ABSTRACT
The alarmone nucleotides guanosine pentaphosphate (pppGpp) and tetraphosphate (ppGpp), collectively referred to as (p)ppGpp, are key regulators of bacterial growth, stress adaptation, antibiotic tolerance and pathogenicity. We have recently shown that the Small Alarmone Synthetase (SAS) RelQ from the Gram-positive pathogen Enterococcus faecalis has an RNA-binding activity (Beljantseva et al. 2017). RelQ's activities as an enzyme and as an RNA-binding protein are mutually incompatible: binding of single-stranded RNA potently inhibits (p)ppGpp synthesis in a sequence-specific manner, and RelQ's enzymatic activity destabilizes the RNA:RelQ complex. RelQ's allosteric regulator, pppGpp, destabilizes RNA binding and activates RelQ's enzymatic activity. Since SAS enzymes are widely distributed in bacteria, and, as has been discovered recently, are also mobilized by phages (Dedrick et al. 2017), RNA binding to SASs could be a widespread mechanism. The initial discovery raises numerous questions regarding RNA-binding function of the SAS enzymes: What is the molecular mechanism underlying the incompatibility of RNA:SAS complex formation with pppGpp binding and (p)ppGpp synthesis? What are the RNA targets in living cells? What is the regulatory output of the system – (p)ppGpp synthesis, modulation of RNA structure and function, or both?
KEYWORDS: ppGpp, RelQ, SAS, RSH, stringent response, RNA, nucleotide, RNA-binding protein, Hfq, CsrA
(p)ppGpp-mediated signaling
Bacterial signaling nucleotides regulate their numerous protein targets either via allosteric (i.e., binding to a dedicated regulatory site) or orthosteric (i.e., competing with the substrate in the active site) mechanisms.1 One of the most well-studied classes of signaling nucleotides is guanosine pentaphosphate (pppGpp) and tetraphosphate (ppGpp), collectively referred to as (p)ppGpp – key regulators of bacterial growth, stress adaptation, pathogenicity and antibiotic tolerance.2,3 In Escherichia coli, (p)ppGpp signaling is orchestrated by two large multi-domain proteins RelA and SpoT, the namesakes of the RelA/SpoT Homolog, RSH, protein family.4 Both RelA5 and SpoT6 synthesize (p)ppGpp from GDP and GTP using ATP as a donor of the pyrophosphate moiety. SpoT – but not RelA – also hydrolyzes pppGpp and ppGpp, yielding GTP and GDP, respectively.7 The hydrolytic function of SpoT is essential since uncontrolled overproduction of (p)ppGpp is fatal for bacteria.6 The enzymatic activities of the two E. coli RSH enzymes are regulated allosterically. Synthesis of (p)ppGpp by RelA is strongly induced by ‘starved’ ribosomal complexes loaded with cognate deacylated tRNA in the A-site that accumulate upon amino acid limitation.8 Activation by ‘starved’ ribosomes is further potentiated by the very product of RelA-catalyzed reaction, (p)ppGpp.9 SpoT is regulated by numerous stress signals, such as fatty acid10 and carbon source6 limitation.
Single-domain RSH enzymes that either synthesize (Small Alarmone Synthetases, SASs11-14) or hydrolyze (Small Alarmone Hydrolases, SAHs15) (p)ppGpp have stepped into the limelight in the last decade. Both classes are widely distributed in bacteria, but are universally absent in the phyla that lack the ‘long’ RSHs i.e., in the Plantomycetes, Verrucomicrobia and Chlamydiales (PVC) superphylum.15,16 Bacterial SAHs are uncharted territory with knowledge about these enzymes limited to mapping out their phylogenetic distribution.15 The biological role and regulation of SAS enzymes are better understood. Alkaline shock and cell wall stresses such as treatment with antibiotics targeting the cell wall (e.g., vancomycin) induce overexpression of SASs, leading to an increase in (p)ppGpp levels, which, in turn, renders bacteria more resilient to stress; genomic disruption of the SAS genes inactivates this adaptive tolerance mechanism.12,14,17
Several allosteric regulatory mechanisms of SAS enzymes have recently been discovered. First, it was shown that the enzymatic activity of the SAS RelQ from Gram-positive pathogen Enterococcus faecalis is activated by pppGpp and, to a lesser extent – ppGpp.18 Subsequent crystallographic analysis of Bacillus subtilis RelQ ortholog YjbM (aka SAS1) by Steinchen and colleagues revealed that the enzyme forms a highly symmetric homotetramer that binds two pppGpp molecules at the interface between subunits, allosterically activating the enzyme's catalytic activity.19 In 2017 using biochemical assays, we discovered that E. faecalis RelQ binds to single-stranded RNA.20 This complex formation leads to the inhibition of RelQ's enzymatic activity in a sequence-specific manner, and the allosteric activator of RelQ, pppGpp, counteracts the effect20 (Fig. 1). This constitutes an example of two archetypical regulatory paradigms combined within a single protein – namely, RNA binding activity and a switch in catalytic activity in response to a secondary nucleotide messenger. In this Point-of-View article, we compare and contrast RelQ with the archetypical bacterial RNA-binding proteins CsrA, Hfq and ProQ and outline the immediate research questions.
