SUMMARY
Mutations of DISC1 (disrupted-in-schizophrenia 1) have been associated with major psychiatric disorders. Despite the hundreds of DISC1-binding proteins reported, almost nothing is known about how DISC1 interacts with other proteins structurally to impact human brain development. Here we solved the high-resolution structure of DISC1 C-terminal tail in complex with its binding domain of Ndel1. Mechanistically, DISC1 regulates Ndel1’s kinetochore attachment, but not its centrosome localization, during mitosis. Functionally, disrupting DISC1/Ndel1 complex formation prolongs mitotic length and interferes with cell cycle progression in human cells, and causes cell cycle deficits of radial glial cells in the embryonic mouse cortex and human forebrain organoids. We also observed similar deficits in organoids derived from schizophrenia patient iPSCs with a DISC1 mutation that disrupts its interaction with Ndel1. Our study uncovers a new mechanism of action for DISC1 based on its structure and has implications for how genetic insults may contribute to psychiatric disorders.
INTRODUCTION
DISC1 (Disrupted-in-schizophrenia 1), originally identified in a large Scottish family suffering from multiple psychiatric disorders due to a chromosomal translocation-induced disruption (Blackwood et al., 2001), has been established as a genetic risk factor for a wide array of psychiatric disorders, including schizophrenia, bipolar disorder, major depression, and autism spectrum disorders (Thomson et al., 2013). Over 200 different proteins with very diverse functions have been reported to interact with DISC1 (Camargo et al., 2007; Soares et al., 2011), although the physiological relevance of most of these protein interactions remains to be verified. Proteins including Ndel1/Nde1, GSK3β, PDE4, FEZ1, ATF4, Kal-7, and Girdin/KIAA1212 are among several functionally well-characterized DISC1 binding partners that are known to play critical roles in neurodevelopment and neuronal signaling in rodent systems (Duan et al., 2007; Enomoto et al., 2009; Hayashi-Takagi et al., 2010; Kang et al., 2011; Kim et al., 2009; Mao et al., 2009; Millar et al., 2005; Soda et al., 2013; Wang et al., 2011). Interestingly, Ndel1/Nde1, PDE4 and GSK3β have been independently identified as genetic risk factors of mental disorders (Blasi et al., 2013; Fatemi et al., 2008; Nicodemus et al., 2010). Thus, DISC1 is hypothesized to function as a major hub protein at the crossroads of neurodevelopment, neuronal signaling, and neurological disorders (Brandon and Sawa, 2011; Ming and Song, 2009; Porteous et al., 2011; Thomson et al., 2013).
Unmatched to the wealth of functional and pathological data on DISC1, biochemical and structural characterizations of DISC1 and its interactions with target proteins are very scarce. No single atomic structure of DISC1 or any of its fragments, either alone or in complex with target proteins, is available. Accordingly, action mechanisms underlying DISC1’s function in brain development and DISC1 mutation-related psychiatric disorders are poorly understood. The reported number of DISC1 binding proteins is very large and many of these proteins co-exist in the same cellular compartments in high abundance. Therefore, it is difficult to understand how the limited amount of DISC1 can possibly be distributed among of such an enormous array of reported binding proteins and impact their functions in the cell.
Ndel1/Nde1, a modulatory component of the dynein complex (Vallee et al., 2012), is one of numerous reported DISC1 binding targets (Brandon et al., 2004). A short C-terminal fragment of DISC1 was identified to be required for Ndel1 binding (Kamiya et al., 2006). The (1; 11)(q42; q14.3) translocation mutation of DISC1, which causes DISC1 C-terminal truncation (deletion of residues 598–854 in humans and residues 595–852 in mice; Figure 1A and S1), disrupts its binding to Ndel1/Nde1 (Brandon et al., 2004). Mutations of Nde1 are known to cause microcephaly both in mice and in humans (Alkuraya et al., 2011; Bakircioglu et al., 2011; Feng and Walsh, 2004). Ndel1 has been shown to epistatically associate with DISC1 in psychiatric disorders (Burdick et al., 2008; Nicodemus et al., 2010). Complete removal of Ndel1 is embryonically lethal in mice (Sasaki et al., 2005), although how it may regulate human brain development remains to be determined. Elucidation of cellular functions of the interaction between DISC1 and Ndel1/Nde1 in brain development has been difficult, as DISC1 may interact with numerous target proteins other than Ndel1/Nde1. Similarly, Ndel1/Nde1 are also scaffold proteins that can interact with several subunits of the cytoplasmic dynein complex, including the dynein heavy chain and Lis1 (Niethammer et al., 2000; Sasaki et al., 2000; Shu et al., 2004). Thus, results derived from loss-of-function approaches on either of DISC1 or Ndel1/Nde1 can be difficult to interpret due to potential compound effects.
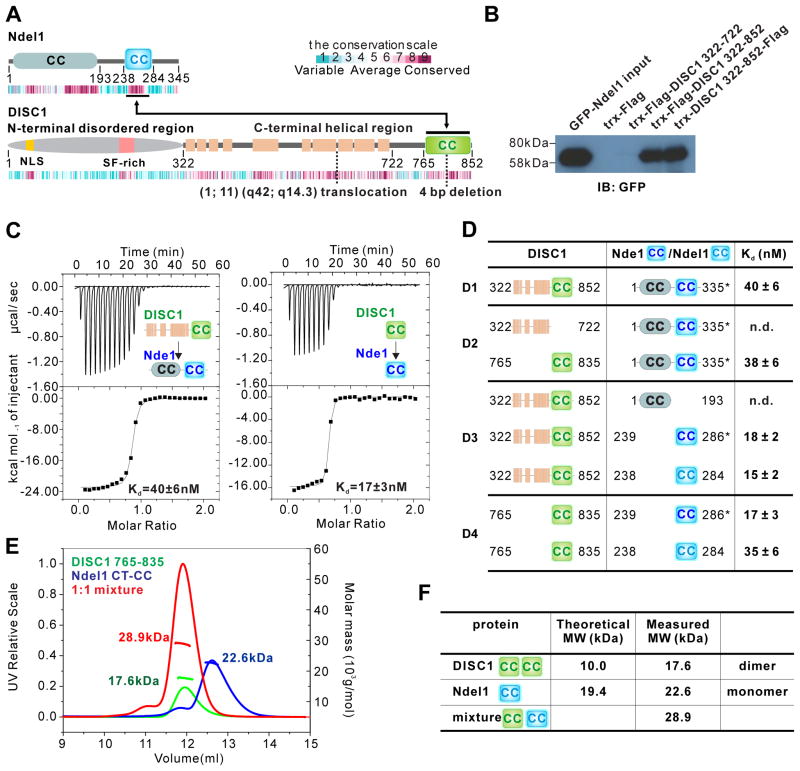
Figure 1. DISC1 765–835 interαcts with Ndel1/Nde1 CT-CC with high αffinity.
(A) Schematic diagram showing the domain organization of Ndel1 and DISC1. The beige-colored rectangles in DISC1 represent predicted α-helices. The two-way arrowed line shows the corresponding regions in the two proteins responsible for their specific interaction. The heat map below each scheme shows the amino acid sequence conservation of each protein throughout the evolution. The relationship between color and conservation is indicated at the upper right corner. CC, predicted coiled-coil region. Positions of translocation break point t(1;11)(q42;q14.3) found in a Scottish family and 4 bp deletion found in American schizophrenia family are highlighted.
(B) Pull-down assay showing the interaction between DISC1 C-terminal helical region (322–852) and full-length Ndel1. Purified DISC1 322–722 or 322–852 with trx and Flag tags were immunoprecipitated with cell lysates from HEK293T cells transfected with the full-length GFP-Ndel1 by anti-Flag beads, and the resulting immunoprecipitates were immunoblotted for GFP-Ndel1.
(C) ITC-based measurements quantifying the binding affinities between DISC1 322–852 and full-length Nde1 (left panel) and DISC1 765–835 and Nde1 CT-CC (right panel). ITC, Isothermal Titration Calorimetry.
(D) ITC-based measurement summarizing the binding affinities between various DISC1 proteins and Nde1/Ndel1 proteins. The mapping results show that DISC1 765–835 and Nde1/Ndel1 CT-CC are the minimal binding regions for these two proteins to interact with high affinity. Nde1 CT-CC is labelled in dark blue, Ndel1 CT-CC in light blue. Due to its superior quality, we used full-length Nde1 indicated by an asterisk (*) when characterizing its interaction with DISC1. n.d., not detected.
(E and F) Analytical gel filtration chromatography analysis coupled with static light scattering analysis of DISC1 765–835 (green line), trx-Ndel1 CT-CC (blue line) and DISC1 765–835/trx-Ndel1 CT-CC complex (red line). The theoretical and measured molecular weights are listed in (F). The results indicate that DISC1 765–835 and Ndel1 CT-CC form a stable 1:1 complex in solution.
See also Figure S1 and S2.
