Abstract
Coenzyme Q (CoQ, ubiquinone) is a redox active lipid produced across all domains of life that functions in electron transport and oxidative phosphorylation and whose deficiency causes human diseases. Yet, CoQ biosynthesis has not been fully defined in any organism. Several proteins with unclear molecular functions facilitate CoQ biosynthesis through unknown means, and multiple steps in the pathway are catalyzed by currently unidentified enzymes. Here we highlight recent progress toward filling these knowledge gaps through both traditional biochemistry and cutting-edge “omics” approaches. To help fill the remaining gaps, we present questions framed by the recently discovered CoQ biosynthetic complex and by putative biophysical barriers. Mapping CoQ biosynthesis, metabolism, and transport pathways has great potential to enhance treatment of numerous human diseases.
Keywords: Coenzyme Q, Ubiquinone, Biosynthesis, Metabolon, Protein Complex, complex Q, CoQ-synthome, Mitochondria, Lipids, Oxidative Phosphorylation, Mitochondrial Disease
Functions of Coenzyme Q and an Overview of its Biosynthesis
Coenzyme Q (CoQ; see Glossary and Box 1) plays a key role in an increasing number of biological processes, human diseases, and therapeutic regimens. While CoQ was discovered 60 years ago in Madison, Wisconsin [1] and Cambridge, England [2], gaps in our knowledge of CoQ biosynthesis, metabolism, and transport continue to limit our understanding of CoQ’s role in human disease and our ability to treat these diseases. Spurred by an influx of new experimental technologies merged with traditional biochemical approaches, such gaps in knowledge are now being filled rapidly, and these efforts have opened promising new therapeutic avenues.
Box 1. Nomenclature of CoQ Biosynthesis.
CoQ8 vs COQ8? The nomenclature of CoQ biochemistry involves subtle, yet meaningful, differences in capitalization.
CoQ
Coenzyme Q (CoQ), also known as ubiquinone (UQ), is the redox active lipid shown in Figure 1A. The names “CoQ” and “UQ” derive from its quinone head group (CoQ and ubiquinone), its function as a coenzyme (CoQ), and its nearly ubiquitous presence across all domains of life (ubiquinone). “CoQ” is the general abbreviation. To specify the number of isoprene subunits in the lipid tail, a subscripted number is included (e.g., “CoQ10”, indicating 10 isoprene subunits). CoQ tail length varies by organism: humans predominantly produce CoQ10; mice, CoQ9; E. coli, CoQ8; and S. cerevisiae, CoQ6. The redox state of the lipid can also be specified: the fully reduced form (the hydroquinone) as “CoQH2” (ubiquinol, UQH2), the singly dehydrogenated radical form (the semiquinone) as “CoQH·” (ubisemiquinone, UQH·), and the oxidized form (the quinone) as “CoQ” (ubiquinone, UQ) (Figure 1B). “Q” can also be found in the literature as an abbreviation for CoQ, but “CoQ” is favored to avoid confusion with the standard abbreviation for the amino acid glutamine (Q).
COQ8
COQ8 is an example of a protein required for CoQ biosynthesis. Eukaryotic genes and proteins required for CoQ biosynthesis are typically given “COQ” or “Coq” names, while their prokaryotic homologs typically have “Ubi” names (for ubiquinone). “COQ” or “Coq” proteins with the same numbers are typically homologs (e.g., human COQ5 and yeast Coq5p are homologs). Capitalization follows the standard format for each species (e.g., human proteins COQ8A and COQ8B, human genes COQ8A and COQ8B, yeast protein Coq8p, yeast gene coq8, E. coli protein UbiB, and E. coli gene ubiB).
Complex Q
The biosynthetic complex that produces CoQ is termed “complex Q” (a name analogous to that of the mitochondrial oxidative phosphorylation complexes I–V) or the “CoQ-synthome”. Complex Q is thought to contain proteins required for the terminal stage of CoQ biosynthesis (COQ3–COQ9), lipids (phospholipids and isoprenoid lipids), small molecule co-factors, and metal ions. However, a complete catalogue of the proteins, lipids, and metabolites that comprise complex Q remains to be determined.
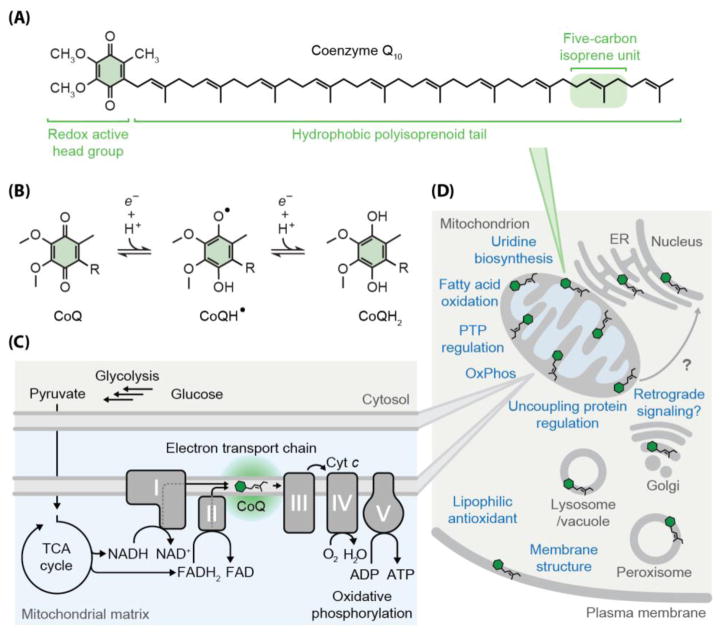
CoQ is a lipid with exceptional biochemical properties (Figure 1). The chemical structure of CoQ includes an extraordinarily long polyisoprenoid lipid tail, which makes it one of the most hydrophobic molecules in life (Figure 1A). This tail is capped by a quinone head group, which is reduction-oxidation (redox) active (Figure 1B) [3, 4]. The redox activity of CoQ allows it to function as a cofactor for numerous enzymes, including those of the mitochondrial electron transport chain (ETC) (Figure 1C). In the ETC, CoQ relays electrons from oxidative phosphorylation (OxPhos) complexes I and II to complex III. The redox chemistry of CoQ also involves protons (Figure 1B), the movement of which is part of the “Q-cycle” [5] that helps generate the proton motive force that drives ATP production via OxPhos.
Figure 1. Chemical and Biological Functions of Coenzyme Q.
(A) Chemical structure of coenzyme Q10 (CoQ10). (B) Reduction-oxidation reactions of the CoQ head group. “R” indicates the polyisoprenoid tail. (C) Cartoon indicating the central role of CoQ in the electron transport chain and mitochondrial oxidative phosphorylation (OxPhos). I–V, OxPhos complexes I–V; Cyt c, cytochrome c; TCA cycle, tricarboxylic acid cycle. (D) Overview of the widespread cellular functions of CoQ. Within mitochondria, CoQ supports and/or regulates the mitochondrial permeability transition pore (PTP) [10], mitochondrial uncoupling proteins [9], uridine biosynthesis [7], fatty acid oxidation [8], and OxPhos. More broadly, CoQ functions as a lipophilic antioxidant [12–14] and a membrane stabilizer [15]. In plants, plastoquinone functions as a retrograde signal from chloroplasts to the nucleus [16], suggesting that mitochondrial CoQ could play an analogous role.
In addition to CoQ’s fundamental role in OxPhos, which it performs across all domains of life [6], broader cellular functions for CoQ are increasingly recognized (Figure 1D). Within mitochondria, CoQ is a critical cofactor for uridine biosynthesis [7], fatty acid oxidation [8], and mitochondrial uncoupling proteins [9], and it is thought to regulate the permeability transition pore [10]. Moreover, an increasing number of extra-mitochondrial functions are coming to light for CoQ, which is found in nearly all eukaryotic cellular membranes [11]. For example, CoQ functions as a lipophilic antioxidant in yeast and in human plasma lipoproteins [12–14] and as a membrane stabilizer [15]. Furthermore, plastoquinone, a plant CoQ analog, can act as a retrograde chloroplast-to-nucleus signal [16], suggesting that mitochondrial CoQ may play an analogous role.
Given its widespread cellular functions, CoQ has powerful and complex effects on health and disease. On one hand, genetic defects in CoQ biosynthesis can cause diseases such as myopathies and ataxias (Box 2), and CoQ abundance decreases with age [17]. On the other hand, genetic disruption of CoQ biosynthesis or dietary depletion of CoQ in C. elegans [18–21] or mice [22] can increase lifespan. The complex mechanisms driving these contrasting effects remain largely undefined [23], in part because of numerous gaps in our understanding of CoQ biochemistry.
Box 2. CoQ Deficiency in the Clinic.
Primary CoQ deficiencies—genetic diseases due to mutations in genes encoding CoQ biosynthesis proteins—are known to be caused by nuclear DNA mutations in COQ2 [145–148], COQ4 [149, 150], COQ6 [151, 152], COQ7 [153], COQ8A (ADCK3) [136, 154–158], COQ8B (ADCK4) [118, 159], COQ9 [160, 161], PDSS1 [162], or PDSS2 [163, 164] (Figure IA). These diseases variably disrupt multiple organ systems. Commonly affected organs include the brain (e.g., encephalopathy, seizures, or cerebellar ataxia), the heart (e.g., hypertrophic cardiomyopathy), the kidneys (e.g., nephrotic syndrome), and skeletal muscles (e.g., myopathy)—organs with high demand for mitochondrial oxidative phosphorylation.
Mutations in some COQ genes (e.g., COQ2) can affect nearly all organ systems. In contrast, other COQ gene mutations have more selective effects. For example, mutations in COQ8A (ADCK3) primarily disrupt brain function, with cerebellar ataxia as the predominant presentation, while mutations in COQ8B (ADCK4) primarily disrupt kidney function, with steroid resistant nephrotic syndrome (SNRS) as the primary feature. Why mutations in different COQ genes have such distinct effects remains unclear. Tissue-specific differences in gene expression or metabolism-regulated protein-protein interactions may play a role [87, 117].
Different mutations within an individual COQ gene can also have widespread clinical presentations with highly variable age of onset—ranging from the day of birth to late adulthood—and differentially affected organ systems. Even the same COQ point mutation can present variably across different patients, potentially due to differences in heteroplasmic mitochondrial DNA (mtDNA) pools, which can affect mitochondrial-nuclear genome interactions. Together, these factors create an extremely complicated scenario for affected families, clinicians, and scientists.
CoQ deficiency can also occur secondary to mutations in genes not directly linked to CoQ biosynthesis such as ETFDH [165], MUT [166], APTX [167], and BRAF [168] (Figure IB), but the underlying mechanisms are unclear. Some drugs, such statins, may also cause secondary CoQ deficiency [169], and gene-drug interactions have been suggested by case reports [170].
Over half of the reported clinical cases of CoQ deficiency lack a molecular diagnosis (an identified causative gene mutation) [171]. Recent biochemical work in model systems nominates additional genes as candidates for currently undefined primary CoQ deficiency syndromes (Figure IC), providing an important foundation for ongoing investigations. Further basic science studies are needed to complete the list of candidate COQ disease genes.
