ABSTRACT
Understanding the mechanisms for mRNA production under normal conditions and in response to cytotoxic stresses has been subject of numerous studies for several decades. The shutdown of canonical mRNA transcription, export and translation is required to have enough free resources for the immediate production of heat shock proteins that act as chaperones to sustain cellular processes. In recent work we uncovered a simple mechanism, in which the export block of regular mRNAs and a fast export of heat shock mRNAs is achieved by deactivation of the nuclear mRNA quality control mediated by the guard proteins. In this point of view we combine long known data with recently gathered information that support this novel model, in which cells omit quality control of stress responsive transcripts to ensure survival.
Keywords: Heat stress, cellular stress, mRNA quality control, nuclear export
Efficient protein production relies on the coordination and correctness of gene expression, transcript processing and translation, which are crucial prerequisites for every cell's growth. In particular, the nuclear regulation of pre-mRNA transcription, maturation, and mRNA export are tightly connected and monitored. Recent work in the model organism Saccharomyces cerevisiae provided novel insights into mRNA quality control and gene expression during stress.
Pre-mRNA maturation
Under normal growth conditions a newly transcribed mRNA undergoes several processing steps before it is exported into the cytoplasm.1-3 The major platform for regulation of mRNA maturation is provided by the C-terminal domain (CTD) of Rpb1, the largest catalytically active component of the RNA polymerase II (RNAP II).4 Changes in the phosphorylation pattern of the CTD's highly conserved YSPTSPS-heptads from the transcription start to the end allow recruitment of proteins at the correct time point.5,6 The first of these steps is the capping reaction, which is performed after approximately 20 nucleotides of the mRNA are synthesized by RNAP II.7 At this point the m7G-cap is added at the 5′ end of the transcript by the Cet1/Ceg1 capping machinery8 and the methyltransferase Abd19 to protect it from 5′−3′ degradation.10,11 Efficiency of this process is supported by a free 5′-end and the immediate recruitment of the capping complex to the CTD, which is at that time highly phosphorylated at Ser5.8,12-14 Once the m7G-cap is synthesized, the cap-binding complex (CBC) can bind and is later involved in nuclear export.15 Co-transcriptional recruitment of the spliceosome follows capping as it requires the presence of the CBC16 and results in excision of introns from the pre-mRNA.17 Splicing is accompanied by binding of the THO/TREX complex to the spliceosome18 and the RNAP II CTD.19,20 The THO complex (Hpr1, Tho2, Mft1, Thp2) forms the TREX (transcription-coupled export) complex by recruitment of Yra1 and Sub2. It supports transcription elongation and links it to the later export of the mRNA.19,21,22 Another complex, which recently was shown to connect transcription with export of the matured mRNA is TREX-2, composed of Sac3, Thp1, Sem1, Sus1 and Cdc31.23 TREX-2 together with the SAGA complex are involved in docking transcribing genes to the NPC in a process called “gene gating” that allows preferential export of these mRNAs.24
To finalize mRNA transcription, the 3′ processing machinery (CF1A, CF1B, CPF) is recruited to the polyadenylation site and after cleavage of the mRNA a 70-90 nucleotide long poly(A) tail is added by the poly(A) polymerase Pap1.25,26
Nuclear mRNA quality control and the guard proteins
During the last years the idea emerged that these maturation processes not only rely on the enzymes and cooperating factors that carry out each processing step, but that the essential steps of mRNA maturation are controlled for their correctness and linked to the recruitment of shuttling adaptor proteins. As such they interact with the mRNA and the export receptor heterodimer Mex67-Mtr2 (TAP-p15 in humans) and accompany the transcript to the cytoplasm. In the bakers yeast S. cerevisiae the most prominent RNA-binding proteins that shuttle with an mRNA from the nucleus to the cytoplasm are the serine/arginine (SR)-rich proteins Npl3, Gbp2 and Hrb1 and the poly(A)-binding protein Nab2. In their function to release the matured mRNA from the nucleus to the cytoplasm by recruitment of Mex67, we will name them from now on the guard proteins. Npl3 is one of the first proteins that contacts a newly emerging mRNA as it interacts with the RNAP II27 as well as with the CBC28 arguing for a very early recruitment. Following that, Npl3 supports efficient splicing by interacting with the early spliceosome.29 Finally, Npl3 recruits Mex67-Mtr2 signaling export competence.30 Correct splicing is subsequently controlled by the guard proteins Gbp2 and Hrb1 that interact with the late spliceosome.31 These two proteins are loaded co-transcriptionally by the THO/TREX complex,32,33 and recruit the export receptor Mex67 in case the mRNA is processed properly. Errors in this step on the other hand result in acquiring the degradation machinery.31 This nuclear removal of faulty RNAs relies on the TRAMP (Trf4/5, Air1/2, Mtr4) complex that marks these RNAs with a short oligo(A) tail for subsequent degradation by the nuclear exosome, which in contrast to its cytoplasmic counterpart contains Rrp6.34-36 It was suggested earlier that preventing export of immature transcripts might be the result of kinetic competitions between factors that facilitate export and others involved in retention and degradation.37,38 Thus, the more an ribonucleoparticle (RNP) differs from being perfectly covered with export receptors, the less likely it is exported39 and rather marked by TRAMP to be degraded by the Rrp6-containing exosome. The proper coverage of large RNA-containing molecules with export receptors is also required for the export of ribosomal subunits, as missing export factors result in the accumulation of subunits in the nucleus.40-46
The last processing step that results in decoration of the mRNA with the guard proteins is the formation of the 3′ end and synthesis of the poly(A) tail. Here the poly(A)-binding protein Nab2, together with its mainly cytoplasmic homolog Pab1,47,48 controls length and quality of the 3′ tail – a process that is antagonized by the exosome component Rrp6.49,50 A fine-tuned interplay between factors that facilitate export and degradation is crucial for cellular functionality. All binding events of the guard proteins suggested to represent quality control checkpoint marks that allow export in case of proper processing or else retain the wrong transcripts and recruit the degradation machinery.
