Abstract
Historical milestones in neuroscience have come in diverse forms, ranging from the resolution of specific biological mysteries via creative experimentation to broad technological advances allowing neuroscientists to ask new kinds of questions. The continuous development of tools is driven with a special necessity by the complexity, fragility, and inaccessibility of intact nervous systems, such that inventive technique development and application drawing upon engineering and the applied sciences has long been essential to neuroscience. Here we highlight recent technological directions in neuroscience spurred by progress in optical, electrical, mechanical, chemical, and biological engineering. These research areas are poised for rapid growth and will likely be central to the practice of neuroscience well into the future.
Introduction
Recent years have witnessed the intriguing and rapidly expanding embodiment of an engineering approach to the study of nervous systems, via influx of ideas, methods, investigators, and scholarly traditions linked to applied and technical fields that were historically far separated from neuroscience. In some ways reminiscent of earlier contributions from theoretical and computational scientists that helped frame aspects of systems neuroscience, we are currently observing a wave of influence from the applied sciences and engineering that is beginning to transform the field. Engineering principles have always been important in neuroscience, but the opportunities today seem greater than ever before due to an especially fertile conceptual and experimental landscape. Because we cannot capture here the full breadth of this ongoing transformation (including the vast realm of biomedical engineering of devices and instrumentation specifically for clinical purposes), we focus instead on specific recent advances and new directions that illustrate how multiple major and distinct fields of engineering are becoming crucial for basic neuroscience research.
Bioengineering
Bioengineering integrates engineering with the life sciences by fusing quantitative and technological approaches with raw materials from the biological domain or focusing on biological applications. Recently, bioengineering principles have found particular traction in neuroscience. For example, although the history of tissue engineering with neural cells has been challenged by the structural complexity of neurons and nervous systems, recent advances have leveraged the self-assembly capabilities of stem or progenitor cells, which, under the right conditions, can form differentiated structures ranging from neural tube-like ellipsoids or neurospheres useful for studying neural stem cell biology (Reynolds and Weiss, 1992) to brain-like organoids that mimic detailed features of brain morphology (Lancaster et al., 2013). Despite (or even building upon) the incomplete stability, consistency, and activity of these artificial structures, it is likely that insights into normal and pathological patterning of nervous systems may result from continued research into such assembly of engineered neural structures in vitro.
Protein engineering (a field of bioengineering in which the raw materials are proteins rather than cells) has exerted a major influence on neuroscience over the past 25 years, exemplified by the process of engineering green fluorescent protein (GFP) and related molecules for improved fluorescence properties via a diverse array of targeted molecular engineering and high-throughput mutation/screening approaches (Heim et al., 1995). This process not only delivered a panel of robust and versatile genetically targetable tools for anatomical and structural investigation of nerve cells and nervous systems but also enabled the development of GFP-based reporters of cellular activity dynamics (Akerboom et al., 2013; Wu et al., 2013b). Various strategies for modification of GFP conferred the ability to report intracellular Ca2+ concentration, allowing tracking of this correlate of neural activity in genetically targetable fashion and culminating over the ensuing 10–15 years in the successful engineering of the GCaMP family of Ca2+ activity probes. These newest Ca2+ indicators cover a range of excitation and emission bands in the visible spectrum and approach single spike detection sensitivity in many neuron types, such as pyramidal cells with relatively low spike rates; resolution of spike timing is presently in the ~10–250 ms range (Akerboom et al., 2013; Ohkura et al., 2012; Wu et al., 2013b).
What do we expect from the future in protein engineering for activity readout? Cognizant that prior efforts have not always considered the dictates of signal detection theory, we note that indicators (for either Ca2+ or voltage dynamics) with ultralow background emissions hold particular importance because background photons often represent the chief impediment to reliable event detection and timing estimation (Wilt et al., 2013). Indicators with ultralow background emission and large signaling dynamic range will also improve the imaging depths that can be attained deep within brain tissue. Likewise, red or near-infrared optical indicators would also improve imaging depths in scattering tissues due to the increased optical attenuation lengths at these wavelengths (Kobat et al., 2009; Lecoq and Schnitzer, 2011; Zhao et al., 2011).
We also anticipate advances in the bioengineering of protein sensors of neuronal transmembrane voltage; sufficient progress in such indicators would permit voltage imaging with single-cell resolution in the living mammalian brain. The latest generation of genetically encoded voltage indicators can now reliably report action potentials in cultured neurons and appear to be on the brink of gaining practical utility in mammalian brain tissue slices (Cao et al., 2013; Gong et al., 2013; Jin et al., 2012; Kralj et al., 2012; Lam et al., 2012). Ideally, improved voltage indicators should dovetail with concurrent advances in targeting proteins to particular cell types or subcellular compartments and would reveal neuronal spiking with millisecond-scale timing resolution, dendritic voltage dynamics, subthreshold inhibition and excitation, and high-frequency oscillations. The improved voltage indicators may well be genetically encoded, but other approaches from chemistry and nanotechnology should also be considered (Alivisatos et al., 2013; Hall et al., 2012; Marshall and Schnitzer, 2013).
While engineered GFP-based tools have transformed neuroscience by enabling the genetically targeted readout of both static anatomy and dynamical activity, experimental strategies to read-in (control) activity dynamics have typically relied on a different class of engineered proteins (Fenno et al., 2011). Devising methods for safely and effectively expressing in neurons members of the microbial opsin gene family, which previously had been studied for many years by physiologists investigating membrane properties of organisms such as algae and archaebacteria (reviewed in Zhang et al., 2011), has opened the door to optical and genetically targetable control of neurons with millisecond resolution within systems as complex as freely behaving mammals. This optogenetic approach, based (as with GFP strategies for imaging) on a single delivered protein component, has likewise benefited enormously from protein engineering (Deisseroth, 2011).
For example, the excitatory channelrhodopsin tools have been engineered to confer many-orders-of-magnitude-increased light sensitivity to neurons (compared with the original wild-type forms) via mutations that selectively lengthen the intrinsic time constant of deactivation of the channelrhodopsin photocurrent (Berndt et al., 2009; Bamann et al., 2010; Yizhar et al., 2011a, 2011b; Mattis et al., 2012). Cells expressing these mutant “step-function” channelrhodopsins become photon integrators, and extraordinarily low-intensity light can be used to increase neural activity in deep-brain genetically targeted cells without penetrating brain tissue with optical hardware (Mattis et al., 2012; Yizhar et al., 2011b). These engineered step-function tools have now found broad application in modulating complex behaviors within systems ranging from flies to worms to mice (Carter et al., 2012; Haikala et al., 2013; Tanaka et al., 2012; Yizhar et al., 2011b; Bepari et al., 2012; Schultheis et al., 2011).