The outstanding knowledge gaps and highly variable gene-phenotype relationships of CoQ deficiencies are an important microcosm of similar multifactorial complexity observed in essentially all mitochondrial diseases [172]. Ongoing research has great potential to help guide treatment strategies (Box 3) and genetic counseling.
Figure I. CoQ Deficiency in Human Health and Disease.
(A) Network of phenotypes and genes associated with primary CoQ deficiency. Lines indicate phenotypes reported to be caused by mutations in the indicate gene(s). (B) Diseases, conditions, drugs, and genes linked to secondary CoQ deficiency. (C) Candidate disease genes for potential primary CoQ deficiencies that have not yet been reported to exist in patients. “(?)” indicates an unproven ortholog relationship to a yeast gene that has been linked to CoQ biosynthesis. Additional uncharacterized (orphan) genes may also be linked to CoQ biosynthesis (e.g., the gene that encodes the unidentified C1 hydroxylase).
CoQ is synthesized endogenously by organisms across all domains of life [6]. Moreover, nearly every mammalian cell produces CoQ, likely because CoQ is poorly absorbed into cells and tissues [24]. The poor absorption of CoQ hinders treatment of CoQ deficiencies in the clinic, but new strategies to bypass this problem are under development (Box 3). Most eukaryotic CoQ is generated at the inner mitochondrial membrane [25], but some CoQ may be produced outside of mitochondria [26–28]. Here, we focus on eukaryotic mitochondrial CoQ biosynthesis, with an emphasis on CoQ production in mammals and in the yeast Saccharomyces cerevisiae, which has been a workhorse model organism for studying CoQ. However, many of the principles discussed also apply to prokaryotic [29, 30] and extra-mitochondrial CoQ production.
Box 3. Therapeutic Strategies for Treating CoQ Deficiency.
Early diagnosis of primary CoQ deficiencies is critical because they are among the very few mitochondrial diseases that can be treatable. Therapy should be initiated as soon as possible to limit irreversible tissue damage [173]. The urgency is such that prior to diagnostic tests, newborn siblings of affected individuals have been treated [148].
The first-line therapy for CoQ deficiency is oral supplementation with high dose exogenous CoQ10. Oral CoQ10 treatment has been at least partially successful in a number of cases of primary CoQ deficiency including those caused by some mutations in COQ2 [145, 173], COQ4 [149], COQ6 [151], COQ7 [153], and COQ8B [118, 159]. However, clinical response to CoQ10 is highly variable, and treatment failure is unfortunately common.
Delivering CoQ to specific cells in need, such as cerebellar Purkinje cells [87], is a major challenge. Exogenous CoQ is efficiently taken up by liver [174, 175], ovarian [176], and brown adipose tissues [177]. However, tissues commonly disrupted by CoQ deficiency, such as the brain and muscle (Box 2), poorly uptake CoQ [178] (Figure IA). Moreover, exogenous CoQ must traverse multiple subcellular compartments to reach the inner mitochondrial membrane (Figure IB). Work in yeast [79, 80, 179] and mouse [174, 175] model systems demonstrates that such CoQ transport is possible, with potential involvement of the endomembrane system [131, 132], but the precise mechanisms are unclear. A recent study showed improved delivery of reduced CoQ (CoQH2) compared to oxidized CoQ [180]. However, uptake is still considered to be inefficient, and new strategies for enhancing CoQ delivery are needed.
Recently, an alternative treatment strategy has emerged that uses CoQ “bypass” precursors to sidestep enzymatic defects. This strategy leverages water soluble CoQ head group precursors, which are predicted to be more bioavailable than CoQ. Mice with defective COQ7 were treated with 2,4-dihydroxybenzoate (2,4-DHB), an unnatural CoQ head group precursor, which was able to bypass the COQ7-catalyzed C6-hydroxylation, elevate CoQ levels, and extend lifespan [181] (Figure IC). 2,4-DHB treatment was similarly effective in human fibroblasts with COQ7 mutations [153] and mice with COQ9 mutations [182]. Building on pioneering work in yeast [89], a similar strategy using vanillic acid or 3,4-dihydroxybenzoate can likewise bypass defects in COQ6 [152]. Importantly, this bypass strategy appears to be limited to COQ mutations that disrupt enzyme activity but maintain complex Q stability, and to select unnatural head group precursors [111].
New strategies for boosting CoQ levels could have widespread impact beyond treatment of primary CoQ deficiencies. CoQ10 supplementation is ubiquitous in the treatment of diverse mitochondrial diseases [183], often as part of a “mitochondrial cocktail” [184], and CoQ10 has been proposed as a therapy for numerous common diseases including Azheimer’s, Parkinson’s, and Huntington’s diseases [185, 186].
Figure I. Strategies for Treatment of CoQ Deficiency.
(A) Efficient uptake of exogenous CoQ into rat tissues after intraperitoneal injection is limited to white blood cells, liver, and spleen. Lesser uptake occurs in other tissues. Data in figure from Bentinger et al [178]. (B) Exogenous CoQ must be transported across multiple lipid membranes and multiple aqueous compartments to reach the mitochondrial inner membrane, but the mechanisms facilitating this CoQ transport are unclear. (C) An example of the “CoQ intermediate bypass strategy” for treating select CoQ deficiencies. 4-hydroxybenzoate (4-HB) is the natural CoQ head group precursor. 2,4-dihydroxybenzoate (2,4-DHB) be used as an alternative head group precursor to bypass a defect in COQ7 [181].
CoQ biosynthesis requires: (i) head group production, (ii) tail production—including isoprene biosynthesis and tail polymerization, (iii) attachment of the tail to the head group, and (iv) a series of head group modifications (Figure 2, Key Figure, and Table 1). In general, CoQ biosynthesis intermediates become increasingly lipophilic as they progress through the pathway, which spans multiple cellular compartments and numerous “omic planes” (e.g., metabolome, lipidome), making it well-suited for investigation with emerging multi-omic technologies (Box 4). Many questions about CoQ biosynthesis remain unanswered, as framed here in the context of existing knowledge about each phase of the pathway.
Figure 2. Current Model for the Eukaryotic CoQ Biosynthesis Pathway.
Scheme of eukaryotic CoQ biosynthesis with currently unidentified enzymes indicated by question marks. The primary CoQ pathway, which is conserved from yeast to humans, is depicted. A secondary CoQ pathway in yeast uses para-aminobenzoate (pABA) as the head group precursor instead of 4-hydroxybenzoate (4-HB) [39, 40], and Coq6p and Coq9p subsequently support C4-deamination via C4-hydroxylation [64, 140]. ‘R’ indicates the polyisoprenoid tail (Figure 1A), which would likely be anchored in the mitochondrial inner membrane. “+” symbol by the arrow from COQ8 and COQ9 indicates a hypothesized supportive role for the indicated reaction. AADAT, mitochondrial alpha-aminoadipate aminotransferase; ALDH3A1, aldehyde dehydrogenase 3A1; FDXR, adrenodoxin reductase; FDX2 (FDX1L), mitochondrial ferredoxin 2 (ferredoxin 1-like); PDSS1, prenyl (decaprenyl) diphosphate synthase subunit 1; TAT, tyrosine aminotransferase; DMAPP, dimethylallyl pyrophosphate; GPP, geranyl pyrophosphate; FPP, farnesyl pyrophosphate; IPP, isopentenyl pyrophosphate; 4-HPP, 4-hydroxyphenylpyruvate; 4-HBz, 4-hydroxybenzaldehyde; PPHB, polyprenyl-hydroxybenzoate; PPDHB, polyprenyl-dihydroxybenzoate; PPVA, polyprenyl-vanillic acid; DDMQ, demethoxy-demethyl-coenzyme Q; DMQ, demethoxy-coenzyme Q; DMeQ, demethyl-coenzyme Q.
Table 1.
Reactions and Enzymes of CoQ Biosynthesis
| Reaction | Eukaryotic Reaction Diagrama | Cofactors in Eukaryotes | Mammalian Enzyme(s)b | Yeast Enzyme | Prokaryotic Enzyme(s)c | Refs |
|---|---|---|---|---|---|---|
| 4-HBz dehydrogenation |

|
NAD+ | ALDH3A1 (?) | Hfd1p | Unidentified | [34,42] |
| Isoprene polymerization |
|
Mg2+ | PDSS1 and PDSS2 | Coq1p | IspA and IspB | [54,163] |
| C1- Hydroxylation |

|
Unidentified | Unidentified | Unidentified | UbiH | [69,110] |
| C1- Decarboxylation | Unidentified | Unidentified | Unidentifiedd | UbiD and UbiX | [96,99] | |
| C2-Methylation |

|
SAM, Mg2+ | COQ5 | Coq5p | UbiE | [75,76,103] |
| C3- Prenylation |

|
Mg2+ | COQ2 | Coq2p | UbiA | [55,145,188] |
| C4- Deamination |

|
FAD and NAD(P)H | N.A. | Coq6p | N.A. | [64] |
| C5- Hydroxylation |

|
FAD and NAD(P)H | COQ6 | Coq6p | UbiI | [89,90] |
| C6- Hydroxylation |

|
Diiron center, NADH | COQ7 | Coq7p | UbiF | [103,106,108] |
| O-Methylations |

|
SAM, Zn2+ | COQ3 | Coq3p | UbiG | [91–93] |
| ATP Hydrolysis or Phosphorylation? |
|
Mg2+ | COQ8A (ADCK3) and COQ8B (ADCK4) | Coq8p | UbiB | [80,82,86,87,96,135] |
| Lipid Binding? | N.A. | COQ9 | Coq9p | UbiJ? | [83,116,139,144] | |
| Lipid Transport? | N.A. | COQ10A and COQ10B | Coq10p | UbiJ? | [141, 142, 144] | |
| Electron Transfers | Fe-S cluster | FDX2 (FDX1L) | Yah1p | Unidentified | [40] | |
| Electron Transfers | NADPH | FDXR | Arh1p | Unidentified | [40] | |
| Head group Reduction? | Unidentified | Unidentified | Coq11p (?) | Unidentified | [115] | |
| Head group Oxidation? | Unidentified | Unidentified | Coq11p (?) | Unidentified | [115] |
“R” indicates polyisoprenoid tail. 4-HB, 4-hydroxybenzoate; 4-HBz, 4-hydroxybenzaldehyde; PPHB, polyprenyl-hydroxybenzoate; PPDHB, polyprenyl-dihydroxybenzoate; PPVA, polyprenyl-vanillic acid; DDMQ, demethoxy-demethyl-coenzyme Q; DMQ, demethoxy-coenzyme Q; DMeQ, demethyl-coenzyme Q.
ALDH3A1, aldehyde dehydrogenase 3A1; FDXR, adrenodoxin reductase; FDX2 (FDX1L), mitochondrial ferredoxin 2 (ferredoxin 1-like); PDSS1, prenyl (decaprenyl) diphosphate synthase subunit 1.
Some, but not all, of these prokaryotic enzymes are sequence homologs of the eukaryotic enzymes.
The Saccharomyces cerevisiae genome, but not the human genome, encodes sequence homologs of E. coli UbiD and UbiX: Fdc1p and Pad1p, respectively. However, whether yeast Pad1p or Fdc1p functions in CoQ biosynthesis remains unclear [101]. Interestingly, a recent large-scale screen showed a significant increase in CoQ in yeast lacking Fdc1p when cultured under respiration conditions (lipidome data available at y3kproject.org) [42].