This serial decoration of the maturing mRNA with the guard proteins and the dependence of their recruitment on the previous step suggested a potential function as control factors that are able to finally allow export by recruitment of the Mex67-export receptor. However, this was not conclusively shown until lately, when an in vivo “leakage assay” was developed in which the retention of false mRNAs was visibly alleviated upon elimination of the guard proteins, Npl3, Gbp2, Hrb1 and Nab2.31,51 Covering the pre-mRNA with the guard proteins might prevent an immediate and independent contact of Mex67 with the premature mRNA, which would seem to be an elegant retention mechanism.
These data together allow describing a model, in which maturing mRNAs bind the guard proteins to prevent Mex67 association until the transcripts are fully processed (Fig. 1). Completed maturation is signaled by Mex67 recruitment and results in their cytoplasmic transfer. Although it seems likely that Npl3 is involved in controlling correctness of 5′ capping, Gbp2 and Hrb1 ensure accurate splicing and Nab2 monitors proper 3′ tailing, it is well possible that the guard proteins have additional functions and are not only loaded once.52,53 Also the existence of other, yet missing guard proteins is conceivable, because a smooth transit of the highly charged mRNA through the hydrophobic interior of the nuclear pore complex (NPC) will probably be more effective in the presence of more Mex67 molecules that shield the charged mRNA and facilitate a smoother passage through the hydrophobic interior of the NPC.
Figure 1.
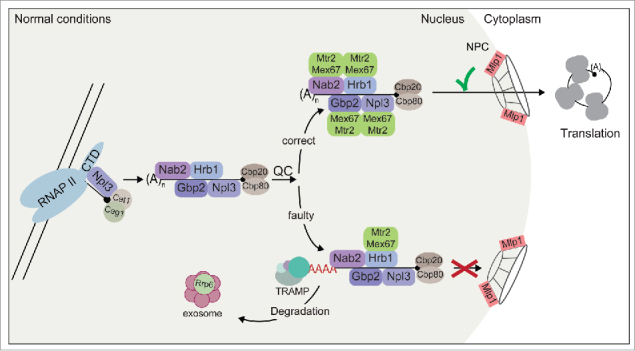
Guard proteins control mRNA maturation and nuclear export. Every maturation step results in association of guard proteins that recruit the export receptor Mex67-Mtr2 to the correct mRNA and support degradation by the TRAMP/exosome pathway for faulty transcripts. Finally, Mlp1 controls proper Mex67 decoration of the guard proteins to allow nuclear export.
Transcripts that do not show the necessary Mex67-decoration of the guard proteins are not only detained because they lack export factors, but also by a last quality control check at the NPC. At the nuclear basket, Mlp1 and the highly homologous nucleoporin Mlp2 are the last nuclear factors involved in retaining erroneous transcripts.54,55 As a final gatekeeper, Mlp1 was shown to interact with the guard proteins,31,55,56 and a model is conceivable that Mlp1 and Mlp2 might control the Mex67-guard-protein interactions, before letting them pass31 and retains the mRNA, if no or insufficient Mex67 is bound to the guard proteins.
This elaborated system ensures efficient translation of correct mRNAs and prevents the translation machinery to deal with an overwhelming number of suboptimal or even defective transcripts. In fact, cytoplasmic non- or wrongly processed mRNAs that are translated into missfunctional proteins threaten homeostasis and have in general a detrimental effect on cellular growth.57 There is growing evidence that not only in yeast but also in mammals mRNA adaptor proteins act as guards and are tightly linked to correctness and thus quality of mRNA expression.58 In the same way as a lack of quality control is harmful to the cell, an overexpression of the guard proteins has the same adverse effect on cellular fitness in yeast, as it possibly even leads to a retention of correct mRNAs.51 In fact, defects or overexpression of human homologues of the guard proteins (ZC3H14, as a human Nab2 ortholog), or the SR-proteins SRSF1, SRSF3 and SRSF7, which also accompany the mRNAs to the cytoplasm and interact with TAP-p15,59 are known to cause several diseases, including cancer and neurodegenerative diseases, cardiovascular diseases or conditions like neuronal dysfunction60-65 and (http://www.cbioportal.org/).