Other forms of protein engineering have (1) accelerated deactivation of photocurrents for improved temporal precision (Gunaydin et al., 2010; Berndt et al., 2011), (2) altered action spectra for red- or blue-shifted control to facilitate the use of multiple bands of the visible-light spectrum in the same experiment (Yizhar et al., 2011b), or (3) enhanced expression of the microbial opsins (Gradinaru et al., 2008, 2010; Zhao et al., 2008; Lin et al., 2009; Wang et al., 2009; Yizhar et al., 2011b; Mattis et al., 2012). The two major protein engineering strategies that led to improved expression have been (1) addition of membrane trafficking tags and (2) chimeric-opsin formation; the first strategy (including addition of tags such as endoplasmic reticulum-export motifs and trafficking signals that guide protein accumulation in axons and dendrites) has enhanced the functionality of every microbial opsin tested, including channelrhodopsins (Yizhar et al., 2011b), chloride pumps (Gradinaru et al., 2008, 2010; Zhao et al., 2008), and proton pumps (Mattis et al., 2012). The resulting many-fold-greater currents also promote application of the most versatile form of optogenetic targeting, “projection targeting,” in which light is delivered to the axon termination field (and the axonally trafficked opsins therein) of a transduced population in order to recruit cells for behavioral control defined by possessing a particular spatially defined projection pattern (Gradinaru et al., 2010); similar trafficking strategies are also reported to have benefited genetically encoded voltage sensors.
The second major protein engineering strategy (thus far particularly successful for the channelrhodopsins) has involved the generation of chimeras by swapping transmembrane helices among various known channelrhodopsins from different microbial genes. This strategy, beginning in 2009 (Lin et al., 2009; Wang et al., 2009), led to the generation of many high-expressing channelrhodopsins, one of which (C1C2, a shortened form of a chimera between the Chlamydomonas reinhardtii channelrhodopsin-1 and channelrhodopsin-2) enabled the 2.3Å crystal structure of channelrhodopsin to be obtained (Kato et al., 2012). Other chimeras were then combined with point mutations for additional optimization, culminating in tools such as CHIEF (with high expression levels, fast kinetics, and reduced desensitization) (Lin et al., 2009) and C1V1 (with high expression, red-light activation, and raster-scanning two-photon optogenetic activation suitability in vivo) (Yizhar et al., 2011b).
What do we expect for the coming years in this realm? The crystal structure (Kato et al., 2012) along with future structures capturing different stages of the photocycle, and in the presence of different permeating or pore-blocking ions, should help drive the directed engineering of opsin genes for new classes of function involving kinetic properties, spectral sensitivity, and ion selectivity; a major goal on this front should be the development of inhibitory channelrhodopsins, which will exceed the utility of the inhibitory pumps by providing decreased membrane resistance as well as hyperpolarization. Molecular dynamics simulations will be of great value, capitalizing on the availability of ever-increasing computational power. Many more opsin genes will be identified in the coming years as numerous genomes are sequenced across the kingdoms of life (Zhang et al., 2011), and protein engineering will be accelerated by the shuffling of motifs among these opsins and other tools (including modulators of biochemical and electrical events) and by development of high-throughput screening methodologies building upon random and combinatorial mutagenesis strategies.
Protein engineering will also bring us other classes of tools for information exchange with nervous systems. Development of nonoptical (e.g., molecular) readouts of neural events will greatly accelerate and will come to include single-neuron transcriptomics, proteomics, and epigenomics, either in cells isolated by high-throughput disassembly strategies or via in situ methods that maintain the assembly of nervous systems while allowing access for molecular and optical interrogation (Chung et al., 2013). Additionally, proteins and particles designed to serve as antennae for sources of information beyond light (e.g., magnetic, acoustic, and thermal energy) will continue to be explored (e.g., Anikeeva et al., 2012). We note that turnkey delivery of these engineered protein tools to arbitrarily defined elements within nervous systems will require in itself future feats of molecular biological engineering; we expect this field to drive, and build heavily upon, major new advances in high-throughput promoter/enhancer screening, viral serotyping and pseudotyping, combinatorial and intersectional access to specific cell types, and genome engineering tools for versatile targeting of endogenous genetic loci (Konermann et al., 2013). And, finally, genetic targeting of protein-based tools will powerfully synergize with spatial targeting of the input stream of information (exemplified by light targeting with increasingly sophisticated optics and photonics, a distinct field of engineering discussed next).
Optics and Photonics
Recent years have witnessed rapid advances in the engineering realms of optics and photonics. Optimal application of these tools to neuroscience demands a holistic view of optical experimentation; the capabilities and limitations of optical hardware in the neuroscience setting should be taken into consideration when developing new optical sensor and control molecules and vice versa, because the collective optical system is what ultimately should be optimized according to the principles of signal detection theory, estimation theory, or other appropriate aspects of theoretical engineering.
Multiple branches of light microscopy have undergone exciting progress. First, there has been rapid development of new methods for superresolution fluorescence imaging (Dani et al., 2010; Testa et al., 2012; Urban et al., 2011; Wilt et al., 2009; Xu et al., 2013). These methods are allowing ultrastructural studies of synaptic content and structure using the light microscope, complementary to traditional electron microscopy methods. A key advantage of using fluorescence to study synaptic ultrastructure is that multiple protein species can be labeled and monitored concurrently (Micheva and Smith, 2007), including in live neurons. We expect rapid advances in this arena, with high-content studies of synaptic molecular organization leveraging new labeling strategies and chemical biology methods.
There has also been dramatic progress in nonlinear optical microscopy. Today, neuroscientists widely appreciate the phenomenon of two-photon excitation, but two-photon effects were once considered esoteric aspects of optical physics. It was the development of solid-state ultrafast lasers, chiefly the development of titanium sapphire laser technology, that propelled the two-photon microscope innovated by Webb and Denk to become broadly usable by biologists and to permeate neuroscience. We expect continued improvement of not just the hardware elements that comprise the two-photon microscope, but also general optical strategies for laser scanning, for scanless approaches to laser illumination, and for other new approaches for imaging faster and deeper into tissue (Kobat et al., 2011; Oron et al., 2005; Quirin et al., 2013; Schrö del et al., 2013).
Beyond the current depths presently attainable by two-photon microscopy, nonlinear optical microscopy modalities relying on multiphoton excitation and long-wavelength, ultrashort-pulsed lasers promise to reveal fundamental features of nervous tissue (Farrar et al., 2011; Horton et al., 2013; Kobat et al., 2011; Mahou et al., 2012). Due to reduced light scattering at longer wavelengths, three-photon excitation with an illumination wavelength of 1.7 μm has been demonstrated in proof-of-concept studies to reach into even the hippocampus in a live mouse (Horton et al., 2013). However, much work remains to make this a practical technique for day-to-day experimental studies of nervous system structure.
