Abstract
We recently identified ten novel SLE susceptibility loci in Asians and uncovered several additional suggestive loci requiring further validation. This study aimed to replicate five of these suggestive loci in a Han Chinese cohort from Hong Kong, followed by meta-analysis (11,656 cases and 23,968 controls) on previously reported Asian and European populations, and to perform bioinformatic analyses on all 82 reported SLE loci to identify shared regulatory signatures. We performed a battery of analyses for these five loci, as well as joint analyses on all 82 SLE loci. All five loci passed genome-wide significance: MYNN (rs10936599, Pmeta = 1.92 × 10−13, OR = 1.14), ATG16L2 (rs11235604, Pmeta = 8.87 × 10 −12, OR = 0.78), CCL22 (rs223881, Pmeta = 5.87 × 10−16, OR = 0.87), ANKS1A (rs2762340, Pmeta = 4.93 × 10−15, OR = 0.87) and RNASEH2C (rs1308020, Pmeta = 2.96 × 10−19, OR = 0.84) and co-located with annotated gene regulatory elements. The novel loci share genetic signatures with other reported SLE loci, including effects on gene expression, transcription factor binding, and epigenetic characteristics. Most (56%) of the correlated (r2 > 0.8) SNPs from the 82 SLE loci were implicated in differential expression (9.81 × 10−198 < P < 5 × 10−3) of cis-genes. Transcription factor binding sites for p53, MEF2A and E2F1 were significantly (P < 0.05) over-represented in SLE loci, consistent with apoptosis playing a critical role in SLE. Enrichment analysis revealed common pathways, gene ontology, protein domains, and cell type-specific expression. In summary, we provide evidence of five novel SLE susceptibility loci. Integrated bioinformatics using all 82 loci revealed that SLE susceptibility loci share many gene regulatory features, suggestive of conserved mechanisms of SLE etiopathogenesis.
Introduction
Systemic Lupus Erythematosus (SLE, ‘lupus’; OMIM 152700) is a common, complex autoimmune disease with profound health consequences across ethnicities. Disease severity, morbidity and mortality are highest in individuals of non-European descent (1). To date, more than ten genome-wide association studies (GWAS) and historical candidate gene studies have identified diverse risk loci (2–4). However, these SLE candidate genes taken together are insufficient to explain the entire heritability of SLE. We recently determined that an aggregate of 47 SLE risk loci explain only ∼24% of SLE heritability (2). Thus, many more SLE loci and genetic variants are yet to be discovered.
We recently reported ten novel loci associated with SLE susceptibility in individuals of Korean, Han Chinese, Japanese and Malaysian Chinese descent (2). Additionally, we uncovered several novel loci that either fell just below genome-wide significance (P < 5×10−8) or were not independently replicated. In this follow-up study, we augmented our previous data with an additional cohort from Hong Kong and performed in silico replication together with published cohorts of European and Chinese descent (4,5). The added statistical power of the meta-analysis with nine cohorts allowed us to independently replicate five candidate loci above genome-wide significance.
In our earlier study, we reported several SLE associated loci, including ATG16L2, MYNN and CCL22, which either failed to pass our genome-wide association threshold or did not replicate in our independent cohorts. Additionally, we reported secondary associations in ANKS1A, RNASEH2C and SIGLEC14 that were statistically independent from reported genes DEF6, PCNXL3 and SIGLEC6, respectively. In this study, we focused on the replication of five loci: ATG16L2 (rs11235604), MYNN (rs10936599), CCL22 (rs223881), ANKS1A (rs2762340) and RNASEH2C (rs1308020).
Among the candidate loci, MYNN encodes the zinc-finger transcription factor myoneurin, which regulates neuromuscular junctions (6) and telomere length (7). ATG16L2 is an autophagy-related gene recently proposed as an SLE risk locus (8), and is associated with multiple sclerosis (MS) (9) and Crohn's disease (10). CCL22 is a pro-inflammatory chemokine (11) secreted by macrophages and dendritic cells, and previously reported as a risk factor in ischemic heart disease (12) and MS (13). CCL22 binds to its receptor CCR4, found on dendritic cells in patients with liver inflammation (14). CCL22/CCR4 is also implicated in T-cell migration to the joints in rheumatoid arthritis (RA), psoriatic arthritis and osteoarthritis (15). RNASEH2C (along with SLE gene IFIH1) has been reported as a risk factor for Aicardi-Goutières syndrome (16), a rare inflammatory disease that clinically overlaps with SLE. RNASEH2C encodes subunit C of the human ribonuclease H2 enzyme complex that trims RNA-DNA duplexes (17). In a recent study, RNASEH2C was associated with SLE (5) through rs494003 (actually in the 3’-UTR of AP5B1, an endocytosis-adaptor protein, instead of RNASEH2C). Our strongest signal for this locus, rs1308020, is in the RNASEH2C 5’-UTR, 44 kb from rs494003. Our fifth candidate locus is lung cancer risk gene (18) ANKS1A, also known as ODIN, a Src kinase that negatively regulates growth factor signaling (19). ANKS1A interacts with and is phosphorylated by Lck (lymphocyte-specific protein tyrosine kinase) (20), a critical component of T-cell activation. rs2820223 of ANKS1A was associated (P = 3.5×10−4, OR (95% CI)=3.14 (1.68–5.87)) with anti-Ro and anti-dsDNA autoantibodies in SLE patients (21). Due to small sample sizes, that study was not sufficiently powered to detect the smaller case-control odds-ratio (OR) that our study found.
To understand how these validated candidate SLE loci fit into the larger scope of SLE pathogenesis, we expanded our analysis to the entire set of known SLE susceptibility genes. We carried out integrated bioinformatic analyses of all 82 SLE loci in the context of gene expression, and identified patterns of regulation within subgroups of SLE susceptibility genes.
Results
Association analysis with five loci
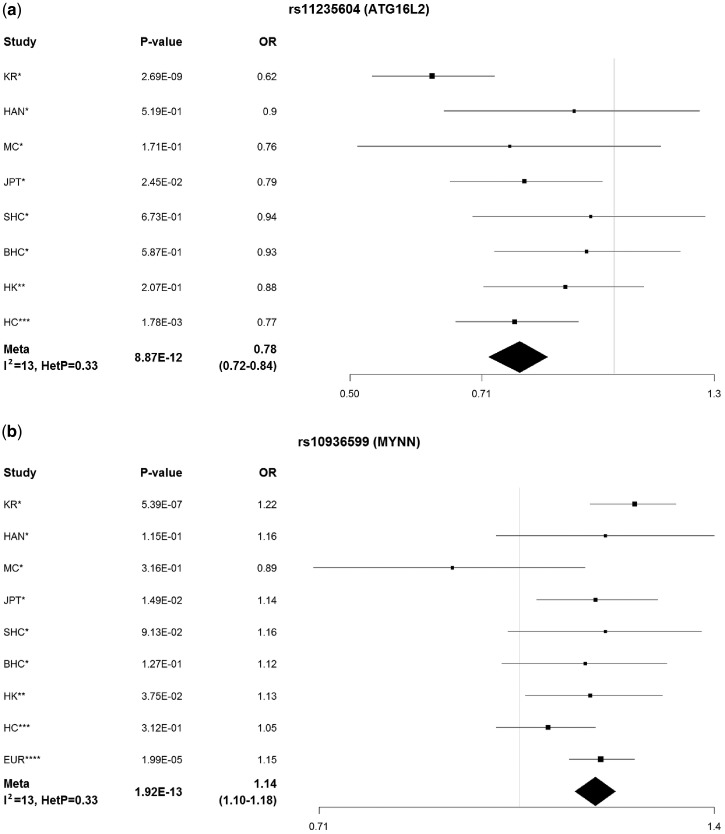
Sample sizes from nine independent cohorts are shown in Table 1. All five SNPs were in Hardy-Weinberg equilibrium. Three SNPs were significantly associated with SLE in the Hong Kong cohort by itself (Table 2): rs10936599 (MYNN, P = 3.75×10−2), rs1308020 (RNASEH2C, P= 2.49×10−3) and rs223881 (CCL22, P = 1.14×10−2). To increase statistical power, we performed meta-analysis with in silico data from published Han Chinese and European cohorts (5). After combining these three cohorts, all five loci became significant (rs223881, P = 5.30×10−9; rs2762340, P = 4.96×10−7; rs1308020, P = 1.40×10−6; rs10936599, P = 4.81×10−6; rs11235604, P = 1.01×10−3).