The stress induced mRNA export block
In situations that are stressful to the cell, such as elevated temperatures or high salt concentrations, a cellular survival response – the heat shock response- is initiated, in which heat shock (HS) proteins are produced to protect other cellular proteins from denaturing. To establish a fast response to survive the hazardous condition, expression, maturation, nuclear export and translation of normal “housekeeping” mRNAs need to be blocked and the export of stress mRNAs has to be of utmost priority.66 That this in fact happens and that mRNAs are blocked in the nucleus upon 42°C heat stress, while HS mRNAs are exported67 and splicing is disrupted68,69 was already described over 20 years ago. Additionally other cellular stress reactions are known, as Npl3 dissociates from mRNAs and is dispensable for HS mRNA export,70,71 and Gbp2 aggregates reversibly upon stress.72 Furthermore, nuclear accumulation of mRNA requires activity of the MAPK kinase Slt2, which is also responsible for the phosphorylation of Nab2 under stress.73 Stress furthermore leads to formation of foci that contain the RNA binding proteins Nab2 and Yra1 as well as the NPC gatekeeper Mlp1.73
However, a systematic analysis of stress in the light of nuclear quality control and nuclear mRNA export was not done until recently. In a systematic approach we examined the behavior of the guard proteins and Mex67 and found that the export block of mature or maturing housekeeping mRNAs appears to be facilitated by a global dissociation of the guard proteins in complex with the exporter Mex6751 (Fig. 2). Even though it is most likely that structural changes or post-translational modifications are responsible for the dissociation of all guard proteins, details still need to be elucidated. Amazingly, inactivation of the guard proteins upon stress not only stops regular mRNA export, but also provides a sophisticated system to prevent the association of guard proteins with newly transcribed HS mRNAs. Thus, production of HS mRNAs is accomplished without any of the guard proteins.51 Certainly the greatest benefit of expressing stress mRNAs without the help of guard proteins is that in this way a time-consuming quality control is omitted and a response to stress is promptly initiated. If formation of nuclear guard protein-containing foci is needed to allow evasion of quality control, or if they form because these proteins are not required and present in excess at that time, remains elusive to date. However, the deactivation of the guard protein-mediated quality control is of particular importance, as the degradation machinery is functional during heat stress and degrades HS RNAs under certain conditions, e.g. when these transcripts are artificially trapped in the nucleus.74,75 Thus, by preventing the guard proteins from binding to HS mRNAs, their potential quality control-mediated degradation is circumvented.
Figure 2.
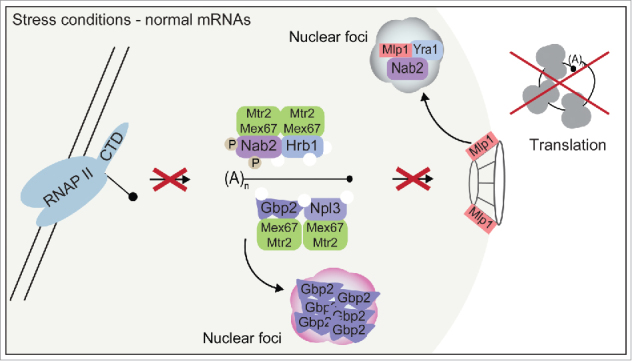
Stress conditions lead to dissociation of Mex67-complexes and a subsequent mRNA export block. Upon stress housekeeping mRNAs are no longer transcribed and Mex67 is dissociated with the guard proteins from mature mRNAs. The recruitment of the guard proteins is prevented by mechanisms, including phosphorylation, aggregation and foci-formation of guard proteins and the NPC gatekeeper Mlp1.
The preferential nuclear export of HS mRNAs
The expression of stress responsive transcripts utilizes a slimmed down pathway. As guard proteins are dispensable, export is very likely promoted by direct binding of the exporter Mex67 to the HS mRNA.51 Not only is Mex67 among the few proteins absolutely essential for the export of stress transcripts,76 but further it was shown to be able to bind, via its loop domain, directly to any kind of RNA including stress mRNAs.46,51 Furthermore, binding of Mex67 to either the guard protein Npl3 or RNA is mutually exclusive,51 arguing for a domain in Mex67 that can either bind to guard proteins or directly to mRNA. This points toward a new view in mRNA export that rather proposes a mechanism in which Mex67 under normal conditions is constantly prevented from binding to the maturing mRNA, by the co-transcriptional decoration of the immature transcript with the guard proteins. Direct binding of the export receptor Mex67-Mtr2 to certain transcripts was also shown for its homolog in higher eukaryotes TAP-p15 where it binds to constitutive transport elements (CTEs) that can be found in viral RNAs as well the TAP pre-mRNA itself77-79 potentially allowing these transcripts to circumvent quality control and being immediately exported.