Here we demonstrate that Ndel1/Nde1 binds to a short, extreme C-terminal fragment of DISC1 with very high specificity and affinity. We reveal the molecular basis governing the specific interaction via solving the atomic structure of this DISC1 C-terminal fragment in complex with its binding sequence of Ndel1/Nde1. The structure of the complex between DISC1 and Ndel1/Nde1 allowed us to design a method to specifically investigate functions of the interaction between DISC1 and Ndel1/Nde1 in vivo without interfering Ndel1/Nde1-mediated dynein complex functions and with minimal obstructing DISC1 binding to other partners. Using this new method, we investigated the role of the interaction between DISC1 and Ndel1/Nde1 in regulating cell cycle in human cells in vitro, in radial glial neural stem cells in the embryonic mouse cortex in vivo and in human forebrain organoids (Qian et al., 2016). In addition, we explored the potential role of this interaction in the context of human psychiatric disorders using patient-derived brain organoids with a specific DISC1 mutation (Chiang et al., 2011). Together, these multifaceted approaches unravel a novel mechanism of action by DISC1 and provide insight into the pathogenesis of psychiatric disorders.
RESULTS
Ndel1/Nde1 binds to a short C-terminal tail of DISC1 with a high affinity
To characterize the property of the interaction between DISC1 and Ndel1/Nde1, we first performed a series of detailed biochemical studies. Using pull-down assay and isothermal titration calorimetry (ITC) analysis, we found that a conformational-homogeneous DISC1 fragment lacking the predicted unstructured N-terminal region (aa 1–321; Figure 1A) binds to the full-length Nde1 (FL-Nde1) at a 1:1 stoichiometry and with a very high affinity (Kd ~40 nM; Figure 1B–C and D1). Further mappings by ITC analyses revealed that a short, predicted coiled-coil region of Nde1 (aa 239–286, termed as “Nde1 CT-CC”) is both necessary and sufficient for DISC1 binding (Figure 1C, 1D3 and S1C3). DISC1 C-terminal helical region is predicted to contain a separate coiled-coil (residue 765–835) that is connected to the upstream helical region with a flexible and variable linker (Figure 1A). Notably, this DISC1 C-terminal region (aa 765–835) is sufficient for binding to both FL-Nde1 and Nde1 CT-CC (Figure 1C, 1D2 and S1C2; we used Nde1 here because FL-Nde1 behaves better for quantitative biochemical studies than Ndel1 does). Additionally, the middle helical region of DISC1 (aa 322–722) has no detectable binding to Nde1 (Figure 1B, 1D2 and S1C2). These ITC data indicate that DISC1 765–835 is solely responsible for binding to Nde1. We also characterized DISC1’s interaction with Ndel1, a paralogue of Nde1. We found that Ndel1 CT-CC (aa 238–284) and Nde1 CT-CC share similar DISC1 affinities (Figure 1D3–D4 and S1C3–C4), a result that is consistent with the very high amino acid sequence identity of the two proteins (Figure S1B). Given that Ndel1 and Nde1 share essentially the identical DISC1 binding property, we refer Ndel1/Nde1 as Ndel1 from here on for simplicity, unless specified otherwise. Gel filtration chromatography analysis revealed that DISC1 765–835 itself can form a weak homodimer, and Ndel1 CT-CC is a monomer. When mixed, DISC1 765–835 and Ndel1 CT-CC can interact with each other and form a stable heterodimer (Figure 1E–F and S2A–B).
Our mapping data also indicates that both DISC1 765–835 and Ndel1 CT-CC function as independent structural units in the context of their respective full-length proteins (Figure 1D). We performed additional biochemical experiments to further support this conclusion, which is important for our functional studies of the DISC1/Ndel1 complex via targeting these specific regions. The purified DISC1 322–852 adopts a highly homogenous tetramer (Figure S3A, D). Deletion of the fragment comprising 723–852, which includes the entire Ndel1 binding region of the protein, does not alter the tetramer structure as well as the stability of DISC1 (Figure S3A, D), indicating that the C-terminal fragment containing residues 723–852 of DISC1 is not structurally coupled with the central tetramerization region of DISC1. The circular dichroism spectrum based urea denaturation profile of DISC1 322–852 and 322–722 are highly similar (Figure S3E–F), indicating similar structure for these two fragments. The NMR spectra of 15N-labeled DISC1 728–852 also revealed that this fragment is largely unstructured in solution (Figure S3G). The full-length Ndel1 forms a stable dimer in solution, and truncation of the C-terminal half of the protein (aa 194–345) does not alter the dimerization property of Ndel1 (Figure S3B–D), supporting an earlier structural study showing that the N-terminal half of Ndel1 forms a coiled-coil dimer and the C-terminal half of the protein is largely unstructured (Derewenda et al., 2007). Together, these results demonstrate that both interacting regions of DISC1 (aa 765–852) and Ndel1 (CT-CC) are independent structural units in each proteins, and they interact with each other to form a highly specific and stoichiometric complex (Figure S3I).
High-resolution solution structure of the DISC1/Ndel1 complex
We next determined the high-resolution complex structure of DISC1 765–852/Ndel1 CT-CC by NMR spectroscopy (Figure S2C, E and Table S1). Structural determination of the DISC1 765–835/Ndel1 CT-CC was aided by fusing Ndel1 CT-CC to the C-terminal end of DISC1 765–852 with a thrombin cleavable linker. The use of the single chain fusion protein allowed us to skip isotope-filtered experiments for identifying inter-molecular nuclear Overhauser enhancement (nOe) signals between DISC1 and Ndel1, and thus simplified the structural determination. The structure of the single chain protein was determined to a high precision due to very high qualities of NMR spectra and thus a large number of structural restraints obtained (~25 restraints/residue in the structured regions, Table S1). The covalent fusion of DISC1 with Ndel1 CT-CC does not alter the structure of the complex as the NMR spectra of the fusion protein with and without thrombin cleavage overlap almost perfectly (Figure S2D). In the complex, DISC1 765–835 contains two helices that form an antiparallel hairpin, and Ndel1 CT-CC adopts a single α-helix that packs with the DISC1 helix hairpin to form a three-helix bundle through canonical coiled-coil interactions (Figure 2A–B). The extension sequence (DISC1 835–852) used to link Ndel1 CT-CC adopts random coil structure, showing that the covalent linking does not alter the structure of the complex.
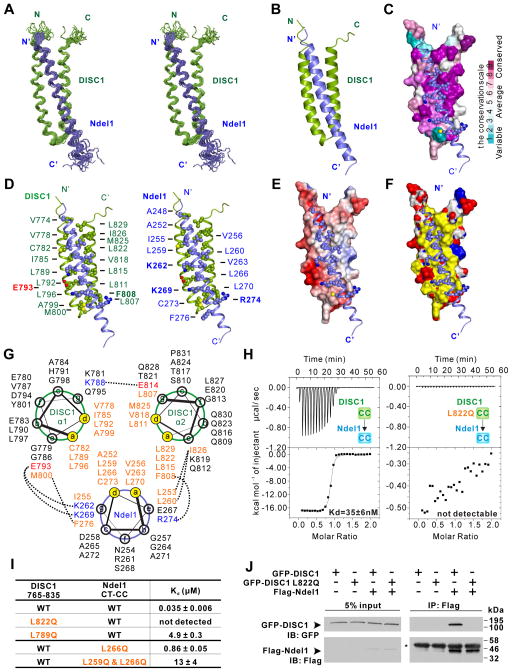
Figure 2. Structure of the DISC1/Ndel1 complex.
(A) Stereo view showing superposition of the backbones of 20 NMR structures of the DISC1 765–835 (green)/Ndel1 CT-CC (blue) complex with the lowest energies.
(B) Ribbon diagram of a representative NMR structure of the DISC1 765–835/Ndel1 CT-CC complex.
(C) Combined surface and ribbon representation showing the conservation map of the DISC1 and Ndel1 binding interface. The residues involved in DISC1/Ndel1 interaction interface are all evolutionarily conserved. In the surface diagram, the highly conserved amino acids are drawn in purple, the less conserved residues in cyan, as indicated in the bar diagram on the right.
(D) Stereo view showing the detailed interaction interface between DISC1 765–835 and Ndel1 CT-CC with combined ribbon and sphere representation. The residues labeled with bold face are involved in charge-charge interactions, whereas others are involved in hydrophobic interactions.
(E) Combined surface and ribbon representation showing the electrostatic potential of DISC1 binding interface for Ndel1. The ± 3-kT/e potential isocontours are shown as blue (positively charged) and red (negatively charged) surfaces, respectively.
(F) Combined surface and ribbon representation showing the hydrophobic interactions dominating the interface between DISC1 and Ndel1. In the surface diagram, hydrophobic residues are colored in yellow, positive charged residues are colored in blue and negative charged residues in red.
(G) Helical wheel representation showing the detailed interactions between the heptad repeats in the DISC1/Ndel1 complex. Residues at the a and d positions forming the hydrophobic core of the coiled coil are highlighted in orange. Residues forming electrostatic interactions are colored with blue for positively charged residues and red for negatively charge residues. Inter-helical interactions between the residues at the e and g (or even b) positions are depicted by dashed lines.
(H) ITC-based measurements quantifying the binding affinities between DISC1 765–835 WT (left panel) and L822Q (right panel) with Ndel1 CT-CC.
(I) ITC-based measurements comparing the binding affinities between DISC1 765–835 (WT or mutants) and Ndel1 CT-CC (WT or mutants). WT, wild type.
(J) Co-IP assay comparing bindings of full-length Ndel1 to the full-length WT DISC1 (or the L822Q mutant). Cell lysates from HEK293T cells transfected with the full-length Flag-Ndel1 and the full-length GFP-DISC1 WT (or L822Q mutant) respectively were mixed and immunoprecipitated by anti-Flag beads. The resulting immunoprecipitates were immunoblotted for DISC1 and Ndel1 as indicated. The heavy chain of Flag antibody on anti-Flag beads is indicated by an asterisk (*).