Figure 1.
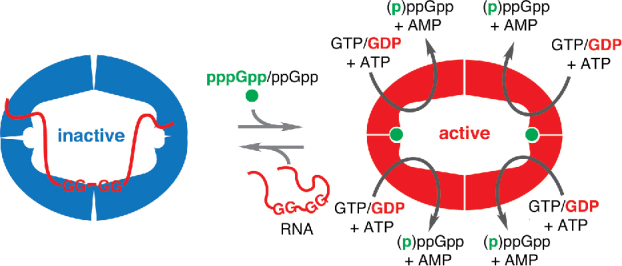
RelQ:RNA interaction as a regulatory switch. RelQ's enzymatic activity is potently inhibited by association with single stranded RNA in a sequence-specific manner with a putative Shine-Dalgarno-like consensus, GGNGG. Association of the primary allosteric regulator pppGpp or, to a lesser extent, the secondary allosteric regulator ppGpp strongly counteracts inhibition by RNA and destabilizes the RNA:RelQ complex. The protective effect is especially strong when the primary allosteric regulator pppGpp synergizes with the preferred substrate, GDP. Both enzymatic activity and mRNA binding could serve as cellular effectors, acting via intracellular concentration of the alarmone and via direct interaction with RNA, respectively. The figure adopted from Beljantseva et al. (2017) with minor modifications.20
Archetypical bacterial RNA-binding proteins: CsrA, Hfq and ProQ
RNA-binding proteins (RBPs) exert their regulatory functions through complex formation with target RNAs.21 Classical examples of this paradigm are the carbon storage regulator CsrA (aka RsmA), broadly distributed across bacterial phyla,22-24 and proteobacterial RNA chaperones Hfq,25,26 and ProQ.27,28 Binding of these RBPs to RNA targets is sequence and structure specific. Hfq uses two RNA-binding interfaces to target single-stranded U-rich (proximal and lateral interfaces) and A-rich (distal interface) sequences.29-32 CsrA binds single-stranded GGA motifs located in the loops of hairpin structures.29,33,34 The binding preferences of ProQ are not as well understood; a transcriptome-wide study mapping its RNA targets (rather than the binding sites per se) identified a diverse set of highly structured small regulatory RNAs (sRNAs) without yielding any obvious sequence consensus.35 Rather, the interaction seems to be structure-driven, with a strong preference for double-stranded RNA over single-stranded.36,37
Broadly speaking, mRNA binding by RBPs regulates gene expression via two strategies. In the case of CsrA, formation of a stable CsrA:RNA complex is the key to regulation. The regulatory GGA element recognized by CsrA is a part of the Shine-Dalgarno motif (AGGAGGU) that directs ribosomal binding to mRNA,38,39 thus CsrA inhibits translation through direct competition with the ribosome.40,41 In the case of RNA chaperones Hfq and ProQ, complex formation is transient and catalyzes structural rearrangements of the target RNA.42,43 The process often involves a third partner – an sRNA – that is complementary to the mRNA target. sRNA binding is the ultimate mRNA regulator, acting through several strategies, including translational repression of the mRNA often followed by degradation by RNase E,44,45 stabilization of mRNA by masking the RNase E cleavage site46 or inhibition of transcription via Rho-dependent termination.47,48
Small Alarmone Synthetase RelQ as an RNA binding protein
In the case of the E. faecalis SAS RelQ, two paradigms – nucleotide-mediated signaling and RNA binding – are combined within a single protein.20 Acting as an enzyme, RelQ increases the levels of the alarmone nucleotide (p)ppGpp, thus increasing tolerance to cell wall stressors such as antibiotics ampicillin and vancomycin.12,14,17 In its role as an RNA-binding protein, RelQ binds single-stranded RNA – and the RNA binding leads to the inhibition of its enzymatic activity in a sequence-specific manner.20 Enzymatic experiments using derivatives of a model mRNA encoding a Met-Phe dipeptide have outlined the specificity of RelQ's inhibition: i) RNA is more potent than DNA, ii), single stranded RNA is more potent than double-stranded, and iii) the two GG elements of the Shine-Dalgarno sequence (GGAGG) are key to the RNA's performance as an inhibitor. (p)ppGpp synthesis and pppGpp binding are mutually incompatible with RelQ:RNA complex formation, suggesting the possibility that the RelQ:RNA interaction acts as a regulatory switch between inactive and active forms of the enzyme (Fig. 1).