For deep studies of nervous system dynamics, further development of red fluorescent sensors of neural activity will be required, which presently lag behind green fluorescent sensors such as the highly successful GCaMP6 Ca2+ sensors. There are also crucial issues of optical aberrations to consider when imaging 1 mm or more into dense brain tissue. Adaptive optical methods may help, offering the possibility of correcting aberrations online, and have already made inroads into neuroscience (particularly for ophthalmic imaging of the retina and visualization of single human photoreceptor cells) (Godara et al., 2010; Hunter et al., 2010). Adaptive optics have also shown utility for two-photon microscopy by improving the resolution of two-photon imaging deep in tissue (Ji et al., 2012). When combined with long-wavelength laser illumination, such as for three-photon excitation, adaptive optics may become even more important.
One-photon fluorescence imaging methods are also making notable progress, including techniques based on selective planar or light-sheet illumination, which has recently been used to scan a plane of fluorescence excitation across the entire zebrafish nervous system at 0.8 Hz (Ahrens et al., 2013). Holographic methods for fluorescence imaging are also emerging, applicable to either one- or two-photon microscopy (Watson et al., 2010). Engineering progress in the spatial light modulators that are a key component for holographic imaging will help drive progress in this area (Quirin et al., 2013). Light-field fluorescence microscopy has now been applied to biology for the first time (L. Grosenick et al., 2013, Society for Neuroscience, abstract), allowing extremely fast three-dimensional image acquisition without scanning (Broxton et al., 2013; A. Andalman et al., 2013, Society for Neuroscience, abstract; L. Grosenick et al., 2013, Society for Neuroscience, abstract). This speed and volumetric information comes at the cost of somewhat reduced lateral resolution but still permits resolution of individual neurons within intact and functioning vertebrate nervous systems (L. Grosenick et al., 2013, Society for Neuroscience, abstract; A. Andalman et al., 2013, Society for Neuroscience, abstract). Going forward, we expect continuous improvement in light-field, holographic, and selective planar illumination methods for improved acquisition rates, resolution, and coverage volume applied to intact nervous systems. We also expect holographic and light-field methods for optogenetic activity manipulation to develop in tandem with corresponding methods for activity imaging.
The resulting large optical data sets require massive improvements in data handling and computational analysis. Optical engineering applied to the nervous system will also continue to benefit from computational methods that improve the capabilities to look through turbid media. In the brain, light attenuation is chiefly due to light scattering (turbidity), rather than light absorption; emerging methods for correcting for effects of light scattering through a combination of computational approaches and optical manipulations (Bertolotti et al., 2012) have yet to have major impacts on neuroscience experimentation, but future years may reveal a role for these computational techniques for imaging deep into turbid brain tissue.
Progress in the engineering of optical hardware continually propels improvements in optical systems. The invention of the charge-coupled device (CCD) camera led to pioneering studies of intracellular Ca2+ dynamics in neurons. Today, scientific-grade cameras routinely monitor neuronal dynamics, but the more recently developed complementary metal oxide semiconductor (CMOS) image sensor has made substantial inroads into experimental terrain previously dominated by the CCD camera. The most recent CMOS image sensors have enabled a new generation of fluorescence imaging experiments.
Commonplace CMOS sensors, such as those in mobile telephones, are now so advanced that they have enabled the engineering and application of miniaturized fluorescence microscopes, small enough to be mounted on the head of a mouse during free behavior (Figure 1) (Ghosh et al., 2011; Ziv et al., 2013). Such CMOS-based miniature microscopes can now provide recordings of up to ~1,200 neurons concurrently during active mouse behavior (Figure 1). This promises to be a useful tool in the study of rodent models of human brain disorders, and perhaps even in primate models. We expect continued progress in camera technology and image sensor chips, leading to larger sensors, faster image-frame acquisition rates, on-chip imaging analyses, wireless imaging, and even capabilities for three-dimensional imaging. Further improvements in tiny light emitting diodes (LEDs) in combination with CMOS image sensors should enable a new generation of devices capable of both optogenetic manipulation and fluorescence imaging concurrently. This need will provide additional impetus for the ongoing engineering of spectrally compatible sets of optogenetic control probes and fluorescence-based sensors of neural activity.
Figure 1. Optics and Protein Engineering Converge for Ca2+ Imaging in >1,200 CA1 Pyramidal Cells in Freely Moving Mice.
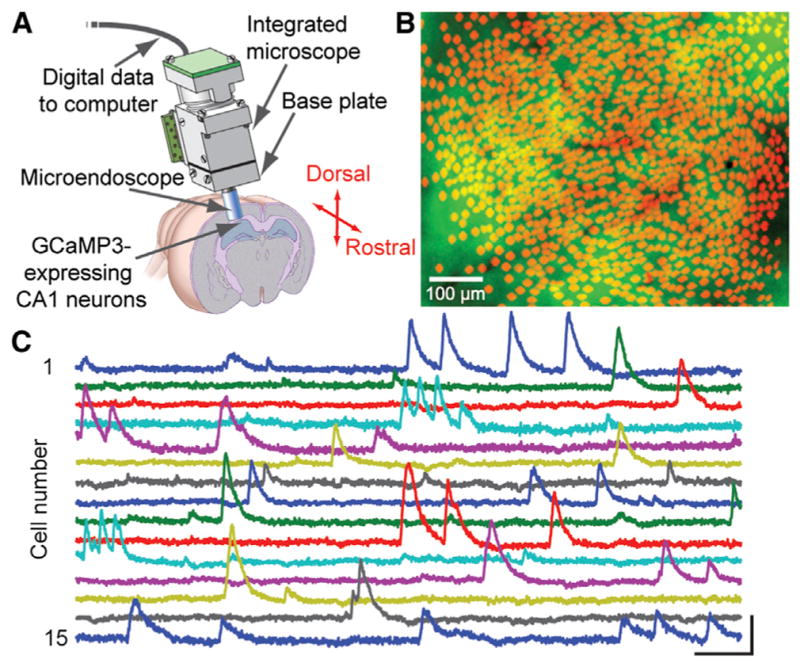
(A) An integrated microscope is equipped with a microendoscope to image CA1 neurons expressing the engineered Ca2+ indicator GCaMP3 under control of the Camk2a promoter. The base plate and microendoscope are fixed to the cranium for repeated access to the same field of view. Republished from Ziv et al., 2013.
(B) Shown are 1,202 CA1 pyramidal cells (red somata) identified by Ca2+ imaging in a freely moving mouse atop a mean fluorescence image (green) of CA1. Vessels appear as dark shadows. Image courtesy of Yaniv Ziv and Lacey Kitch, Stanford University.