See also Figure S1, S2 and S3.
Design of a highly specific inhibitory peptide based on the complex structure
The structure of the DISC1/Ndel1 complex reveals the biochemical mechanism governing the specific interaction between DISC1 and Ndel1. Hydrophobic residues located at the a and d positions of the three helices form the hydrophobic core of the complex via the typical ‘knobs into holes’ packing mode in coiled-coil structures (Lupas, 1996) (Figure 2C–G). These hydrophobic residues are highly conserved in both DISC1 and Ndel1 (Figure 2C and S1A–B). Substitutions of each of these hydrophobic amino acids (e.g. L789 and L822 in DISC1, and L259 and L266 Ndel1) with polar ones significantly weaken or even completely disrupt the complex formation (Figure 2H–I and S1D). Apart from the hydrophobic interactions, charge-charge interactions formed by residues at the e and g positions along the heptad repeats also contribute to the affinity and specificity of the DISC1/Ndel1 interaction (Figure 2D–E, G).
The structure of the DISC1 765–835/Ndel1 CT-CC complex allowed us to develop specific tools for studying cellular functions of the DISC1/Ndel1 interaction. Based on our biochemical and structural results, introduction of the DISC1 765–835 peptide into living cells is expected to specifically block the interaction between DISC1 and Ndel1/Nde1. This DISC1 peptide should not interfere with any of the cellular functions of DISC1 mediated by the rest of the protein as the DISC1 tail (i.e. aa765–852) is not structurally coupled to the rest of the protein (Figure S3E–F and I). Additionally, the DISC1 765–835 peptide is expected to only disrupt the cellular functions of both Ndel1 and Nde1 mediated by their short DISC1 binding segment (i.e. the 47-residue Ndel1 CT-CC) and leave the rest of Ndel1/Nde1’s functions, for example as a dynein regulatory subunit, intact. This strategy is particularly advantageous for studying specific functions of the interaction between DISC1 and Ndel1, two multi-domain scaffold proteins with very broad cellular functions and diverse binding partners. To ensure the specificity of our experimental approach used below for investigating functions of the DISC1/Ndel1 interaction, we used a single point mutation of the DISC1 765–835 peptide (DISC1 765–835 L822Q), which has no detectable binding to Ndel1 and thus should not interfere cellular functions of the DISC1/Ndel1 interaction, as the control (Figure 2H–J).
Disrupting the DISC1/Ndel1 interaction causes mitotic delay in heterologous cells
Since Ndel1 is known to play critical roles in cell cycle control, presumably by recruiting the dynein complex to various cell cycle apparatuses, including centrosomes and kinetochores (Bakircioglu et al., 2011; Liang et al., 2007; Raaijmakers et al., 2013; Vallee et al., 2012), we first asked whether the DISC1/Ndel1 interaction regulates cell cycle progression in general using Hela cells as a model. Time-lapse live imaging analysis revealed that over-expression of the GFP-DISC1 765–835 peptide, but not control GFP-DISC1 765–835 L822Q peptide, significantly prolongs the duration of the mitotic phase of cell cycle (Figure 3A). Further analysis revealed that the DISC1 peptide-induced mitotic delay is mainly due to lengthening metaphase to anaphase duration, whereas the duration of prophase to metaphase transition was not altered (Figure 3B–D and S4A–B). In cells expressing GFP-DISC1 765–835, the sister chromatids appeared to exhibit difficulty to separate after onset of the metaphase and the cell cycle was trapped at the metaphase to anaphase stage for a prolonged duration (Figure S4B).
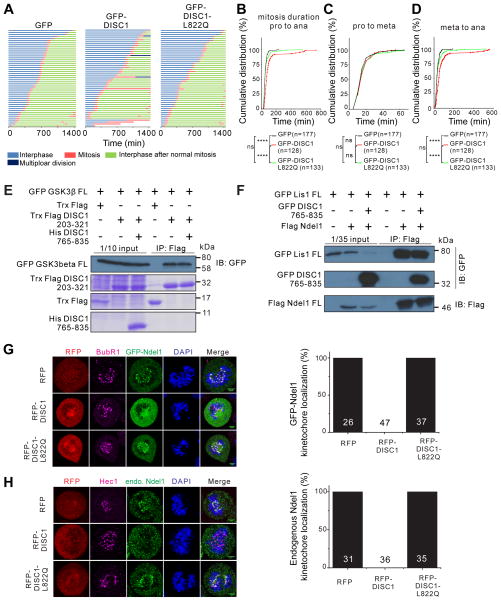
Figure 3. DISC1/Nde1l interaction regulates cell cycle progression during mitosis by controlling Ndel1’s kinetochore localization.
(A) Time-lapse imaging analysis showing that the DISC1 765–835 peptide can prolong the duration of mitosis. Hela cells expressing histone H2B-RFP were transfected with GFP control, GFP-DISC1 765–835 or GFP-DISC1 765–835 L822Q respectively. After 24 hours, the Hela cells were tracked using time-lapse microscopy for 24 hours. Each horizontal bar represents one cell (n = 50). Various colors corresponding to different periods in cell cycle are indicated below the diagram.
(B) Cumulative distributions of the entire mitosis duration time from prophase (pro) to anaphase (ana) of cells tracked in (A).
(C) Cumulative distributions of time durations from prophase to metaphase (meta).
(D) Cumulative distributions of time durations from metaphase to anaphase of mitosis. Three independent experiment results were pooled for statistical analysis. The total number “n” is indicated for each group. The statistical analysis in (B–D) was carried out by Kolmogorov-Smirnov test (****P < 0.0001; ns, not significant).
(E) Pull-down-based competition experiment showing the interaction between DISC1 and GSK3β is not affected by DISC1 C-terminal 765–835 peptide. We used purified Trx-Flag-tagged DISC1 (203–321) for the binding assay, and the purified proteins are shown using Coomassie Blue staining. The input lanes are bacterial cell lysates expressing corresponding proteins and also stained with Coomassie Blue
(F) Pull-down-based competition experiment showing the interaction between Ndel1 and LIS1 is not affected by DISC1 C-terminal 765–835 peptide.
(G and H) Representative images showing that the DISC1 peptide can disrupt the exogenously expressed Ndel1 (G) and endogenous Ndel1 (H) kinetochore localization in Hela cells. Cells were synchronized at early S phase by double thymidine block procedure. During the interval of two thymidine treatments, cells were transfected with RFP control, RFP DISC1 765–835, RFP DISC1 765–835 L822Q and GFP-full-length Ndel1. Cells in prometaphase to metaphase are chosen for the Ndel1 kinetochore localization analysis. Anti-BubR1 and Anti-Hec1 (magenta) are used to mark kinetochore. Scale bar, 10 μm. Quantifications of the impact of DISC1 765–835 on exogenously expressed Ndel1 (derived from 6 independent experiments) or endogenous Ndel1 (derived from 3 independent experiments) kinetochore localization during mitosis. “n” stands for number of cells analyzed.
See also Figure S4
Previous studies have shown that both DISC1 and Nde1l can regulate cell cycle by interacting with GSK3β (Mao et al., 2009) and Lis1/dynein, respectively (Derewenda et al., 2007; Moon et al., 2014; Niethammer et al., 2000; Shu et al., 2004; Tarricone et al., 2004; Zylkiewicz et al., 2011). To ensure that the cell cycle progression deficit caused by overexpression of DISC1 765–835 peptide is not due to the disruption of the DISC1/GSK3β complex or the Ndel1/Lis1 complex, we assayed the potential impact of the DISC1 765–835 peptide on DISC1’s binding to GSK3β as well as Ndel1’s binding to Lis1. We confirmed that the N-terminal fragment preceding the tetramerization domain of DISC1 (i.e. residue 1–321) is responsible for binding to GSK3β (Figure S3I) (Mao et al., 2009). We further mapped the GSK3β-binding region to residues 203–321 of DISC1 (Figure 3E). As expected, the DISC1/GSK3β interaction was not affected by the presence of excess amount of the DISC1 765–835 peptide, as the GSK3β-binding region on DISC1 is far away from the DISC1 C-terminal tail (Figure 3E). Similarly, the interaction between Ndel1 and Lis1 was not affected by the DISC1 765–835 peptide (Figure 3F). This result is also expected as the Lis1 binding region is in the N-terminal dimerization domain of Ndel1 and is away from the Ndel1 CT-CC (Derewenda et al., 2007; Tarricone et al., 2004). Together, our results indicate that specific interaction between DISC1 and Ndel1 regulates cell cycle progression, particularly at the metaphase-to-anaphase transition of mitosis, in the heterologous cell culture model.
DISC1 regulates the kinetochore localization of Ndel1 during mitosis
To understand the mechanism underlying the mitotic delay caused by disrupting the interaction between DISC1 and Ndel1, we examined the dynamics of cellular localizations of Ndel1 during cell cycle. Ndel1 mainly localizes at three regions, namely centrosomes, kinetochores and the nuclear envelope, in dividing cells (Figure S4C) (Alkuraya et al., 2011; Hebbar et al., 2008; Hu et al., 2013; Liang et al., 2007). Ndel1 persistently co-localized to centrosomes throughout the entire cell cycle. When cells entered the prophase, Ndel1 became concentrated on the nuclear envelope to regulate its breakdown process. After nuclear envelope broke down, Ndel1 became concentrated at kinetochores and centrosomes (Figure S4C), a process believed to be important for dynein-mediated separation of sister chromatids during mitosis (Liang et al., 2007; Vergnolle and Taylor, 2007). Nde1 showed a similar cellular localization pattern as Ndel1 did during cell cycle (Figure S4D).
