Abstract
Background: The mammalian gut microbiota is a highly abundant and diverse microbial community that resides in the gastrointestinal tract. One major benefit that the gut microbiota provides to its host is colonization resistance—the ability to prevent colonization by foreign microbes, including diarrheal pathogens such as Clostridium difficile, Salmonella enterica serovar Typhimurium and diarrheagenic Escherichia coli.
Methods: We conducted a literature review of the effects of the gut microbiota on infection by diarrheal pathogens. We used PubMed to search for relevant articles published before July 2016, as well as incorporated data from our laboratory.
Results: The gut microbiota provides protection from diarrheal infections both by direct inhibition of pathogens and by indirect effects on host functions. Direct effects of the microbiota on diarrheal pathogens include competing for nutrients and producing metabolites that inhibit pathogen growth or virulence. Indirect effects of the gut microbiota include promoting maintenance of the gut mucosal barrier and stimulating innate and adaptive immunity.
Conclusions: Human epidemiological studies and experimental infections of laboratory animals both demonstrate that antibiotic treatment can alter the gut microbial community and thereby reduce colonization resistance against diarrheal pathogens. Further research might lead to the development of next-generation probiotics that could be used to bolster colonization resistance and thus prevent travellers’ diarrheal.
Introduction
Keywords: gut microbiota, antibiotics, bacterial pathogens, diarrhea
All animals that have a gastrointestinal tract also have a gut microbiota— the community of microbes (bacteria, archaea, viruses and eukaryotic microbes) that inhabits the gut. These microbes are highly abundant; a recent estimate suggests that the ratio of bacterial cells to human cells in or on the human body is ∼1:1, with the vast majority of bacteria residing in the colon.1 Both the density and the diversity of microbes increases from the proximal to the distal end of the gastrointestinal tract, shaped by the concentration of such factors as stomach acid, nutrients, oxygen and antimicrobial peptides.2 Due to its long co-evolution with its host species, the gut microbiota influences numerous aspects of host physiology including digestion, metabolism, immunity and even behaviour.3 In this review, we describe a key role of the gut microbiota: providing the host with colonization resistance, which is the ability to prevent incursion of foreign microbes (including but not limited to pathogens) into the host gut.4,5 In particular, we will focus on how the gut microbiota protects its host from the well-studied diarrheal pathogens Clostridium difficile, Salmonella enterica and diarrheagenic Escherichia coli.
Evidence that the Gut Microbiota Protects against Infection
Strong evidence that a healthy gut microbiota provides colonization resistance against diarrheal pathogens comes from studies of germ-free animals (which completely lack a gut microbiota) and antibiotic-treated animals (whose gut microbial community has been disrupted) (Yurist-Doutsch et al.5 and Keeney et al.6 and references therein).
Germ-free animals are extremely susceptible to diarrheal infections, including those caused by S. enterica serovars Typhimurium and Enteritidis— both of which cause gastroenteritis in humans— and Citrobacter rodentium, a natural mouse pathogen that is used as a model for the human-specific intestinal pathogens enterohemorrhagic E. coli (EHEC) and enteropathogenic E. coli (EPEC). In the case of Salmonella, oral infection of germ-free mice with as few as 10 colony-forming units (CFU) causes a lethal infection, whereas doses of 103 –109 CFU are required to kill 50% of mice with an intact gut microbiota.7,8 Specific pathogen-free (SPF) mice (which possess a gut microbiota) infected with C. rodentium develop self-limiting colitis, with clearance of the pathogen three to 4 weeks post-infection. While C. rodentium infections are not lethal in germ-free mice, the animals remain heavily colonized for at least 6 weeks post-infection (the maximum infection length assessed in the study), demonstrating that the gut microbiota is essential for clearance of this pathogen.9 Since germ-free animals possess immature immune responses,10 it is likely that the increased susceptibility of these animals to diarrheal pathogens results from both defects in the host response to infection and a lack of ecological competition from commensal microbes.