Table 1.
Samples used in this study. This study used samples from our previous analysis as the discovery set, and added a genotyped cohort as well as two published in silico sources as the replication sets.
| Data and Source populations | Ethnicity | Case | Control | Total | |
|---|---|---|---|---|---|
| Discovery (Sun et al. 2016) (2) | Korean | East Asian | 1,710 | 6,836 | 8,546 |
| Han Chinese | East Asian | 490 | 493 | 983 | |
| Malaysian Chinese | East Asian | 285 | 287 | 572 | |
| Japanese | East Asian | 891 | 3,384 | 4,275 | |
| Shanghai Han Chinese | East Asian | 501 | 622 | 1,123 | |
| Beijing Han Chinese | East Asian | 601 | 1,034 | 1,635 | |
| Replication (Genotyped) (Morris et al. 2016) (5) (Bentham et al. 2015) (4) | Hong-Kong | East Asian | 1,456 | 955 | 2,411 |
| Han Chinese (5) | East Asian | 1,659 | 3,398 | 5,057 | |
| European (4,5) | European | 4,063 | 6,959 | 11,022 | |
| Meta-analysis | Total | 11,656 | 23,968 | 35,624 | |
Table 2.
Association of five novel SLE loci replicated across eight Asian cohorts and a European cohort. KR: Korean, HC: Han Chinese, MC: Malaysian Chinese; JPT: Japanese; SHC: Shanghai Han Chinese; BHC: Beijing Han Chinese; HK: Hong Kong Chinese; European Ancestry cohort (Bentham et al. 2016); Han Chinese replication cohort (Morris et al. 2016); Overall: Meta-analysis of all eight Asian and European cohorts; OR: odds ratio; CI: 95% confidence interval for odds. In the Han Chinese replication cohort for rs11235604, we used MAF and P-value for the proxy SNP rs77971648 which is in LD (D'=1; r2=0.88) with rs11235604. MAF: Minor allele frequency for cases/controls
| SNP (Alleles) | Nearest Gene | MAF/P-value for the discovery cohorts |
Meta P-value OR (95% CI) | MAF/P-value for the replication cohorts |
Overall P-value OR (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KR | HC | MC | JPT | SHC | BHC | HK | European | Han Chinese | ||||
| rs11235604 (T/C) | ATG16L2 | 0.067/0.099 | 0.092/0.100 | 0.093/0.119 | 0.072/0.088 | 0.083/0.087 | 0.093/0.099 | 1.90E-09 | 0.081/0.091 | monomorphic | 0.076/0.097 | 8.87E-12 |
| 2.69E-09 | 5.19E-01 | 1.71E-01 | 2.45E-02 | 6.98E-01 | 5.87E-01 | 0.76 (0.69-0.84) | 2.07E-01 | 1.78E-3 | 0.78(0.71-0.85) | |||
| rs10936599 (C/T) | MYNN | 0.435/0.389 | 0.492/0.460 | 0.456/0.484 | 0.394/0.363 | 0.489/0.455 | 0.482/0.455 | 4.93E-09 | 0.483/0.452 | 0.783/0.754 | 0.465/0.457 | 1.92E-13 |
| 5.39E-07 | 1.15E-01 | 3.16E-01 | 1.49E-02 | 9.19E-02 | 1.27E-01 | 1.16 (1.10-1.22) | 3.75E-02 | 1.99E-05 | 3.12E-01 | 1.14(1.10-1.18) | ||
| rs223881 (C/T) | CCL22 | 0.455/0.492 | 0.457/0.528 | 0.509/0.587 | 0.362/0.402 | 0.536/0.534 | 0.497/0.506 | 2.06E-08 | 0.538/0.579 | 0.741/0.765 | 0.533/0.567 | 5.87E-16 |
| 9.31E-05 | 1.29E-03 | 7.23E-03 | 2.60E-03 | 9.08E-01 | 5.94E-01 | 0.87 (0.82-0.91) | 1.14E-02 | 6.88E-06 | 5.51E-03 | 0.87(0.84-0.90) | ||
| rs2762340 (G/A) | ANKS1A | 0.341/0.381 | 0.278/0.339 | 0.235/0.279 | 0.348/0.391 | 0.282/0.268 | 0.255/0.291 | 5.05E-07 | 0.234/0.235 | 0.474/0.499 | 0.253/0.277 | 4.93E-15 |
| 3.15E-05 | 7.19E-03 | 1.30E-01 | 6.43E-04 | 4.63E-01 | 2.50E-02 | 0.85 (0.81-0.90) | 9.81E-01 | 3.19E-04 | 1.84E-05 | 0.87(0.84-0.90) | ||
| rs1308020 (T/C) | RNASEH2C | 0.227/0.269 | 0.231/0.235 | 0.210/0.275 | 0.229/0.258 | 0.187/0.262 | 0.217/0.291 | 1.77E-09 | 0.223/0.262 | 0.327/0.344 | 0.219/0.252 | 2.96E-19 |
| 1.17E-07 | 8.80E-01 | 6.45E-03 | 1.34E-02 | 3.46E-05 | 1.23E-05 | 0.78 (0.73-0.83) | 2.49E-03 | 2.32E-03 | 8.28E-03 | 0.84(0.81-0.88) | ||
Finally, we performed a meta-analysis of these SNPs by adding data from our discovery cohorts (Table 1), which further increased significance levels, confirming association with SLE. All signals were replicated and passed genome-wide significance: MYNN (rs10936599 Pmeta = 1.92×10−13, I2 = 12.8, PHeterogeneity =0.33), ATG16L2 (using rs77971648 (D' = 1; r2 = 0.88) as a proxy for rs11235604, Pmeta = 8.87×10−12, I2 = 12.6, PHeterogeneity =0.33), CCL22 (rs223881, Pmeta = 5.87×10−16, I2 = 28.3, PHeterogeneity = 0.19), ANKS1A (rs2762340, Pmeta = 4.93×10−15, I2 = 42.9, PHeterogeneity =0.08) and RNASEH2C (rs1308020, Pmeta = 2.96×10−19, I2= 64.9, PHeterogeneity = 3.60×10−3) (Table 2). ANKS1A and RNASEH2C were identified by conditioning within the association regions of the DEF6 and PCNXL3 signals (Fig. 1).
Figure 1.