This instant binding of Mex67 to newly transcribed HS mRNAs seems to happen already at the site of transcription as the exporter interacts with the RNAP II component Rpb1 and the transcription factor Hsf1 when heat stress conditions are applied51 (Fig. 3). The transcription factor Hsf1 is the master factor of heat stress gene induction as it regulates their expression by binding to the heat-shock-element (HSE) in the gene's promoter.80
Figure 3.
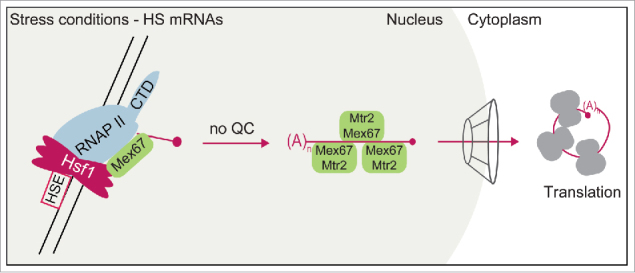
Export of HS RNAs. Direct co-transcriptional recruitment of Mex67 to HS mRNAs allows omission of quality control and fast HS mRNA export.
Overall, the expression of stress responsive transcripts appears to rely on other requirements than normal gene expression. Thus, transcriptional activation by Hsf1 does not require the presence of general transcriptional activators like TFIIA,81 Taf9 (a component of TFIID and SAGA)82,83 or the TFIIH kinase Kin28.84,85 Even the CTD of RNAP II, which usually mediates most regulatory processes is dispensable upon stress.85 On the contrary, Hsf1 on its own is able to recruit the Mediator complex86 that tightly regulates stress gene expression.87 Whether “gene gating” supports HS mRNA export and involves for instance TREX-2 is currently unknown, but might be an attractive way of supporting fast nuclear export.
Being expressed under the control of Hsf1 appears to change the fate of an mRNA dramatically. Thus, housekeeping transcripts can be converted into heat stress responsive mRNAs that are not quality controlled by only inserting an HSE in their promoter.51 Therefore, the HSE resembles an express ticket to the cytoplasm. Furthermore, it allows a preferential translation, as glucose starvation was shown to induce an Hsf1-controlled transcription and translation priority for heat shock proteins, while mRNAs controlled by other stress transcription factors (Msn2/4) are indeed produced, but directly stored in stress granules for later use.88 Bypassing quality control appears to be a drastic course of action when facing cellular stress, but the gain of time in responding to a threat potentially exceeds the drawbacks of translating a couple of faulty proteins - at least for a limited time. Moreover, the overall cellular mRNA export block relieves the competing situation for the many mRNAs at the translation machinery, as in comparison only a small amount of HS RNAs needs to be translated so that it is not necessary to be very efficient in producing correct proteins. Consistently, cells can survive a period of severe heat stress without guard proteins and quality control factors, but not without the general exporter Mex67 or heat shock proteins, as shown in growth analyses.51
Not only changes in the nucleus contribute to efficient HS mRNA expression, but also cytoplasmic processes adapt to ensure a fast heat shock protein production. In response to stress the general translation is inhibited and polysomes can no longer be detected.89,90 Depending on the type of stress, initiation of translation is inhibited either by disturbing formation of the closed loop structure in an Eap1/Caf20-dependent way, or by titrating GTP-bound eIF2 out of the ternary complex.91,92 All cellular responses in the end lead to the preferential translation of HS mRNA.
Concluding remarks and open questions
In summary, the mechanisms, by which normal and HS mRNA expression is performed are significantly different, as the guard protein mediated quality control is deactivated during stress. The export receptor heterodimer Mex67-Mtr2 alone is sufficient to ensure a fast transport to the cytoplasm. The important role of the guard proteins lies in normal mRNA transport by regulation and control of maturation and the stepwise approval through Mex67 recruitment. Future studies have to address how the guard proteins detect defects and initiate degradation of false transcripts. It might be a combination of the timely detection of the successively associating complexes and a competition with the degradation machinery. Also post-translational modifications may play a role. De-phosphorylation of Npl3 was indeed shown to lead to the recruitment of Mex67,30 but what triggers de-phosphorylation is unknown. Furthermore, it is unclear if there are additional guard proteins and how exactly Mlp1 at the NPC detects the proper Mex67-coverage of the guards. Many questions are also unanswered for the particular situation during cellular stress, such as what triggers the dissociation or aggregation of the guard proteins and which complexes and subcomplexes that contribute to a smooth mRNA production under normal conditions (like THO, TREX, TREX-2 or SAGA) are also involved in HS mRNA transcription. Finally, it remains to be understood how HS mRNA translation differs from normal translation. Comparing the molecular basics of gene expression under normal and challenging conditions may help to unravel fundamental principles in science and get a better understanding of diseases based on defects in mRNA surveillance factors.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank the Krebber laboratory for comments on the manuscript.
Funding
This work was funded by grants of the Deutsche Forschungsgemeinschaft (DFG) and the SFB860 awarded to H.K.