(C) Example traces of [Ca2+]i dynamics from 15 cells. Scale bars denote 5% ΔF/F (vertical) and 10 s (horizontal).
Even as next-generation optical tools offer increasingly sophisticated technological capabilities, the practice of systems neuroscience will have to remain grounded in rigorous, clever, and insightful behavioral paradigms. Here, digital imaging may help advance the field, as many emerging opportunities exist for high-throughput and high-resolution video tracking of animal behavior. To maximally leverage the newfound capabilities for optically monitoring individual cells over many weeks in the live brain, new behavioral assays should be compatible with long-term tracking and quantification of behavior. Machine-learning approaches to scoring digital image sequences of animal behavior (Kabra et al., 2013) might facilitate the combined automation of both brain imaging and behavioral data analyses.
Finally, we note that for in vivo animal experimentation, the demands of small animal surgery often remain a limiting factor on the rate of experimental progress. In recent years there has been exploration of laser surgical methods to perform highly precise surgeries. One candidate approach involves the use of regenerative laser amplifiers that emit high-energy ultrashort pulses of light for highly precise tissue ablation (down to the sub-micron scale, to cut or ablate individual axons, neurons, and even organelles) (Jeong et al., 2012; Samara et al., 2010). However, the fine spatial scale of the cutting action is a limiting factor for performing dissections over broad tissue regions. An alternative approach is to make use of ultraviolet lasers, such as those commonly used in clinical ophthalmology for reshaping the cornea (Sinha et al., 2013). Ultraviolet excimer lasers can cut precision holes down to the sub-10-μm scale, with clean-cut edges straight to <1 μm, and at much faster cutting rates than the regenerative laser amplifiers. These properties enable automated forms of laser surgery in insects, nematodes, and even rodents on the seconds timescale, offering intriguing possibilities for increasing the throughput of neuroscience experimentation (Sinha et al., 2013).
Electrical Engineering
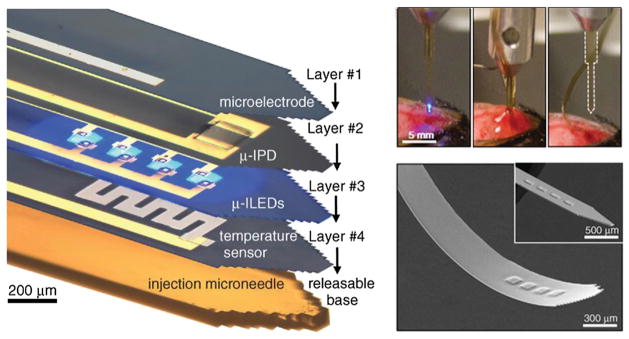
Electrical engineering has long influenced neuroscience, dating back to the contributions of cable theory and radio electronics on the pioneering research by Cole, Curtis, Hodgkin, and Huxley on the squid giant axon. This influence has persisted, as in the advancement of low-noise electronic amplifiers driving improved electrophysiological instrumentation for single channel biophysics. Recent years have seen an acceleration of technologies for performing large-scale multielectrode recordings—not just expanding arrays of electrodes to numbers and densities beyond those previously feasible, but also novel surface electrodes and mechanically flexible recording devices that can be bent to match the brain’s curvature and could be used to monitor dynamics and help detect phenomena such as those involved in epilepsy at the neocortical surface (Figure 2) (Viventi et al., 2011).
Figure 2. Materials Science and Optoelectronics Converge for Neural Interface Research.
Left: exploded-view layout of injectable semiconductor device for integrated stimulation/sensing/actuation, highlighting distinct layers for electrophysiological measurement (1), optical measurement (2), optical stimulation (3, micro-ILED array), and temperature sensing (4), all bonded to a releasable base for injection with a microneedle. Top right: injection and release of the microneedle. After insertion (left), artificial cerebrospinal fluid dissolves an external silk-based adhesive (middle) and the microneedle can be removed (right), leaving the active device in the brain. Bottom right: SEM of an injectable micro-LED array, 8.5 μm thick; flexible and rigid forms shown. Adapted with permission from Kim et al., 2013.
We expect continued progress in the development of large-scale and flexible electronic technology platforms (Kim et al., 2012, 2013). Active electrodes or smart electronics will be internally incorporated to permit in situ signal amplification, reducing the impact of noise and allowing immediate extraction of specific physiological signals. It may become commonplace to incorporate closed-loop capabilities within devices that allow both measurement and manipulation—the latter being electrical, optical, or pharmacologic. Such capabilities could have an important clinical impact as well as an impact in basic science; for example, early detection of epileptic episodes could trigger immediate preventive action, perhaps taken by the same device.
We expect continued progress in the area of hybrid probes such as optrodes (Gradinaru et al., 2007, 2008), which allow optogenetic stimulation of neurons along with electrical recordings from the very same cells. Advanced forms of optrodes have enabled the recording of neural circuit dynamics simultaneously with high-speed optical control and behavior (Anikeeva et al., 2012; Wu et al., 2013a; Ozden et al., 2013; Kim et al., 2013) and will also facilitate the identification or tagging of spikes from cells that express opsins. Integration of electrical and optical capabilities in the same devices will continue to improve; for example, flexible electronics will be combined with high-density multicolor miniature light sources and optical detectors, and optrodes will become smaller, easier to fabricate, and better integrated for more ready implementation in behaving animals. Unconventional optrode designs such as new types of metal-coated, tapered fiberoptics may likewise serve to facilitate combined electrophysiological measurement and light delivery (Dufour et al., 2013; Ozden et al., 2013). More generally, we foresee an expansion of new types of multifaceted probes for electrophysiological recording and stimulation that might incorporate not only capabilities for light detection or delivery, but also drug delivery or microfluidic sampling.
Another major area in which electrical engineering is exerting a strong influence on neuroscience concerns brain-machine interfaces. An established class of such interfaces concerns sensory perception, with the cochlear implant as a paradigmatic example. Likewise, there has been sustained progress toward retinal prosthetics for restoring vision (Mathieson et al., 2012) and toward motor prosthetics for achieving artificial-limb control using neural signals sent from the brain and transduced into electronic commands. Recent progress has conferred the ability to control a computer cursor or robotic arm by motor-impaired patients (Hochberg et al., 2012). This realm of prosthesis engineering is building heavily upon concepts from computational and analytical aspects of electrical engineering and computer science, including dynamical systems modeling, state space analysis, dimensionality reduction, and adaptive filtering (Dangi et al., 2013; Gilja et al., 2011; Shenoy et al., 2013). We note that the notion of a neural prosthetic is conceptually broad, and nonelectrical prosthetics (e.g., optical or magnetic) might be developed to augment or correct aspects of cognition or behavior. For basic neuroscience experimentation, all-optical approaches to brain-machine interfaces should also be feasible (optical readouts combined with optical manipulation of neural dynamics). We expect to see increased complexity in this prosthetics-focused fusion of engineering and systems neuroscience, as the needs and opportunities are enormous.