Meta-analysis of all studies for five novel loci. We carried out meta-analysis for our five novel loci on nine independent cohorts. P-values and odds ratios (ORs) are presented for each cohort, as well as heterogeneity statistics for each locus. *Sun et al. 2016; **this study; ***Morris et al. 2016; ****Bentham et al. 2016. (a) ATG16L2; (b) MYNN; (c)CCL22; (d)ANKS1A; (e) RNASEH2C.
Bioinformatic analysis of five SLE susceptibility loci
We next conducted a series of bioinformatic analyses to assess the functional consequences of the five SNPs individually and in concert with previously reported SNPs. To identify possible contributions of these SNPs to SLE pathogenesis, we assessed co-localization of these SNPs with functional genomic elements. Two SNPs (rs11235604 and rs10936599) are exonic: SNP rs11235604 (ATG16L2) changes residue 114 to tryptophan (Arg114Trp), while rs10936599 (MYNN) is synonymous (His4His). rs2762340 (ANKS1A) is intronic, and the two remaining SNPs are intergenic. Since only one SNP produced a coding mutation, we next assessed their effect on gene regulation and expression.
To understand how these SNPs affect gene expression, we queried them against external expression quantitative trail loci (eQTL) databases. In whole blood data (22), SNPs in CCL22, ANKS1A and RNASEH2C are eQTLs for multiple targeted genes, including SLE locus DEF6 (P = 1.69×10−7), the largest subunit of RNA polymerase II (POLR2C) (P = 1.08×10−10) and SCYL1 (P= 9.81×10−198) (Supplementary Material, Table S1A). Additionally, among cell-specific expression data (23), rs10936599 is an eQTL for MYNN in testis (P = 1.90×10−7) and for LRRC34 in transformed fibroblasts (P = 6.60×10−6). rs223881 is an eQTL for cytokine-induced apoptosis inhibitor CIAPIN1 in testis (P = 1.70×10−7). rs1308020 affects expression of SIPA1 (a key modulator of T-cell activation (24)) and MAP3K11 (part of the NF-κB signaling pathway) in esophageal mucosa (P = 1.40×10−5), arterial tissues (P = 4.80×10−6) and thyroid (3.20×10−12) (Supplementary Material, Table S1B).
To assess other effects on gene function, we first examined ENCODE annotations of the loci. rs223881 (CCL22) is predicted to be in a weak enhancer sequence of the GM12878 lymphoblastoid cell line; rs11235604 (ATG16L2) is in a GM12878 ENCODE weak promoter and transcription start site. Next, we analyzed allele-specific disruption of transcription factor binding sites (TFBSs). The lead SNPs for the five candidate loci directly and strongly disrupted 24 TFBSs. rs223881 disrupts binding of SLE risk gene STAT5A (25,26), which contributes to B-cell response to cytokines and glucocorticoid receptor NR3C1 (27). rs1308020 (RNASEH2C) disrupts binding of NR3C1 itself. Variants highly correlated to the five lead SNPs (LD r2 > 0.8 in Asians: 75 SNPs) yielded 350 more strongly disrupted TFBSs (Supplementary Material, Table S2). This set of correlated, disrupted TFBSs includes those for known SLE risk genes such as ETS1 (28), NF-κB (29), STAT2 and STAT3 (48). Immune-system TFs including NRF1, TFAP4, SMAD4, p53, pre-B-cell leukemia transcription factor (PBX) proteins, interferon regulatory factors (IRFs), nuclear factor of activated T-cell (NFAT) proteins, B-cell lymphoma (BCL) proteins, and T-cell-specific (TCF) proteins were all highly represented in the set of strongly disrupted allele-specific TFBSs (Supplementary Material, Table S2). In summary, the five replicated loci have functional involvement in transcription and gene expression, particularly in B- and T-cells.
We also investigated whether these loci were pleiotropic, as is the case with many other autoimmune disease (AD)-related loci (2). MYNN, CCL22 and ATG16L2 have previous reported association with AD. SNP rs10936599 (MYNN) has been reportedly associated with celiac disease (30) and chronic lymphocytic leukemia (31), while CCL22 has been related to MS (13) and RA (15). ATG16L2 was recently linked to SLE (8) through a correlated SNP (rs11235667) located between FCHSD2 and P2RY2, and also associated with MS (9) and Crohn's disease (10) (Supplementary Material, Table S3).
Bioinformatic analysis of 82 SLE susceptibility loci
To understand how these five loci fit within the published set of SLE loci, we continued our bioinformatic analysis on the expanded set of 82 SLE loci (five novel and 77 previously known SLE loci) (Supplementary Material, Table S4). For a comprehensive evaluation, we used a set of all 1,825 correlated SNPs (r2 > 0.8) from the 82 loci and explored their potential as functional variants. Only 28 SNPs (1.5%) are coding (missense or synonymous). 1,134 (62%) of the correlated SNPs fall within gene regions (including untranscribed regions). Most correlated SNPs are located within ENCODE-annotated functional elements, such as enhancers (1,206 SNPs; 66%) and promoters (492 SNPs; 27%) (Supplementary Material, Table S4). Using all correlated SNPs, we identified significant (2×10−6 < P < 5×10−2) enrichment of enhancers in T-, B- and natural killer cells, together constituting the 16 most enriched cell types (Supplementary Material, Table S5).
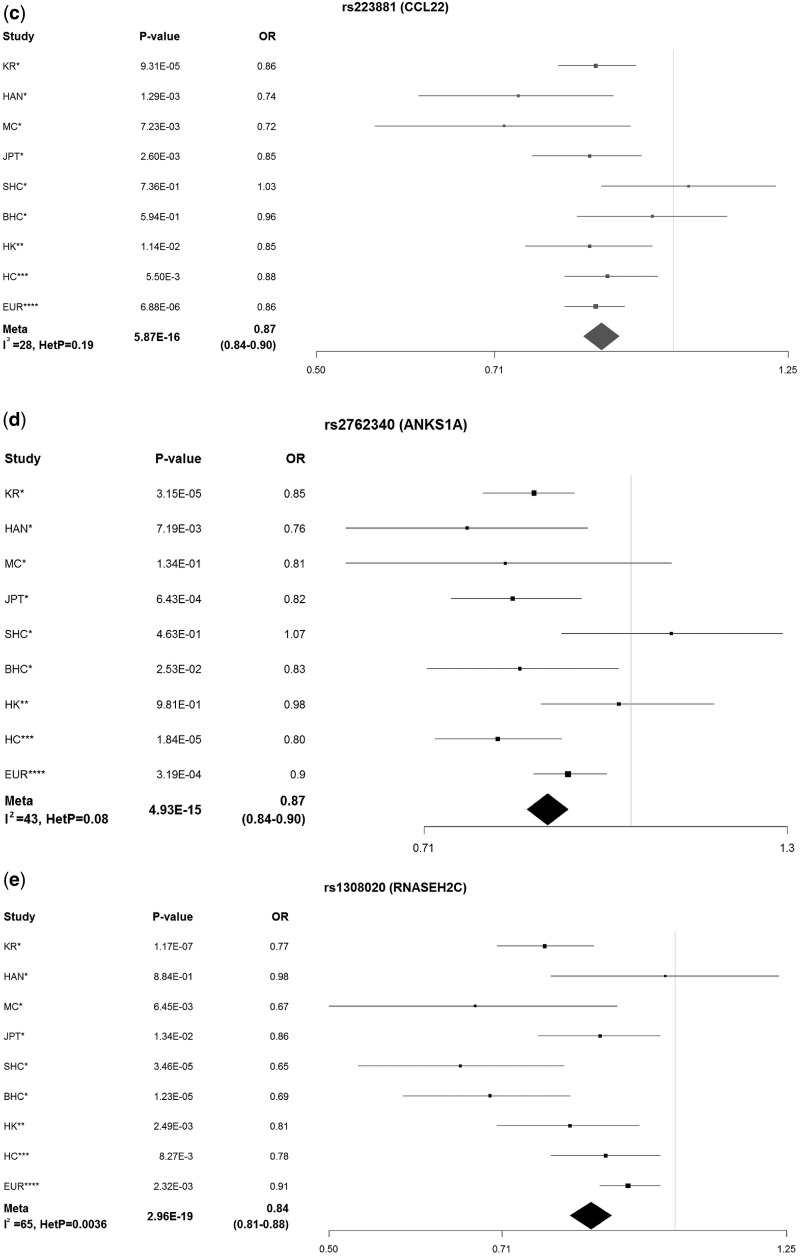
Given that SLE loci were enriched in epigenetic functional elements, we sought to identify an enriched set of bound transcription factors (TFs). Variant set enrichment (VSE) analysis identified 7 TFs significantly enriched (P < 5×10−5) in the expanded variant set in lymphoblastoid cells (Fig. 2). The most enriched TF was nuclear factor IC (Padj = 2.54×10−8), followed by EBF1 (P = 1.29×10−8). EBF1 binds to SLE locus ITGAM (32), and contributes to B-cell development, activation and proliferation (33). TATA-box binding protein (TBP) was also enriched, consistent with reports of its association with SLE (34) (Fig. 2). Other significantly enriched TFs were lupus-related BCL3 and RELA, as well as immune-system genes RUNX3, MTA3, WRNIP1, POLR2A and FOXM1 (Fig. 2). Previously reported TFs MXI1 and IKZF1, although nominally suggestive, did not pass our predefined (P < 5×10−8) significance threshold.