Studies with antibiotic-treated animals have demonstrated that the gut microbiota provides protection against diarrheal infections even in immune-competent hosts. In SPF mice untreated with antibiotics, S. enterica serovar Typhimurium causes a systemic disease similar to human typhoid fever, with little Salmonella colonization in the gut. However, treating these animals with streptomycin 24 h prior to infection leads to increased Salmonella colonization and inflammation in the large intestine and increased faecal shedding of Salmonella relative to untreated animals.11 Antibiotic-treated mice remain susceptible to Salmonella gastroenteritis up to 21 days post-withdrawal of antibiotics, even though the total number of microbiota in the gut has returned to normal levels by this time.12 Susceptibility to Salmonella gut colonization can be induced by multiple antibiotics, including streptomycin, vancomycin and metronidazole, even though these antibiotics have differing effects on the composition of the microbiota community,13 suggesting that it is a disruption of the stable gut community rather than changes in abundance of specific organisms that enables Salmonella to colonize the mouse gut. Antibiotic treatment is known to affect the metabolic activities of the microbiota14; the resulting increase or decrease in abundance of certain host or bacterial metabolites could be one of the factors underlying the loss of colonization resistance. The relevance of these mouse studies to human disease is underscored by epidemiological evidence that humans are also more susceptible to Salmonella gastroenteritis following antibiotic therapy.15
Perhaps the best-known antibiotic-associated pathogen in humans is C. difficile, the majority of cases of which are associated with previous antibiotic treatment, particularly fluoroquinolones, penicillins, cephalosporins and clindamycin.16 These effects can be recapitulated in mice; oral treatment of mice with cefoperazone induces susceptibility to C. difficile-mediated disease 2 days post-treatment.17 Mice that are treated with both cefoperazone and clindamycin remain susceptible to C. difficile for up to 6 weeks after antibiotic treatment is ceased.17 As further evidence that C. difficile infection requires a breach in colonization resistance, recurrent C. difficile infections in humans have successfully been treated by restoring microbial communities using either stool transplants or synthetic bacterial communities, thereby reestablishing colonization resistance.18,19
Although antibiotic treatment induces susceptibility to numerous diarrheal pathogens in mice, it is important to note that the effects of antibiotics can be pathogen-specific. For example, as described above, streptomycin pretreatment increases Salmonella colonization, leading to inflammation in the large intestine,11 while streptomycin treatment does not affect C. rodentium colonization or gut inflammation at 6 days post-infection.20 In contrast, metronidazole pretreatment increases Salmonella gut colonization without increasing Salmonella-mediated inflammation,13 which may be related to the direct anti-inflammatory effects of metronidazole21; however, the same antibiotic increases C. rodentium-induced colitis without affecting C. rodentium gut colonization.20 Although it is clear that the gut microbiota provides protection from numerous diarrheal infections, these studies illustrate the difficulties in predicting what effect antibiotic disruption of the microbiota may have on host susceptibility to specific infections.
Although much is known about how the microbiota affects susceptibility to diarrheal pathogens such as Salmonella and C. difficile, less is known about its effects on common causes of travellers’ diarrhea, such as enterotoxigenic E. coli (ETEC). However, the necessity of streptomycin pretreatment in a mouse model of ETEC infection22 strongly suggests that the gut microbiota provides colonization resistance against this pathogen similarly to those described above. In addition, a recent study examining the microbiota of human volunteers before ETEC infection identified a biomarker panel of 32 microbiota taxa that could predict with 76% accuracy whether the subject would develop diarrhea after ETEC exposure,23 suggesting that the composition of the gut microbiota has a major effect on ETEC’s ability to cause disease in humans.