For imaging the human brain, engineering and physics have long played key roles; for example, magnetic resonance imaging (MRI) arose from nuclear magnetic resonance spectroscopy. We expect continued major progress in the realm of MRI, with new computational approaches and instrumentation allowing unprecedented levels of detail to be revealed concerning the human brain and cognition. This will include not just instrumentation advances such as higher magnetic field strengths, but also improved computational approaches for registration of brain anatomy across different individuals and new methods for interpreting with high confidence the nature of the signals seen, as with diffusion tractography. And for controlling human nervous systems, there has been recent engineering progress in the design and development of optogenetic interfaces that may be useful for bidirectional modulation of activity, such as for major peripheral nerves (Liske et al., 2013).
Finally, we take note of miniaturization, which involves electrical, mechanical, and materials engineering, among other domains. Major industries such as telecommunications, consumer electronics, and defense will continue to drive rapid innovation in miniaturization of optics, electronics, wireless technology, and computational elements, all of which have contributed to superior instrumentation for neuroscience experimentation. Major virtues of miniaturized systems for use in freely moving animals include compatibility with behavioral assays that have already been deployed and validated over decades of neuroscience research. Akin to EEG and EMG telemetry systems in present usage, wireless and miniaturized brain imaging systems may come to permit around-the-clock studies of brain activity, e.g., for monitoring neural activity and brain states across sleeping, eating, and other behaviors, in substantial numbers of animals (e.g., for large behavioral cohorts in basic neuroscience laboratory investigations or in drug screening) without constant human supervision.
Chemical and Materials Engineering
The chemistry- and physics-based engineering of materials has accelerated several exciting and important technologies for neuroscience research (beyond miniaturization and electrode design, already discussed above). Here we touch on only two of many categories of chemical engineering that seem well poised to grow with neuroscience into the future: (1) the engineering of materials into which organisms and cells are placed and (2) the engineering of materials from within intact organisms.
Small organisms such as nematodes, fruit flies, and mammalian embryos could be amenable to high-throughput investigations of nervous system development, structure, physiology, and behavior. However, only recently have technologies been developed to allow high-throughput positioning and interrogation of small, intact organisms. Microfabrication and microfluidics, often with computer-aided design (CAD) molding, and soft lithography with an elastomer such as polydimethylsiloxane (PDMS), which is poured or spun into the micropatterned mold, have been applied to the positioning of Caenorhabditis elegans and mouse embryos (Albrecht and Bargmann, 2011; Chung et al., 2011a, 2011b). While zebrafish are too large for typical high-throughput microfabricated devices, approaches based on multiple well plates are coming of age (Chang et al., 2012).
Chemical engineering and applied chemistry efforts have led to the development of materials, nanoparticles, and polymers for the study of central nervous system regeneration and repair (Tam et al., 2013), delivery of small interfering RNAs for causal testing of specific transcripts (Chan et al., 2013), and construction of hydrogel environments into which nervous system cells (or stem/progenitor cells) may be seeded to study proliferation, differentiation, survival, and other properties (Cha et al., 2012; Ferreira et al., 2007; Owen et al., 2013; Tibbitt and Anseth, 2012). Going forward, we expect growth areas in this domain to include increasingly sophisticated and modular engineering of these scaffolds and environments, for example using versatile click chemistry as well as novel conductive or transparent polymers for compatibility with optical or electrical interrogation of the resulting construct (Chung and Deisseroth, 2013; Keplinger et al., 2013). This general approach may also synergize with the studies of the development and assembly of neural structures beginning from the other direction, with biology rather than chemistry, as in the stem cell/brain organoid (Lancaster et al., 2013) approach described above.
A newly emerged concept at the interface of neuroscience and chemical engineering, CLARITY (Figure 3), involves the chemical construction of new physical forms from within biological systems such as the brain (Chung and Deisseroth, 2013; Chung et al., 2013). For example, hydrogel infrastructures can be constructed from within intact brains to covalently stabilize native proteins and nucleic acids in preparation for stringent removal of membrane phospholipids with strong ionic detergents and active electrophoresis of the entire brain. This lipid removal, in turn, allows interrogation of the intact brain with photons (which no longer scatter heavily due to removal of the lipid-aqueous interfaces) and macromolecules (such as antibodies and oligonucleotide probes, which can at that point penetrate the tissue without interference from intact plasma membranes). We expect this kind of approach to find utility in mapping volumetric anatomical features from animal models as well as clinical samples; moreover, many kinds of gels and scaffolds could be constructed in this way with a range of passive and active properties for a broad range of different kinds of structural and functional studies of nervous systems. Finally, distinct from gel and scaffold diversity, there also exists a broad diversity of macromolecular probe type that can be used to interrogate the resulting nanoporous hybrid structures, including functionalized proteins and active enzymes.
Figure 3. Chemical Engineering, Bioengineering, and Photonics Give Rise to Circuit-Probing Hardware and Wetware.
Top: construction of a hydrogel from within tissue (CLARITY) creates a transparent mammalian brain for intact-system anatomical analysis; adapted from Chung et al., 2013. Bottom left: 2.3Å crystal structure of the channel-rhodopsin optogenetic control tool enables directed protein engineering for enhanced interventional functionality; adapted from Kato et al., 2012. Bottom right: optogenetic neural interfaces deliver light from laser diodes or advanced LEDs; flexible fiberoptic control in freely moving mouse shown. Photo credit Inbal Goshen and Karl Deisseroth.
Outlook
As exciting as these domains of neuroscience have become, the future may hold even greater opportunities—for example, via combinations of multiple engineering subdisciplines (e.g., computer science with chemical engineering, or optical instrumentation with bioengineering, for applications to increasingly sophisticated questions in increasingly complex nervous systems; Figure 3). CLARITY is already being used in human tissue, and advanced electrical and optical interfaces have already been designed for human and nonhuman primate applications.
Emerging optical methods may bring among the most exciting synergistic possibilities for integrative studies of neural circuit dynamics, connectivity, cytoarchitecture, and molecular composition. Specifically, in vivo optical recordings of neural activity and optogenetic manipulations in cells defined genetically or by anatomical projections can be naturally combined and registered with technologically advanced studies of circuitry, synaptic structure, and other macromolecular information (e.g., using CLARITY). By comparison, due to the challenges inherent to large-scale in vivo electrical recordings regarding unambiguous assignments of cells’ types and postmortem registration of their identities, circuit reconstructions in the very same cell ensembles recorded electrically are likely to be far more challenging.