Figure 2.
Variant set enrichment analysis of 76 transcription factors (TFs) in GM12878. (a) Number of SLE loci enriched in each TF (x-axis). (b) Enrichment score for each TF, boxplots for all permutations of enrichment analysis. Dots represent enrichment score for each TF. Empty circles represent all TFs that passed 5 × 10−8 enrichment P-value.
We next assessed how gene-level expression correlates with SLE-associated loci through eQTLs, and identified 51 SLE loci altering expression of 168 genes (103 cis, 65 trans) (Supplementary Material, Table S1A and C). Of note, among the five novel genes, CCL22 affected POLR2C, COQ9 and DOK4; and RNASEH2C affected SCYL1, FIBP and other seven genes (Supplementary Material, Table S1A).
To identify common genetic signatures among SLE loci, we carried out gene-level enrichment analyses. The most significantly enriched pathways (KEGG annotation) included stimulus-related immune pathways such as the RIG-I-like receptor signaling pathway (P = 6.62×10−5; NFKBIA; IFIH1; IL12B; IL12A; IKBKE). The five novel loci share membership with other SLE susceptibility loci in the cytokine-cytokine receptor interaction (P = 1.93×10−2: IL10, CCL22, TNFSF4, IL12A/B) and chemokine signaling (P = 4.80×10−3; LYN, NFKBIA, CCL22, PRKCB) pathways (Supplementary Material, Table S6). Our enrichment results further cement these five novel loci within the current understanding of SLE susceptibility and pathogenesis, while further providing a functional context for them.
In order to cluster the functions of our novel SLE loci with the set of known SLE genes, we performed an enrichment analysis of their protein domain types. Our novel loci share protein domain types (Pfam), although not significantly enriched, such as the zinc finger Znf_C2H2 domain (MYNN, IKZF1, PRDM1, RNASEH2C), as well as the SH3 domain (LYN, NCF2), suggesting common modes of action among SLE genes. Furthermore, to test for commonalities among modes of transcriptional regulation, we performed enrichment analysis of the TFBSs. Four new loci were enriched in E2F1 (P = 2.64×10−4; which regulates p53-dependent/independent apoptosis (35)) TFBSs, as did 21 other SLE loci (Supplementary Material, Table S7A). p53 itself is the second-most enriched TFBS. Sites for B-/T-cell apoptotic factors RELA, MEF2A (36), PU.1/SPI-1 (37), BCL6 (38) and ETS1 (39) were similarly enriched (Supplementary Material, Table S7B), as was glucocorticoid receptor NR3C1. Taken together, these results highlight the importance of apoptosis (particularly for B- and T-cells) and glucocorticoid signaling in the pathogenesis of SLE.
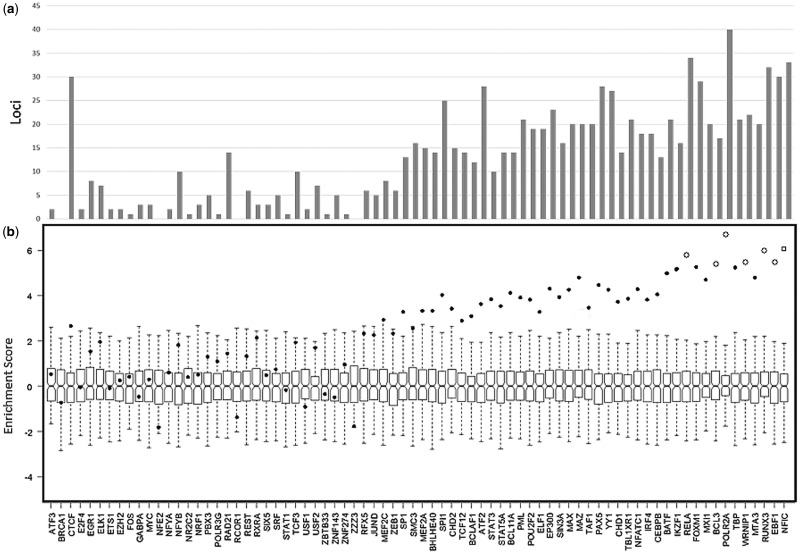
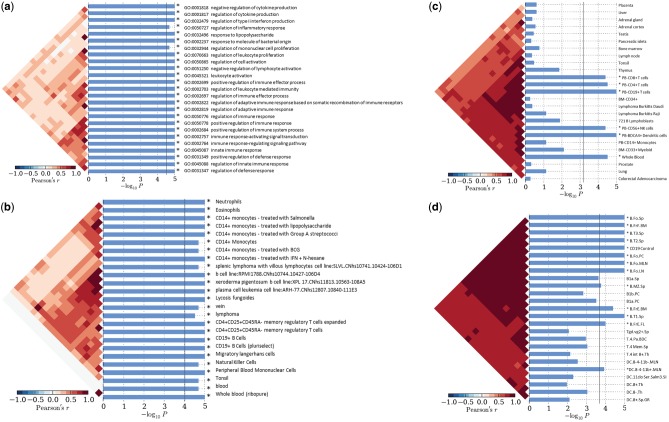
In order to understand how SLE loci fit together in the context of function and cell-specific expression, we performed enrichment analysis (40). In our cell-specific enrichment analyses, the expanded set of SLE loci was enriched in 32 ontology categories; including GO:0001817 (P = 1.00×10−5; regulation of cytokine production) and GO:0002252 (P = 2.00×10−5; immune effector processes); CCL22 and ATG16L2 both contribute (Fig. 3A). Additionally, SLE gene expression was enriched in dendritic cells (P = 3.0×10−5) and lymphoblasts (P = 6.5×10−4), and in migratory Langerhans cells (P = 1.0×10−5) (Fig. 3B and C). B-cells (P = 1×10−5) and dendritic cells (P = 1.20×10−4) were also enriched in SLE gene expression in a mouse model (ImmVar) (Fig. 3D). These results corroborate previously observed immune cell expression patterns (2,41).