Box 4. Multi-omic Investigation of Mitochondrial Pathways.
A grand challenge in mitochondrial biology is to completely characterize the functions of all ~1,200 mammalian mitochondrial proteins, ~20% of which are currently “mitochondrial uncharacterized (x) proteins” (MXPs) [117]. Accomplishing this goal would enhance understanding of how mitochondria impact human health and disease. Proven approaches for characterizing MXPs include both traditional biochemical methods and cutting-edge “omics” methods (Figure I).
As complex organelles composed of dynamic and intertwined populations of proteins, lipids, and soluble metabolites, mitochondria are also prime targets for emerging “multi-omic” (“transomic”) approaches. Multi-omic investigations integrate data across multiple “omic planes” (Figure I), which can provide deeper and more confident insight than that of an individual omic method. Large-scale multi-omic approaches were recently used to discover previously unknown facets of mitochondrial biochemistry in mice [187] and yeast [42].
The CoQ biosynthesis pathway provides a salient example of a multi-omic mitochondrial process. CoQ-related molecules span numerous omic planes (Figure I). Early CoQ precursors are small soluble metabolites, the terminal CoQ intermediates are lipids, and COQ proteins (e.g., the enzymes in the pathway) form a protein complex (complex Q) and are encoded byCOQ mRNAs and COQ genes. As such, the outstanding gaps in knowledge about CoQ biosynthesis are aptly positioned for study using multi-omics.
A large-scale multi-omic project in yeast recently uncovered a role for the enzyme Hfd1p in producing the CoQ head group precursor 4-hydroxybenzoate (4-HB) (Figure I) [42]. Notably, this function of Hfd1p was independently validated and co-discovered via an orthogonal genetic approach [34]. The discovery via multi-omics hinged on integration of both metabolomics, which uncovered diagnostic changes in the metabolites 4-HB and 4-hydroxybenzaldehyde, and lipidomics, which revealed telltale changes in lipid CoQ intermediates. The large-scale nature of this work also helped highlight the biological importance of a statistically significant, but small in magnitude, decrease in 4-HB observed in yeast that lack Hfd1p because this change was unique across all strains in the study. We hypothesize that similar multi-omic approaches will be central to filling the remaining knowledge gaps in CoQ biosynthesis and, more broadly, key for completely mapping mitochondrial biology.
Figure I. A Multi-Omic Approach to Mitochondrial Biochemistry.
Overview of methods used for multi-omic investigation of mitochondrial pathways, using CoQ biosynthesis as a key example. The cartoon indicates gaps in knowledge (?) that were recently filled using a multi-omic approach (Hfd1p) [34, 42] or that are targets for linking a currently undefined mitochondrial uncharacterized protein (MXP) to CoQ biosynthesis.
CoQ Head Group Biosynthesis
In mammals, the CoQ head group is derived from the essential amino acid phenylalanine, which is converted into tyrosine (Tyr) and, subsequently, 4-hydroxybenzoate (4-HB) [31–33], but the responsible enzymes are unidentified (Figure 2). A Tyr-to-4-HB pathway also operates in yeast [34]. However, unlike mammals, bacteria and yeast can produce 4-HB de novo through the shikimate pathway [30, 35–38]. Yeast can also use p-aminobenzoate (pABA) as an alternative head group precursor [39, 40]. Because typical yeast culture media contain either 4-HB (in rich media) or pABA (in synthetic media), the Tyr-to-4-HB pathway is often not required for growth under laboratory conditions. It is likely for this reason that the responsible genes have been bypassed in many genetic screens. Recognizing this technical barrier and using distinct methods, two independent studies recently identified the first and last enzymes in the yeast Tyr-to-4-HB pathway [34, 41, 42].
Tyrosine Transamination by Aro8p or Aro9p
The yeast aromatic amino acid transaminases Aro8p and Aro9p were recently linked to the first step of the Tyr-to-4-HB pathway, initially through either hypothesis-driven heavy atom tracing studies [34] or unbiased global protein co-expression studies [42] (Figure 2). The analogous mammalian enzymes remain unidentified, but the Aro8p/Aro9p homolog AADAT and the tyrosine aminotransferase TAT are candidates.
4-Hydroxybenzaldehyde Dehydrogenation by Hfd1p
While steps after the tyrosine transamination remain unclear, Hfd1p was recently discovered to catalyze the final step in the Tyr-to-4-HB pathway—dehydrogenation of 4-hydroxybenzaldehyde (4-HBz) to generate 4-HB (Figure 2) [34, 42]. One of four human co-orthologs of Hfd1p, ALDH3A1, was also shown to catalyze 4-HB production in vitro and to rescue the growth defect of yeast that lack Hfd1p. However, whether ALDH3A1 participates in endogenous CoQ biosynthesis in mammalian cells remains unknown. The independent and complementary approaches used to discover this activity for Hfd1p and ALDH3A1 are summarized in Box 4.
Because Hfd1p is localized to the mitochondrial outer membrane [43] and exogenous 4-HB can rescue CoQ production in yeast lacking Hfd1p, we predict that 4-HB is the head group precursor that is transported across the inner mitochondrial membrane into the mitochondrial matrix, but this has not yet been demonstrated. Moreover, the putative mitochondrial membrane transporter for the head group precursor remains unidentified (Figure 2).
Biosynthesis and Attachment of the Polyisoprenoid Tail
In mammals, the isoprene subunits for CoQ biosynthesis are generated through the mevalonate pathway [32, 44–46], stitched together by head-to-tail polymerization [47, 48], and attached to the head group via an electrophilic aromatic substitution [49] (Figure 2). The responsible prenyltransferases, Coq1p (PDSS1 and PDSS2) and Coq2p (COQ2) (see Glossary and Box 1 for “COQ” nomenclature) are localized to the inner mitochondrial membrane [25, 50–59], a key point marking the switch from cytosolic precursor generation to mitochondrial CoQ biosynthesis. Coq1p, which independently determines the CoQ tail length [60], is peripherally associated with the matrix face of the inner mitochondrial membrane [56], while Coq2p is an integral membrane protein [59, 61].
The mechanism by which the highly polar isopentenyl pyrophosphate (IPP) molecule is transported from the cytosol to the mitochondrial matrix is unknown. We hypothesize that an IPP transporter exists in the inner mitochondrial membrane (Figure 2), but it remains unidentified. An allylic pyrophosphate is also needed for the isoprene polymerization reactions. In vitro evidence suggests that Coq1p can use dimethylallyl pyrophosphate (DMAPP), geranyl pyrophosphate (GPP), or farnesyl pyrophosphate (FPP) [51], but which is used in vivo is unclear. DMAPP could potentially be formed by IPP isomerase activity, which has been inferred to exist in mitochondria [25] (Figure 2), but the presence and identity of this enzyme is unclear. If an IPP isomerase is not active in the mitochondrial matrix, then a transporter for the allylic pyrophosphate is also likely to exist.
The Terminal Stage of CoQ Biosynthesis – Head Group Modifications
In the terminal stage of CoQ biosynthesis, the head group is modified by a decarboxylation and a series of hydroxylations and methylations (Figure 2). In bacteria and in yeast, the added methyl groups and hydroxyl groups derive from S-adenosyl methionine (SAM) [62] and O2 [63, 64], respectively, and the same is predicted to be true in mammals. The proposed order of these reactions is based on isolation of CoQ intermediates from various strains of bacteria [65–69] and yeast [52, 53, 70–72].
Hypothesized Chemical Logic
The proposed sequence of CoQ head group modifications (Figure 2) follows the chemical logic of electrophilic aromatic substitution (EAS) reactions. First, the C4-hydroxyl of 4-HB acts as an ortho-directing group to enhance EAS at C3 and C5. Second, the C4-hydoxyl acts as a para-directing group to enhance EAS at C1. Third, the newly installed C1-hydroxyl group acts as an ortho-directing group to enhance EAS at C2 and C6. This EAS chemistry, combined with substrate orientation in enzyme active sites, likely affords the regioselectivity needed to produce CoQ, yet these hypotheses are untested in vitro. A precise understanding of this chemistry could assist design of new CoQ intermediate “bypass precursors” for treating primary CoQ deficiencies (Box 3).
Sub-cellular Location of COQ Proteins
The proteins required for the terminal phase of CoQ production localize to the mitochondrial matrix. Mammalian COQ3–COQ9 were localized to mitochondria in the “MitoCarta” study [57], and COQ3, COQ5, COQ6, COQ8A/B, and COQ9 were localized to the mitochondrial matrix via an “APEX” study [58]. Biochemical fractionation studies localized yeast Coq3p–Coq9p to the mitochondrial matrix and, importantly, showed that they are peripherally associated with the matrix face of the inner membrane [59, 73–84]. An exception to this peripheral association is COQ8A, which has a transmembrane helix [85–87], although its yeast ortholog Coq8p may be peripherally associated [59, 81].
COQ enzymes might also co-localize to extra-mitochondrial locations. A nuclear form of COQ7 was suggested to function in mitochondrial-nuclear retrograde signaling [88], but this remains debated [21]. Additionally, the COQ2 homolog UBIAD1 localizes to the Golgi membrane, where it supports Golgi CoQ production [28]. Whether the downstream CoQ biosynthesis proteins (COQ3–9) also localize to the Golgi is unclear, but, interestingly, Golgi dysmorphology was observed in mice that lack COQ8A [87].
C5-Hydroxylation by COQ6
The C5-hydroxylation is catalyzed by COQ6 in eukaryotes [89] (Figure 2) and UbiI in prokaryotes [90]. Coq6p is a predicted flavin-dependent monooxygenase that uses flavin adenine dinucleotide (FAD) as a cofactor and NAD(P)H as a coenzyme for reduction of FAD [89]. A structure of truncated UbiI revealed a Rossman-like fold—a widespread structural fold that can bind nucleotide cofactors—with a FAD binding site [90], but no in vitro enzymology was reported.
In yeast, the C5-hydroxylation requires the ferredoxin Yah1p—an essential iron-sulfur protein that mediates electron transfers in numerous pathways—and the ferredoxin reductase Arh1p [40], which are predicted to provide electrons to Coq6p (Figure 2). FDXR and FDX2 are the mammalian orthologs of Arh1p and Yah1p, respectively, but whether they function in mammalian CoQ biosynthesis is unknown.
O-Methylation by COQ3
The O-methylations are catalyzed by COQ3 in eukaryotes [70, 91, 92] and UbiG in prokaryotes [73, 93–95]. Purified UbiG or mitochondrial extracts containing COQ3 can catalyze both CoQ head group O-methylations in vitro in a reaction that depends on SAM and a divalent cation (e.g., Zn2+ or Co2+) [73, 92, 94, 95]. The molecular basis for the regioselectivity of COQ3 for each of its reactions (e.g., selective methylation of the DMeQ C6-hydroxyl over the C1-hydroxyl, see Figure 2) is unknown.