Consistent with the observation that the DISC1 peptide did not alter the onset of and the prophase of mitosis, Ndel1’s nuclear envelope localization during the prophase was not perturbed by DISC1 binding (Figure S4E). We also found that the DISC1 peptide does not alter the centrosome localization of the endogenous Ndel1 throughout the cell cycle (Figure S4E), indicating that DISC1 binding does not alter Ndel1’s function on centrosomes. This observation is consistent with our biochemical finding that the DISC peptide does not affect the Ndel1/Lis1 interaction, which is known to be critical for Ndel1-mediated dynein complex assembly and functions (Moon et al., 2014; Niethammer et al., 2000; Sasaki et al., 2000; Shu et al., 2004; Zylkiewicz et al., 2011). Notably, the DISC1 peptide, but not the control peptide, completely disrupted the kinetochore localization of over-expressed as well as endogenous Ndel1 (Figure 3G–H). Likewise, overexpression of the DISC1 peptide, but not the L822Q control peptide, completely disrupted kinetochore localization of Nde1 (Figure S4F). Taken together, our results suggest that interaction between DISC1 and Ndel1 regulates the cell cycle progression by specifically controlling the Ndel1’s kinetochore localization.
DISC1/Nde1l interaction regulates proliferation of radial glial cells during embryonic mouse neocortical development
The neocortex is responsible for higher brain functions such as cognitive and emotional processing, which are impaired in patients with psychiatric disorders. Radial glial cells (RGCs) in the ventricular zone (VZ) function as neural stem cells in the embryonic cortex (Taverna et al., 2014). To investigate the function of the DISC1/Ndel1 interaction in regulating RGCs in vivo, we performed in utero electroporations with vectors expressing GFP (GFP), or co-expressing GFP and the DISC1 765–835 peptide (GFP-DISC1), or the DISC1 L822Q control peptide (GFP-DISC1-L822Q), into the E13.5 neocortex (Yoon et al., 2014). To label the RGCs during the S phase of cell cycle, we injected EdU into the pregnant dams 6 hr prior to collection (Figure S5A). We first examined the interkinetic nuclear migration (INM) of GFP+EdU+ RGCs in VZ. During INM, the nucleus of each RGCs migrates away from the ventricular surface through the basal process of the cell in G1, progresses through the S-phase, and then returns to the ventricular surface during G2 for the mitotic division (Taverna et al., 2014). Consistent with our results from the eterologous system, we did not observe any difference in the distribution of GFP+EdU+ cells in VZ upon expression of the DISC1 765–835 peptide, indicating that progressions from S to G2, and G2 to M phases are not affected by the disruption of the DISC1/Ndel1 interaction (Figure S5B).
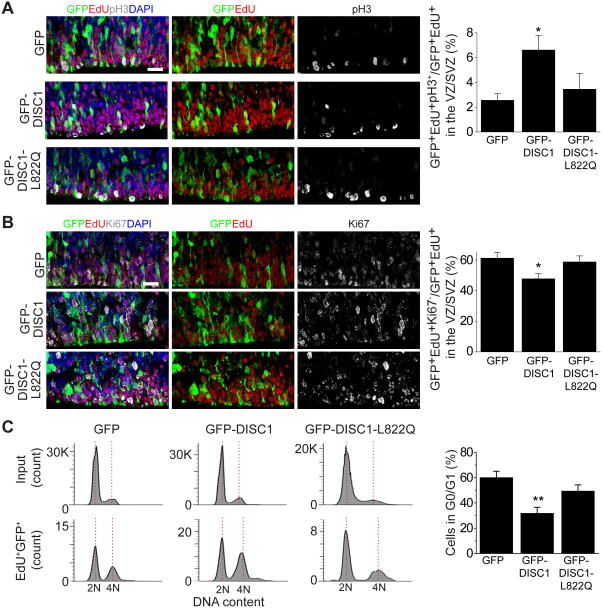
Next, we examined cell cycle progression during mitosis by analyzing the percentage of cells with the mitotic marker, pH3, among all GFP+EdU+ cell in VZ and subventricular zone (SVZ). We observed an increased percentage of pH3+ cells upon expression of the DISC1 765–835 peptide compared to the empty vector control, while the DISC1 L822Q control peptide had no effect (Figure 4A). We further assessed cell cycle exit by measuring the percentage of GFP+ cells labeled in the S-phase (GFP+EdU+) that exited the cell cycle (Ki67−) 24 hr later. We found that expression of DISC1 765–835 peptide, but not DISC1 L822Q control peptide, caused substantially decreased cell cycle exit (Figure 4B). We next performed fluorescence-activated cell sorting (FACS) analysis with dissociated cortex after in utero electroporation and EdU treatment as described above (Yoon et al., 2017a) (Figure S5C). The FACS analysis revealed that progression from S/G2/M to G0/G1 was significantly reduced by the expression of the DISC1 765–835 peptide, but not by the DISC1 L822Q control peptide (Figure 4C). Together, these results indicate that the DISC1/Ndel1 interaction is important for proper cell cycle progression of RGCs in the developing mouse cortex in vivo.
Figure 4. The interaction between DISC1 and Ndel1 regulates the cell cycle progression of RGCs in developing mouse cortex in vivo.
(A and B) Expression of DISC1 765–835 (GFP-DISC1), but not GFP-DISC1 765–835 L822Q (GFP-DISC1-L822Q), leads to delayed cell cycle progression and results in increased mitotic index and decreased cell cycle exit index of RGCs. Shown on left are sample confocal images of brain sections immunostained with GFP, pH3, EdU, and DAPI (A) or GFP, Ki67, EdU, and DAPI (B). White arrows indicate GFP+EdU+pH3+ cells (A) and GFP+EdU+Ki67- cells (B). Quantifications are shown on the right for each panel. Scale bars: 20 μm. Numbers associated with each bar graph refer to the total number of brain sections analyzed under each condition. Values represent mean ± SEM (*P < 0.05, One-way ANOVA).
(C) Shown on the left are histograms for FACS analysis of dissociated embryonic cortical cells after electroporation. Bar graph depicts the percentage of GFP positive cells in G0/G1. Number associated with each bar graph refers to the number of replicates of flow cytometry under each condition. Values represent mean ± SEM (**P < 0.01, One-way ANOVA). See also Figure S5.
DISC1/Ndel1 interaction is critical for neurogenesis in human forebrain organoids
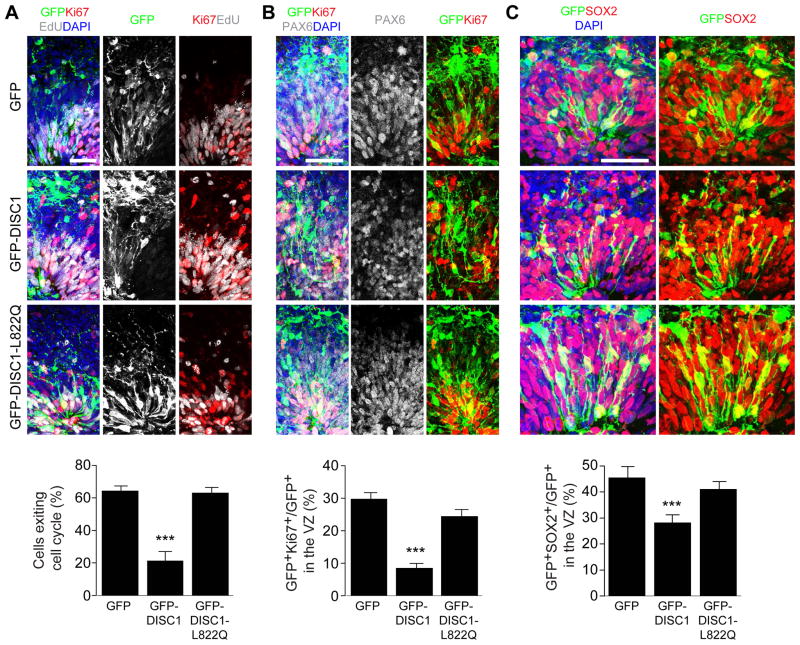
To determine the potential functional impact of disrupting the DISC1 and Ndel1 complex during human brain development, we used forebrain-specific organoids derived from human induced pluripotent stem cells (iPSCs) (Qian et al., 2016; Xu et al., 2016). At day 45–47, NESTIN+ RGCs in forebrain organoids proliferate robustly as revealed by mitotic NSC marker p-VIMENTINE (Figure S6A). To genetically manipulate and label RGCs in organoids, we delivered plasmids expressing GFP, GFP-DISC1 765–835, or GFP-DISC1 765–835 L822Q into organoids by electroporation (Figure S6B) (Yoon et al., 2017b). At 5 days post electroporation (dpe), we observed GFP-labeled Ki67+PAX6+ proliferating RGCs in the ventricular zone-like structure (VZ, defined by PAX6 labeling; Figure S6C). We pulsed organoids with EdU for 2 hr at 4 dpe, followed by analysis 24 hr later (Figure S6B). We found that the cell cycle index was significantly decreased upon expression of GFP-DISC1 765–835, but not control peptide, when compared to the GFP control (Figure 5A). An increase in the percentage of EdU-labeled cells in the cell cycle can be due to either delayed cell cycle progression or accelerated re-entrance to the next cell cycle. We therefore examined the number of proliferating RGCs in the VZ. The portion of Ki67+GFP+ proliferating cells among total GFP+ RGCs in VZ was dramatically reduced in the organoids expressing GFP-DISC1 765–835 peptide, but not the control peptide (Figure 5B). Moreover, the number of SOX2+ GFP+ RGCs in the VZ was significantly decreased in the organoids expressing GFP-DISC1 765–835 peptide (Figure 5C). Together, these sets of experiments suggest that specific disruption of DISC1 and Ndel1 interaction results in reduced proliferation of neural stem cells in the VZ due to a delayed cell-cycle progression.