References
- 1.Meinel DM, Sträßer K. Co-transcriptional mRNP formation is coordinated within a molecular mRNP packaging station in S. cerevisiae. Bioessays 2015; 37:666-77; PMID:25801414; https://doi.org/ 10.1002/bies.201400220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niño CA, Hérissant L, Babour A, Dargemont C. mRNA nuclear export in yeast. Chem Rev 2013; 113:8523-45; PMID:23731471; https://doi.org/ 10.1021/cr400002g [DOI] [PubMed] [Google Scholar]
- 3.Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat Rev 2011; 12:377-84; PMID:21602906; https://doi.org/ 10.1038/nrm3119 [DOI] [PubMed] [Google Scholar]
- 4.Hsin J-p, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 2012; 26:2119-37; PMID:23028141; https://doi.org/ 10.1101/gad.200303.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bataille AR, Jeronimo C, Jacques PÉ, Laramée L, Fortin MÈ, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 2012; 45:158-70; PMID:22284676; https://doi.org/ 10.1016/j.molcel.2011.11.024 [DOI] [PubMed] [Google Scholar]
- 6.Bentley DL. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 2005; 17:251-6; PMID:15901493; https://doi.org/ 10.1016/j.ceb.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 7.Coppola JA, Field AS, Luse DS. Promoter-proximal pausing by RNA polymerase II in vitro: Transcripts shorter than 20 nucleotides are not capped. Proc Natl Acad Sci U S A 1983; 80:1251-5; PMID:6572384; https://doi.org/ 10.1073/pnas.80.5.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu M, Rajashankar KR, Lima CD. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure 2010; 18:216-27; PMID:20159466; https://doi.org/ 10.1016/j.str.2009.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwer B, Saha N, Mao X, Chen HW, Shuman S. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics 2000; 155:1561-76; PMID:10924457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 2010; 467:608-11; PMID:20802481; https://doi.org/ 10.1038/nature09338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwer B, Mao X, Shuman S. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res 1998; 26:2050-7; PMID:9547258; https://doi.org/ 10.1093/nar/26.9.2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev 1997; 11:3319-26; PMID:9407025; https://doi.org/ 10.1101/gad.11.24.3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suh MH, Meyer PA, Gu M, Ye P, Zhang M, Kaplan CD, Lima CD, Fu J. A dual interface determines the recognition of RNA polymerase II by RNA capping enzyme. J Biol Chem 2010; 285:34027-38; PMID:20720002; https://doi.org/ 10.1074/jbc.M110.145110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Rucobo FW, Kohler R, van de Waterbeemd M, Heck AJR, Hemann M, Herzog F, Stark H, Cramer P. Molecular basis of transcription-coupled Pre-mRNA capping. Mol Cell 2015; 58:1079-89; PMID:25959396; https://doi.org/ 10.1016/j.molcel.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 15.Lewis JD, Izauralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem 1997; 247:461-9; PMID:9266685; https://doi.org/ 10.1111/j.1432-1033.1997.00461.x [DOI] [PubMed] [Google Scholar]
- 16.Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 2005; 19:53-63; PMID:15989964; https://doi.org/ 10.1016/j.molcel.2005.05.007 [DOI] [PubMed] [Google Scholar]
- 17.Will CL, Lührmann R. 13 spliceosome structure and function. Cold Spring Harb Monogr Arch 2006; 43:369-400; PMID:21441581; https://doi.org/ 10.1101/087969739.43.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chanarat S, Seizl M, Sträßer K. The prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev 2011; 25:1147-58; PMID:21576257; https://doi.org/ 10.1101/gad.623411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J 2004; 23:2620-31; PMID:15192704; https://doi.org/ 10.1038/sj.emboj.7600261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meinel DM, Burkert-Kautzsch C, Kieser A, O'Duibhir E, Siebert M, Mayer A, Cramer P, Söding J, Holstege FC, Sträßer K. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLoS Genet 2013; 9:e1003914; PMID:24244187; https://doi.org/ 10.1371/journal.pgen.1003914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saguez C, Gonzales FA, Schmid M, Bøggild A, Latrick CM, Malagon F, Putnam A, Sanderson L, Jankowsky E, Brodersen DE, et al.. Mutational analysis of the yeast RNA helicase Sub2p reveals conserved domains required for growth, mRNA export, and genomic stability. RNA 2013; 19:1363-71; PMID:23962665; https://doi.org/ 10.1261/rna.040048.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, et al.. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002; 417:304-8; PMID:11979277; https://doi.org/ 10.1038/nature746 [DOI] [PubMed] [Google Scholar]