An Unidentified Decarboxylase
UbiD catalyzes the decarboxylation in prokaryotes [96], but the eukaryotic decarboxylase remains unidentified. Evidence for the mammalian decarboxylation was demonstrated by lack of radioactivity in CoQ when [carbonyl-C14]-4-HBz was used as a precursor [97]. Polyprenyl-vanillic acid (Figure 2) has been isolated from a mutant yeast strain [71], suggesting that C5-methoxy group installation occurs before the decarboxylation. A second prokaryotic enzyme, UbiX, was recently shown to support the decarboxylation by generating a previously unknown prenylated-FMN (prFMN) cofactor used by UbiD [98–100]. Whether prFMN functions similarly in eukaryotes is unclear [101]. No sequence homolog of UbiD or UbiX exists in humans and, thus, the decarboxylation appears to be a mechanistic deviation between prokaryotes and eukaryotes. A possibility to consider is that the eukaryotic C1-hydroxylation could be part of the decarboxylation mechanism.
An Unidentified Hydroxylase
UbiH catalyzes the C1-hydroxylation in Escherichia coli [69], but the eukaryotic enzyme has not been identified. Unlike the C5-hydroxylation described above, the eukaryotic C1-hydroxylation does not require Arh1p and Yah1p and may not require FAD [40], suggesting a different enzyme mechanism. In contrast, prokaryotic UbiH is likely a flavin-dependent monooxygenase, as it shares 30% sequence identity with UbiI, the C5-hydroxylase described above [90].
C2-Methylation by COQ5
The C2-methylation is catalyzed by COQ5 in eukaryotes [75–77, 102] and UbiE in prokaryotes [103, 104]. Lysates containing Coq5p or UbiE can catalyze demethoxy-demethyl-CoQ (DDMQ) C2-methylation in vitro in a reaction that depends on SAM and NADH [75]. Why NADH enhances this reaction is unclear, but we hypothesize that NADH helps keep the head group in the reduced state, which would enhance electrophilic aromatic substitution. Recently, X-ray structures of Coq5p revealed a Rossmann-like fold with a SAM-dependent methyltransferase domain [105], but no in vitro activity was reported.
C6-Hydroxylation by COQ7
The C6-hydroxylation is catalyzed by COQ7 in eukaryotes [19, 79, 106, 107] and UbiF in prokaryotes [103]. COQ7 is a carboxylate-bridged diiron hydroxylase [107], and its mechanism involves NADH passing electrons through demethoxy-CoQ (DMQ) to the diiron center [108, 109]. In contrast, E. coli UbiF is predicted to be a flavin-dependent monooxygenase similar to UbiI and UbiH, the C5- and C1- hydroxylases described above [90]. Why prokaryotes use highly similar enzymes for all three CoQ head group hydroxylations [110], but eukaryotes use different enzyme chemistries, is unknown.
The CoQ Biosynthetic Complex
The enzymes in the terminal phase of CoQ biosynthesis form a biosynthetic complex termed complex Q (or the “CoQ-synthome”), which is located on the matrix face of the inner mitochondrial membrane.
The presence of a yeast CoQ biosynthetic complex was initially suggested by genetic studies that showed interdependence between Coq proteins: deleting any one of the genes coq3–9 caused depletion of numerous Coq proteins and accumulation of the same CoQ precursor, PPHB [72, 111, 112] (Figure 2). Subsequently, physical evidence for complex Q was uncovered through protein-protein interaction studies and native gel electrophoresis experiments [59, 84, 113–115]. These studies also demonstrated that lipids, including CoQ and CoQ intermediates, are part of complex Q [115].
Recently, evidence for a mammalian complex Q has also been revealed. Global mass spectrometry proteomics studies demonstrated that deletion of Coq8a or Coq9 in mice causes selective and significant depletion of numerous COQ proteins [87, 116], not just that encoded by the deleted gene. Furthermore, a mitochondria-focused affinity enrichment mass spectrometry (AE-MS) study revealed a dynamic network of protein-protein interactions between COQ3–COQ9 [117]. Numerous complex Q interactions were also confirmed by cell-free protein expression and purification studies [117], and by immunoprecipitation-immunoblot studies of COQ5 [77], COQ8A [87], and COQ8B [118]. Additionally, native gel electrophoresis studies tracking COQ5 suggested the presence of a high molecular weight complex Q in human cells [102]. Select lipids and small molecules have also been shown to bind to individual complex Q proteins, such as COQ9 and COQ8A [87, 116], suggesting that mammalian complex Q contains nonproteinacious components. Together, these studies provide robust support for complex Q in mammals.
Similar “metabolons”, defined as “supramolecular complexes of sequential metabolic enzymes and cellular structural elements” [119], have been observed in other systems, such as the tricarboxylic acid cycle [120], the purinosome [121], dhurrin biosynthesis [122], and steroid biosynthesis [123]. Identifying the functions of such metabolon organization remains an active area of research with multiple hypotheses. First, metabolons are thought to enhance flux via enzyme proximity, local concentration of substrates, or substrate channeling. Second, metabolons may sequester potentially toxic biosynthetic intermediates (e.g., redox active CoQ intermediates) or help keep potentially poisonous enzymes (e.g., the hydroxylase COQ7) from escaping and promiscuously disrupting other processes. Third, metabolons may be key for regulation via allosteric enzyme activation, post-translational modifications, or organizing assembly in time and space. Finally, metabolons could also help solve biophysical problems, such as the barriers to CoQ biosynthesis and transport discussed in the next section.
While numerous exciting questions about complex Q components, structure, assembly, regulation, and activity remain unanswered (Figure 3 and Outstanding Questions), the discovery of this CoQ metabolon has dramatically altered the way that we think about each individual COQ protein and the biosynthetic process as a whole.
Figure 3. Models Framing Outstanding Questions and Knowledge Gaps in CoQ Biochemistry.
(A) Model framing questions about complex Q assembly, regulation of CoQ biosynthesis, and mechanisms for CoQ transport. (B) Model for a biophysical barrier to accessing lipophilic CoQ intermediates, highlighting the currently unclear mechanism for solving this problem. CoQ and hydrophobic CoQ intermediates are predicted to reside near the lipid bilayer midplane (i) with some movement (gray arrows) toward the bilayer surface (ii), but not beyond the glycerol backbones [129, 130]. Biochemical mechanisms for moving CoQ intermediates past the layer of glycerol backbones and polar lipid head groups are unclear. (C) Models framing unanswered questions about complex Q biochemistry. (D) Summary of the currently incomplete state of complex Q structural biology. X-ray structures have been reported for Coq5p (PDB IDs 4OBW and 4OBX) [105], COQ8A (ADCK3) (PDB IDs 4PED and 5I35) [86] [87], and COQ9 (PBD ID 4RHP) [116].
Outstanding Questions Box.
What enzymes catalyze the undefined steps in the Tyr-to-4-HB pathway in yeast and in mammals?
How are the CoQ biosynthesis precursors—4-HB and isoprenoid subunits—transported into mitochondria?
What eukaryotic enzymes catalyze the C1 decarboxylation and the C1 hydroxylation of CoQ biosynthesis? In eukaryotes, does the C1 hydroxylation occur before, after, or in concert with the C1 decarboxylation?
What are the precise biochemical functions of the MXPs COQ4, COQ8, COQ9, COQ10, and Coq11p in CoQ biosynthesis?
How are lipophilic CoQ intermediates extracted from the membrane to be acted upon by COQ enzymes?
What proteins, lipids, and metabolites constitute complex Q? What is their stoichiometry?
What is the three-dimensional structure of complex Q?
How is complex Q anchored to the mitochondrial membrane?
Can a functional complex Q be reconstituted in vitro?
Does complex Q activity require its endogenous context on the inner mitochondrial membrane?
What is the complex Q assembly pathway? Are assembly factors required? How is complex Q assembly seeded?
What benefit does organizing the COQ proteins in a biosynthetic complex provide? Does complex Q enable substrate channeling? Does complex Q sequester CoQ intermediates or promiscuous COQ enzymes?
Does complex Q localize to a specific region of the inner mitochondrial membrane?
Does complex Q interact with any of the oxidative phosphorylation complexes?
How is complex Q activity regulated and coordinated with other mitochondrial processes, such as mitochondrial biogenesis and OxPhos?
How is CoQ abundance regulated through biosynthesis and turnover?
How are COQ proteins regulated transcriptionally, post-transcriptionally, and post-translationally?
Do canonical COQ proteins function in extra-mitochondrial CoQ biosynthesis?
How is CoQ transported from the inner mitochondrial membrane to the outer mitochondrial membrane?
How is CoQ transported from mitochondria to other organelles throughout the cell?
Biophysical Barriers in CoQ Biosynthesis
Transmembrane Transport of Polar Precursors and Proteins
The precursors for building CoQ are negatively charged small molecules that cannot passively cross the inner mitochondrial membrane (Figure 2). How these polar CoQ precursors are transported from their site of production outside of mitochondria into the mitochondrial matrix is unclear.
The COQ proteins, which are encoded by nuclear DNA, must also be transported into the mitochondrial matrix. Mitochondrial protein import and processing is a complex process that can help coordinate and regulate biochemical pathways [124]. Although some transcriptional regulators [125] and post-transcriptional coordinators [126] of CoQ biosynthesis have been identified, the specific mitochondrial import and processing pathways of COQ proteins are incompletely defined (Figure 3A). As leads for future work, the import of yeast Coq2p is mediated by the Tim9p-Tim10p complex [127], the import of mouse COQ7 requires a mitochondrial targeting sequence and ATP, but not the mitochondrial membrane potential [128], and a recent large-scale study linked the mitochondrial intermediate peptidase Oct1p to CoQ biosynthesis [42].
Lipophilic Quinone Inaccessibility
CoQ resides primarily in the midplane of the lipid bilayer [129, 130], between the distal ends of the phospholipid tails (Figure 3B). This creates a biophysical problem for its transport to membranes outside of the mitochondria, where it is known to be present [11]. We predict that a series of CoQ lipid transport pathways exist (Figure 3A), likely involving both specific lipid transport proteins and the endomembrane system [131, 132]. These CoQ transport pathways are also likely to function in the reverse process—transport of extracellular CoQ into mitochondria—which has important implications for treatment of CoQ deficiency via delivery of exogenous CoQ (see Box 3 for further discussion of this point and evidence for CoQ transport).
We predict that the hydrophobic quinone-containing CoQ intermediates (Figure 2) are also localized to the lipid bilayer midplane (Figure 3B). In contrast, CoQ biosynthesis enzymes are primarily peripheral membrane proteins, located at the membrane surface. How these peripheral membrane enzymes access substrates located deep in the midplane of the lipid bilayer is unclear (Figure 3B). One or more of the biochemically uncharacterized COQ proteins could potentially help solve these biophysical problems.
Uncharacterized Proteins in CoQ Biosynthesis
A number of genes that support CoQ biosynthesis encode proteins of unknown molecular function. These CoQ-related mitochondrial uncharacterized (x) proteins (MXPs) include COQ4, COQ8A (ADCK3), COQ8B (ADCK4), COQ9, COQ10A, COQ10B, and Coq11p.
COQ4 – Complex Q Scaffold?