However, all of the experiments so far were performed with purified protein in a test tube. There are more questions than answers: What are the RNA targets of RelQ in live cells and what is the biologic role of the interaction? What is the molecular mechanism of E. faecalis RelQ SAS inhibition by RNA and why is pppGpp binding and (p)ppGpp synthesis incompatible with RNA binding? To determine the biologic relevance of RNA binding to RelQ, one has to demonstrate the connection between the biochemistry and the cellular function, as manifested in an observable phenotype, e.g. cell wall stress sensitivity of strains lacking the SAS enzymes.12,14,17
Before we outline the possible roadmap for assessing the in vivo relevance of RelQ:RNA interactions, let us dissect RelQ as a potential RBP. Could it be a bona fide RBP given the biochemical data? If so, what kind of RBP – an RNA chaperone (like Hfq) or a regulator working via formation of a stable complex with its target RNA (like CsrA)? RelQ's affinity to the tested RNA oligonucleotides (EC50 of 1–2 μM20) is modest in comparison with RBPs CsrA, Hfq and ProQ that all have characteristic equilibrium affinities, Kd, in the range of 1–50 nM.33,34,36,37,43,49 In the case of Pseudomonas bacteria, Hfq co-acts with another protein – a Pseudomonas-specific Catabolite repression control, Crc – which, while lacking affinity for RNA itself, greatly increases the affinity of Hfq for RNA.50,51 Given the low affinity of RelQ for RNA, one could imagine that it may have a similar protein partner, but there is no experimental evidence for this exotic possibility. Therefore, RelQ's affinity to RNA is seemingly too low to be physiologically relevant if one were to think within the standard RBP:RNA regulatory framework.
However, in the absence of pppGpp, RNA is a dramatically efficient inhibitor of RelQ's synthetic activity: 100 nM single-strand RNA completely abolishes the ppGpp synthesis by 250 nM RelQ (62.5 nM homotetramer).20 Therefore, it is likely that the regulatory output of the system is not modulation of RNA structure and function, but (p)ppGpp synthesis by RelQ. In this case, it is the RNA that is in control of the signaling system, just like deacylated tRNA in the ribosomal A-site is the ultimate activator of the enzymatic activity of ‘long’ RSHs Rel52 and RelA.8
By shifting the focus to inhibition of RelQ's enzymatic activity – rather than RNA binding per se – binding and inhibition should be considered separately when it comes to determining the functional consensus RNA sequence: the best binder is not necessarily the best inhibitor. Our understanding of the sequence specificity of RelQ inhibition is based on the enzymatic assays that identified two GG motifs in the model single-stranded RNA as the key sequence elements for inhibition of ppGpp synthesis. Binding specificity per se has been studied less extensively, limited to electrophoretic mobility shift assays demonstrating that RelQ virtually does not bind double-stranded RNA and DNA.20 As was recently done for Hfq,29,30 and CsrA,29 one can apply genome-wide approaches to identify RelQ's RNA targets in the cell. This suite of related methods relies on UV-induced crosslinking, isolation of RNA:protein complexes and deep sequencing of the bound RNA fragments.53 Note that these kind of experiments select for the best RNA binders, rather than the best inhibitors. However, there can be no inhibition without binding, so the results of such genomic analyses would give a list of candidate RNA regulators that can be tested for inhibitory activity in vitro.