- 23.Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R, Pugh BF, Clausen T, Köhler A. The nuclear pore-associated TREX-2 complex employs mediator to regulate gene expression. Cell 2015; 162:1016-28; PMID:26317468; https://doi.org/ 10.1016/j.cell.2015.07.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev 2007; 8:761-73; PMID:17786152; https://doi.org/ 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- 25.Chan S, Choi E-AA, Shi Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA 2011; 2:321-35; PMID:21957020; https://doi.org/ 10.1002/wrna.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proudfoot N, O'Sullivan J. Polyadenylation: A tail of two complexes. Curr Biol 2002; 12:R855-7; PMID:12498707; https://doi.org/ 10.1016/S0960-9822(02)01353-2 [DOI] [PubMed] [Google Scholar]
- 27.Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 2001; 15:1771-82; PMID:11459827; https://doi.org/ 10.1101/gad.892401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen EC, Stage-Zimmermann T, Chui P, Silver PA. 7The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J Biol Chem 2000; 275:23718-24; PMID:10823828; https://doi.org/ 10.1074/jbc.M002312200 [DOI] [PubMed] [Google Scholar]
- 29.Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell 2008; 32:727-34; PMID:19061647; https://doi.org/ 10.1016/j.molcel.2008.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert W, Guthrie C. The glc7p nuclear phosphatase promotes mRNA export by facilitating association of mex67p with mRNA. Mol Cell 2004; 13:201-12; PMID:14759366; https://doi.org/ 10.1016/S1097-2765(04)00030-9 [DOI] [PubMed] [Google Scholar]
- 31.Hackmann A, Wu H, Schneider UM, Meyer K, Jung K, Krebber H. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat Commun 2014; 5:3123; PMID:24452287; https://doi.org/ 10.1038/ncomms4123 [DOI] [PubMed] [Google Scholar]
- 32.Hacker S, Krebber P. Differential export requirements for shuttling serine/arginine-type mRNA-binding proteins. J Biol Chem 2004; 279:5049-52; PMID:14676199; https://doi.org/ 10.1074/jbc.C300522200 [DOI] [PubMed] [Google Scholar]
- 33.Hurt E, Luo M-J, Röther S, Reed R, Strässer K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A 2004; 101:1858-62; PMID:14769921; https://doi.org/ 10.1073/pnas.0308663100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem 2010; 285:3540-7; PMID:19955569; https://doi.org/ 10.1074/jbc.M109.058396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fasken MB, Corbett AH. Mechanisms of nuclear mRNA quality control. RNA Biol 2009; 6:237-41; PMID:19574733; https://doi.org/ 10.4161/rna.6.3.8330 [DOI] [PubMed] [Google Scholar]
- 36.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell 2009; 136:763-76; PMID:19239894; https://doi.org/ 10.1016/j.cell.2009.01.019 [DOI] [PubMed] [Google Scholar]
- 37.Doma MK, Parker R. RNA quality control in eukaryotes. Cell 2007; 131:660-8; PMID:18022361; https://doi.org/ 10.1016/j.cell.2007.10.041 [DOI] [PubMed] [Google Scholar]
- 38.Mühlemann O, Jensen TH. mRNP quality control goes regulatory. Trends Genet 2012; 28:70-7; PMID:22154474; https://doi.org/ 10.1016/j.tig.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 39.Soheilypour M, Mofrad MRK. Regulation of the affinity between RNA-binding proteins and the export receptor enables uclear basket proteins to distinguish and retain aberrant mRNAs; Sci Rep [Internet] 2016. doi: 10.1038/srep35380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG. Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes. PLoS Genet 2012; 8:e1002915; PMID:22956913; https://doi.org/ 10.1371/journal.pgen.1002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 2001; 21:3405-15; PMID:11313466; https://doi.org/ 10.1128/MCB.21.10.3405-3415.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerhardy S, Menet AM, Pena C, Petkowski JJ, Panse VG. Assembly and nuclear export of pre-ribosomal particles in budding yeast. Chromosoma 2014; 123:327-44; PMID:24817020; https://doi.org/ 10.1007/s00412-014-0463-z [DOI] [PubMed] [Google Scholar]
- 43.Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 2000; 151:1057-66; PMID:11086007; https://doi.org/ 10.1083/jcb.151.5.1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moy TI, Silver PA. Requirements for the nuclear export of the small ribosomal subunit. J Cell Sci 2002; 115(14):2985-95; PMID:12082158 [DOI] [PubMed] [Google Scholar]
- 45.Neumann B, Wu H, Hackmann A, Krebber H. Nuclear Export of pre-ribosomal subunits requires Dbp5, but not as an RNA-Helicase as for mRNA export. PLoS One 2016; 11:e0149571; PMID:26872259; https://doi.org/ 10.1371/journal.pone.0149571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao W, Roser D, Köhler A, Bradatsch B, Baßler J, Hurt E. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell 2007; 26:51-62; PMID:17434126; https://doi.org/ 10.1016/j.molcel.2007.02.018 [DOI] [PubMed] [Google Scholar]
- 47.Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly (A) -binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA 2005; 11:517-31; PMID:15769879; https://doi.org/ 10.1261/rna.7291205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev 2005; 19:90-103; PMID:15630021; https://doi.org/ 10.1101/gad.1267005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmid M, Olszewski P, Pelechano V, Gupta I, Steinmetz LM, Jensen TH. The nuclear PolyA-binding protein Nab2p is essential for mRNA production. Cell Rep 2015; 12:128-39; PMID:26119729; https://doi.org/ 10.1016/j.celrep.2015.06.008 [DOI] [PubMed] [Google Scholar]