Integrated optical studies in larger brains exacerbate the “big data” problem, which is already becoming a notable challenge in multiple subareas of neuroscience. Collaborations between neuroscientists and computer scientists will become increasingly important, and even essential, for the challenges of the next 25 years—not only for generating testable hypotheses arising from models of brain dynamics or machine learning research, but also for storing, handling, processing, and making accessible these vast data streams concurrent with the emergence of integrated and computational optical approaches. For example, large-scale Ca2+ recordings in mice will come to produce gigabytes per second of data, while CLARITY data sets for individual whole rodent brains can be ~1–10 terabytes in size, depending on the number of color channels (Figures 1 and 3). These optical data sets will soon grow to the ~10 petabyte scale and beyond, especially when larger brains including those of humans are examined at high resolution. However, conventional “cloud storage” approaches for large data sets are in many ways suboptimal for the kinds of data encountered in neuroscience, and computational/analytical methods will have to be profoundly accelerated simply to keep pace with the exhilarating new rate of data acquisition in neuroscience.
Lastly, we close with some remarks on how engineers and neuroscientists might fruitfully interact in the coming years. Traditionally, there often have not been conventional career paths, at least in academics, for engineers playing critical supporting roles in neuroscience research. In many cases, engineering departments might not view such activity as breaking sufficient ground in the engineering realm, whereas biology departments might not appreciate the crucial but underlying links to biological discovery. As the engineering challenges become increasingly severe for neuroscientists in the years ahead, with an upcoming deluge of sophisticated instrumentation and massive data sets, the neuroscience community will need to consider carefully how best to engage and retain the best, brightest, and most ambitious engineers.
Both the engineering and neuroscience communities might be well served by further appreciation of each other’s intellectual traditions and modus operandi. Engineers are typically motivated to address wide sets of problems that share central features, permitting common tools and approaches. Biologists are usually motivated to solve specific mysteries in detail. These are distinct intellectual mind sets, and the two communities can sometimes talk past each other. Engineers are generally well served by learning which classes of problems are the true stumbling blocks in biological science, rather than looking for suitable puzzles to solve after inventing a new widget. Biologists can benefit from enhanced appreciation of the intellectual potency of simultaneously examining all problems of a given category, an approach that has yielded many technologies that form the bedrock of modern biological research practice and infrastructure. In the coming years, neuroscientists and engineers will need (and want) to work more closely together than ever before, making “cross-cultural” exchange of ideas and working modes increasingly important for, and part of, the natural fabric of neuroscience.
Acknowledgments
K.D. acknowledges support from the Wiegers Family Fund, NIMH, NIDA, NSF, the DARPA REPAIR Program, and the Gatsby Charitable Foundation. M.J.S. acknowledges support from NIMH, NSF, the Paul Allen Family Foundation, DARPA, the Ellison Foundation, the Keck Foundation, NIDA, and NIBIB. M.J.S. is a cofounder and consults scientifically for Inscopix Inc., which has commercialized the miniature integrated microscope technology of Figure 1. K.D. is a cofounder and consults for Circuit Therapeutics Inc., which is using optogenetics to screen for medications and build devices for treating diseases in the peripheral nervous system; optogenetics tools, training, and protocols are freely available (http://www.optogenetics.org).
References
- Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- Akerboom J, Carreras Calderón N, Tian L, Wabnig S, Prigge M, Tolö J, Gordus A, Orger MB, Severi KE, Macklin JJ, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht DR, Bargmann CI. High-content behavioral analysis of Caenorhabditis elegans in precise spatiotemporal chemical environments. Nat Methods. 2011;8:599–605. doi: 10.1038/nmeth.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivisatos AP, Andrews AM, Boyden ES, Chun M, Church GM, Deisseroth K, Donoghue JP, Fraser SE, Lippincott-Schwartz J, Looger LL, et al. Nanotools for neuroscience and brain activity mapping. ACS Nano. 2013;7:1850–1866. doi: 10.1021/nn4012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anikeeva P, Andalman AS, Witten I, Warden M, Goshen I, Grosenick L, Gunaydin LA, Frank LM, Deisseroth K. Optetrode: a multi-channel readout for optogenetic control in freely moving mice. Nat Neurosci. 2012;15:163–170. doi: 10.1038/nn.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- Bepari AK, Sano H, Tamamaki N, Nambu A, Tanaka KF, Takebayashi H. Identification of optogenetically activated striatal medium spiny neurons by Npas4 expression. PLoS ONE. 2012;7:e52783. doi: 10.1371/journal.pone.0052783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- Berndt A, Schoenenberger P, Mattis J, Tye KM, Deisseroth K, Hegemann P, Oertner TG. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc Natl Acad Sci USA. 2011;108:7595–7600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti J, van Putten EG, Blum C, Lagendijk A, Vos WL, Mosk AP. Non-invasive imaging through opaque scattering layers. Nature. 2012;491:232–234. doi: 10.1038/nature11578. [DOI] [PubMed] [Google Scholar]
- Broxton M, Grosenick L, Yang S, Cohen N, Andalman A, Deisseroth K, Levoy M. Wave optics theory and 3-D deconvolution for the light field microscope. Opt Express. 2013;21:25418–25439. doi: 10.1364/OE.21.025418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Platisa J, Pieribone VA, Raccuglia D, Kunst M, Nitabach MN. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, Brill J, Bonnavion P, Huguenard JR, Huerta R, de Lecea L. Mechanism for Hypocretin-mediated sleep-to-wake transitions. Proc Natl Acad Sci USA. 2012;109:E2635–E2644. doi: 10.1073/pnas.1202526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha C, Liechty WB, Khademhosseini A, Peppas NA. Designing biomaterials to direct stem cell fate. ACS Nano. 2012;6:9353–9358. doi: 10.1021/nn304773b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DP, Deleavey GF, Owen SC, Damha MJ, Shoichet MS. Click conjugated polymeric immunonanoparticles for targeted siRNA and antisense oligonucleotide delivery. Biomaterials. 2013;34:8408–8415. doi: 10.1016/j.biomaterials.