Figure 3.
Cell-specific enrichment was carried out in SNPsea. Enrichment P-value is presented as –log10P, significantly enriched categories pass the multiple testing threshold (vertical line). Correlation between categories is presented to the left of each plot. (a) Gene ontology; (b) FANTOM5; (c) Gene Atlas; (d) ImmVar.
We did not find significant heterogeneity of odds-ratios at most of the risk loci between Asian and European cohorts, consistent with published results (5,42). To understand the contribution of the loci to SLE inheritance, we estimated the percentage of explained heritability of 78 loci (four loci were monomorphic in Asians). The proportion of explainable heritability increased from 26 to 28% (Supplementary Material, Table S8).
Discussion
In this study, we aimed not only to replicate five novel SLE loci, but also to identify common regulatory modes of these associated variants together with 77 previously identified SLE GWAS loci. We deployed a series of bioinformatic analyses to identify transcriptional modes of regulation, and to cluster SLE loci by function and expression profiles.
We replicated five previously identified suggestive SLE loci in eight independent Asian cohorts and one European cohort. As a result, we increased the total number of SLE susceptibility loci to 82. One locus (rs11235604 in ATG16L2) alters the protein sequence, while all loci modulate transcription and/or gene expression. As with other reported inherited autoimmune diseases, three of the new loci are located in intergenic regions of the genome. The genetic function of the loci identified in this and other studies can be partly explained by epigenetic changes in gene activation and expression, e.g. TFBSs, enhancer activity, and eQTL effects on other genes.
Recently, Morris et al. (5) identified rs494003 as an SLE susceptibility locus tagging RNASEH2C; we localized the strongest signal to rs1308020, 9.1 kb 5' of RNASEH2C and 44.7 kb from rs494003. The SNPs are uncorrelated (D'= 0.99, r2 = 0.05 in East Asians and D'= 0.99, r2 = 0.14 in Western Europeans) and independently associated with SLE. rs494003 actually lies in the 3'-UTR region of AP5B1 (43) and should be considered a separate signal from RNASEH2C.
Three of the five novel loci we identified have known immune functions, and have been previously associated with autoimmune disease. CCL22 is a cytokine associated with RA (15) and MS (13). ATG16L2 is involved in apoptosis and physically interacts with SLE locus ATG5. MYNN has been previously linked to celiac disease susceptibility through immune cell gene expression differences (44).
There have been several genome-wide association studies together reporting 77 SLE susceptibility loci (2–4,45) (Supplementary Material, Table S8). Few loci have identified coding variants. Most identified loci have predicted effects on gene regulation through non-coding epigenetic effects (Supplementary Material, Table S4) such as enhancers, promoters or eQTLs. The five loci we replicated here show similar modes of gene regulation. Moreover, we determined that 51 of the 82 loci affected gene expression of 115 non-SLE genes, which could be tested further for candidate loci for SLE (Supplementary Material, Table S1A–C). Expression of these 82 SLE loci is enriched in immune cell types (33 enriched cell lines), especially in various B-cell types (e.g. CD19+), consistent with our previous (2) observations (Fig. 3B).
Our novel loci also share pathway membership, gene ontology and cell-type expression patterns with other SLE loci, such as CCL22 mentioned above. In fact, pathways identified in this study, which contain multiple known SLE loci, could be used as follow-up targets for further studies. Perhaps most interesting is apoptosis, which has been previously reported as a model for SLE pathogenesis (46).
We determined in our enrichment analyses that the majority of our SLE loci are enriched in enhancer and promoter marks (H3K27Ac and H3K4me3), as well as in immune response gene ontologies. These characteristics, shared among SLE susceptibility loci, identify SLE regulatory signatures that will aid in understanding disease pathobiology and identifying further candidate loci. Consistent with our epigenetic mark enrichment analyses, we identified multiple TFs linked to SLE (e.g. EBF1, TBP, BCL3, MXI1, RELA and IKZF1). The most strongly enriched TFs highlight the link of T-/B-cell biology to SLE, consistent with our allele-specific analysis.
In our analysis, we identified multiple regulatory commonalities among SLE loci. These susceptibility loci are located primarily in epigenetic regulatory regions that favor enhancers. Further studies into SLE loci would benefit greatly from investigating all enhancer targets for these loci, especially if they share membership with SLE loci in immune-related pathways.
Interestingly, in four of the five new loci (except CCL22), the highly conserved ancestral alleles increase SLE risk. Furthermore, in four of five loci (except ATG16L2, where the protective allele is extremely rare), risk allele frequency in European populations is significantly different from that in Asians. Thus, it seems that Asian populations are experiencing different selective pressure at these immune loci from Europeans.
It is important to experimentally validate predicted loci and SNPs when possible. As we have done before (47), testing of promoter/enhancer fragments (differing only at the predicted risk/protective SNP) in reporter assays can reveal profound differences in activity. Such results can be explored further with electrophoretic mobility shift assays (EMSAs) (47), bound proteins in shifted bands can be identified by mass spectrometry, and finally bands can be ‘super-shifted’ with antibodies to bound proteins. Such experiments can definitively identify SNPs ‘causal’ for association signals. The associated SNPs are predicted to disrupt many conserved enhancers and TFBSs; it is likely that reporter assays could reveal function at many of these sites. It is intriguing to consider the possibility that the enriched, pro-apoptotic TFs could be common elements in the activity of the associated enhancers and the effects of the SNPs.
In conclusion, we have added five novel loci of SLE susceptibility that share common functional traits with currently established loci, bringing the total to 82. In our analysis we were able to identify common signatures of gene regulation through variants that directly affect expression, as well as variants that disrupt the binding of key transcription factors. Among enriched TFBSs, many have well-documented immune-related roles in apoptosis of B- and T-cells, including p53, E2F1, MEF2A, PU.1, BCL6, ETS1 and RELA. With this evidence, we have increased current knowledge of the drivers of SLE susceptibility, and have identified pathways critical to SLE pathogenesis.
Materials and Methods
Samples and available data
The data used in this study came from three sources. In the first set, we recruited 1,456 SLE patients from five hospitals in Hong Kong: Queen Mary Hospital, Tuen Mun Hospital, Queen Elizabeth Hospital, Pamela Youde Nethersole Eastern Hospital, and Princess Margaret Hospital. The 955 controls were healthy blood donors collected from the Hong Kong Red Cross. All recruited patients and controls filled an informed consent form and the use of their data was approved by the Institutional Review Board of the involved institutes. We genotyped five SNPs (rs10936599, rs11235604, rs223881, rs2762340 and rs1308020) on all 2,411 individuals (1,456 cases and 955 controls) using the TaqMan assay.
The second set of data (in silico replication) for 5,057 Han Chinese (1,659 cases and 3,398 controls) and 11,022 Europeans (4,063 cases and 6,959 controls) was obtained as summary statistics from Morris etal. (5). The third set of data was obtained from our previously published study of the SLE association in Asians (2), and included 17,140 individuals (4,478 cases and 12,656 controls) from six East Asian cohorts (Table 2).
Patients in all the cohorts used in this study fulfilled at least four of 11 American College of Rheumatology classification criteria for SLE (48). Patient, control recruitment and sample collection were done in compliance with the Institutional Review Board of both institutions. All individuals provided informed consent.