Coq4p stabilizes complex Q and is required for efficient CoQ biosynthesis [113, 133], but the underlying mechanism is undefined. Coq4p has homologs in some proteobacteria, which share a motif predicted to bind a divalent cation [133], but the specific biochemical functions of Coq4p remain unclear. The current model is that COQ4 is a complex Q scaffold protein that binds both proteins and lipids, as suggested by an X-ray structure of a COQ4 homolog that co-crystallized with geranylgeranyl monophosphate (PDB 3KB4).
COQ8 – ATPase or Kinase?
The ancient UbiB family of enzymes [134], which includes COQ8, is found across all domains of life. E. coli UbiB [96, 135], yeast Coq8p [80], and mammalian COQ8A (ADCK3) [87, 136] and COQ8B (ADCK4) [118] each enhance CoQ biosynthesis, but their precise biochemical activities remain obscure. Coq8p is required for complex Q integrity, and Coq8p overexpression has a remarkable ability to rescue complex Q stability in various mutant yeast strains that would otherwise be complex Q deficient [59, 111]. Yet, how Coq8p stabilizes complex Q is unknown. A physical and functional interaction between COQ8 and COQ5 may be particularly important, as suggested by robust physical interactions between COQ8A and COQ5 [87, 117], and the common occurrence of prokaryotic operons containing both ubiB and ubiE [30], the COQ8 and COQ5 orthologs, respectively.
Coq8p and its UbiB family members fit within the protein kinase-like (PKL) superfamily [137], which includes protein kinases, lipid kinases, and ATPases. Initially, Coq8p was hypothesized to be a protein kinase. Indeed, phosphorylation of Coq3p, Coq5p, and Coq7p is altered in Δcoq8 yeast [81, 82], but whether this effect is caused directly by the absence of Cop8p is unclear. More recently, an X-ray structure of COQ8A revealed a PKL fold with unique features that inhibit protein kinase activity and afford an unusual preference for binding ADP over ATP [86]. Furthermore, COQ8A was shown to lack canonical protein kinase activity in trans, and to instead bind lipid CoQ intermediates and have ATPase activity that is enhanced by binding to cardiolipin-containing liposomes [87, 138]. Based on these findings, new models for COQ8A function were presented [87], including the possibility that it acts primarily as an ATPase, or that its ATPase activity is indicative of small molecule or lipid kinase activity against an undiscovered substrate. Yet, precisely how COQ8’s ATPase activity (or kinase activity) and lipid binding function might stabilize complex Q to enhance CoQ biosynthesis remains an open question.
COQ9 – Lipid Sensor or Precursor Presenter?
Coq9p and COQ9 stabilize complex Q through an unclear mechanism [83, 84, 116, 139]. Recently, an X-ray structure of COQ9 revealed a lipid-binding pocket that is important for function in vivo [116], but the endogenous lipid ligands remain undefined. Coq9p interacts physically and functionally with Coq7p to enhance the C6-hydroxylation reaction [116]. Coq9p also appears to enhance Coq6p activity and the deamination of CoQ intermediates derived from pABA [64, 140]. Our current model is that COQ9 helps extract hydrophobic CoQ intermediates from the membrane and presents them to other COQ enzymes, such as COQ7. An alternative model is that COQ9 is a lipid sensor that allosterically regulates complex Q upon ligand binding.
COQ10 – Lipid Transporter?
Coq10p, COQ10A, and COQ10B localize to mitochondria [57, 141]. Coq10p contains a conserved lipid-binding domain that binds CoQ and CoQ intermediates in vitro [141, 142]. CoQ biosynthesis is less efficient in yeast with coq10 mutations, and the CoQ produced is used less efficiently for OxPhos [142], but the molecular basis for these effects is unclear. Neither Coq10p nor Coq9p has a known homolog in E. coli. However, the recently discovered E. coli CoQ biosynthesis proteins UbiJ and UbiK, which physically bind each other and a lipid [143, 144], could be functional homologs of Coq10p or Coq9p.
Coq11p – Dehydrogenase or Reductase?
Yeast Coq11p (YLR290c) was recently linked to CoQ biosynthesis via immunoprecipitation studies of complex Q [115]. Unlike most coq gene deletion yeast strains, yeast lacking Coq11p retain respiratory growth and are not completely CoQ deficient [42, 115]. The primary sequence of Coq11p places it in the short chain dehydrogenase/reductase (SDR) superfamily [115], but its biochemical activity is unclear.
We hope that the specific questions and knowledge gaps highlighted by this review, especially those related to complex Q function and biophysical barriers (Figure 3), will help guide ongoing research to definitively characterize the molecular functions of these CoQ-related MXPs.
Concluding Remarks and Future Perspectives
In recent years, the known functions of CoQ in human health and disease have expanded, rekindling the drive to fully define its biosynthesis. CoQ now has numerous defined functions outside of its primary role in mitochondrial OxPhos, including regulation of mitochondrial and possibly extra-mitochondrial functions (Figure 1). Likewise, human diseases with links to CoQ biosynthesis have expanded to include common diseases such as Parkinson’s disease, rare diseases such as a cerebellar ataxia, and even the aging process itself (Box 2), elevating the potential impact of therapies that boost CoQ levels (Box 3).
Given the importance of CoQ in human biology and the fact that it was discovered 60 years ago [1, 2], it may be surprising that we still do not fully understand its biosynthesis. This apparent disconnect highlights the challenges associated with studying CoQ biosynthesis, which include unstable membrane-bound proteins, fleeting biosynthetic intermediates, highly hydrophobic lipids, and numerous undefined biochemical components. Fortunately, advances in protein and lipid biochemistry, X-ray crystallography, mass spectrometry, and multi-omic systems biology (Box 4) are rising to meet the challenge. Yet, numerous knowledge gaps still exist (Figures 2 and 3 and Outstanding Questions). We hope that this review—by defining gaps in knowledge about CoQ biosynthesis at the biochemical level—will catalyze further progress toward understanding CoQ in human health and disease.
Trends Box.
A biosynthetic complex for producing coenzyme Q (CoQ) was recently revealed in yeast and mammals. This complex likely contains proteins, lipids, and polar small molecules, but its precise composition, structure, and activities remain largely unknown.
Multiple mitochondrial uncharacterized proteins (MXPs) have been linked to CoQ biosynthesis, and multiple steps in the pathway are enabled by currently unidentified proteins.
Recent progress was made toward understanding the biochemistry of a dehydrogenase, a deaminase, a lipid-binding protein, and a protein kinase-like enzyme in the CoQ pathway.
CoQ biosynthesis spans multiple “omic” planes—the metabolome, the lipidome, and the proteome—and thus has proven to be a prime target for investigation with emerging multi-omic technologies.
Mapping CoQ biochemistry recently spurred new therapeutic strategies.
Acknowledgments
We thank all members of the Pagliarini lab, especially fellow “Q Branch” colleagues Andrew Reidenbach and Danielle Lohman, for helpful discussions. This work was supported by National Institutes of Health (NIH) grants R01GM112057 and R01GM115591 (to D.J.P.); and Ruth L. Kirschstein NRSA F30AG043282 (to J.A.S.).
Glossary
- CoQ
Coenzyme Q (CoQ) is the redox-active lipid product of the biosynthetic pathway discussed here. See Box 1 for further explanations of CoQ nomenclature.
- COQ5
COQ5 is an example of a human enzyme required for CoQ biosynthesis.
- Coq5p
Coq5p is an example of a yeast enzyme required for CoQ biosynthesis.
- Complex Q (CoQ-synthome)
Complex Q, also known as the CoQ-synthome, is the biosynthetic complex that produces CoQ.
- MXP
A mitochondrial uncharacterized (x) protein (MXP) is a protein that localizes to mitochondria but lacks a well-defined molecular or cellular function in mitochondria.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crane FL, et al. Isolation of a quinone from beef heart mitochondria. Biochim Biophys Acta. 1957;25:220–221. doi: 10.1016/0006-3002(57)90457-2. [DOI] [PubMed] [Google Scholar]
- 2.Morton RA. Ubiquinone. Nature. 1958;182:1764–1767. doi: 10.1038/1821764a0. [DOI] [PubMed] [Google Scholar]
- 3.Lester RL, et al. Coenzyme Q: a new group of quinones. Journal of the American Chemical Society. 1958;80:4751–4752. [Google Scholar]
- 4.Wolf DEHCH, Trenner NR, Arison BH, Shunk CH, Linn BO, McPherson JF, Folkers K. Coenzyme Q. I. Structure Studies on the Coenzyme Q Group. J Am Chem Soc. 1958;80:4752. [Google Scholar]
- 5.Mitchell P. Protonmotive redox mechanism of the cytochrome b-c1 complex in the respiratory chain: protonmotive ubiquinone cycle. FEBS Lett. 1975;56:1–6. doi: 10.1016/0014-5793(75)80098-6. [DOI] [PubMed] [Google Scholar]
- 6.Lester RL, Crane FL. The natural occurrence of coenzyme Q and related compounds. J Biol Chem. 1959;234:2169–2175. [PubMed] [Google Scholar]
- 7.Jones ME. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu Rev Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- 8.Frerman FE. Acyl-CoA dehydrogenases, electron transfer flavoprotein and electron transfer flavoprotein dehydrogenase. Biochem Soc Trans. 1988;16:416–418. doi: 10.1042/bst0160416. [DOI] [PubMed] [Google Scholar]
- 9.Echtay KS, et al. Coenzyme Q is an obligatory cofactor for uncoupling protein function. Nature. 2000;408:609–613. doi: 10.1038/35046114. [DOI] [PubMed] [Google Scholar]
- 10.Fontaine E, et al. A ubiquinone-binding site regulates the mitochondrial permeability transition pore. J Biol Chem. 1998;273:25734–25740. doi: 10.1074/jbc.273.40.25734. [DOI] [PubMed] [Google Scholar]
- 11.Sastry SP, et al. Distribution of coenzyme Q in rat liver cell fractions. Nature. 1961;189:577. doi: 10.1038/189577a0. [DOI] [PubMed] [Google Scholar]
- 12.Do TQ, et al. Enhanced sensitivity of ubiquinone-deficient mutants of Saccharomyces cerevisiae to products of autoxidized polyunsaturated fatty acids. Proc Natl Acad Sci U S A. 1996;93:7534–7539. doi: 10.1073/pnas.93.15.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stocker R, et al. Ubiquinol-10 protects human low density lipoprotein more efficiently against lipid peroxidation than does alpha-tocopherol. Proc Natl Acad Sci U S A. 1991;88:1646–1650. doi: 10.1073/pnas.88.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frei B, et al. Ubiquinol-10 is an effective lipid-soluble antioxidant at physiological concentrations. Proc Natl Acad Sci U S A. 1990;87:4879–4883. doi: 10.1073/pnas.87.12.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sevin DC, Sauer U. Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat Chem Biol. 2014;10:266–272. doi: 10.1038/nchembio.1437. [DOI] [PubMed] [Google Scholar]
- 16.Petrillo E, et al. A chloroplast retrograde signal regulates nuclear alternative splicing. Science. 2014;344:427–430. doi: 10.1126/science.1250322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalen A, et al. Age-related changes in the lipid compositions of rat and human tissues. Lipids. 1989;24:579–584. doi: 10.1007/BF02535072. [DOI] [PubMed] [Google Scholar]