Even more poorly understood is the interaction between the gut microbiota and non-bacterial enteric pathogens. In the case of the protozoal parasite Giardia lamblia, the gut microbiota appears to play a protective role. Mice purchased from two different suppliers differ in their susceptibility to G. lamblia infection; however, the normally susceptible mice can be made resistant to G. lamblia via co-housing with the resistant mice.24 Since this resistance to G. lamblia infection can be abolished by treating the mice with the antibiotic neomycin,24 resistance is likely due to transfer of microbiota from the resistant to the susceptible mice. Interestingly, some studies indicate that the gut microbiota might facilitate, rather than inhibit, norovirus infection. Antibiotic-treated mice infected with murine norovirus have a reduced viral titer compared with untreated mice, and certain gut microbes, including Enterobacter cloacae, stimulate the ability of human norovirus to infect human B cells in vitro.25 However, it remains unclear whether differences in microbiota composition between humans might predispose or protect individuals from norovirus infection.
Mechanisms of Microbiota-Mediated Protection
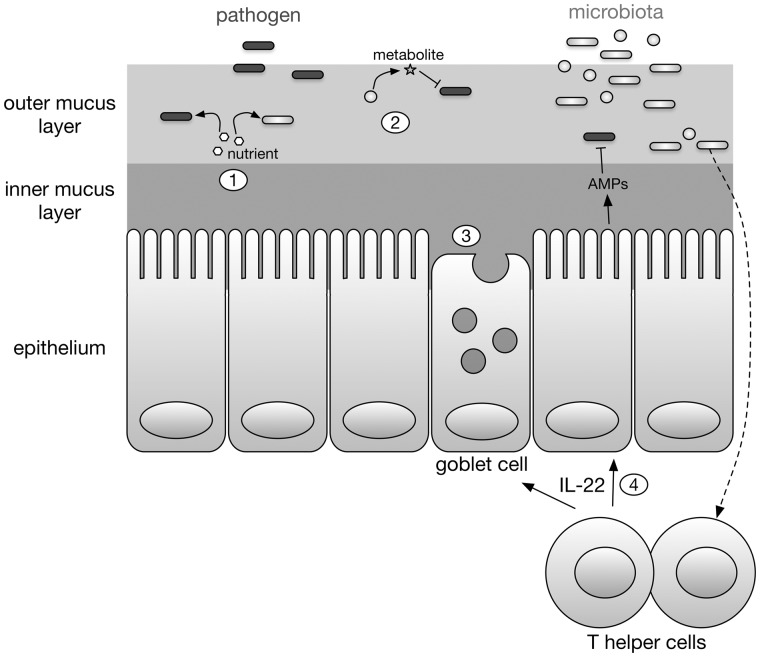
There are numerous mechanisms by which the gut microbiota provides colonization resistance (summarized in Figure 1). These can be divided into two broad categories: direct inhibition of pathogens, and indirect inhibition of pathogens via effects on the host.26,27
Figure 1.
Inhibition of diarrheal pathogens by the gut microbiota. The gut microbiota inhibits diarrheal pathogens both directly (1 and 2) and indirectly (3 and 4). (1) The gut microbiota competes with pathogens for nutrients such as free sialic acid. (2) Certain gut microbiota species produce metabolites that inhibit the growth or virulence of pathogens. (3) The gut microbiota promotes maintenance of the mucosal barrier, including secretion of mucin by goblet cells. (4) The gut microbiota also stimulate innate and adaptive immunity, including production of the cytokine IL-22, which acts on epithelial cells to increase production of antimicrobial peptides (AMPs).
One major way in which the microbiota directly inhibits diarrheal pathogens is by competition for nutrients. Many gut-dwelling bacteria use host-produced carbohydrates, such as the glycans covalently linked to the colonic mucin protein MUC2, as a carbon source.28 In order to do so, some microbiota species secrete sialidases that cleave terminal sialic acid residues from mucin glycans. 29 The liberated sialic acid can then be consumed by any nearby bacteria that possess genes for sialic acid transport and catabolism, including those organisms that do not produce sialidase on their own. Interestingly, the amount of free sialic acid (i.e. not covalently attached to mucus) in the mouse cecum drastically increases 1 day post-treatment with antibiotics.30 At this timepoint, mice are also more susceptible to infection with Salmonella and C. difficile, both of which are capable of consuming sialic acid but lack the sialidase needed to release sialic acid from mucus.30,31 Ng et al.30 showed that mutants of Salmonella and C. difficile that are incapable of consuming sialic acid have a reduced ability to colonize the guts of antibiotic-treated mice. These findings suggest that antibiotic treatment kills many sialic acid-consuming microbiota but leaves some of the sialidase producers alive, such that there is less competition for free sialic acid. Thus, one way that antibiotic treatment fuels the growth of pathogens such as Salmonella and C. difficile is by reducing competition for nutrients including sialic acid.
