tissue specific transcription factors changing the epigenetic landmarks of their target genes.23,24 Interestingly, the polycomb complex associates with histone deacetylases and two other HDAC complex members (Rbbp4 and 7); this interaction further strengthens its repressive function by favouring the removal of histone acetyl groups.25 This appears to be a common theme; in fact, other HDAC complex members, such as SAP18, were also found at a polycomb regulated gene in drosophila,9 suggesting that there is a strong network of chromatin modifiers that act together to remodel the epigenetic landscape and regulate gene expression. As observed here, HDAC complexes are also present at Repo-Man targeted genes and Repo-Man interacts with members of HDAC complexes (Mass Spectrometry, SPLIT Bio-ID and PLA). We could therefore hypothesize that this network would indeed favour a rapid transition between K27ac and K27me as suggested before and it would be interesting to explore this idea in the context of stress signalling.
In fact, it has been shown that H3S28Ph is present at 50% of all stress-induced genes. Moreover, MSK1/2-mediated phosphorylation of H3S28 at stress-responsive promoters contributes to the dissociation of HDAC corepressor complexes and promotes local histone acetylation and subsequent transcriptional activation of stress-induced genes.19 We have also shown that the binding of Repo-Man to chromatin is inhibited by H3S28 phosphorylation and that depletion of Repo-Man alleviates the repressive marks at some polycomb repressed genes.4 Taken these datasets together, it is becoming clear that H3S28Ph mark plays a key role in the activation of mammalian genes in response to MAP and stress activated kinases. A cooperation between HDACs and phosphatases in this context could represent a safeguard mechanism to ensure the maintenance of a specific chromatin landscape but also a mean to revert to the default situation once the stress stimuli are over.
Several HDAC complex members seem to be able to interact with Repo-Man; this could be due either to their participation in different HDAC complexes or to the presence of multi-complexes at a given gene. The latter possibility seems to occur in some cases; for example, coREST and Sin3a can be found together in REST regulated sites7,8 as well as Sin3, HDACs and NCor are targeted by given receptors to their target genes (reviewed in 26). Therefore, albeit the view is that HDAC complexes bind transcription factors/receptors according to tissue specificity or signalling cues, it can also be the case that, in certain genes HDACs are brought together with other chromatin modifiers (e.g. protein phosphatases) to reinforce the status of repression.
Future direction
Here we have just explored the tip of the iceberg and we have shown that Repo-Man sits at a crossroad of important interactions with chromatin modifiers.
These findings add an extra view to the complexity of chromatin regulations and place phosphatase complexes in the inner circle of epigenetic modifiers that contribute to maintain and regulate gene expression.
It will be important to investigate the functional consequences of these interactions for the epigenetic and transcriptional status of Repo-Man bound genes upon different kind of stimuli. Moreover, the identification of the specific networks in terms of dependencies and recruitment of the different HDACs to a particular chromatin domain will clarify how the system works and highlight possible overlaps and redundancies.
Future investigations will shed light into this and other potential interactions with chromatin regulators that might be important for the normal functioning of the cell.
Materials and methods
Cell culture, cloning, and transfections
HeLa cells were grown in DMEM supplemented with 10% FBS and 1% Penicillin-Streptomycin (Invitrogen Gibco) at 37ºC with 5% CO2.
Transient transfections were performed using Polyplus jetPRIME (PEQLAB, Southampton, UK) with 1μg of plasmid DNA and incubated for 24 hours before fixation as per manufacturer's instructions.
Immunofluorescence microscopy
Cells were fixed in 4% PFA and processed as previously described.27 SAP18 antibody (abcam) was used at a concentration of 1:200 and fluorescence-labeled secondary antibodies were applied at 1:200 (Jackson ImmunoResearch). 3D data sets were acquired using a wide-field microscope (NIKON Ti-E super research Live Cell imaging system) with a NA 1.45 Plan Apochromat lens. The datasets were deconvolved with NIS Elements AR analysis software (NIKON). Three-dimensional datasets were converted to Maximum Projection in the NIS software, exported as TIFF files, and imported into Adobe Photoshop for final presentation.