Figure 5. Interaction between DISC1 and Ndel1 regulates the cell cycle of RGCs in human forebrain-specific organoids.
(A) Expression of DISC1 765–835 peptide leads to delayed cell cycle progression of RGCs in human forebrain organoids. Human forebrain organoids generated from human iPSC line C3-1 were electroporated with GFP (GFP), GFP-DISC1 765–835 (GFP-DISC1) or GFP-DISC1 765–835 L822Q (GFP-DISC1-L822Q) plasmids at day 45–47 in culture, and further cultured for 5 days before analysis. Shown are representative sample confocal images of organoids under different conditions pulsed with EdU for 2 hr at 4 days after electroporation and immunostained with GFP, Ki67, EdU and DAPI. Cell exit index is calculated by GFP+EdU+Ki67-/GFP+EdU+ cells.
(B) Effect of DISC1 765–835 peptide expression on NSC proliferation. Shown are representative sample confocal images of organoids under different conditions immunostained with GFP, Ki67, PAX6 and DAPI. Quantification of proliferating RGCs (Ki67+) is shown at the bottom.
(C) Effect of DISC1 765–835 peptide expression on the maintenance of the RGC population. Shown are representative sample confocal images of organoids under different conditions immunostained with GFP, SOX2 and DAPI. Quantification of total SOX2+ RGCs is shown at the bottom. Scale bars: 50 μm. Numbers associated with each bar graph refer to the total number of neural tube structures analyzed under each condition. Values represent mean ± SEM (***P < 0.001, One-way ANOVA).
See also Figure S6.
Defective neurogenesis in human brain organoids derived from a schizophrenia patient with a DISC1 mutation
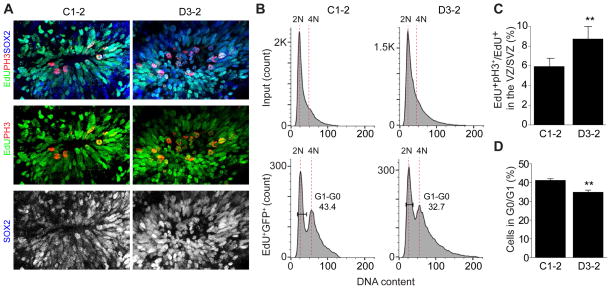
The characterization of the structure of the DISC1/Ndel1 complex prompted us to examine the potential impact of DISC1 mutations founds in patients with major psychiatric disorders. A (1; 11)(q42; q14.3) translocation found in the Scottish family leads to a truncation of DISC1 with a large part of the C-terminal tail (aa 598–854) removed (Blackwood et al., 2001). This mutation would cause a total loss of DISC1’s binding to Ndel1. A 4 bp deletion of DISC1 discovered in an American family with psychiatric disorders leads to truncation of DISC1 at I808 (Figure 1A and S1A) (Sachs et al., 2005). This frame-shift mutation completely eliminates the DISC1 α2 helix and will therefore abolish its interaction with Ndel1, in view that single residue-substitution in the same helix can disrupt DISC1’s binding to Ndel1 (Figure 2J). To evaluate the potential impact of DISC1 mutations in regulating proliferation of neural stem cells, we generated forebrain organoids from previously characterized iPSCs derived from a schizophrenia patient with the DISC1 mutation (D3–2) and from an unrelated healthy individual as a control (C1–2) (Chiang et al., 2011; Wen et al., 2014). To minimize potential accumulated neurodevelopmental deficits caused by the DISC1 mutation, we analyzed forebrain organoids at as early as 20 days when VZ-like structures is already well established (Qian et al., 2016). We labeled cells in the S phase of cell cycle with EdU followed by analysis 16 hr later. Quantification showed an increased in the percentage of pH3+ cells among all SOX2+EdU+ cells in D3–2 compared to C1–2 groups (Figure 6A, C). FACS analysis further revealed that the progression from S/G2/M phase to G0/G1 was significantly reduced in D3–2 compared to C1–2 groups (Figure 6B, D). Together, these results indicate delayed cell cycle progression during mitosis of RGCs in human forebrain organoids carrying a DISC1 mutation with a defect in its interaction with Nde1/Nde1.
Figure 6. Disrupted cell cycle progression of RGCs during mitosis in forebrain organoids derived from a schizophrenia patent with a DISC1 mutation.
(A) Cell cycle progression is delayed and results in increased mitotic index of RGCs in D3-2 forebrain organoids. Shown on top are sample confocal images of organoid sections immunostained with, pH3, EdU, Sox2 and DAPI. Scale bars, 20 μm
(B) Shown histograms for FACS analysis of dissociated forebrain organoids at day 20.
(C) Quantification of (A). Numbers associated with each bar graph refer to the total number of neural tube structures analyzed under each condition. Values represent mean ± SEM (**P < 0.01, t-test).
(D) Bar graph depicting the percentage of EdU positive cells in G0/G1. Number associated with each bar graph refers to the number of replicates of flow cytometry under each condition. Values represent mean ± SEM (** P < 0.01, t-test).
See also Figure S6.
DISCUSSION
In this study, we have characterized the binding properties between DISC1 and Ndel1 and determined the atomic structure of the complex formed between DISC1 (765–835) and Ndel1-CT (Figure 1 and 2). This structural insight of the DISC1/Ndel1 complex allowed us to develop a highly specific approach to study the function of their interaction by designing a highly specific peptide, which only inhibits the formation of the DISC1/Ndel1 complex, but does not interfere with the bindings of these two proteins to many other target proteins. Using this method, we uncovered the specific function of the DISC1/Ndel1 interaction in regulating cell cycle progression during mitosis by controlling kinetochore localization of Ndel1 and Nde1 in a heterologous system (Figure 3). We further demonstrated a critical role of the DISC1/Ndel1 interaction in regulating mitosis of RGCs both in the developing mouse cortex in vivo and in a human forebrain organoid system (Figure 4 and 5). Finally, based on the structural and mechanistic findings, we tested a potential impact of the DISC1 mutation found in an American family with a 4 bp deletion mutation in the C-terminal tail of the DISC1 coding region on brain development using human forebrain organoids derived from the patient iPSCs, and found cell cycle progression deficits during mitosis of the DISC1 mutation bearing neural stem cells (Figure 6). Our study provides an example of how interaction of two genetic risk factors for complex human disorders can be effectively investigated by integrating biochemical and structural studies with different model systems from heterologous cells to mouse model in vivo and human brain organoids from normal subjects and from patients.
Both DISC1 and Ndel1 are genetic risk factors for psychiatric disorders and have attracted extensive attentions in the past. Since both DISC1 and Ndel1 are multi-functional scaffold proteins, removal or over-expression of either of the proteins is undoubtedly going to cause pleotropic impacts on brain development and functions. Additionally, it is often difficult to dissect and trace the mechanistic origins of functional alterations caused by manipulations of either of the full-length proteins. For example, alterations of Ndel1 will undoubtedly impact the cellular functions of the dynein complex and therefore cause perturbations to many cellular processes in developing neurons. Similarly, manipulation of the full-length DISC1 will change the interaction landscape of many DISC1 targets, including GSK3β and PDE4, which are a master signaling molecules in brain development and functions (Mao et al., 2009; Millar et al., 2005; Soda et al., 2013). In this study, we present the first atomic-level structural description of DISC1/Ndel1 interaction. The DISC1/Ndel1 complex structure allowed us to design a very specific tool to investigate the functional role of DISC1 and Ndel1 interaction while leaving the rest of the protein interactions of these two scaffold proteins intact. Therefore, our study provides an example of an effective strategy to investigate the functional interaction in a specific manner based on the structure.