The gut microbiota can also directly inhibit the growth of diarrheal pathogens. There are many ways in which this inhibition can occur, including secretion of bacteriocins (antimicrobial peptides)32 or small-molecule metabolites such as secondary bile acids,33 or via cell contact-dependent inhibitory structures such as the type VI secretion system.34 In addition, microbiota species can also produce molecules that reduce pathogens’ virulence. As an example, when Salmonella is grown in the presence of organic compounds from human faeces, the expression of its type III secretion genes, which are essential for its ability to colonize the gut, is reduced.35 Correspondingly, Salmonella’s ability to invade cultured human epithelial cells is diminished in the presence of human faecal extract.35 The authors demonstrated that metabolites from lab-grown cultures of several microbiota species were capable of repressing Salmonella type III secretion, with Clostridium citroniae being particularly effective.35 The microbiota may have evolved to produce such antivirulence metabolites in order to protect their intestinal niche against intruders like Salmonella. Alternatively, Salmonella could have evolved the ability to sense gut microbiota metabolites as a way of ensuring that virulence genes are expressed in the correct time and place during infection.
The microbiota also provides protection from diarrheal pathogens via its effects on the host. One important effect of the microbiota on the host is promoting gut barrier maintenance. The colonic epithelium is protected by a mucus layer, consisting of two distinct regions (reviewed in Johansson et al.28). The inner mucus layer (nearest to the epithelial surface) is composed of dense mucin polymers and is essentially free of microbes. In contrast, the outer mucus layer (nearer to the lumen of the colon) is more loosely packed and inhabited by a variety of microbiota species. In addition to physically occluding pathogens from the epithelial surface, the mucus layer also protects the epithelium by retaining antimicrobial factors such as antimicrobial peptides and secretory IgA antibodies.36 Signals from the microbiota play an important role in maintenance of this mucus layer,37 as evidenced by the increased penetrability of colonic mucus in germ-free mice.38 Mice treated with the antibiotic metronidazole develop a thinner inner mucus layer compared with untreated animals or those treated with streptomycin.20 When metronidazole-treated mice are subsequently infected with C. rodentium, they develop more severe colitis than untreated or streptomycin-treated mice.20 Since C. rodentium localizes close to the epithelial surface earlier during the infection and penetrates deeper into colonic crypts in metronidazole-treated mice, thinning of the protective inner mucus layer is likely responsible for the more severe inflammation observed in these animals.20
The gut microbiota is also known to stimulate both the innate and the adaptive immune system,39,40 thereby altering the susceptibility of the host to diarrheal pathogens. One such example originates from a study examining the outcome of C. rodentium infection in different strains of mice. In most strains of mice (including NIH Swiss), C. rodentium causes self-limiting colitis that resolves within three to 4 weeks. However, in several strains (such as C3H/HeJ), C. rodentium infections are fatal.41 Since the resistant and susceptible strains of mice harbour substantially different gut microbiota communities,42 a microbiota transplantation study was performed in order to determine whether differences in microbiota composition contribute to differing susceptibility to C. rodentium. When C3H/HeJ (susceptible) mice received faecal microbiota from NIH Swiss (resistant) mice, they had significantly higher survival rates when subsequently challenged with C. rodentium compared with mock-treated mice that received faeces from susceptible mice.42 The authors then examined the effect of microbiota transplantation on expression of the cytokine IL-22, which was previously known to be stimulated by the microbiota and to be important for protection against C. rodentium.43,44 They found that IL-22 levels were higher in the guts of resistant mice than in susceptible mice, and that levels of IL-22 were increased by transplantation of resistant microbiota, but not by mock transplantation.42 Importantly, immunoneutralizing IL-22 by injecting mice with an anti-IL-22 antibody significantly reduced the ability of resistant microbiota transplantation to protect susceptible mice from C. rodentium lethality,42 demonstrating that the induction of immune responses mediated by IL-22 is one mechanism by which the gut microbiota can protect against diarrheal infection.