- 18.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 19.Ewbank JJ, et al. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- 20.Larsen PL, Clarke CF. Extension of life-span in Caenorhabditis elegans by a diet lacking coenzyme Q. Science. 2002;295:120–123. doi: 10.1126/science.1064653. [DOI] [PubMed] [Google Scholar]
- 21.Liu JL, et al. A single biochemical activity underlies the pleiotropy of the aging-related protein CLK-1. Scientific Reports. 2017;7:859. doi: 10.1038/s41598-017-00754-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, et al. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Hekimi S. Understanding Ubiquinone. Trends Cell Biol. 2016;26:367–378. doi: 10.1016/j.tcb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Miles MV. The uptake and distribution of coenzyme Q10. Mitochondrion. 2007;7(Suppl):S72–77. doi: 10.1016/j.mito.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Momose K, Rudney H. 3-Polyprenyl-4-hydroxybenzoate synthesis in the inner membrane of mitochondria from p-hydroxybenzoate and isopentenylpyrophosphate. A demonstration of isoprenoid synthesis in rat liver mitochondria. J Biol Chem. 1972;247:3930–3940. [PubMed] [Google Scholar]
- 26.Kalen A, et al. Nonaprenyl-4-hydroxybenzoate transferase, an enzyme involved in ubiquinone biosynthesis, in the endoplasmic reticulum-Golgi system of rat liver. J Biol Chem. 1990;265:1158–1164. [PubMed] [Google Scholar]
- 27.Kalen A, et al. Ubiquinone biosynthesis by the microsomal fraction from rat liver. Biochim Biophys Acta. 1987;926:70–78. doi: 10.1016/0304-4165(87)90183-8. [DOI] [PubMed] [Google Scholar]
- 28.Mugoni V, et al. Ubiad1 is an antioxidant enzyme that regulates eNOS activity by CoQ10 synthesis. Cell. 2013;152:504–518. doi: 10.1016/j.cell.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke CF. New advances in coenzyme Q biosynthesis. Protoplasma. 2000;213:134–147. [Google Scholar]
- 30.Aussel L, et al. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta. 2014;1837:1004–1011. doi: 10.1016/j.bbabio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Bentley RR, VG, Springer CM, Dialameh GH, Olson RE. The Origin of the Benzoquinone Ring of Coenzyme Q9 in the Rat. Biochem Biophys Res Commun. 1961;5:443–446. [Google Scholar]
- 32.Olson RE, et al. Benzoate Derivatives as Intermediates in the Biosynthesis of Coenzyme Q in the Rat. J Biol Chem. 1963;238:3146–3148. [PubMed] [Google Scholar]
- 33.Booth ANM, MS, Robbins DJ, Emerson OH, Jones FT, Deeds F. Urinary Phenolic Acid Metabolites of Tyrosine. J Biol Chem. 1960;235:2649–2652. [Google Scholar]
- 34.Payet LA, et al. Mechanistic Details of Early Steps in Coenzyme Q Biosynthesis Pathway in Yeast. Cell Chem Biol. 2016;23:1241–1250. doi: 10.1016/j.chembiol.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Gibson MIGF, Doy CH, Morgan P. The Branch Point in the Biosynthesis of the Aromatic Amino-Acids. Nature. 1962;195:1173–1175. [Google Scholar]
- 36.Morgan PNG, MI, Gibson F. Conversion of Shikimic Acid to Aromatic Compounds. Nature. 1962;194:1239–1241. [Google Scholar]
- 37.Cox GB, Gibson F. Biosynthesis of Vitamin K and Ubiquinone. Relation to the Shikimic Acid Pathway in Escherichia Coli. Biochim Biophys Acta. 1964;93:204–206. doi: 10.1016/0304-4165(64)90285-5. [DOI] [PubMed] [Google Scholar]
- 38.Lawrence J, et al. Biosynthesis of ubiquinone in Escherichia coli K-12: biochemical and genetic characterization of a mutant unable to convert chorismate into 4-hydroxybenzoate. J Bacteriol. 1974;118:41–45. doi: 10.1128/jb.118.1.41-45.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marbois B, et al. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J Biol Chem. 2010;285:27827–27838. doi: 10.1074/jbc.M110.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierrel F, et al. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem Biol. 2010;17:449–459. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 41.Enriquez JA, et al. Hypothesis Driven versus Hypothesis-free: Filling the Gaps in CoQ Biosynthesis. Cell Metab. 2016;24:525–526. doi: 10.1016/j.cmet.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 42.Stefely JA, et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat Biotechnol. 2016;34:1191–1197. doi: 10.1038/nbt.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zahedi RP, et al. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell. 2006;17:1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gloor U, Wiss O. On the biosynthesis of ubiquinone (50) Arch Biochem Biophys. 1959;83:216–222. doi: 10.1016/0003-9861(59)90026-8. [DOI] [PubMed] [Google Scholar]
- 45.Olson RE, et al. Studies on Coenzyme Q. Pattern of Labeling in Coenzyme Q9 after Administration of Isotopic Acetate and Aromatic Amino Acids to Rats. J Biol Chem. 1965;240:514–523. [PubMed] [Google Scholar]
- 46.Gold PH, Olson RE. Studies on coenzyme Q. The biosynthesis of coenzyme Q9 in rat tissue slices. J Biol Chem. 1966;241:3507–3516. [PubMed] [Google Scholar]
- 47.Chaykin S, et al. Phosphorylated Intermediates in the Synthesis of Squalene. Proc Natl Acad Sci U S A. 1958;44:998–1004. doi: 10.1073/pnas.44.10.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lynen FEH, Henning U, Kessel I. Farnesyl-pyrophosphat und 3-Methyl-d3-butenyl-1-pyrophosphat, die biologischen Vorstufen des Squalens. Angewandte Chemie. 1958;70:738–742. [Google Scholar]
- 49.Lynen FA, BW, Eggerer H, Henning U, Moslein EM. g,g-Dimethyl-allyl-pyrophosphat und Geranyl-pyrophosphat, biologische Vorstufen des Squalens. Angewandte Chemie. 1959;71:657–684. [Google Scholar]
- 50.Winrow MJ, Rudney H. The incorporation of p-hydroxybenzoic acid and isopentenyl pyrophphate into ubiquinone precursors by cell-free preparations of rat tissues. Biochem Biophys Res Commun. 1969;37:833–840. doi: 10.1016/0006-291x(69)90967-x. [DOI] [PubMed] [Google Scholar]
- 51.Casey J, Threlfall DR. Formation of 3-hexaprenyl-4-hydroxybenzoate by matrix-free mitochondrial membrane-rich preparations of yeast. Biochim Biophys Acta. 1978;530:487–502. doi: 10.1016/0005-2760(78)90168-6. [DOI] [PubMed] [Google Scholar]
- 52.Tzagoloff A, et al. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem. 1975;250:8228–8235. [PubMed] [Google Scholar]
- 53.Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashby MN, Edwards PA. Elucidation of the deficiency in two yeast coenzyme Q mutants. Characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J Biol Chem. 1990;265:13157–13164. [PubMed] [Google Scholar]
- 55.Ashby MN, et al. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J Biol Chem. 1992;267:4128–4136. [PubMed] [Google Scholar]
- 56.Gin P, Clarke CF. Genetic evidence for a multi-subunit complex in coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase. J Biol Chem. 2005;280:2676–2681. doi: 10.1074/jbc.M411527200. [DOI] [PubMed] [Google Scholar]
- 57.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhee HW, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He CH, et al. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim Biophys Acta. 2014;1841:630–644. doi: 10.1016/j.bbalip.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okada K, et al. Polyprenyl diphosphate synthase essentially defines the length of the side chain of ubiquinone. Biochim Biophys Acta. 1996;1302:217–223. doi: 10.1016/0005-2760(96)00064-1. [DOI] [PubMed] [Google Scholar]
- 61.Cheng W, Li W. Structural insights into ubiquinone biosynthesis in membranes. Science. 2014;343:878–881. doi: 10.1126/science.1246774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackman LM, et al. Methionine as the source of methyl groups for ubiquinone and vitamin K: a study using nuclear magnetic resonance and mass spectrometry. Biochim Biophys Acta. 1967;141:1–7. doi: 10.1016/0304-4165(67)90239-5. [DOI] [PubMed] [Google Scholar]
- 63.Alexander K, Young IG. Three hydroxylations incorporating molecular oxygen in the aerobic biosynthesis of ubiquinone in Escherichia coli. Biochemistry. 1978;17:4745–4750. doi: 10.1021/bi00615a023. [DOI] [PubMed] [Google Scholar]
- 64.Ozeir M, et al. Coq6 is responsible for the C4-deamination reaction in coenzyme Q biosynthesis in Saccharomyces cerevisiae. J Biol Chem. 2015;290:24140–24151. doi: 10.1074/jbc.M115.675744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olsen RKS, JL, Daves GD, Moore HW, Folkers K, Parson WW, Rudney H. 2-Decaprenylphenol, Biosynthetic Precursor of Ubiquinone-10. J Am Chem Soc. 1965;87:2298–2300. doi: 10.1021/ja01088a045. [DOI] [PubMed] [Google Scholar]
- 66.Friis PD, GD, Folkers K. Complete Sequence of Biosynthesis from p-Hydroxybenzoic Acid to Ubiquinone. J Am Chem Soc. 1966;88:4754–4756. [Google Scholar]
- 67.Olsen RKD, GD, Moore HW, Folkers K, Parson WW, Rudney H. 2-Multiprenylphenols and 2-Decaprenyl-6-methoxyphenol, Biosynthetic Precursors of Ubiquinones. J Am Chem Soc. 1966;88:5919–5923. doi: 10.1021/ja00976a036. [DOI] [PubMed] [Google Scholar]
- 68.Cox GB, et al. Mutant strains of Escherichia coli K-12 unable to form ubiquinone. J Bacteriol. 1968;95:1591–1598. doi: 10.1128/jb.95.5.1591-1598.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young IG, et al. Pathway for ubiquinone biosynthesis in Escherichia coli K-12: gene-enzyme relationships and intermediates. J Bacteriol. 1973;114:42–52. doi: 10.1128/jb.114.1.42-52.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goewert RR, et al. Identification of 3,4-dihydroxy-5-hexaprenylbenzoic acid as an intermediate in the biosynthesis of ubiquinone-6 by Saccharomyces cerevisiae. Biochemistry. 1981;20:4217–4223. doi: 10.1021/bi00517a041. [DOI] [PubMed] [Google Scholar]
- 71.Goewert RR, et al. Identification of 3-methoxy-4-hydroxy-5-hexaprenylbenzoic acid as a new intermediate in ubiquinone biosynthesis by Saccharomyces cerevisiae. Biochemistry. 1981;20:5611–5616. doi: 10.1021/bi00522a040. [DOI] [PubMed] [Google Scholar]
- 72.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7(Suppl):S62–71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hsu AY, et al. Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry. 1996;35:9797–9806. doi: 10.1021/bi9602932. [DOI] [PubMed] [Google Scholar]
- 74.Belogrudov GI, et al. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch Biochem Biophys. 2001;392:48–58. doi: 10.1006/abbi.2001.2448. [DOI] [PubMed] [Google Scholar]
- 75.Barkovich RJ, et al. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J Biol Chem. 1997;272:9182–9188. doi: 10.1074/jbc.272.14.9182. [DOI] [PubMed] [Google Scholar]
- 76.Dibrov E, et al. The COQ5 gene encodes a yeast mitochondrial protein necessary for ubiquinone biosynthesis and the assembly of the respiratory chain. J Biol Chem. 1997;272:9175–9181. doi: 10.1074/jbc.272.14.9175. [DOI] [PubMed] [Google Scholar]
- 77.Nguyen TP, et al. Molecular characterization of the human COQ5 C-methyltransferase in coenzyme Q10 biosynthesis. Biochim Biophys Acta. 2014;1841:1628–1638. doi: 10.1016/j.bbalip.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gin P, et al. The Saccharomyces cerevisiae COQ6 gene encodes a mitochondrial flavin-dependent monooxygenase required for coenzyme Q biosynthesis. J Biol Chem. 2003;278:25308–25316. doi: 10.1074/jbc.M303234200. [DOI] [PubMed] [Google Scholar]
- 79.Jonassen T, et al. Yeast Clk-1 homologue (Coq7/Cat5) is a mitochondrial protein in coenzyme Q synthesis. J Biol Chem. 1998;273:3351–3357. doi: 10.1074/jbc.273.6.3351. [DOI] [PubMed] [Google Scholar]