The first step toward uncovering the molecular mechanism of RelQ's co-regulation by RNA and pppGpp is by mapping the RNA-binding site of the protein. In addition to solving the structure of the RNA:RelQ complex directly with X-ray or NMR, there are two promising experimental approaches. The first is protein cross-linking coupled with mass spectrometry, XL-MS, using purified SAS complexed with a chemically synthesized RNA oligonucleotide. In the XL-MS workflow the UV-irradiated protein:RNA complexes are specifically digested by proteases, the resultant peptides resolved on liquid chromatography, and analyzed by mass spectrometry yielding the location of the crosslinked amino acid residues.54 The second approach is hydrogen deuterium exchange mass spectrometry, HDX.55 This method has been successfully applied to study nucleotide substrate binding to B. subtilis RelQ (aka SAS1 or YjbM).19 In the HDX workflow RelQ in the presence and absence of the RNA ligand is mixed with D2O-containing buffer, and the kinetics of hydrogen exchange to deuterium in amide groups of the protein backbone – a proxy for their mobility and solvent accessibility – is followed by mass spectrometry upon quenching the reaction by lowering the pH. The structural data can be directly validated by mutagenesis and enzymology, yielding RNA-insensitive RelQ variants. These mutants, in turn, can be tested in vivo for cell wall stress phenotypes and used as specificity controls for the genome-wide RNA fishing approaches described above.
Evolutionary diversity of bacterial and phage SAS enzymes
The discovery of E. faecalis RelQ's regulation by (p)ppGpp and RNA is the first glimpse into a mechanism that is likely to be used by other bacterial SAS enzymes. Phylogenetic analyses have uncovered the evolutionary diversity of bacterial SAS enzymes constituting 12 subfamilies.15 These can now be tested for inhibition by RNA, e.g., one can test whether there are functional differences between B. subtilis RelQ and closely related RelP: Are both proteins equally sensitive to inhibition by RNA and activation by pppGpp (Fig. 2)? And if so; do all SAS enzymes share the same sequence/structure specificity for the RNA targets?
Figure 2.
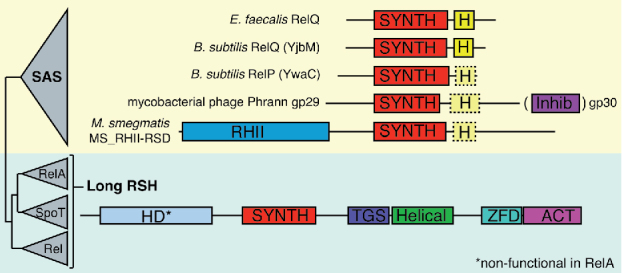
Examples of the evolutionary diversity of bacterial and phage SAS enzymes. The B. subtilis genome encodes two SAS enzymes: RelQ (aka SAS1 or YjbM) and RelP (aka SAS2 or YwaC).12 Mycobacterial phage Phrann encodes a toxic SAS gp29 that is neutralized by a putative inhibitor, gp30, encoded as a downstream gene.57 In M. smegmatis SAS MS_RHII-RSD possesses an RNase HII domain.56 Schematic RSH phylogeny and domain nomenclature are as per Atkinson et al. (2011),15 with minor modifications: HD: (p)ppGpp hydrolase; SYNTH: (p)ppGpp synthetase; H: SAS-specific helix 5α that mediates tetramerisation20 (shown with a dotted outline where this function is unconfirmed but sequence homology is present); Inhib: putative gp29 inhibitor, gp30; RHII: RNase HII; TGS: Threonyl-tRNA Synthetase, GTPase, and SpoT domain; Helical: conserved helix domain; ZFD: Zn finger domain; ACT: Aspartate kinase, Chorismate mutase and TyrA domain.
Another interesting experimental target is M. smegmatis SAS MS_RHII-RSD: in addition to a ppGpp synthetase domain, this SAS contains an RNase HII domain, and is implicated in the R-loop-induced stress response.56 Finally, Dedrick and colleagues have recently demonstrated mobilization of SAS enzymes by mycobacterial Phrann and related viruses.57 Surprisingly, the phage-encoded SAS gp29 is highly cytotoxic, and the effect is countered by co-expression of an adjacent phage gene, gp30.57 In this respect these two proteins behave like a toxin-antitoxin (TA) pair58 – a widespread mechanism mediating both phage addiction and phage immunity.59 Neither the mechanism of gp29 cytotoxicity (overproduction of (p)ppGpp? RNA binding?) and its inhibition by gp30 (direct inhibition via gp29:pg30 complex formation?) are known.