Conclusions and Outlook
It is now clear that the outcome of infection with a diarrheal pathogen depends not only on the pathogen’s intrinsic characteristics, but also on the health and immune status of the host and the level of colonization resistance provided by the gut microbiota. Currently, we cannot accurately predict the degree to which an individual’s microbiota will provide colonization resistance against diarrheal pathogens. However, in the future, it may be possible to identify missing taxa or functions in an individual’s microbiota and prescribe probiotics, prebiotics, or dietary modifications to strengthen protection against diarrheal infections. For example, a probiotic organism or cocktail that is exceptionally efficient at catabolizing free sialic acid might enhance colonization resistance against Salmonella and C. difficile.45 As another example, Pop et al. identified a number of microbiota species whose presence predicted resistance to ETEC, including Sutterella sp., Prevotella copri and Bacteroides vulgatus.23 If further studies confirm that these organisms are protective against ETEC, travellers could undergo faecal microbiota analysis prior to travel to predict their level of colonization resistance to ETEC; if resistance is predicted to be low, physicians may be able to recommend custom prebiotics, probiotics, or even faecal transplants (in severe cases) to help prevent travellers’ diarrhea. Since there is no evidence that current probiotics can prevent travellers’ diarrhea,46 such next-generation probiotics could be a major advance in maintaining travellers’ health.
Funding
S.L.V. is supported by a Killam Postdoctoral Research Fellowship, a Canadian Institutes of Health Research (CIHR) Postdoctoral Fellowship, and a Michael Smith Foundation for Health Research (MSFHR) Postdoctoral Fellowship. Work in B.B.F.’s laboratory is supported by CIHR (grant MOP-130290). B.B.F. is the University of British Columbia Peter Wall Distinguished Professor.
Conflict of interest: None declared.
References
- 1. Sender R, Fuchs S, Milo R.. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016; 164: 337–40. [DOI] [PubMed] [Google Scholar]
- 2. Donaldson GP, Lee SM, Mazmanian SK.. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 2016; 14: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sommer F, Bäckhed F.. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol 2013; 11: 227–38. [DOI] [PubMed] [Google Scholar]
- 4. Lawley TD, Walker AW.. Intestinal colonization resistance. Immunology 2013; 138: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yurist-Doutsch S, Arrieta MC, Vogt SL, Finlay BB.. Gastrointestinal microbiota-mediated control of enteric pathogens. Annu Rev Genet 2014; 48: 361–82. [DOI] [PubMed] [Google Scholar]
- 6. Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB.. Effect of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 2014; 68: 217–35. [DOI] [PubMed] [Google Scholar]
- 7. Nardi RM, Silva ME, Vieira EC. et al. Intragastric infection of germfree and conventional mice with Salmonella typhimurium. Braz J Med Biol Res 1989; 22: 1389–92. [PubMed] [Google Scholar]
- 8. Collins FM, Carter PB.. Growth of salmonellae in orally infected germfree mice. Infect Immun 1978; 21: 41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kamada N, Kim YG, Sham HP. et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 2012; 336: 1325–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Round JL, Mazmanian SK.. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barthel M, Hapfelmeier S, Quintanilla-Martínez L. et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 2003; 71: 2839–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Croswell A, Amir E, Teggatz P. et al. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun 2009; 77: 2741–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferreira RB, Gill N, Willing BP. et al. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One 2011; 6: e20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pérez-Cobas AE, Gosalbes MJ, Friedrichs A. et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 2013; 62: 1591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crum-Cianflone NF. Salmonellosis and the gastrointestinal tract: more than just peanut butter. Curr Gastroenterol Rep 2008; 10: 424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Britton RA, Young VB.. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol 2012; 20: 313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reeves AE, Theriot CM, Bergin IL. et al. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2011; 2: 145–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee CH, Steiner T, Petrof EO. et al. Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: a randomized clinical trial. JAMA 2016; 315: 142–9. [DOI] [PubMed] [Google Scholar]