Proximity ligation assay (PLA)
PLA was performed according to the manufacturer's protocol (Sigma). HeLa cells were fixed, permeabilised and blocked with BSA as previously described.27 GFP (Thermo Scientific) and Repo-Man27 antibodies were used at a concentration of 1:500 and 1:300, respectively. PLA probes were added and ligation was performed following manufacturer instructions (Sigma). Coverslips were mounted on DAPI and observed on the previously mentioned wide-field NIKON microscope. Spots were counted using object detection by thresholding (ROI) on the NIKON software.
Bioinformatics
The Repo-Man dataset was extracted from the publically available data (Accession: GSE84035) as described in de Castro et al. (2017) except that chrX regions were included, giving a total of 643 regions. The datasets for the HDAC complex components were obtained from ENCODE (doi: 10.1038/nature11247). The p-values (p) used to generate the heat map were obtained empirically using , where m is the number of times that o < e, where o is the number of Repo-Man binding regions that overlap (and/or are within ± 10 Kb of) HDAC complex component binding sites and e is the number of randomly generated Repo-Man regions that overlap (and/or are within ± 10 Kb of) HDAC complex component binding sites and n is the number of repeats (n = 10,000). Each randomly generated dataset was identical to the Repo-Man dataset except that the starting position of each region was a random integer between x and 1 + y - w, where x is 1, y is the maximum length of the chromosome to which that region was mapped to and w is the width of that region. The maximum length of each chromosome was assumed to be the value listed in the “All scaffolds” column of the “Total lengths” table from Human Genome Assembly GRCh38.p10 (doi: 10.1093/nar/gkv1290). For chromosomes 13, 14, 15, 21 and 22, x was equated to the value listed in the “Start” column of the “Modeled [sic] centromeres and heterochromatin regions” table under GRCh38.p10 (doi: 10.1093/nar/gkv1290). RM_RefSeq are Repo-Man binding sites dataset that overlap RefSeq and RM_No_RefSeq are all the rest. RefSeq is a dataset obtained from UCSC table browser (genome = “Human”, assembly = “Dec. 2013 (GRCh38/hg38)”, group = “Genes and Gene Predictions”, track = “RefSeq Genes”, table = “refGene”) and subsetted such that only regions with the NM_ accession prefix remained.
Data availability
The microscopy data are available from the corresponding author upon request and will be released via figshare.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the BBSRC grant BB/K017632/1 to PV.
References
- [1].Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176-80. doi: 10.1038/nature04254. PMID:16222244 [DOI] [PubMed] [Google Scholar]
- [2].Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116-22. doi: 10.1038/nature04219. PMID:16222246 [DOI] [PubMed] [Google Scholar]
- [3].Varier RA, Outchkourov NS, de Graaf P, van Schaik FM, Ensing HJ, Wang F, Higgins JM, Kops GJ, Timmers HT. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. Embo J. 2010;29:3967-78. doi: 10.1038/emboj.2010.261. PMID:20953165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].de Castro IJ, Budzak J, Di Giacinto ML, Ligammari L, Gokhan E, Spanos C, Moralli D, Richardson C, de Las Heras JI, Salatino S, et al.. Repo-Man/PP1 regulates heterochromatin formation in interphase. Nat Commun. 2017;8:14048. doi: 10.1038/ncomms14048. PMID:28091603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Qian J, Lesage B, Beullens M, Van Eynde A, Bollen M. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr Biol. 2011;21:766-73. doi: 10.1016/j.cub.2011.03.047. PMID:21514157 [DOI] [PubMed] [Google Scholar]
- [6].Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans. 2013;41:741-9. doi: 10.1042/BST20130010. PMID:23697933 [DOI] [PubMed] [Google Scholar]
- [7].Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544-54. doi: 10.1038/nrg2100. PMID:17572692 [DOI] [PubMed] [Google Scholar]