Meta-analysis of all nine cohorts was performed in METAL (49), adjusting for the effects of each cohort sample size. Confidence intervals for meta-analysis were estimated using the standard error model.
Independent SNP analysis
In our previous study, we performed conditional analysis to identify independent SNPs within two SLE associated regions (PCNXL3 and DEF6). Our approach used a logistic regression model choosing the strongest associated SNP as an iterative process starting with the strongest SNP and adding additional SNPs to the model until no more variants passed the P < 5×10−5 threshold of significance (2).
In order to confirm the results from conditional analysis, we performed a stepwise model selection (implemented in GCTA (50)) on the summary statistics of our previous study. We used genotypes for 207 Han Chinese and Japanese from the 1000Genomes Project v3 as the reference panel for the estimation of linkage disequilibrium. Hardy-Weinberg equilibrium (HWE) and allelic association were evaluated using PLINK (51).
Meta-analysis
We used METAL (49) to perform sample size-corrected meta-analysis with nine cohorts (Table 2). We included a total of 7,595 cases and 17,013 controls of Asian descent, and 4,063 cases and 6,959 controls of European descent. We estimated I2 and its associated P-value as a measure of the heterogeneity among the effect sizes.
SNP-based bioinformatics analysis
The expanded set of 82 SLE loci and their correlated SNPs (r2 > 0.8) were annotated for co-location of epigenetic features and SNP enhancer enrichment using Haploreg (52).
To assess if the candidate loci in this study alter gene expression, we queried previously published information from BloodeQTL (22). BloodeQTL is a study of expression quantitative loci (eQTLs) performed on non-transformed peripheral blood from 8,086 European patients. In order to account for unobserved variants correlated with the candidate SNP, we expanded our search to variants with linkage disequilibrium (LD) r2 > 0.8 in Asian populations (HapMap CHB, Han Chinese; and JPT, Japanese). To identify if the five novel loci were differentially expressed in a given tissue, we used the Genotype-Tissue Expression (GTEx) database, which establishes relationships between SNPs and gene expression in 52 different tissues (53).
To identify functional or regulatory effects of our candidate SNPs, we performed a series of bioinformatic analyses, including annotating the candidate variants (as well as their correlated SNPs r2 > 0.8), for transcription factor binding sites (TFBSs). Tested TFBSs were obtained from the Factorbook (54), Homer (55) and Hocomoco (56) databases. Allele-specific changes in TFBS affinity were scored with the MotifbreakR (57) library in R. Strongly disrupted TFBSs are presented.
In order to identify cell type-specific enrichment in gene expression, we used normalized expression data obtained from the GeneAtlas (58), ImmVar (59) and FANTOM (60) databases, as well as the GeneOntology Atlas, as implemented in SNPSEA (40). Altogether, we assessed enrichment of gene expression in over 500 cell lines for all 82 SLE loci.
In order to assess the regulatory features tagged by identified susceptibility loci, we performed variant set enrichment (VSE) (61) on the expanded set of 82 loci, together with all strongly correlated SNPs (r2 > 0.8, and within ±500 bp) in East Asian 1000Genomes populations (CHB, CHD, JPT and KHV), as well as in the combination of African (ACB, ASW, ESN, GWD, LWK, MSL and YRI) and European (CEU, FIN, GBR, IBS and TSI) 1000Genomes populations. The expanded set of SNPs in LD with our 82 loci was obtained through the rAggr database. The East Asian LD expanded set contained 2,096 SNPs, while the combined population LD set contained 4,992 SNPs. With these variant sets, we expect to capture ‘true’ functional variants tagged by SLE susceptibility loci. We tested VSE of transcription factor binding sites (TFBSs) for both SNP sets. Since the East Asian SNP set did not contain all SLE loci it was left out of this analysis. TFBS data used for VSE were obtained from the ChIP-Seq database version 3 from the ENCODE Project (wgEncodeRegTfbsClusteredV3). We focused on cell line GM12878 and 76 TFs. VSE was implemented in the R package VSE, and used 200 random SNP sets to set the expected distribution of TF sites.
To further understand regulation within SLE loci, we annotated each locus (as well as all highly correlated SNPs: r2 > 0.8 in CHB + JPT) for the presence of epigenetic marks (Histone marks [H3K27Ac, H3K4me3]; DNAse I hypersensitivity sites; and enhancer marks) using Enlight (62) and rSNPbase (63).
We performed an enrichment analysis on the expanded set of SLE correlated SNPs to identify the cell lines enriched in enhancers as annotated by the core 15-state model from ChromHMM, and by peaks from H3K27ac and H3K9ac (as enhancers and promoters), as implemented in Haploreg (52).
Proportion of heritability was estimated for the 78 non-monomorphic (in Asians) loci using the variance in a liability model explained in So and Sham (64).
Gene set-based bioinformatics analysis
In order to further assess changes in gene expression related to our candidate loci, we performed a gene-based association test of the imputed expression profiles (11,553 autosomal genes) in whole blood. By doing this, we expected to identify expression changes related to our candidate loci with greater precision. For this purpose, we used MetaXcan, an extension of PrediXcan (65), to impute the expression profile of GTEx whole-blood expression data on the summary statistics of our Korean samples, since it was the largest cohort. Genes within 1 Mb of each SLE locus were included to account for neighboring gene involvement.
We carried out enrichment analysis and over-representation analysis using the expanded set of 82 known SLE loci (as gene sets) to identify genetic signatures of SLE loci, through shared regulation of function, or shared functional group membership. Enrichment analysis was implemented in the Enrichr tool (66), which includes analysis within pathway, transcription factor binding, and gene ontology databases. Over-representation analysis was performed using ConsensuspathDB (67), a tool that also aggregates multiple databases of pathways, gene ontology and protein complex databases. Confirmatory enrichment analysis was performed using the GREAT database (68), PANTHER (69) and gPROFILER (70).
URLs: ENCODE project https://genome.ucsc.edu/ENCODE/; VSE https://cran.r-project.org/web/packages/VSE; rAggr http://raggr.usc.edu; BloodeQTL http://genenetwork.nl/bloodeqtlbrowser/; gPROFILER http://biit.cs.ut.ee/gprofiler/; Enlight http://enlight.usc.edu/; rSNPbase http://rsnp.psych.ac.cn/; Enrichr http://amp.pharm.mssm.edu/Enrichr/; GREAT great.stanford.edu/; PANTHER http://pantherdb.org/; MetaXcan https://github.com/hakyimlab/MetaXcan; www.insidegen.com; date last accessed August 15, 2016.
All websites were last accessed on January 24, 2017.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank affected and unaffected individuals who participated in this study; as well as research assistants, coordinators and physicians who helped in the recruitment of subjects, including the individuals in the coordinating projects.
Conflict of Interest statement. None declared.
Funding
The US National Institutes of Health (AR060366, MD007909, AI107176), a grant from the Korea Healthcare Technology R&D Project (HI15C2958), Ministry for Health and Welfare, Republic of Korea, and Basic Science Research Program (NRF-2015R1 C1A1A02036527) through the National Research Foundation of Korea (NRF), Ministry of Education, Republic of Korea.