Our structure and biochemical studies also provide a model for future testing of mechanistic differences of psychiatric diseases caused by the two DISC1 mutations. Based on our data (Figure 1 and 2), the DISC1 mutation in the American family patients is predicted to selectively disrupt the DISC1/Ndel1 interaction and spares the rest of the interaction mediated by DISC1 regions other than short C-terminal. As such the majority of the DISC1 interactions (e.g. the interactions mediated by the central tetramerization domain of the protein and the N-terminal GSK3β-binding region) are predicted not to be affected by the mutation (Figure S3I). However, since the DISC1 mutation carriers are almost invariably heterozygous, the short C-terminal truncated DISC1 mutant protein generated by the mutant allele, if is translated into proteins, will form hetero-tetramer with the WT protein. Therefore, the mutant DISC1 protein in the American family patients may display certain additional functional properties due to the formation of DISC1WT/DISC1mutant hetero-tetramer. Consistent with this model, the expression of DISC1mutant , both in iPSCs derived from patients and in HEK293 heterologous cells, can cause stability decrease of WT DISC1 (Wen et al., 2014). In contrast, the (1; 11)(q42; q14.3) translocation-induced mutation deletes a large part of the tetramerization domain in addition to the Ndel1 binding region. We found that any further truncations of DISC1 from its C-terminal end beyond residue 722, including residue 598 at the Chr1.11 translocation point, resulted in expression of insoluble proteins (Figure S3H), suggesting mutation-induced misfolding of the tetramerization domain. Therefore, the Chr1.11 translocation mutation is likely to generate one copy of functionally null allele DISC1, and decrease the effective dosage of DISC1 in patients even if the mutant transcript is translated. An alternative explanation is that the (1; 11)(q42; q14.3) translocation causes the mutant allele of DISC1 transcript to be unstable and results haploinsufficiency for DISC1 in the mutation carriers (Millar et al., 2005).
Our DISC1 (765–835) peptide targets both Ndel1 and Nde1, and cannot differentiate the two dynein regulators due to their same binding properties. As such, our assay would be observing the combined impacts of the perturbations on the interactions between DISC1 and Ndel1/Nde1. Nde1 and Ndel1 paralogues have been previously studied and show similar behaviors in cell cycle in non-neuronal cells (Stehman et al., 2007). Recently, it was found Nde1 and Ndel1 can substitute each other in many aspects of neurogenesis and neuronal migration (Bradshaw and Hayashi, 2017) . However, Nde1 also displays its unique roles in the cell cycle progression of radial glial progenitors (Doobin et al., 2016). It is interesting to note that ndel1 and nde1 show different expression pattern during the mouse neocortical development (Allen Developing Mouse Brain Atlas, http://developingmouse.brin-map.org/). While Nde1 mRNA is more highly detected in the region with high proliferation, such as VZ, ndel1 is more strongly expressed in the cortical plate, where neurons are localized as result of neuronal migration at E13.5. Therefore, it is likely that NDE1 plays more crucial role in regulating cell cycle progression of neural stem cells in VZ. Future studies are needed to elucidate their potential differential function in vivo.
The human brain organoid system confers a unique platform to investigate neural development in a genetically manipulable and disease-relevant brain region (Nguyen et al., 2016; Qian et al., 2017). We have developed a forebrain-specific organoid model for major mental disorders including schizophrenia at the cellular level with a highly penetrant and disease-relevant genotype. Our finding from studying human forebrain organoids derived from a patient iPSCs suggests a model in which susceptibility gene for major psychiatric disorders could affect neural development via dysregulation of RGC proliferation. Our results delineate a potential mechanistic bridge in patient forebrain organoids for a prominent hypothesis of complex psychiatric disorders, namely aberrant neurodevelopment. Formation of the cerebral cortex requires a tight control of neural stem cell proliferation and differentiation to generate proper number of neurons. RGCs in VZ are precisely regulated to maintain their own stem cell population while generating intermediate progenitors which migrate and form the SVZ and neurons that form the upper layers of cortex (Molyneaux et al., 2007). Therefore, proper control of RGC proliferation is a fundamental and critical process of cortex. Using forebrain organoids generated from human iPSCs, we further show that the specific interaction between DISC1 and Ndel1 plays an important role in maintaining the neural stem cell population during human forebrain development (Figure 5C). Our study provides an example that genetic risk factors for psychiatric disorders can lead to specific cellular abnormalities of biological processes implicated in the neurodevelopmental origins of these disorders.
In summary, based on structural insights we identified a function of DISC1/Ndel1 interaction in regulating cell cycle progression of RGCs in in vivo animal model and in human forebrain organoids. Our study from patient-derived forebrain organoids provides a potential mechanistic understanding of how DISC1 mutation with its C-term deletion can affect neural developmental processes. Together, our multifaceted approaches provide insight into mechanisms of pathogenesis of complex psychiatric disorders.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Mingjie Zhang (mzhang@ust.hk).
EXPERIMENTAL MODELS AND SUBJECTS DETAILS
Human iPSC lines
All studies were performed with approved protocols of Johns Hopkins University School of Medicine. All human iPSC lines were previously characterized (Chiang et al., 2011; Wen et al., 2014)
Animals
All animal experimental procedures were performed in accordance with the protocols approved by the Institutional Animal Care and Use Committee at The Johns Hopkins University School of Medicine. Embryos from pregnant CD1 mice (Charles River) were used for the experiment without determining their sex. All mice were housed in a dedicated animal facility with a standard 12-hour light/dark cycle.
HeLa Cells and HEK293T Cells Culture
HeLa and HEK293T cells (both from ATCC) were cultured in DMEM media supported by fetal bovine serum. The cell lines used in this study were not further authenticated and not found to be on the list of commonly misidentified cell lines (International Cell Line Authentication Committee). Cells were tested negative for mycoplasma contamination by cytoplasmic DAPI staining.
METHODS DETAILS
Constructs and Protein Expression
Genes encoding various lengths of mouse DISC1, Ndel1 and human Nde1 were amplified by PCR and cloned into a modified pET32a vector. Residue numbers indicated in the study refer to the full-length DISC1 (NP_777279.2; 852 residues), Ndel1 (NP_076157.2; 345 residues) and Nde1 (NP_001137451.1; 335 residues). Recombinant proteins were expressed in BL21-Codon Plus (DE3) Escherichia coli cells. The N-terminal His6-tagged proteins were purified using an Ni2+-nitrilotriacetic acid agarose column followed by a step of size-exclusion chromatography (Superdex 200 column from GE Healthcare) in the final buffer of 50 mM Tris-HCl (pH 7.8), 100 mM NaCl, 1 mM DTT and 1 mM EDTA. GST-fused proteins were purified by GSH-Sepharose affinity chromatography, followed by a step of size-exclusion chromatography. For NMR structure determination, Ndel1 238–284 was fused to the C-terminus of DISC1 (765–852) with a 6-residue thrombin cleavable linker (“LVPRGS”).
Isothermal Titration Calorimetry Assay
ITC measurements were carried out on a MicroCal VP-ITC calorimeter at 25 °C. Titration buffer contained 50 mM Tris-HCl pH 7.8, 100 mM NaCl, 1 mM EDTA and 1 mM DTT. Each titration point was performed by injecting a 10 μl aliquot of a protein sample from a syringe into a protein sample in the cell at a time interval of 120 s to ensure that the titration peak returned to the baseline. The titration data were analyzed by Origin7.0 and fitted by a one-site binding model.
Analytical Gel Filtration Chromatography Coupled with Static Light Scattering
This analysis was performed on a fast protein liquid chromatography (FPLC) system coupled with a static light-scattering detector (miniDAWN; Wyatt) and a differential refractive index detector (Optilab; Wyatt). Protein samples (100 μl, concentration of 200μM) were loaded to a Superose 12 10/300 GL column (GE Healthcare) pre-equilibrated with an assay buffer containing 50 mM Tris-HCl pH 7.8, 100 mM NaCl, 1 mM EDTA and 1 mM DTT on an AKTA FPLC system (GE Healthcare). Data were analyzed with Astra 6 (Wyatt).
NMR spectroscopy
NMR samples contained 0.8 mM of the DISC1 (765–852) fused with Ndel1 (CT-CC) protein in 100 mM potassium phosphate at pH6.5. NMR spectra were acquired at 30 °C on Varian Inova 750- and 800-MHz spectrometers each equipped with an actively z-gradient shielded triple resonance probe. Backbone and side-chain resonance assignments were achieved by the standard heteronuclear correlation experiments.
NMR structural calculation
Structural determination of the DISC1 765–835/Ndel1 CT-CC was aided by fusing Ndel1 CT-CC to the C-terminal end of DISC1 765–852 with a thrombin cleavable linker. Single chain fusion protein has simplified the NOE assignments, as we can treat the complex as a single chain protein. The use of the single chain fusion protein is validated by the near identical backbone HSQC spectra of the protein before and after thrombin cleavage (Figure S2D). Inter-proton distance restraints were obtained from three-dimensional, 13C-and 15N-separated NOESY experiments using a mixing time of 80 ms. Hydrogen bonding restraints were generated from the standard secondary structure of the protein based on the NOE patterns (3 and angles) were derived from the chemical shift analysis program TALOS. A total of 2098 NOE structural restrains are obtained and classified as short, medium, and long range (Table S1). Structures were calculated using the program CNS. The 20 lowest energy conformations were obtained. The program Procheck was used to assess the overall quality of the structures. Figures were generated using PYMOL (http://pymol.sourceforge.nt/) and MOLMOL. Ramachandran statistics for the final ensemble of structures for residues containing secondary structure (residue 773–801, 805–830 for DISC1 and residue 247–281 for Ndel1) show that 96.5% of residues are in the most favored region, 2.7% of the residues are in the additionally allowed region, and 0.8% of the residues are in the generally allowed region. None of the structures exhibit distance violations greater than 0.3 Å or dihedral angle violations greater than 4°.
Co-immunoprecipitation and GST Pull-Down Assays
For the co-immunoprecipitation (Co-IP) assay, cell lysates from HEK293T cells transfected with the full-length Flag-Ndel1, the full-length GFP-Lis1 and GFP DISC1 765–835 respectively were mixed for 1 hour at 4 °C. Then the cell lysate mixtures were incubated with 30 μl of Anti-Flag M2 magnetic beads (Sigma-Aldrich, M8823) for another 30 mins at 4 °C. The captured proteins were eluted by boiling, resolved by 15% SDS PAGE, and immunoblotted with the anti-Flag antibody or with anti-GFP antibody (abcam, ab6658).