- [8].Seki M, Masaki H, Arauchi T, Nakauchi H, Sugano S, Suzuki Y. A comparison of the rest complex binding patterns in embryonic stem cells and epiblast stem cells. PloS One. 2014;9:e95374. doi: 10.1371/journal.pone.0095374. PMID:24752154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Canudas S, Perez S, Fanti L, Pimpinelli S, Singh N, Hanes SD, Azorin F, Espinas ML. dSAP18 and dHDAC1 contribute to the functional regulation of the Drosophila Fab-7 element. Nucleic Acids Res. 2005;33:4857-64. doi: 10.1093/nar/gki776. PMID:16135462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jones B. Gene expression: layers of gene regulation. Nat Rev Genet. 2015;16:128-9. doi: 10.1038/nrg3918. PMID:25690392 [DOI] [PubMed] [Google Scholar]
- [11].Nuytten M, Beke L, Van Eynde A Ceulemans H, Beullens M, Van Hummelen P Fuks F, Bollen M. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene. 2008;27:1449-60. doi: 10.1038/sj.onc.1210774. PMID:17724462 [DOI] [PubMed] [Google Scholar]
- [12].Petruk S, Black KL, Kovermann SK, Brock HW, Mazo A. Stepwise histone modifications are mediated by multiple enzymes that rapidly associate with nascent DNA during replication. Nat Commun. 2013;4:2841. doi: 10.1038/ncomms3841. PMID:24276476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].De Munter S Gornemann J, Derua R, Lesage B, Qian J, Heroes E, Waelkens E, Van Eynde A Beullens M, Bollen M. Split-BioID: a proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett. 2017;591:415-424. doi: 10.1002/1873-3468.12548. PMID:28032891 [DOI] [PubMed] [Google Scholar]
- [14].de Castro IJ, Budzak J, Di Giacinto ML, Ligammari L, Gokhan E, Spanos C, Moralli D, Richardson C, de Las Heras JI, Salatino S, et al.. Repo-Man/PP1 regulates heterochromatin formation in interphase. Nat Commun. 2017;8:14048. doi: 10.1038/ncomms14048. PMID:28091603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vagnarelli P, Ribeiro S, Sennels L, Sanchez-Pulido L, de Lima Alves F, Verheyen T, Kelly DA, Ponting CP, Rappsilber J, Earnshaw WC. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev Cell. 2011;21:328-42. doi: 10.1016/j.devcel.2011.06.020. PMID:21820363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].de Castro IJ Gokhan E, Vagnarelli P. Resetting a functional G1 nucleus after mitosis. Chromosoma. 2016;125:607-19. doi: 10.1007/s00412-015-0561-6. PMID:26728621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lau PN, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci U S A. 2011;108:2801-6. doi: 10.1073/pnas.1012798108. PMID:21282660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell. 2010;39:886-900. doi: 10.1016/j.molcel.2010.08.020. PMID:20864036 [DOI] [PubMed] [Google Scholar]
- [19].Sawicka A, Hartl D, Goiser M, Pusch O, Stocsits RR, Tamir IM, Mechtler K, Seiser C. H3S28 phosphorylation is a hallmark of the transcriptional response to cellular stress. Genome Res. 2014;24:1808-20. doi: 10.1101/gr.176255.114. PMID:25135956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J Biol Chem. 2009;284:7970-6. doi: 10.1074/jbc.M808220200. PMID:19155214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. PMID:22414492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, et al.. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660-72. doi: 10.1016/j.cell.2009.05.050. PMID:19703393 [DOI] [PubMed] [Google Scholar]
- [23].Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25:10338-51. doi: 10.1128/MCB.25.23.10338-10351.2005. PMID:16287849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol. 2007;27:1407-24. doi: 10.1128/MCB.00944-06. PMID:17145766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275-86 PMID:11124122 [DOI] [PubMed] [Google Scholar]
- [26].McDonel P, Costello I, Hendrich B. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. The international journal of biochemistry & cell biology. 2009;41:108-16. doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vagnarelli P, Hudson DF, Ribeiro SA, Trinkle-Mulcahy L, Spence JM, Lai F, Farr CJ, Lamond AI, Earnshaw WC. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol. 2006;8:1133-42. doi: 10.1038/ncb1475. PMID:16998479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The microscopy data are available from the corresponding author upon request and will be released via figshare.