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Chang TY, Pardo-Martin C, Allalou A, Wählby C, Yanik MF. Fully automated cellular-resolution vertebrate screening platform with parallel animal processing. Lab Chip. 2012;12:711–716. doi: 10.1039/c1lc20849g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat Methods. 2013;10:508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- Chung K, Kim Y, Kanodia JS, Gong E, Shvartsman SY, Lu H. A microfluidic array for large-scale ordering and orientation of embryos. Nat Methods. 2011a;8:171–176. doi: 10.1038/nmeth.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Zhan M, Srinivasan J, Sternberg PW, Gong E, Schroeder FC, Lu H. Microfluidic chamber arrays for whole-organism behavior-based chemical screening. Lab Chip. 2011b;11:3689–3697. doi: 10.1039/c1lc20400a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi S, Orsborn AL, Moorman HG, Carmena JM. Design and analysis of closed-loop decoder adaptation algorithms for brain-machine interfaces. Neural Comput. 2013;25:1693–1731. doi: 10.1162/NECO_a_00460. [DOI] [PubMed] [Google Scholar]
- Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour S, Lavertu G, Dufour-Beauséjour S, Juneau-Fecteau A, Calakos N, Deschênes M, Vallée R, De Koninck Y. A multimodal micro-optrode combining field and single unit recording, multispectral detection and photolabeling capabilities. PLoS ONE. 2013;8:e57703. doi: 10.1371/journal.pone.0057703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar MJ, Wise FW, Fetcho JR, Schaffer CB. In vivo imaging of myelin in the vertebrate central nervous system using third harmonic generation microscopy. Biophys J. 2011;100:1362–1371. doi: 10.1016/j.bpj.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LS, Gerecht S, Fuller J, Shieh HF, Vunjak-Novakovic G, Langer R. Bioactive hydrogel scaffolds for controllable vascular differentiation of human embryonic stem cells. Biomaterials. 2007;28:2706–2717. doi: 10.1016/j.biomaterials.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AE, Schnitzer MJ. Miniaturized integration of a fluorescence microscope. Nat Methods. 2011;8:871–878. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilja V, Chestek CA, Diester I, Henderson JM, Deisseroth K, Shenoy KV. Challenges and opportunities for next-generation intracortically based neural prostheses. IEEE Trans Biomed Eng. 2011;58:1891–1899. doi: 10.1109/TBME.2011.2107553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godara P, Dubis AM, Roorda A, Duncan JL, Carroll J. Adaptive optics retinal imaging: emerging clinical applications. Optom Vis Sci. 2010;87:930–941. doi: 10.1097/OPX.0b013e3181ff9a8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Li JZ, Schnitzer MJ. Enhanced Archaerhodopsin Fluorescent Protein Voltage Indicators. PLoS ONE. 2013;8:e66959. doi: 10.1371/journal.pone.0066959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, Schneider MB, Deisseroth K. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J Neurosci. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- Haikala V, Joesch M, Borst A, Mauss AS. Optogenetic control of fly optomotor responses. J Neurosci. 2013;33:13927–13934. doi: 10.1523/JNEUROSCI.0340-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LT, Beart GC, Thomas EA, Simpson DA, McGuinness LP, Cole JH, Manton JH, Scholten RE, Jelezko F, Wrachtrup J, et al. High spatial and temporal resolution widefield imaging of neuron activity using quantum NV-diamond. Sci Rep. 2012;2:401. doi: 10.1038/srep00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Hochberg LR, Bacher D, Jarosiewicz B, Masse NY, Simeral JD, Vogel J, Haddadin S, Liu J, Cash SS, van der Smagt P, Donoghue JP. Reach and grasp by people with tetraplegia using a neurally controlled robotic arm. Nature. 2012;485:372–375. doi: 10.1038/nature11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton NG, Wang K, Kobat D, Clark CG, Wise FW, Schaffer CB, Xu C. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photonics. 2013;7:205–209. doi: 10.1038/nphoton.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JJ, Masella B, Dubra A, Sharma R, Yin L, Merigan WH, Palczewska G, Palczewski K, Williams DR. Images of photoreceptors in living primate eyes using adaptive optics two-photon ophthalmoscopy. Biomed Opt Express. 2010;2:139–148. doi: 10.1364/BOE.2.000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DC, Tsai PS, Kleinfeld D. Prospect for feedback guided surgery with ultra-short pulsed laser light. Curr Opin Neurobiol. 2012;22:24–33. doi: 10.1016/j.conb.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N, Sato TR, Betzig E. Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc Natl Acad Sci USA. 2012;109:22–27. doi: 10.1073/pnas.1109202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Han Z, Platisa J, Wooltorton JR, Cohen LB, Pieribone VA. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabra M, Robie AA, Rivera-Alba M, Branson S, Branson K. JAABA: interactive machine learning for automatic annotation of animal behavior. Nat Methods. 2013;10:64–67. doi: 10.1038/nmeth.2281. [DOI] [PubMed] [Google Scholar]
- Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, Hirata K, Ito J, Aita Y, Tsukazaki T, Hayashi S, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keplinger C, Sun JY, Foo CC, Rothemund P, Whitesides GM, Suo Z. Stretchable, transparent, ionic conductors. Science. 2013;341:984–987. doi: 10.1126/science.1240228. [DOI] [PubMed] [Google Scholar]
- Kim DH, Ghaffari R, Lu N, Rogers JA. Flexible and stretchable electronics for biointegrated devices. Annu Rev Biomed Eng. 2012;14:113–128. doi: 10.1146/annurev-bioeng-071811-150018. [DOI] [PubMed] [Google Scholar]
- Kim TI, McCall JG, Jung YH, Huang X, Siuda ER, Li Y, Song J, Song YM, Pao HA, Kim RH, et al. Injectable, cellular-scale opto-electronics with applications for wireless optogenetics. Science. 2013;340:211–216. doi: 10.1126/science.1232437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobat D, Durst ME, Nishimura N, Wong AW, Schaffer CB, Xu C. Deep tissue multiphoton microscopy using longer wavelength excitation. Opt Express. 2009;17:13354–13364. doi: 10.1364/oe.17.013354. [DOI] [PubMed] [Google Scholar]
- Kobat D, Horton NG, Xu C. In vivo two-photon microscopy to 1.6-mm depth in mouse cortex. J Biomed Opt. 2011;16:106014. doi: 10.1117/1.3646209. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq J, Schnitzer MJ. An infrared fluorescent protein for deeper imaging. Nat Biotechnol. 2011;29:715–716. doi: 10.1038/nbt.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liske H, Towne C, Anikeeva P, Zhao S, Feng G, Deisseroth K, Delp S. Optical inhibition of motor nerve and muscle activity in vivo. Muscle Nerve. 2013;47:916–921. doi: 10.1002/mus.23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahou P, Zimmerley M, Loulier K, Matho KS, Labroille G, Morin X, Supatto W, Livet J, Débarre D, Beaurepaire E. Multicolor two-photon tissue imaging by wavelength mixing. Nat Methods. 2012;9:815–818. doi: 10.1038/nmeth.2098. [DOI] [PubMed] [Google Scholar]