- 50.Schmid M, Poulsen MB, Olszewski P, Pelechano V, Saguez C, Gupta I, Steinmetz LM, Moore C, Jensen TH. Rrp6p controls mRNA poly(A) tail length and its decoration with poly(A) binding proteins. Mol Cell 2012; 47:267-80; PMID:22683267; https://doi.org/ 10.1016/j.molcel.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zander G, Hackmann A, Bender L, Becker D, Lingner T, Salinas G, Krebber H. mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature 2016; 540(7634):593–596;PMID:27951587; https://doi.org/ 10.1038/nature20572 [DOI] [PubMed] [Google Scholar]
- 52.Baejen C, Torkler P, Gressel S, Essig K, Soding J, Cramer P. Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol Cell 2014; 55:745-57; PMID:25192364; https://doi.org/ 10.1016/j.molcel.2014.08.005 [DOI] [PubMed] [Google Scholar]
- 53.Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 2013; 154:996-1009; PMID:23993093; https://doi.org/ 10.1016/j.cell.2013.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 2004; 116:63-73; PMID:14718167; https://doi.org/ 10.1016/S0092-8674(03)01026-2 [DOI] [PubMed] [Google Scholar]
- 55.Green DM, Johnson CP, Hagan H, Corbett AH. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci U S A 2003; 100:1010-5; PMID:12531921; https://doi.org/ 10.1073/pnas.0336594100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fasken MB, Stewart M, Corbett AH. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J Biol Chem 2008; 283:27130-43; PMID:18682389; https://doi.org/ 10.1074/jbc.M803649200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kallehauge TB, Robert MC, Bertrand E, Jensen TH. Nuclear retention prevents premature cytoplasmic appearance of mRNA. Mol Cell 2012; 48:145-52; PMID:22921936; https://doi.org/ 10.1016/j.molcel.2012.07.022 [DOI] [PubMed] [Google Scholar]
- 58.Müller-McNicoll M, Botti V, de Jesus Domingues AM, Brandl H, Schwich OD, Steiner MC, Curk T, Poser I, Zarnack K, Neugebauer KM. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev 2016; 30:553-66; PMID:26944680; https://doi.org/ 10.1101/gad.276477.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang Y, Steitz JA. SRprises along a messenger's journey. Mol Cell 2005; 17:613-5; PMID:15749011; https://doi.org/ 10.1016/j.molcel.2005.02.020 [DOI] [PubMed] [Google Scholar]
- 60.Corbo C, Orru S, Salvatore F. SRp20: An overview of its role in human diseases. Biochem Biophys Res Commun 2013; 436:1-5; PMID:23685143; https://doi.org/ 10.1016/j.bbrc.2013.05.027 [DOI] [PubMed] [Google Scholar]
- 61.Das S, Krainer AR. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 2014; 12:1195-204; PMID:24807918; https://doi.org/ 10.1158/1541-7786.MCR-14-0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Bruin RG, Rabelink TJ, van Zonneveld AJ, van der Veer EP. Emerging roles for RNA-binding proteins as effectors and regulators of cardiovascular disease. Eur Heart J 2017; 38:1380-8; PMID:28064149; https://doi.org/ 10.1093/eurheartj/ehw567 [DOI] [PubMed] [Google Scholar]
- 63.Fasken MB, Corbett AH. Links between mRNA splicing, mRNA quality control, and intellectual disability. RNA Dis 2016; 3(4): e1448; PMID:27868086; https://doi.org/ 10.14800/RD.1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Huang B, Xiao Y, Xiong HM, Li J, Feng DQ, Chen XM, Zhang HB, Wang XZ. Aberrant expression of splicing factors in newly diagnosed acute myeloid leukemia. Onkologie 2012; 35:335-40; PMID:22722453; https://doi.org/ 10.1159/000338941 [DOI] [PubMed] [Google Scholar]
- 65.Wong J, Garner B, Halliday GM, Kwok JB. Srp20 regulates TrkB pre-mRNA splicing to generate TrkB-Shc transcripts with implications for Alzheimer's disease. J Neurochem 2012; 123:159-71; PMID:22788679; https://doi.org/ 10.1111/j.1471-4159.2012.07873.x [DOI] [PubMed] [Google Scholar]
- 66.Bond U. Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res 2006; 6:160-70; PMID:16487339; https://doi.org/ 10.1111/j.1567-1364.2006.00032.x [DOI] [PubMed] [Google Scholar]
- 67.Saavedra C, Tung KS, Amberg DC, Hopper AK, Cole CN. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev 1996; 10:1608-20; PMID:8682292; https://doi.org/ 10.1101/gad.10.13.1608 [DOI] [PubMed] [Google Scholar]
- 68.Yost HJ, Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell 1986; 45:185-93; PMID:2421918; https://doi.org/ 10.1016/0092-8674(86)90382-X [DOI] [PubMed] [Google Scholar]