Concluding remarks
Recent structural and biochemical investigations of SAS enzymes have uncovered unexpected interactions with ligands that activate (in the case of pppGpp18,19) or inhibit (in the case of RNA20) their enzymatic activity. The biological significance of either of these interactions is unclear, and concerted microbiological, genomic and structure-functional investigations are necessary to bridge the gap. We have even less information about SAH enzymes and it might well be that these humble single-domain enzymes also hold exciting surprises.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are thankful to Gert Bange, Jose Lemos, Erik Holmqvist, Yasuhiko Irie, Victoria Shingler, Victoriia Murina and Liis Andresen for their comments on the manuscript.
Funding
This work was supported by the Swedish Research Council (Vetenskapsrådet) under Grants 2013–4680 (VH) and 2015–04746 (GCA).
References
- 1.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr Opin Microbiol. 2009;12:170–6. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinchen W, Bange G. The magic dance of the alarmones (p)ppGpp. Mol Microbiol. 2016;101:531–44. doi: 10.1111/mmi.13412. [DOI] [PubMed] [Google Scholar]
- 4.Mittenhuber G. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J Mol Microbiol Biotechnol. 2001;3:585–600. [PubMed] [Google Scholar]
- 5.Haseltine WA, Block R, Gilbert W, Weber K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature. 1972;238:381–4. doi: 10.1038/238381a0. [DOI] [PubMed] [Google Scholar]
- 6.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–90. [PubMed] [Google Scholar]
- 7.Sy J. In vitro degradation of guanosine 5'-diphosphate, 3'-diphosphate. Proc Natl Acad Sci U S A. 1977;74:5529–33. doi: 10.1073/pnas.74.12.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci U S A. 1973;70:1564–8. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shyp V, Tankov S, Ermakov A, Kudrin P, English BP, Ehrenberg M, Tenson T, Elf J, Hauryliuk V. Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep. 2012;13:835–9. doi: 10.1038/embor.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seyfzadeh M, Keener J, Nomura M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:11004–8. doi: 10.1073/pnas.90.23.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol. 2007;65:1568–81. doi: 10.1111/j.1365-2958.2007.05897.x. [DOI] [PubMed] [Google Scholar]
- 12.Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- 13.Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abranches J, Martinez AR, Kajfasz JK, Chavez V, Garsin DA, Lemos JA. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol. 2009;191:2248–56. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuerst JA. The PVC superphylum: Exceptions to the bacterial definition? Antonie Van Leeuwenhoek. 2013;104:451–66. doi: 10.1007/s10482-013-9986-1. [DOI] [PubMed] [Google Scholar]
- 17.Geiger T, Kastle B, Gratani FL, Goerke C, Wolz C. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol. 2014;196:894–902. doi: 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol. 2015;197:1146–56. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinchen W, Schuhmacher JS, Altegoer F, Fage CD, Srinivasan V, Linne U, Marahiel MA, Bange G. Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc Natl Acad Sci U S A. 2015;112:13348–53. doi: 10.1073/pnas.1505271112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beljantseva J, Kudrin P, Andresen L, Shingler V, Atkinson GC, Tenson T, Hauryliuk V. Negative allosteric regulation of Enterococcus faecalis Small Alarmone Synthetase RelQ by single stranded RNA. Proc Natl Acad Sci U S A. 2017;114(14):3726–3731. doi: 10.1073/pnas.1617868114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Assche E Van Puyvelde S, Vanderleyden J, Steenackers HP. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front Microbiol. 2015;6:141. doi: 10.3389/fmicb.2015.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev. 2015;79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–85. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol Microbiol. 2011;82:447–61. doi: 10.1111/j.1365-2958.2011.07822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Updegrove TB, Zhang A, Storz G. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol. 2016;30:133–8. doi: 10.1016/j.mib.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–71. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Attaiech L, Glover JN, Charpentier X. RNA Chaperones Step Out of Hfq's Shadow. Trends Microbiol. 2017;25:247–9. doi: 10.1016/j.tim.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Attaiech L, Boughammoura A, Brochier-Armanet C, Allatif O, Peillard-Fiorente F, Edwards RA, Omar AR, MacMillan AM, Glover M, Charpentier X. Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. Proc Natl Acad Sci U S A. 2016;113:8813–8. doi: 10.1073/pnas.1601626113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016;35:991–1011. doi: 10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell. 2014;55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murina V, Lekontseva N, Nikulin A. Hfq binds ribonucleotides in three different RNA-binding sites. Acta Crystallogr D Biol Crystallogr. 2013;69:1504–13. doi: 10.1107/S090744491301010X. [DOI] [PubMed] [Google Scholar]