For Flag pull-down assay, purified trx-Flag (2 nmol), trx-Flag-tagged DISC1 203–321 (2 nmol) or his-tagged DISC1 765–835 (10 nmol) was incubated with 100 μl of HEK293 cell lysate expressing full-length GFP-GSK3β for 1 hour at 4 °C. The mixtures were incubated with 60 μl of Anti-Flag M2 magnetic beads (Sigma-Aldrich, M8823) for 30 minutes at 4 °C. After three times washing with TBST buffer, the captured proteins were eluted by boiling, resolved by 15% SDS PAGE, and detected by immunoblotting with anti-GFP antibody (abcam, ab6658).
Live cell imaging
HeLa cells stably expressing H2B-RFP were seeded onto 24-well culture plates and imaged using a Ti-E inverted fluorescent microscope (Nikon, Tokyo, Japan) equipped with a 10X objective lens, a SPOT BOOST EMCCD camera (Diagnostic Instrument, Sterling Heights, MI, USA) and an INU-NI-F1 temperature, humidity, and CO2 control system (Tokai Hit, Shizuoka, Japan). Data were acquired with MetaMorph software (Molecular Devices) for up to 24 hrs at 5 minute/frame. Images were processed with ImageJ software (NIH).
Kinetochore localization assay
HeLa cells were cultured in DMEM containing 10% fetal bovine serum in 10% CO2. Cells were synchronized at early S phase by double thymidine (at 2 mM) blocks. During the interval of two thymidine treatments, cells were transiently transfected with 1 μg of each plasmid per dish with a Lipofectamine PLUS Kit (Invitrogen) in 35-mm dishes. At 9 hours after second release from thymidine, cells were fixed by 4% paraformaldehyde with 4% sucrose and 0.5% triton in PBS at room temperature for 15 min. After permeabilization with 0.2% Triton X-100 in PBS, cells were stained with primary antibodies at 4 °C overnight. After extensive washing with 0.05% Tween-20 in PBS, secondary antibodies coupled with Alexa-488 or Alexa-594 or Alexa-647 (Invitrogen) were used for staining for another 1 hr at room temperature. Nuclear DNA was stained with 10 μg/ml DAPI. Fluorescent images were acquired with a Leica sp8 confocal microcopy. Mouse Hec1 primary antibody is from Abcam (ab3613). Rabbit Ndel1 primary antibody was provided by Shinji Hirotsune at Osaka City University (Sasaki et al., 2000).
Culture of human iPSC-derived forebrain organoids
Human iPSC lines (C3-1; female), C1–2 (male), D3-2 (male) used in the current study were previously fully characterized (Chiang et al., 2011; Wen et al., 2014). Forebrain-specific organoids were generated as previously described (Qian et al., 2016). Briefly, human iPSC colonies were detached 7 days after passage with Collagenase Type IV, washed with fresh stem cell medium in a 15ml conical tube. On day 1, detached iPSC colonies were transferred to an Ultra-Low attachment 6-well plate (Corning Costar), containing 3 ml of stem cell medium (without FGF-2), plus 2 μM Dorsomorphine (Sigma) and 2 μM A83-01 (Tocris). On days 5–6, half of the medium was replaced with induction medium consisting of DMEM:F12, 1X N2 Supplement (Invitrogen), 1X Penicillin/Streptomycin, 1X Non-essential Amino Acids, 1X Glutamax, 4 ng/ml WNT-3A (R&D Systems), 1 μM CHIR99021 (Cellagentech), and 1 μM SB-431542 (Cellagentech). On day 7, organoids were embedded in Matrigel (Corning) and continued to grow in induction medium for 6 more days. On day 14, embedded organoids were mechanically dissociated from Matrigel by pipetting up and down onto the plate with a 5 ml pipette tip. Typically, 10 – 20 organoids were transferred to each well of a 12-well spinning bioreactor (Spin3) containing differentiation medium, consisting of DMEM:F12, 1X N2 and B27 Supplements (Invitrogen), 1X Penicillin/Streptomycin, 1X 2-Mercaptoenthanol, 1X Non-essential Amino Acids, 2.5 μg/ml Insulin (Sigma).
In utero electroporation and EdU pulsing of mouse
Plasmids expressing GFP, or the DISC1 765–835 peptide or the DISC1 765–835 L822Q peptide mixed with EGFP expressing plasmid pSUbGW) (~ 2μg/μl) were delivered to ventricular zone of embryo brain by in utero electroporation at E13.5 as previously described (Yoon et al., 2014; Yoon et al., 2017b). Briefly, DNA was injected using a beveled and calibrated micropipette with a ~10 μm opening at 15 psi, then five pulses (43 V, 50 ms in duration with a 950 ms interval) were delivered with tweezer electrodes (CUY650-5, Nepa Gene) by a CUY21SC electroporator (Nepa Gene). 50 mg/kg of EdU was injected to a mom 24 hours after electroporation and embryos were sacrificed and fixed with 4% PFA 6 hours after EdU injection. All animal procedures were performed in accordance with the protocol approved by the Institutional Animal Care and Use Committee.
Electroporation and EdU labeling of organoids
At day 45~47, organoids were transferred into Petri dishes containing 37°C PBS, and 3 μl of plasmids expressing GFP, orDISC1 765–835 peptide or DISC1 765–835 L822Q peptide in cFUGW vector mixed with EGFP expressing plasmid (pSUbGW) and 0.01% fast green diluted in sterile PBS was injected into 4–5 ventricle-like cavities of neural tube structures in forebrain organoids at 5 psi, using a beveled and calibrated micropipette with a ~20 μm diameter opening. Five pulses (43 V, 50 ms in duration with a 950 ms interval) were delivered with tweezer electrodes (CUY650–5, Nepa Gene) by a CUY21SC electroporator (Nepa Gene) as previously described (Yoon et al., 2017a; Yoon et al., 2017b). Electroporated organoids were transferred back to Spin3 bioreactor and cultured until fixation. 4 days after electroporation, organoids were pulsed with 10 μM EdU for 2 hr. The media was then replaced and organoids were washed 3 times with fresh media.
Immunohistochemistry and quantitative analysis of mouse embryo and organoids
At 5-day post electroporation, organoids were fixed and sectioned at 30 μm using a cryostat for immunohistochemistry as previously described (Qian et al., 2016). Brains were sectioned at 20 μm using a cryostat. The following primary antibodies were used: GFP (goat, 1:250, Rockland), GFP (chicken, 1:250, Aves), PAX6 (mouse, 1:250, BD), KI67 (rabbit, 1:250, ThermoFisher), and SOX2 (goat, 1:250, Santa Cruz), p-VIMENTINE (mouse, 1:1000, MBL) and NESTIN (Chick, 1:300; Aves). EdU detection was done using Click-iT® EdU Alexa Fluor® 594/647 Imaging Kit (ThermoFisher C10339) according to the manufacturer’s manual, followed by immunostaining for GFP and Ki67. Images were acquired on a Zeiss LSM 710 confocal system (Carl Zeiss, Thornwood, NY, USA) using a multi-track configuration at 40x magnification. All quantification was performed by counting the number of GFP+ cells within the Pax6+ layer (defined as VZ) using Zeiss LSM Image browser (Version 4.2.0.121, Carl Zeiss). Quantification was conducted by investigators blind to manipulation conditions. Cell-cycle exit index was calculated as percentage of GFP+EdU+Ki67− cells/GFP+EdU+ cells in the VZ.
Flow cytometric analysis of cell cycle
Cortex of electroporated brain were dissociated using MACS® Neural Tissue Dissociation Kit (P) (Miltenyl Biotec) and fixed with 4%PFA for 20 min. For EdU detection, Click-iT™ EdU Alexa Fluor™ 647 Flow Cytometry Assay Kit (ThermoFisher C10424) was used, followed by GFP and Vybrant DyeCycle Violet (ThermoFisher V35003) staining. For sorting, 50,000 cells were sorted with Vybrant DyeCycle Violet (405nm), EdU (640 nm), and GFP (488nm) using FACSJazz Cell Sorter (BD). Each sample from cortex from 2~3mice and sorting was repeated four to six times. Analyses were done using FlowJo_v10.3 (FlowJo, LLC) (Yoon et al., 2017a).
QUANTIFICATION AND STATISTICAL ANALYSIS
All data represent Mean ± SEM. For two independent data comparisons, unpaired t-test and Kolmogorov–Smirnov tests were used to determine statistical significance. For multiple comparisons, ANOVAs were used as indicated in the text. *P < 0.05, **P < 0.01, ***P < 0.001. n,s; not significant. The “n” are indicated in the summary quantification plots of each experiments. All statistical analyses were performed using Origin software (OriginLab). All experiments related to cell cultures and imaging studies were performed in blinded fashion.
DATA AND SOFTWARE AVAILABILITY
Data Resources
The atomic coordinates of the DISC1/Ndel1 complex are deposited to the Protein Data Bank under the accession codes PDB: 5YI4.