References
- 1. Alarcon G.S., McGwin G. Jr., Petri M., Reveille J.D., Ramsey-Goldman R., Kimberly R.P. and PROFILE Study Group (2002) Baseline characteristics of a multiethnic lupus cohort: PROFILE. Lupus, 11, 95–101. [DOI] [PubMed] [Google Scholar]
- 2. Sun C., Molineros J.E., Looger L.L., Zhou X.J., Kim K., Okada Y., Ma J., Qi Y.Y., Kim-Howard X., Motghare P., et al. (2016) High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet., 48, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. International Consortium for Systemic Lupus Erythematosus, G., Harley J.B., Alarcon-Riquelme M.E., Criswell L.A., Jacob C.O., Kimberly R.P., Moser K.L., Tsao B.P., Vyse T.J., Langefeld C.D., et al. (2008) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet., 40, 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bentham J., Morris D.L., Cunninghame Graham D.S., Pinder C.L., Tombleson P., Behrens T.W., Martin J., Fairfax B.P., Knight J.C., Chen L., et al. (2015) Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet., 47, 1457–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morris D.L., Sheng Y., Zhang Y., Wang Y.F., Zhu Z., Tombleson P., Chen L., Cunninghame Graham D.S., Bentham J., Roberts A.L., et al. (2016) Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet., 48,940–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cifuentes-Diaz C., Bitoun M., Goudou D., Seddiqi N., Romero N., Rieger F., Perin J.P., Alliel P.M. (2004) Neuromuscular expression of the BTB/POZ and zinc finger protein myoneurin. Muscle Nerve, 29, 59–65. [DOI] [PubMed] [Google Scholar]
- 7. Do S.K., Yoo S.S., Choi Y.Y., Choi J.E., Jeon H.S., Lee W.K., Lee S.Y., Lee J., Cha S.I., Kim C.H., et al. (2015) Replication of the results of genome-wide and candidate gene association studies on telomere length in a Korean population. Korean J. Intern. Med., 30, 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lessard C.J., Sajuthi S., Zhao J., Kim K., Ice J.A., Li H., Ainsworth H., Rasmussen A., Kelly J.A., Marion M., et al. (2016) Identification of a Systemic Lupus Erythematosus Risk Locus Spanning ATG16L2, FCHSD2, and P2RY2 in Koreans. Arthritis Rheumatol., 68, 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Igci M., Baysan M., Yigiter R., Ulasli M., Geyik S., Bayraktar R., Bozgeyik I., Bozgeyik E., Bayram A., Cakmak E.A. (2016) Gene expression profiles of autophagy-related genes in multiple sclerosis. Gene, 588, 38–46. [DOI] [PubMed] [Google Scholar]
- 10. Yang S.K., Hong M., Zhao W., Jung Y., Baek J., Tayebi N., Kim K.M., Ye B.D., Kim K.J., Park S.H., et al. (2014) Genome-wide association study of Crohn's disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut, 63, 80–87. [DOI] [PubMed] [Google Scholar]
- 11. Godiska R., Chantry D., Raport C.J., Sozzani S., Allavena P., Leviten D., Mantovani A., Gray P.W. (1997) Human macrophage-derived chemokine (MDC), a novel chemoattractant for monocytes, monocyte-derived dendritic cells, and natural killer cells. J. Exp. Med., 185, 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Safa A., Rashidinejad H.R., Khalili M., Dabiri S., Nemati M., Mohammadi M.M., Jafarzadeh A. (2016) Higher circulating levels of chemokines CXCL10, CCL20 and CCL22 in patients with ischemic heart disease. Cytokine, 83, 147–157. [DOI] [PubMed] [Google Scholar]
- 13. Galimberti D., Fenoglio C., Comi C., Scalabrini D., De Riz M., Leone M., Venturelli E., Cortini F., Piola M., Monaco F., et al. (2008) MDC/CCL22 intrathecal levels in patients with multiple sclerosis. Mult. Scler., 14, 547–549. [DOI] [PubMed] [Google Scholar]
- 14. Oo Y.H., Weston C.J., Lalor P.F., Curbishley S.M., Withers D.R., Reynolds G.M., Shetty S., Harki J., Shaw J.C., Eksteen B., et al. (2010) Distinct roles for CCR4 and CXCR3 in the recruitment and positioning of regulatory T cells in the inflamed human liver. J. Immunol., 184, 2886–2898. [DOI] [PubMed] [Google Scholar]
- 15. Flytlie H.A., Hvid M., Lindgreen E., Kofod-Olsen E., Petersen E.L., Jorgensen A., Deleuran M., Vestergaard C., Deleuran B. (2010) Expression of MDC/CCL22 and its receptor CCR4 in rheumatoid arthritis, psoriatic arthritis and osteoarthritis. Cytokine, 49, 24–29. [DOI] [PubMed] [Google Scholar]
- 16. Crow Y.J., Chase D.S., Lowenstein Schmidt J., Szynkiewicz M., Forte G.M., Gornall H.L., Oojageer A., Anderson B., Pizzino A., Helman G., et al. (2015) Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. A, 167A, 296–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., et al. (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat. Genet., 38, 910–916. [DOI] [PubMed] [Google Scholar]
- 18. Lee Y., Yoon K.A., Joo J., Lee D., Bae K., Han J.Y., Lee J.S. (2013) Prognostic implications of genetic variants in advanced non-small cell lung cancer: a genome-wide association study. Carcinogenesis, 34, 307–313. [DOI] [PubMed] [Google Scholar]
- 19. Pandey A., Blagoev B., Kratchmarova I., Fernandez M., Nielsen M., Kristiansen T.Z., Ohara O., Podtelejnikov A.V., Roche S., Lodish H.F., et al. (2002) Cloning of a novel phosphotyrosine binding domain containing molecule, Odin, involved in signaling by receptor tyrosine kinases. Oncogene, 21, 8029–8036. [DOI] [PubMed] [Google Scholar]
- 20. Emaduddin M., Edelmann M.J., Kessler B.M., Feller S.M. (2008) Odin (ANKS1A) is a Src family kinase target in colorectal cancer cells. Cell Commun. Signal, 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kariuki S.N., Franek B.S., Kumar A.A., Arrington J., Mikolaitis R.A., Utset T.O., Jolly M., Crow M.K., Skol A.D., Niewold T.B. (2010) Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis Res. Ther., 12, R151.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet., 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mele M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., et al. (2015) Human genomics. The human transcriptome across tissues and individuals. Science, 348, 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katagiri K., Hattori M., Minato N., Kinashi T. (2002) Rap1 functions as a key regulator of T-cell and antigen-presenting cell interactions and modulates T-cell responses. Mol. Cell Biol., 22, 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meshaal S., El Refai R., El Saie A., El Hawary R. (2016) Signal transducer and activator of transcription 5 is implicated in disease activity in adult and juvenile onset systemic lupus erythematosus. Clin. Rheumatol., 35, 1515–1520. [DOI] [PubMed] [Google Scholar]
- 26. Goropevsek A., Holcar M., Avcin T. (2016) The role of STAT signaling pathways in the pathogenesis of systemic Lupus Erythematosus. Clin. Rev. Allergy Immunol., in press. [DOI] [PubMed] [Google Scholar]
- 27. Tsuruyama T., Hiratsuka T., Wulamujiang A., Nakamura T. (2016) STAT5A Modulates Chemokine Receptor CCR6 Expression and Enhances Pre-B Cell Growth in a CCL20-Dependent Manner. J. Cell Biochem., 117,2630–2642. [DOI] [PubMed] [Google Scholar]
- 28. Yang W., Shen N., Ye D.Q., Liu Q., Zhang Y., Qian X.X., Hirankarn N., Ying D., Pan H.F., Mok C.C., et al. (2010) Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet., 6, e1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wong H.K., Kammer G.M., Dennis G., Tsokos G.C. (1999) Abnormal NF-kappa B activity in T lymphocytes from patients with systemic lupus erythematosus is associated with decreased p65-RelA protein expression. J. Immunol., 163, 1682–1689. [PubMed] [Google Scholar]