- 32.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A. 2009;106:19292–7. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–87. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapouge K, Perozzo R, Iwaszkiewicz J, Bertelli C, Zoete V, Michielin O, Scapozza L, Haas D. RNA pentaloop structures as effective targets of regulators belonging to the RsmA/CsrA protein family. RNA Biol. 2013;10:1031–41. doi: 10.4161/rna.24771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smirnov A, Forstner KU, Holmqvist E, Otto A, Gunster R, Becher D, Reinhardt R, Vogel J. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci U S A. 2016;113:11591–6. doi: 10.1073/pnas.1609981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaulk SG, Smith Frieday MN, Arthur DC, Culham DE, Edwards RA, Soo P, Soo P, Frost LS, Keates RA, Glover JN, et al. ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry. 2011;50:3095–106. doi: 10.1021/bi101683a. [DOI] [PubMed] [Google Scholar]
- 37.Arthur DC, Ghetu AF, Gubbins MJ, Edwards RA, Frost LS, Glover JN. FinO is an RNA chaperone that facilitates sense-antisense RNA interactions. EMBO J. 2003;22:6346–55. doi: 10.1093/emboj/cdg607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shine J, Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974;71:1342–6. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–7. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol. 2002;44:1599–610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 41.Baker CS, Eory LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol. 2007;189:5472–81. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hopkins JF, Panja S, Woodson SA. Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res. 2011;39:5193–202. doi: 10.1093/nar/gkr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–6. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smirnov A, Wang C, Drewry LL, Vogel J. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J. 2017;36:1029–45. doi: 10.15252/embj.201696127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol. 2011;79:419–32. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- 46.Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, Romeo T, Babitzke P. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol. 2013;87:851–66. doi: 10.1111/mmi.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabhi M, Espeli O, Schwartz A, Cayrol B, Rahmouni AR, Arluison V, Boudvillain M. The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 2011;30:2805–16. doi: 10.1038/emboj.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci U S A. 2011;108:13059–64. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henderson CA, Vincent HA, Casamento A, Stone CM, Phillips JO, Cary PD, Sobott F, Gowers DM, Taylor JE, Callaghan AJ. Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS. RNA. 2013;19:1089–104. doi: 10.1261/rna.034595.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milojevic T, Sonnleitner E, Romeo A, Djinovic-Carugo K, Blasi U. False positive RNA binding activities after Ni-affinity purification from Escherichia coli. RNA Biol. 2013;10:1066–9. doi: 10.4161/rna.25195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno R, Hernandez-Arranz S, La Rosa R, Yuste L, Madhushani A, Shingler V, Rojo F. The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ Microbiol. 2015;17:105–18. doi: 10.1111/1462-2920.12499. [DOI] [PubMed] [Google Scholar]
- 52.Avarbock A, Avarbock D, Teh JS, Buckstein M, Wang ZM, Rubin H. Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry. 2005;44:9913–23. doi: 10.1021/bi0505316. [DOI] [PubMed] [Google Scholar]
- 53.Li X, Song J, Yi C. Genome-wide mapping of cellular protein-RNA interactions enabled by chemical crosslinking. Genomics Proteomics Bioinformatics. 2014;12:72–8. doi: 10.1016/j.gpb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kramer K, Sachsenberg T, Beckmann BM, Qamar S, Boon KL, Hentze MW, Kohlbacher O, Urlaub H. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat Methods. 2014;11:1064–70. doi: 10.1038/nmeth.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev Proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnan S, Petchiappan A, Singh A, Bhatt A, Chatterji D. R-loop induced stress response by second (p)ppGpp synthetase in Mycobacterium smegmatis: functional and domain interdependence. Mol Microbiol. 2016;102:168–82. doi: 10.1111/mmi.13453. [DOI] [PubMed] [Google Scholar]
- 57.Dedrick RM, Jacobs-Sera D, Bustamante CA, Garlena RA, Mavrich TN, Pope WH, Reyes JC, Russell DA, Adair T, Alvey R, et al. Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol. 2017;2:16251. doi: 10.1038/nmicrobiol.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 59.Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: Defeating bacterial defences. Nat Rev Microbiol. 2013;11:675–87. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]