- Marshall JD, Schnitzer MJ. Optical strategies for sensing neuronal voltage using quantum dots and other semiconductor nanocrystals. ACS Nano. 2013;7:4601–4609. doi: 10.1021/nn401410k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson K, Loudin J, Goetz G, Huie P, Wang L, Kamins TI, Galambos L, Smith R, Harris JS, Sher A, Palanker D. Photovoltaic Retinal Prosthesis with High Pixel Density. Nat Photonics. 2012;6:391–397. doi: 10.1038/nphoton.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O’Shea DJ, Prakash R, Gunaydin LA, Hyun M, Fenno LE, Gradinaru V, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura M, Sasaki T, Sadakari J, Gengyo-Ando K, Kagawa-Nagamura Y, Kobayashi C, Ikegaya Y, Nakai J. Genetically encoded green fluorescent Ca2+ indicators with improved detectability for neuronal Ca2+ signals. PLoS ONE. 2012;7:e51286. doi: 10.1371/journal.pone.0051286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron D, Tal E, Silberberg Y. Scanningless depth-resolved microscopy. Opt Express. 2005;13:1468–1476. doi: 10.1364/opex.13.001468. [DOI] [PubMed] [Google Scholar]
- Owen SC, Fisher SA, Tam RY, Nimmo CM, Shoichet MS. Hyaluronic acid click hydrogels emulate the extracellular matrix. Langmuir. 2013;29:7393–7400. doi: 10.1021/la305000w. [DOI] [PubMed] [Google Scholar]
- Ozden I, Wang J, Lu Y, May T, Lee J, Goo W, O’Shea DJ, Kalanithi P, Diester I, Diagne M, et al. A coaxial optrode as multifunction write-read probe for optogenetic studies in non-human primates. J Neurosci Methods. 2013;219:142–154. doi: 10.1016/j.jneumeth.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirin S, Peterka DS, Yuste R. Instantaneous three-dimensional sensing using spatial light modulator illumination with extended depth of field imaging. Opt Express. 2013;21:16007–16021. doi: 10.1364/OE.21.016007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Samara C, Rohde CB, Gilleland CL, Norton S, Haggarty SJ, Yanik MF. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc Natl Acad Sci USA. 2010;107:18342–18347. doi: 10.1073/pnas.1005372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödel T, Prevedel R, Aumayr K, Zimmer M, Vaziri A. Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light. Nat Methods. 2013;10:1013–1020. doi: 10.1038/nmeth.2637. [DOI] [PubMed] [Google Scholar]
- Schultheis C, Liewald JF, Bamberg E, Nagel G, Gottschalk A. Optogenetic long-term manipulation of behavior and animal development. PLoS ONE. 2011;6:e18766. doi: 10.1371/journal.pone.0018766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy KV, Sahani M, Churchland MM. Cortical control of arm movements: a dynamical systems perspective. Annu Rev Neurosci. 2013;36:337–359. doi: 10.1146/annurev-neuro-062111-150509. [DOI] [PubMed] [Google Scholar]
- Sinha S, Liang L, Ho ETW, Urbanek KE, Luo L, Baer TM, Schnitzer MJ. High-speed laser microsurgery of alert fruit flies for fluorescence imaging of neural activity. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1216287110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam RY, Fuehrmann T, Mitrousis N, Shoichet MS. Regenerative Therapies for Central Nervous System Diseases: a Biomaterials Approach. Neuropsychopharmacology. 2013 doi: 10.1038/npp.2013.237. in press Published online September 4, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KF, Matsui K, Sasaki T, Sano H, Sugio S, Fan K, Hen R, Nakai J, Yanagawa Y, Hasuwa H, et al. Expanding the repertoire of optogenetically targeted cells with an enhanced gene expression system. Cell Rep. 2012;2:397–406. doi: 10.1016/j.celrep.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Testa I, Urban NT, Jakobs S, Eggeling C, Willig KI, Hell SW. Nanoscopy of living brain slices with low light levels. Neuron. 2012;75:992–1000. doi: 10.1016/j.neuron.2012.07.028. [DOI] [PubMed] [Google Scholar]
- Tibbitt MW, Anseth KS. Dynamic microenvironments: the fourth dimension. Sci Transl Med. 2012;4:60ps24. doi: 10.1126/scitranslmed.3004804. [DOI] [PubMed] [Google Scholar]
- Urban NT, Willig KI, Hell SW, Nägerl UV. STED nanoscopy of actin dynamics in synapses deep inside living brain slices. Biophys J. 2011;101:1277–1284. doi: 10.1016/j.bpj.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viventi J, Kim DH, Vigeland L, Frechette ES, Blanco JA, Kim YS, Avrin AE, Tiruvadi VR, Hwang SW, Vanleer AC, et al. Flexible, foldable, actively multiplexed, high-density electrode array for mapping brain activity in vivo. Nat Neurosci. 2011;14:1599–1605. doi: 10.1038/nn.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sugiyama Y, Hikima T, Sugano E, Tomita H, Takahashi T, Ishizuka T, Yawo H. Molecular determinants differentiating photo-current properties of two channelrhodopsins from chlamydomonas. J Biol Chem. 2009;284:5685–5696. doi: 10.1074/jbc.M807632200. [DOI] [PubMed] [Google Scholar]
- Watson BO, Nikolenko V, Araya R, Peterka DS, Woodruff A, Yuste R. Two-photon microscopy with diffractive optical elements and spatial light modulators. Front Neurosci. 2010:4. doi: 10.3389/fnins.2010.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt BA, Burns LD, Wei Ho ET, Ghosh KK, Mukamel EA, Schnitzer MJ. Advances in light microscopy for neuroscience. Annu Rev Neurosci. 2009;32:435–506. doi: 10.1146/annurev.neuro.051508.135540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt BA, Fitzgerald JE, Schnitzer MJ. Photon shot noise limits on optical detection of neuronal spikes and estimation of spike timing. Biophys J. 2013;104:51–62. doi: 10.1016/j.bpj.2012.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Stark E, Im M, Cho IJ, Yoon ES, Buzsáki G, Wise KD, Yoon E. An implantable neural probe with monolithically integrated dielectric waveguide and recording electrodes for optogenetics applications. J Neural Eng. 2013a;10:056012. doi: 10.1088/1741-2560/10/5/056012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liu L, Matsuda T, Zhao Y, Rebane A, Drobizhev M, Chang YF, Araki S, Arai Y, March K, et al. Improved orange and red Ca2+ indicators and photophysical considerations for optogenetic applications. ACS Chem Neurosci. 2013b;4:963–972. doi: 10.1021/cn400012b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011a;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011b;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, Prigge M, Berndt A, Cushman J, Polle J, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Cunha C, Zhang F, Liu Q, Gloss B, Deisseroth K, Augustine GJ, Feng G. Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain Cell Biol. 2008;36:141–154. doi: 10.1007/s11068-008-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci. 2013;16:264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]