Supplementary Material
Acknowledgments
We thank S. Hirotsune for anti-Ndel1 antibody, and members of Ming, Song and Zhang laboratories for comments. This work was supported by grants from RGC of Hong Kong (664113, 16103614, AoE-M09-12, and T13-607/12R) and a 973 program grant from the Minister of Science and Technology of China (2014CB910204 to M.Z.), NIH (R35NS097370, R01MH105128 and U19AI131130 to G-l.M.; R37NS047344 and U19 MH106434 to H.S.), the Simons Foundation (SFARI grant 308988 to H.S. and SFARI grant 401625 to G.-l.M.), and the NARSAD young investigator award (to E.K.). M.Z. is a Kerry Holdings Professor in Science and a Senior Fellow of IAS at HKUST.
Footnotes
AUTHOR CONTRIBUTIONS
F.Y., Chuan Y., Cong Y., M.M., R.Y.C.P. contributed to structural analysis, E.K. performed functional analysis of mouse and organoid development, X.Q. and F.J. helped with organoid analyses. J.K. contributed to data collection. F.Y., E.K., H.S., G-l.M. and M.Z. conceived the project and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkuraya FS, Cai X, Emery C, Mochida GH, Al-Dosari MS, Felie JM, Hill RS, Barry BJ, Partlow JN, Gascon GG, et al. Human mutations in NDE1 cause extreme microcephaly with lissencephaly [corrected] American journal of human genetics. 2011;88:536–547. doi: 10.1016/j.ajhg.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakircioglu M, Carvalho OP, Khurshid M, Cox JJ, Tuysuz B, Barak T, Yilmaz S, Caglayan O, Dincer A, Nicholas AK, et al. The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. American journal of human genetics. 2011;88:523–535. doi: 10.1016/j.ajhg.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. American journal of human genetics. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi G, Napolitano F, Ursini G, Di Giorgio A, Caforio G, Taurisano P, Fazio L, Gelao B, Attrotto MT, Colagiorgio L, et al. Association of GSK-3beta genetic variation with GSK-3beta expression, prefrontal cortical thickness, prefrontal physiology, and schizophrenia. The American journal of psychiatry. 2013;170:868–876. doi: 10.1176/appi.ajp.2012.12070908. [DOI] [PubMed] [Google Scholar]
- Bradshaw NJ, Hayashi MA. NDE1 and NDEL1 from genes to (mal)functions: parallel but distinct roles impacting on neurodevelopmental disorders and psychiatric illness. Cell Mol Life Sci. 2017;74:1191–1210. doi: 10.1007/s00018-016-2395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran-Jimeniz B, Camargo LM, Oliver KR, Beher D, Shearman MS, et al. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Molecular and cellular neurosciences. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nature reviews Neuroscience. 2011;12:707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick KE, Kamiya A, Hodgkinson CA, Lencz T, DeRosse P, Ishizuka K, Elashvili S, Arai H, Goldman D, Sawa A, et al. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Human molecular genetics. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, Bonnert TP, Whiting PJ, Brandon NJ. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Molecular psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, Ming GL. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011;16:358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda U, Tarricone C, Choi WC, Cooper DR, Lukasik S, Perrina F, Tripathy A, Kim MH, Cafiso DS, Musacchio A, et al. The structure of the coiled-coil domain of Ndel1 and the basis of its interaction with Lis1, the causal protein of Miller-Dieker lissencephaly. Structure. 2007;15:1467–1481. doi: 10.1016/j.str.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Doobin DJ, Kemal S, Dantas TJ, Vallee RB. Severe NDE1-mediated microcephaly results from neural progenitor cell cycle arrests at multiple specific stages. Nature communications. 2016;7:12551. doi: 10.1038/ncomms12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A, Asai N, Namba T, Wang Y, Kato T, Tanaka M, Tatsumi H, Taya S, Tsuboi D, Kuroda K, et al. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, King DP, Reutiman TJ, Folsom TD, Laurence JA, Lee S, Fan YT, Paciga SA, Conti M, Menniti FS. PDE4B polymorphisms and decreased PDE4B expression are associated with schizophrenia. Schizophrenia research. 2008;101:36–49. doi: 10.1016/j.schres.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nature neuroscience. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbar S, Mesngon MT, Guillotte AM, Desai B, Ayala R, Smith DS. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. The Journal of cell biology. 2008;182:1063–1071. doi: 10.1083/jcb.200803071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DJ, Baffet AD, Nayak T, Akhmanova A, Doye V, Vallee RB. Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell. 2013;154:1300–1313. doi: 10.1016/j.cell.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Tomoda T, Chang J, Takaki M, Zhan C, Morita M, Cascio MB, Elashvili S, Koizumi H, Takanezawa Y, et al. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Human molecular genetics. 2006;15:3313–3323. doi: 10.1093/hmg/ddl407. [DOI] [PubMed] [Google Scholar]
- Kang E, Burdick KE, Kim JY, Duan X, Guo JU, Sailor KA, Jung DE, Ganesan S, Choi S, Pradhan D, et al. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72:559–571. doi: 10.1016/j.neuron.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-anpongkul N, Kang E, Song H, Ming GL. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Yu W, Li Y, Yu L, Zhang Q, Wang F, Yang Z, Du J, Huang Q, Yao X, et al. Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase. Molecular biology of the cell. 2007;18:2656–2666. doi: 10.1091/mbc.E06-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends in biochemical sciences. 1996;21:375–382. [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, Tassa C, Berry EM, Soda T, Singh KK, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. DISC1 partners with GSK3beta in neurogenesis. Cell. 2009;136:990–992. doi: 10.1016/j.cell.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nature reviews Neuroscience. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Moon HM, Youn YH, Pemble H, Yingling J, Wittmann T, Wynshaw-Boris A. LIS1 controls mitosis and mitotic spindle organization via the LIS1-NDEL1-dynein complex. Human molecular genetics. 2014;23:449–466. doi: 10.1093/hmg/ddt436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HN, Song H, Ming G. Enginerring human pluripotet stem cell-derived 3D brain tissues for drug discovery. Journal of Translational Neuroscience. 2016;1:38–48. [Google Scholar]
- Nicodemus KK, Callicott JH, Higier RG, Luna A, Nixon DC, Lipska BK, Vakkalanka R, Giegling I, Rujescu D, St Clair D, et al. Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Human genetics. 2010;127:441–452. doi: 10.1007/s00439-009-0782-y. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Porteous DJ, Millar JK, Brandon NJ, Sawa A. DISC1 at 10: connecting psychiatric genetics and neuroscience. Trends in molecular medicine. 2011;17:699–706. doi: 10.1016/j.molmed.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Jacob F, Song H, Ming GL. Using brain organoids to understand Zika virus-induced microcephaly. Development. 2017;144:952–957. doi: 10.1242/dev.140707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Medema RH. Systematic dissection of dynein regulators in mitosis. The Journal of cell biology. 2013;201:201–215. doi: 10.1083/jcb.201208098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs NA, Sawa A, Holmes SE, Ross CA, DeLisi LE, Margolis RL. A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Molecular psychiatry. 2005;10:758–764. doi: 10.1038/sj.mp.4001667. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, Muramatsu M, Miyagawa S, Hiraiwa N, Yoshiki A, Wynshaw-Boris A, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Molecular and cellular biology. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Soares DC, Carlyle BC, Bradshaw NJ, Porteous DJ. DISC1: Structure, Function, and Therapeutic Potential for Major Mental Illness. ACS chemical neuroscience. 2011;2:609–632. doi: 10.1021/cn200062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda T, Frank C, Ishizuka K, Baccarella A, Park YU, Flood Z, Park SK, Sawa A, Tsai LH. DISC1-ATF4 transcriptional repression complex: dual regulation of the cAMP-PDE4 cascade by DISC1. Molecular psychiatry. 2013;18:898–908. doi: 10.1038/mp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehman SA, Chen Y, McKenney RJ, Vallee RB. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. The Journal of cell biology. 2007;178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarricone C, Perrina F, Monzani S, Massimiliano L, Kim MH, Derewenda ZS, Knapp S, Tsai LH, Musacchio A. Coupling PAF signaling to dynein regulation: structure of LIS1 in complex with PAF-acetylhydrolase. Neuron. 2004;44:809–821. doi: 10.1016/j.neuron.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Taverna E, Gotz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Malavasi EL, Grunewald E, Soares DC, Borkowska M, Millar JK. DISC1 genetics, biology and psychiatric illness. Front Biol (Beijing) 2013;8:1–31. doi: 10.1007/s11515-012-1254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nature cell biology. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- Vergnolle MA, Taylor SS. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Current biology : CB. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- Wang Q, Charych EI, Pulito VL, Lee JB, Graziane NM, Crozier RA, Revilla-Sanchez R, Kelly MP, Dunlop AJ, Murdoch H, et al. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Molecular psychiatry. 2011;16:1006–1023. doi: 10.1038/mp.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, Makri G, Nauen D, Shin JH, Park Y, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15:79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell. 2017a doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Song G, Qian X, Pan J, Xu D, Rho HS, Kim NS, Habela C, Zheng L, Jacob F, et al. Zika-Virus-Encoded NS2A Disrupts Mammalian Cortical Neurogenesis by Degrading Adherens Junction Proteins. Cell Stem Cell. 2017b doi: 10.1016/j.stem.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylkiewicz E, Kijanska M, Choi WC, Derewenda U, Derewenda ZS, Stukenberg PT. The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. The Journal of cell biology. 2011;192:433–445. doi: 10.1083/jcb.201011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.