- 69.Yost HJ, Lindquist S. Heat shock proteins affect RNA processing during the heat shock response of Saccharomyces cerevisiae. Mol Cell Biol 1991; 11:1062-8; PMID:1899282; https://doi.org/ 10.1128/MCB.11.2.1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krebber H, Taura T, Lee MS, Silver PA. Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes Dev 1999; 13:1994-2004; PMID:10444597; https://doi.org/ 10.1101/gad.13.15.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rollenhagen C, Hodge CA, Cole CN. Following temperature stress, export of heat shock mRNA occurs efficiently in cells with mutations in genes normally important for mRNA export. Eukaryot Cell 2007; 6:505-13; PMID:17259545; https://doi.org/ 10.1128/EC.00317-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wallace EWJ, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al.. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 2015; 162:1286-98; PMID:26359986; https://doi.org/ 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carmody SR, Tran EJ, Apponi LH, Corbett AH, Wente SR. The mitogen-activated protein kinase Slt2 regulates nuclear retention of non-heat shock mRNAs during heat shock-induced stress. Mol Cell Biol 2010; 30:5168-79; PMID:20823268; https://doi.org/ 10.1128/MCB.00735-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 2001; 413:538-42; PMID:11586364; https://doi.org/ 10.1038/35097110 [DOI] [PubMed] [Google Scholar]
- 75.Thomsen R, Libri D, Boulay J, Rosbash M, Jensen TH. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 2003; 9:1049-57; PMID:12923254; https://doi.org/ 10.1261/rna.5170303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hurt E, Sträßer K, Segref A, Bailer S, Schlaich N, Presutti C, Tollervey D, Jansen R. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J Biol Chem 2000; 275:8361-8; PMID:10722667; https://doi.org/ 10.1074/jbc.275.12.8361 [DOI] [PubMed] [Google Scholar]
- 77.Braun IC, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J 1999; 18:1953-65; PMID:10202158; https://doi.org/ 10.1093/emboj/18.7.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, Hammarskjöld ML. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci U S A 1994; 91:1256-60; PMID:8108397; https://doi.org/ 10.1073/pnas.91.4.1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y, Bor Y-C, Misawa Y, Xue Y, Rekosh D, Hammarskjöld M-L. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature 2006; 443:234-7; PMID:16971948; https://doi.org/ 10.1038/nature05107 [DOI] [PubMed] [Google Scholar]
- 80.Sorger PK, Pelham HR. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J 1987; 6:3035-41; PMID:3319580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chou S, Chatterjee S, Lee M, Struhl K. Transcriptional activation in yeast cells lacking transcription factor IIA. Genetics 1999; 153(4):1573-81; PMID:10581267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Apone LM, Virbasius CA, Holstege FC, Wang J, Young RA, Green MR. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell 1998; 2:653-61; PMID:9844637; https://doi.org/ 10.1016/S1097-2765(00)80163-X [DOI] [PubMed] [Google Scholar]
- 83.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell 1998; 2:675-82; PMID:9844639; https://doi.org/ 10.1016/S1097-2765(00)80165-3 [DOI] [PubMed] [Google Scholar]
- 84.Lee D, Lis JT. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature 1998; 393:389-92; PMID:9620805; https://doi.org/ 10.1038/30770 [DOI] [PubMed] [Google Scholar]
- 85.McNeil JB, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev 1998; 12:2510-21; PMID:9716404; https://doi.org/ 10.1101/gad.12.16.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim S, Gross DS. Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and mediator tail subunits Med15 and Med16. J Biol Chem 2013; 288:12197-213; PMID:23447536; https://doi.org/ 10.1074/jbc.M112.449553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singh H, Erkine AM, Kremer SB, Duttweiler HM, Davis DA, Iqbal J, Gross RR, Gross DS. A functional module of yeast mediator that governs the dynamic range of heat-shock gene expression. Genetics 2006; 172:2169-84; PMID:16452140; https://doi.org/ 10.1534/genetics.105.052738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zid BM, O'Shea EK. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. TL - 514. Nature 2014; 514:117-21; PMID:25119046; https://doi.org/ 10.1038/nature13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005; 310:486-9; PMID:16141371; https://doi.org/ 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, Burge CB. Widespread regulation of translation by elongation pausing in heat shock. Mol Cell 2013; 49:439-52; PMID:23290915; https://doi.org/ 10.1016/j.molcel.2012.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Costello J, Castelli LM, Rowe W, Kershaw CJ, Talavera D, Mohammad-Qureshi SS, Sims PF, Grant CM, Pavitt GD, Hubbard SJ, et al.. Global mRNA selection mechanisms for translation initiation. TL - 16. Genome Biol 2015; 16:10; PMID:25650959; https://doi.org/ 10.1186/s13059-014-0559-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simpson CE, Ashe MP. Adaptation to stress in yeast: To translate or not? Biochem Soc Trans 2012; 40:794-9; PMID:22817736; https://doi.org/ 10.1042/BST20120078 [DOI] [PubMed] [Google Scholar]