- 80.Do TQ, et al. A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. J Biol Chem. 2001;276:18161–18168. doi: 10.1074/jbc.M100952200. [DOI] [PubMed] [Google Scholar]
- 81.Tauche A, et al. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 2008;8:1263–1275. doi: 10.1111/j.1567-1364.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 82.Xie LX, et al. Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim Biophys Acta. 2011;1811:348–360. doi: 10.1016/j.bbalip.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johnson A, et al. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J Biol Chem. 2005;280:31397–31404. doi: 10.1074/jbc.M503277200. [DOI] [PubMed] [Google Scholar]
- 84.Hsieh EJ, et al. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch Biochem Biophys. 2007;463:19–26. doi: 10.1016/j.abb.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Khadria AS, et al. A Gly-zipper motif mediates homodimerization of the transmembrane domain of the mitochondrial kinase ADCK3. J Am Chem Soc. 2014;136:14068–14077. doi: 10.1021/ja505017f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stefely JA, et al. Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Mol Cell. 2015;57:83–94. doi: 10.1016/j.molcel.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stefely JA, et al. Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity. Mol Cell. 2016;63:608–620. doi: 10.1016/j.molcel.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Monaghan RM, et al. A nuclear role for the respiratory enzyme CLK-1 in regulating mitochondrial stress responses and longevity. Nat Cell Biol. 2015;17:782–792. doi: 10.1038/ncb3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ozeir M, et al. Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem Biol. 2011;18:1134–1142. doi: 10.1016/j.chembiol.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 90.Hajj Chehade M, et al. ubiI, a new gene in Escherichia coli coenzyme Q biosynthesis, is involved in aerobic C5-hydroxylation. J Biol Chem. 2013;288:20085–20092. doi: 10.1074/jbc.M113.480368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clarke CF, et al. Ubiquinone biosynthesis in Saccharomyces cerevisiae. Isolation and sequence of COQ3, the 3,4-dihydroxy-5-hexaprenylbenzoate methyltransferase gene. J Biol Chem. 1991;266:16636–16644. [PubMed] [Google Scholar]
- 92.Jonassen T, Clarke CF. Isolation and functional expression of human COQ3, a gene encoding a methyltransferase required for ubiquinone biosynthesis. J Biol Chem. 2000;275:12381–12387. doi: 10.1074/jbc.275.17.12381. [DOI] [PubMed] [Google Scholar]
- 93.Stroobant P, et al. Mutants of Escherichia coli K-12 blocked in the final reaction of ubiquinone biosynthesis: characterization and genetic analysis. J Bacteriol. 1972;109:134–139. doi: 10.1128/jb.109.1.134-139.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leppik RA, et al. Membrane-associated reactions in ubiquinone biosynthesis. 2-Octaprenyl-3-methyl-5-hydroxy-6-methoxy-1,4-benzoquinone methyltransferase. Biochim Biophys Acta. 1976;428:146–156. doi: 10.1016/0304-4165(76)90116-1. [DOI] [PubMed] [Google Scholar]
- 95.Poon WW, et al. Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J Biol Chem. 1999;274:21665–21672. doi: 10.1074/jbc.274.31.21665. [DOI] [PubMed] [Google Scholar]
- 96.Cox GB, et al. Biosynthesis of ubiquinone in Escherichia coli K-12: location of genes affecting the metabolism of 3-octaprenyl-4-hydroxybenzoic acid and 2-octaprenylphenol. J Bacteriol. 1969;99:450–458. doi: 10.1128/jb.99.2.450-458.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rudney H, Parson WW. The Conversion of P-Hydroxybenzaldehyde to the Benzoquinone Ring of Ubiquinone in Rhodospirillum Rubrum. J Biol Chem. 1963;238:3137–3138. [PubMed] [Google Scholar]
- 98.Gulmezian M, et al. The role of UbiX in Escherichia coli coenzyme Q biosynthesis. Arch Biochem Biophys. 2007;467:144–153. doi: 10.1016/j.abb.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.White MD, et al. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature. 2015;522:502–506. doi: 10.1038/nature14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Payne KA, et al. New cofactor supports alpha,beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature. 2015;522:497–501. doi: 10.1038/nature14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Clarke CF, Allan CM. Biochemistry: Unexpected role for vitamin B2. Nature. 2015;522:427–428. doi: 10.1038/nature14536. [DOI] [PubMed] [Google Scholar]
- 102.Yen HC, et al. Disruption of the human COQ5-containing protein complex is associated with diminished coenzyme Q10 levels under two different conditions of mitochondrial energy deficiency. Biochim Biophys Acta. 2016;1860:1864–1876. doi: 10.1016/j.bbagen.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 103.Young IGM, LM, Stroobant P, Gibson F. Chacaterization and Genetic Analysis of Mutant Strains of Escherichia coli K-12 Accumulating the Ubiquinone Precursors 2-Octaprenyl-6-Methoxy-1,4-Benzoquinone and 2-Octaprenyl-3-Methyl-6-Methoxy-1,4-Benzoquinone. J Bacteriol. 1971;105:769–778. doi: 10.1128/jb.105.3.769-778.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee PT, et al. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J Bacteriol. 1997;179:1748–1754. doi: 10.1128/jb.179.5.1748-1754.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dai YN, et al. Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway. Acta Crystallogr D Biol Crystallogr. 2014;70:2085–2092. doi: 10.1107/S1399004714011559. [DOI] [PubMed] [Google Scholar]
- 106.Marbois BN, Clarke CF. The COQ7 gene encodes a protein in saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J Biol Chem. 1996;271:2995–3004. doi: 10.1074/jbc.271.6.2995. [DOI] [PubMed] [Google Scholar]
- 107.Stenmark P, et al. A new member of the family of di-iron carboxylate proteins. Coq7 (clk-1), a membrane-bound hydroxylase involved in ubiquinone biosynthesis. J Biol Chem. 2001;276:33297–33300. doi: 10.1074/jbc.C100346200. [DOI] [PubMed] [Google Scholar]
- 108.Behan RK, Lippard SJ. The aging-associated enzyme CLK-1 is a member of the carboxylate-bridged diiron family of proteins. Biochemistry. 2010;49:9679–9681. doi: 10.1021/bi101475z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lu TT, et al. Aging-associated enzyme human clock-1: substrate-mediated reduction of the diiron center for 5-demethoxyubiquinone hydroxylation. Biochemistry. 2013;52:2236–2244. doi: 10.1021/bi301674p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pelosi L, et al. Evolution of Ubiquinone Biosynthesis: Multiple Proteobacterial Enzymes with Various Regioselectivities To Catalyze Three Contiguous Aromatic Hydroxylation Reactions. mSystems. 2016:1. doi: 10.1128/mSystems.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xie LX, et al. Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J Biol Chem. 2012;287:23571–23581. doi: 10.1074/jbc.M112.360354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Poon WW, et al. 3-Hexaprenyl-4-hydroxybenzoic acid forms a predominant intermediate pool in ubiquinone biosynthesis in Saccharomyces cerevisiae. Arch Biochem Biophys. 1995;320:305–314. doi: 10.1016/0003-9861(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 113.Marbois B, et al. Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J Biol Chem. 2005;280:20231–20238. doi: 10.1074/jbc.M501315200. [DOI] [PubMed] [Google Scholar]
- 114.Tran UC, et al. Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide: two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis. J Biol Chem. 2006;281:16401–16409. doi: 10.1074/jbc.M513267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Allan CM, et al. Identification of Coq11, a new coenzyme Q biosynthetic protein in the CoQ-synthome in Saccharomyces cerevisiae. J Biol Chem. 2015;290:7517–7534. doi: 10.1074/jbc.M114.633131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lohman DC, et al. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc Natl Acad Sci U S A. 2014;111:E4697–4705. doi: 10.1073/pnas.1413128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Floyd BJ, et al. Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function. Mol Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ashraf S, et al. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest. 2013;123:5179–5189. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Srere PA. The Metabolon. Trends in Biochem Sci. 1985:109–110. [Google Scholar]
- 120.Barnes SJ, Weitzman PD. Organization of citric acid cycle enzymes into a multienzyme cluster. FEBS Lett. 1986;201:267–270. doi: 10.1016/0014-5793(86)80621-4. [DOI] [PubMed] [Google Scholar]
- 121.Pedley AM, Benkovic SJ. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem Sci. 2017;42:141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Laursen T, et al. Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science. 2016;354:890–893. doi: 10.1126/science.aag2347. [DOI] [PubMed] [Google Scholar]
- 123.Midzak A, Papadopoulos V. Adrenal Mitochondria and Steroidogenesis: From Individual Proteins to Functional Protein Assemblies. Front Endocrinol (Lausanne) 2016;7:106. doi: 10.3389/fendo.2016.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schmidt O, et al. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 125.Gonzalez-Mariscal I, et al. The regulation of coenzyme q biosynthesis in eukaryotic cells: all that yeast can tell us. Mol Syndromol. 2014;5:107–118. doi: 10.1159/000362897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lapointe CP, et al. Post-Transcriptional Control of Coenzyme Q Biosynthesis Revealed by Transomic Analysis of the RNA-Binding Protein Puf3p. 2017 bioRxiv 146985. [Google Scholar]
- 127.Leuenberger D, et al. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 1999;18:4816–4822. doi: 10.1093/emboj/18.17.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiang N, et al. Mouse CLK-1 is imported into mitochondria by an unusual process that requires a leader sequence but no membrane potential. J Biol Chem. 2001;276:29218–29225. doi: 10.1074/jbc.M103686200. [DOI] [PubMed] [Google Scholar]
- 129.Ulrich EL, et al. Location and mobility of ubiquinones of different chain lengths in artificial membrane vesicles. Biochemistry. 1985;24:2501–2508. doi: 10.1021/bi00331a016. [DOI] [PubMed] [Google Scholar]
- 130.Metz GH, KP, van Liemt WBS, Prestegard JH, Lugtenburg J, Smith SO. NMR Studies of Ubiquinone Location in Oriented Model Membranes: Evidence for a Single Motionally-Averaged Population. J Am Chem Soc. 1995;117:564–565. [Google Scholar]