- 19. Petrof EO, Gloor GB, Vanner SJ. et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ′RePOOPulating′ the gut. Microbiome 2013; 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wlodarska M, Willing B, Keeney KM. et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun 2011; 79: 1536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shakir L, Javeed A, Ashraf M, Riaz A.. Metronidazole and the immune system. Pharmazie 2011; 66: 393–8. [PubMed] [Google Scholar]
- 22. Allen KP, Randolph MM, Fleckenstein JM.. Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect Immun 2006; 74: 869–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pop M, Paulson JN, Chakraborty S. et al. Individual-specific changes in the human gut microbiota after challenge with enterotoxigenic Escherichia coli and subsequent ciprofloxacin treatment. BMC Genomics 2016; 17: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singer SM, Nash TE.. The role of normal flora in Giardia lamblia infections in mice. J Infect Dis 2000; 181: 1510–2. [DOI] [PubMed] [Google Scholar]
- 25. Jones MK, Watanabe M, Zhu S. et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014; 346: 755–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamada N, Chen GY, Inohara N, Núñez G.. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013; 14: 685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McKenney PT, Pamer EG.. From hype to hope: The gut microbiota in enteric infectious disease. Cell 2015; 163: 1326–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johansson ME, Sjövall H, Hansson GC.. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol 2013; 10: 352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lewis AL, Lewis WG.. Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell Microbiol 2012; 14: 1174–82. [DOI] [PubMed] [Google Scholar]
- 30. Ng KM, Ferreyra JA, Higginbottom SK. et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013; 502: 96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoyer LL, Hamilton AC, Steenbergen SM, Vimr ER.. Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol Microbiol 1992; 6: 873–84. [DOI] [PubMed] [Google Scholar]
- 32. Kommineni S, Bretl DJ, Lam V. et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 2015; 526: 719–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buffie CG, Bucci V, Stein RR. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517: 205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Russell AB, Wexler AG, Harding BN. et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 2014; 16: 227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Antunes LC, McDonald JA, Schroeter K. et al. Antivirulence activity of the human gut metabolome. MBio 2014; 5: e01183–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McGuckin MA, Lindén SK, Sutton P, Florin TH.. Mucin dynamics and enteric pathogens. Nat Rev Microbiol 2011; 9: 265–78. [DOI] [PubMed] [Google Scholar]
- 37. Johansson ME, Hansson GC.. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol 2016; 16: 639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johansson ME, Jakobsson HE, Holmén-Larsson J. et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 2015; 18: 582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Honda K, Littman DR.. The microbiota in adaptive immune homeostasis and disease. Nature 2016; 535: 75–84. [DOI] [PubMed] [Google Scholar]
- 40. Thaiss CA, Zmora N, Levy M, Elinav E.. The microbiome and innate immunity. Nature 2016; 535: 65–74. [DOI] [PubMed] [Google Scholar]
- 41. Vallance BA, Deng W, Jacobson K, Finlay BB.. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect Immun 2003; 71: 3443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Willing BP, Vacharaksa A, Croxen M. et al. Altering host resistance to infections through microbial transplantation. PLoS One 2011; 6: e26988.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S. et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 2008; 29: 958–70. [DOI] [PubMed] [Google Scholar]
- 44. Zheng Y, Valdez PA, Danilenko DM. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 2008; 14: 282–9. [DOI] [PubMed] [Google Scholar]
- 45. Ley RE. Harnessing microbiota to kill a pathogen: the sweet tooth of Clostridium difficile. Nat Med 2014; 20: 248–9. [DOI] [PubMed] [Google Scholar]
- 46. Ritchie ML, Romanuk TN.. A meta-analysis of probiotic efficacy for gastrointestinal diseases. PLoS One 2012; 7: e34938.. [DOI] [PMC free article] [PubMed] [Google Scholar]