- 30. Sharma A., Liu X., Hadley D., Hagopian W., Liu E., Chen W.M., Onengut-Gumuscu S., Simell V., Rewers M., Ziegler A.G., et al. (2016) Identification of Non-HLA Genes Associated with Celiac Disease and Country-Specific Differences in a Large, International Pediatric Cohort. PLoS One, 11, e0152476.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Speedy H.E., Di Bernardo M.C., Sava G.P., Dyer M.J., Holroyd A., Wang Y., Sunter N.J., Mansouri L., Juliusson G., Smedby K.E., et al. (2014) A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat. Genet., 46, 56–60. [DOI] [PubMed] [Google Scholar]
- 32. Maiti A.K., Kim-Howard X., Motghare P., Pradhan V., Chua K.H., Sun C., Arango-Guerrero M.T., Ghosh K., Niewold T.B., Harley J.B., et al. (2014) Combined protein- and nucleic acid-level effects of rs1143679 (R77H), a lupus-predisposing variant within ITGAM. Hum. Mol. Genet., 23, 4161–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo S., Liu Y., Liang G., Zhao M., Wu H., Liang Y., Qiu X., Tan Y., Dai Y., Yung S., et al. (2015) The role of microRNA-1246 in the regulation of B cell activation and the pathogenesis of systemic lupus erythematosus. Clin Epigenetics, 7, 24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chauhan R., Handa R., Das T.P., Pati U. (2004) Over-expression of TATA binding protein (TBP) and p53 and autoantibodies to these antigens are features of systemic sclerosis, systemic lupus erythematosus and overlap syndromes. Clin. Exp. Immunol., 136, 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Korotayev K., Ginsberg D. (2008) Many pathways to apoptosis: E2F1 regulates splicing of apoptotic genes. Cell Death Differ., 15, 1813–1814. [DOI] [PubMed] [Google Scholar]
- 36. Wang M., Windgassen D., Papoutsakis E.T. (2008) A global transcriptional view of apoptosis in human T-cell activation. BMC Med. Genomics, 1, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamada T., Kondoh N., Matsumoto M., Yoshida M., Maekawa A., Oikawa T. (1997) Overexpression of PU.1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood, 89, 1383–1393. [PubMed] [Google Scholar]
- 38. Phan R.T., Dalla-Favera R. (2004) The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature, 432, 635–639. [DOI] [PubMed] [Google Scholar]
- 39. Bories J.C., Willerford D.M., Grevin D., Davidson L., Camus A., Martin P., Stehelin D., Alt F.W. (1995) Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature, 377, 635–638. [DOI] [PubMed] [Google Scholar]
- 40. Slowikowski K., Hu X., Raychaudhuri S. (2014) SNPsea: an algorithm to identify cell types, tissues and pathways affected by risk loci. Bioinformatics, 30, 2496–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu L., Yin X., Wen L., Yang C., Sheng Y., Lin Y., Zhu Z., Shen C., Shi Y., Zheng Y., et al. (2016) Several Critical Cell Types, Tissues, and Pathways Are Implicated in Genome-Wide Association Studies for Systemic Lupus Erythematosus. G3 (Bethesda), 6, 1503–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang C., Ahlford A., Jarvinen T.M., Nordmark G., Eloranta M.L., Gunnarsson I., Svenungsson E., Padyukov L., Sturfelt G., Jonsen A., et al. (2013) Genes identified in Asian SLE GWASs are also associated with SLE in Caucasian populations. Eur. J. Hum. Genet., 21, 994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zuo X., Sun L., Yin X., Gao J., Sheng Y., Xu J., Zhang J., He C., Qiu Y., Wen G., et al. (2015) Whole-exome SNP array identifies 15 new susceptibility loci for psoriasis. Nat. Commun., 6, 6793.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dubois P.C., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A., Adany R., Aromaa A., et al. (2010) Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet., 42, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Okada Y., Shimane K., Kochi Y., Tahira T., Suzuki A., Higasa K., Takahashi A., Horita T., Atsumi T., Ishii T., et al. (2012) A genome-wide association study identified AFF1 as a susceptibility locus for systemic lupus eyrthematosus in Japanese. PLoS Genet., 8, e1002455.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mahajan A., Herrmann M., Munoz L.E. (2016) Clearance Deficiency and Cell Death Pathways: A Model for the Pathogenesis of SLE. Front. Immunol., 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Molineros J.E., Maiti A.K., Sun C., Looger L.L., Han S., Kim-Howard X., Glenn S., Adler A., Kelly J.A., Niewold T.B., et al. (2013) Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS Genet., 9, e1003222.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hochberg M.C. (1997) Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum., 40, 1725.. [DOI] [PubMed] [Google Scholar]
- 49. Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang J., Ferreira T., Morris A.P., Medland S.E., Genetic Investigation of A.T.C., Replication D.I.G., Meta-analysis C., Madden P.A., Heath A.C., Martin N.G., et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet., 44, 369–375. S361-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ward L.D., Kellis M. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Consortium G.T. (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang J., Zhuang J., Iyer S., Lin X., Whitfield T.W., Greven M.C., Pierce B.G., Dong X., Kundaje A., Cheng Y., et al. (2012) Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res., 22, 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kulakovskiy I.V., Medvedeva Y.A., Schaefer U., Kasianov A.S., Vorontsov I.E., Bajic V.B., Makeev V.J. (2013) HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res., 41, D195–D202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Coetzee S.G., Coetzee G.A., Hazelett D.J. (2015) motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics, 31, 3847–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. U S A., 101, 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hyatt G., Melamed R., Park R., Seguritan R., Laplace C., Poirot L., Zucchelli S., Obst R., Matos M., Venanzi E., et al. (2006) Gene expression microarrays: glimpses of the immunological genome. Nat. Immunol., 7, 686–691. [DOI] [PubMed] [Google Scholar]
- 60. Lizio M., Harshbarger J., Shimoji H., Severin J., Kasukawa T., Sahin S., Abugessaisa I., Fukuda S., Hori F., Ishikawa-Kato S., et al. (2015) Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol., 16, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cowper-Sal lari R., Zhang X., Wright J.B., Bailey S.D., Cole M.D., Eeckhoute J., Moore J.H., Lupien M. (2012) Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet., 44, 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guo Y., Conti D.V., Wang K. (2015) Enlight: web-based integration of GWAS results with biological annotations. Bioinformatics, 31, 275–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guo L., Du Y., Chang S., Zhang K., Wang J. (2014) rSNPBase: a database for curated regulatory SNPs. Nucleic Acids Res., 42, D1033–103D1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. So H.C., Gui A.H., Cherny S.S., Sham P.C. (2011) Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet. Epidemiol., 35, 310–317. [DOI] [PubMed] [Google Scholar]
- 65. Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Consortium G.T., Nicolae D.L., et al. (2015) A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet., 47, 1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 14, 128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kamburov A., Wierling C., Lehrach H., Herwig R. (2009) ConsensusPathDB–a database for integrating human functional interaction networks. Nucleic Acids Res., 37, D623–D628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol., 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D. (2016) PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res., 44, D336–3D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H., Vilo J. (2016) g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res., 44 (W1), W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.