- 131.Padilla-Lopez S, et al. Genetic evidence for the requirement of the endocytic pathway in the uptake of coenzyme Q6 in Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1788:1238–1248. doi: 10.1016/j.bbamem.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fernandez-Ayala DJ, et al. Coenzyme Q distribution in HL-60 human cells depends on the endomembrane system. Biochim Biophys Acta. 2005;1713:129–137. doi: 10.1016/j.bbamem.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 133.Marbois B, et al. The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim Biophys Acta. 2009;1791:69–75. doi: 10.1016/j.bbalip.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Leonard CJ, et al. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome research. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- 135.Poon WW, et al. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol. 2000;182:5139–5146. doi: 10.1128/jb.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lagier-Tourenne C, et al. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am J Hum Genet. 2008;82:661–672. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kannan N, et al. Structural and functional diversity of the microbial kinome. PLoS biology. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reidenbach A, et al. Conserved lipid and small molecule modulation of COQ8 reveals regulation of the ancient UbiB family. 2017 doi: 10.1016/j.chembiol.2017.11.001. bioRxiv 149823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Garcia-Corzo L, et al. Dysfunctional Coq9 protein causes predominant encephalomyopathy associated with CoQ deficiency. Hum Mol Genet. 2013;22:1233–1248. doi: 10.1093/hmg/dds530. [DOI] [PubMed] [Google Scholar]
- 140.He CH, et al. Yeast Coq9 controls deamination of coenzyme Q intermediates that derive from para-aminobenzoic acid. Biochim Biophys Acta. 2015;1851:1227–1239. doi: 10.1016/j.bbalip.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Barros MH, et al. The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J Biol Chem. 2005;280:42627–42635. doi: 10.1074/jbc.M510768200. [DOI] [PubMed] [Google Scholar]
- 142.Allan CM, et al. A conserved START domain coenzyme Q-binding polypeptide is required for efficient Q biosynthesis, respiratory electron transport, and antioxidant function in Saccharomyces cerevisiae. Biochim Biophys Acta. 2013;1831:776–791. doi: 10.1016/j.bbalip.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Loiseau L, et al. The UbiK protein is an accessory factor necessary for bacterial ubiquinone (UQ) biosynthesis and forms a complex with the UQ biogenesis factor UbiJ. Journal of Biological Chemistry. 2017 doi: 10.1074/jbc.M117.789164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Aussel L, et al. ubiJ, a new gene required for aerobic growth and proliferation in macrophage, is involved in coenzyme Q biosynthesis in Escherichia coli and Salmonella enterica serovar Typhimurium. J Bacteriol. 2014;196:70–79. doi: 10.1128/JB.01065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Quinzii C, et al. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet. 2006;78:345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jakobs BS, et al. A novel mutation in COQ2 leading to fatal infantile multisystem disease. J Neurol Sci. 2013;326:24–28. doi: 10.1016/j.jns.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 147.Diomedi-Camassei F, et al. COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. J Am Soc Nephrol. 2007;18:2773–2780. doi: 10.1681/ASN.2006080833. [DOI] [PubMed] [Google Scholar]
- 148.Desbats MA, et al. Primary coenzyme Q10 deficiency presenting as fatal neonatal multiorgan failure. Eur J Hum Genet. 2015;23:1254–1258. doi: 10.1038/ejhg.2014.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Salviati L, et al. Haploinsufficiency of COQ4 causes coenzyme Q10 deficiency. J Med Genet. 2012;49:187–191. doi: 10.1136/jmedgenet-2011-100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brea-Calvo G, et al. COQ4 mutations cause a broad spectrum of mitochondrial disorders associated with CoQ10 deficiency. Am J Hum Genet. 2015;96:309–317. doi: 10.1016/j.ajhg.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Heeringa SF, et al. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Doimo M, et al. Effect of vanillic acid on COQ6 mutants identified in patients with coenzyme Q10 deficiency. Biochim Biophys Acta. 2014;1842:1–6. doi: 10.1016/j.bbadis.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Freyer C, et al. Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4-dihydroxybensoic acid. J Med Genet. 2015 doi: 10.1136/jmedgenet-2015-102986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mollet J, et al. CABC1 gene mutations cause ubiquinone deficiency with cerebellar ataxia and seizures. Am J Hum Genet. 2008;82:623–630. doi: 10.1016/j.ajhg.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu YT, et al. Autosomal-recessive cerebellar ataxia caused by a novel ADCK3 mutation that elongates the protein: clinical, genetic and biochemical characterisation. J Neurol Neurosurg Psychiatry. 2014;85:493–498. doi: 10.1136/jnnp-2013-306483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Blumkin L, et al. Heterozygous Mutations in the ADCK3 Gene in Siblings with Cerebellar Atrophy and Extreme Phenotypic Variability. JIMD Rep. 2014;12:103–107. doi: 10.1007/8904_2013_251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gerards M, et al. Nonsense mutations in CABC1/ADCK3 cause progressive cerebellar ataxia and atrophy. Mitochondrion. 2010;10:510–515. doi: 10.1016/j.mito.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 158.Horvath R, et al. Adult-onset cerebellar ataxia due to mutations in CABC1/ADCK3. J Neurol Neurosurg Psychiatry. 2012;83:174–178. doi: 10.1136/jnnp-2011-301258. [DOI] [PubMed] [Google Scholar]
- 159.Korkmaz E, et al. ADCK4-Associated Glomerulopathy Causes Adolescence-Onset FSGS. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2014121240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Duncan AJ, et al. A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary coenzyme Q10 deficiency: a potentially treatable form of mitochondrial disease. Am J Hum Genet. 2009;84:558–566. doi: 10.1016/j.ajhg.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Danhauser K, et al. Fatal neonatal encephalopathy and lactic acidosis caused by a homozygous loss-of-function variant in COQ9. Eur J Hum Genet. 2015 doi: 10.1038/ejhg.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mollet J, et al. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J Clin Invest. 2007;117:765–772. doi: 10.1172/JCI29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lopez LC, et al. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet. 2006;79:1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gasser DL, et al. Focal segmental glomerulosclerosis is associated with a PDSS2 haplotype and, independently, with a decreased content of coenzyme Q10. Am J Physiol Renal Physiol. 2013;305:F1228–1238. doi: 10.1152/ajprenal.00143.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gempel K, et al. The myopathic form of coenzyme Q10 deficiency is caused by mutations in the electron-transferring-flavoprotein dehydrogenase (ETFDH) gene. Brain. 2007;130:2037–2044. doi: 10.1093/brain/awm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Haas D, et al. Coenzyme Q(10) is decreased in fibroblasts of patients with methylmalonic aciduria but not in mevalonic aciduria. J Inherit Metab Dis. 2009;32:570–575. doi: 10.1007/s10545-009-1150-8. [DOI] [PubMed] [Google Scholar]
- 167.Quinzii CM, et al. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology. 2005;64:539–541. doi: 10.1212/01.WNL.0000150588.75281.58. [DOI] [PubMed] [Google Scholar]
- 168.Aeby A, et al. Cardiofaciocutaneous (CFC) syndrome associated with muscular coenzyme Q10 deficiency. J Inherit Metab Dis. 2007;30:827. doi: 10.1007/s10545-007-0612-0. [DOI] [PubMed] [Google Scholar]
- 169.Marcoff L, Thompson PD. The role of coenzyme Q10 in statin-associated myopathy: a systematic review. J Am Coll Cardiol. 2007;49:2231–2237. doi: 10.1016/j.jacc.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 170.Kassardjian CD, et al. Myopathy during treatment with the antianginal drug ranolazine. J Neurol Sci. 2014;347:380–382. doi: 10.1016/j.jns.2014.10.037. [DOI] [PubMed] [Google Scholar]
- 171.Emmanuele V, et al. Heterogeneity of coenzyme Q10 deficiency: patient study and literature review. Arch Neurol. 2012;69:978–983. doi: 10.1001/archneurol.2012.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 173.Montini G, et al. Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. N Engl J Med. 2008;358:2849–2850. doi: 10.1056/NEJMc0800582. [DOI] [PubMed] [Google Scholar]
- 174.Wang Y, Hekimi S. Mitochondrial respiration without ubiquinone biosynthesis. Hum Mol Genet. 2013;22:4768–4783. doi: 10.1093/hmg/ddt330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lapointe J, et al. The submitochondrial distribution of ubiquinone affects respiration in long-lived Mclk1+/- mice. J Cell Biol. 2012;199:215–224. doi: 10.1083/jcb.201203090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ben-Meir A, et al. Coenzyme Q10 restores oocyte mitochondrial function and fertility during reproductive aging. Aging Cell. 2015;14:887–895. doi: 10.1111/acel.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Anderson CM, et al. Dependence of brown adipose tissue function on CD36-mediated coenzyme Q uptake. Cell Rep. 2015;10:505–515. doi: 10.1016/j.celrep.2014.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bentinger M, et al. Distribution and breakdown of labeled coenzyme Q10 in rat. Free Radic Biol Med. 2003;34:563–575. doi: 10.1016/s0891-5849(02)01357-6. [DOI] [PubMed] [Google Scholar]
- 179.Santos-Ocana C, et al. Uptake of exogenous coenzyme Q and transport to mitochondria is required for bc1 complex stability in yeast coq mutants. J Biol Chem. 2002;277:10973–10981. doi: 10.1074/jbc.M112222200. [DOI] [PubMed] [Google Scholar]
- 180.Garcia-Corzo L, et al. Ubiquinol-10 ameliorates mitochondrial encephalopathy associated with CoQ deficiency. Biochim Biophys Acta. 2014;1842:893–901. doi: 10.1016/j.bbadis.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 181.Wang Y, et al. Mitochondrial function and lifespan of mice with controlled ubiquinone biosynthesis. Nat Commun. 2015;6:6393. doi: 10.1038/ncomms7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Luna-Sanchez M, et al. The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol Med. 2015;7:670–687. doi: 10.15252/emmm.201404632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Parikh S, et al. Practice patterns of mitochondrial disease physicians in North America. Part 2: treatment, care and management. Mitochondrion. 2013;13:681–687. doi: 10.1016/j.mito.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 184.Parikh S, et al. A modern approach to the treatment of mitochondrial disease. Curr Treat Options Neurol. 2009;11:414–430. doi: 10.1007/s11940-009-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chaturvedi RK, Beal MF. Mitochondria targeted therapeutic approaches in Parkinson’s and Huntington’s diseases. Mol Cell Neurosci. 2013;55:101–114. doi: 10.1016/j.mcn.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 186.Dumont M, et al. Coenzyme Q10 decreases amyloid pathology and improves behavior in a transgenic mouse model of Alzheimer’s disease. J Alzheimers Dis. 2011;27:211–223. doi: 10.3233/JAD-2011-110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Williams EG, et al. Systems proteomics of liver mitochondria function. Science. 2016;352:aad0189. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Young IG, et al. Biochemical and genetic studies on ubiquinone biosynthesis in Escherichia coli K-12:4-hydroxybenzoate octaprenyltransferase. J Bacteriol. 1972;110:18–25. doi: 10.1128/jb.110.1.18-25.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]