ABSTRACT
Epithelial-mesenchymal interactions are required to coordinate cell proliferation, patterning, and functional differentiation of multiple cell types in a developing organ. This exquisite coordination is dependent on various secreted molecules that provide developmental signals to mediate these tissue interactions. Recently, it was reported that mature mesenchymal-derived microRNAs (miRNAs) in the fetal mouse salivary gland are loaded into exosomes, and transported to the epithelium where they influence progenitor cell proliferation. The exosomal miRNAs regulated epithelial expression of genes involved in DNA methylation in progenitor cells to influence morphogenesis. Thus, exosomal miRNAs are mobile genetic signals that cross tissue boundaries within an organ. These findings raise many questions about how miRNA signals are initiated to coordinate organogenesis and whether they are master regulators of epithelial-mesenchymal interactions. The development of therapeutic applications using exosomal miRNAs for the regeneration of damaged adult organs is a promising area of research.
KEYWORDS: KIT, epithelial-mesenchymal interaction, exosomes, miR-133b-3p, microRNA, salivary gland
Intercellular communication with RNAs
Intercellular communication is an essential feature required for multicellular organisms.1 The communication often involves various proteins, such as soluble factors, their receptors, and secreted extracellular matrix.1 RNA was initially not considered a mediator of intercellular communication because of its instability and rapid degradation by endogenous RNases.2 However, since the late 1990s RNA was discovered to have functional roles as a mobile signal across cell and/or tissue boundaries in multicellular organisms,3-5 including plant, nematode, and insect cells.1,2,6-8 In plants, RNA transport was demonstrated by grafting tissue from a genetically modified plant into a control plant, allowing the transgene signals to be tracked over long distances through phloem.4,9 The report of RNA-transport and RNA-interference (RNAi) in Caenorhabditis elegans (C. elegans) were awarded the 2006 Nobel Prize in Physiology or Medicine.5 C. elegans expresses SID-1, an RNA channel, to import double-stranded RNA (dsRNA) into cells,8,10,11 and the dsRNA is also transported from cell to cell throughout the whole body and surprisingly from one generation to the next.8,12
An important question is whether RNA transfer has been implicated in vertebrate organs. In 1974, experiments were performed to examine whether RNA is transferred between lung tissues in embryonic chick and mouse.13 Isolated lung mesenchyme was incubated with ribonucleosides labeled by stable heavy isotopes. Then the mesenchyme was cultured with unlabeled-epithelium separated with a filter. Subsequent equilibrium centrifugation of RNA from the epithelium showed no detectable transfer of macromolecular RNA such as messenger mRNAs (mRNAs).13 However the extensive transfer of labeled ribonucleosides was detected from mesenchyme to epithelium with subsequent incorporation into epithelial RNA, in spite of thoroughly washing the mesenchyme to remove the remaining labeled ribonucleosides.13 The authors speculated that this radioactivity was noise, comprised of heterogeneous RNA including some small RNAs.13 Therefore the “noise” detected in the unlabeled-epithelium may have been functional small RNAs transported from the mesenchyme in the lung.
Similarly, a report in 2012 may have implied that microRNAs (miRNAs) were transported between tissues in the mouse mammary gland.14 miRNAs are small noncoding regulatory RNAs that involved in various biologic processes and function by silencing mRNA expression at post-transcriptional levels.15-17 The authors eliminated the miR-17/92 cluster from genomic DNA specifically in mammary epithelial cells and assessed the functional roles of this miRNA cluster during mammary gland development. Unfortunately, the mammary epithelium showed no defects during development from prepuberty to lactation. Therefore, the authors concluded that the miR-17/92 cluster was dispensable for mammary gland development, and suggested that other miRNA paralogues may compensate for the miR-17/92 cluster in the mammary epithelium.14 In this study, it is interesting to note that miRNAs in the miR-17/92 cluster were still detected in the mammary epithelial cells, contrary to an expectation that they were specifically deleted in the mammary epithelium.14 The authors argued that the unexpected miRNA detection was due to other cells contaminating the epithelial cell purification.14 However, it may be possible that the miRNAs were transported to the mammary epithelium from surrounding tissues such as mesenchyme and fat pad, although the authors did not suggest the possibility. Taken together, these studies using lung and mammary gland, in hindsight, suggest small RNA transfer may occur between different tissues within vertebrate organs.
RNAs in extracellular vesicles
Last decade, a major advance in understanding intercellular communication in mammalian cells was the discovery that miRNAs are involved. This showed that cells in vitro secreted extracellular vesicles (EV) containing functional miRNAs in a stable form as a cargo and that the EV were delivered to other cells.18,19 Later studies reported that miRNAs packaged in EV were also found in body fluids in vivo, which was promising for clinical applications, such as liquid biopsies.1,19,20 EV are categorized into three groups: apoptotic bodies of 1–4 µm in diameter, which are generated from plasma membrane when apoptosis occurs in damaged cells21,22; microvesicles of 200 nm-1 µm in diameter,23,24 which are formed by outward blebbing of plasma membrane; and exosomes (30 to 100 nm in diameter), which are formed by invagination and budding of late endosome membranes and released by fusion of multivesicular bodies with the plasma membrane.25 To date, an open source resource for EVs catalogs RNAs and other biomolecules that detected in these EVs (Vesiclepedia; http://www.microvesicles.org).26 As of July 2017, there were 4,934 entries for miRNAs, 27,642 entries for mRNAs, 584 entries for lipids, and 92,897 entries for proteins including developmental signals such as WNTs and Hedgehog proteins.27 Among these EVs, exosomes have been of particular interest since the first discovery that RNAs are “signaling molecules” in cell-to-cell communication via exosomal secretion and uptake.18
Epithelial-mesenchymal interactions in embryonic salivary gland
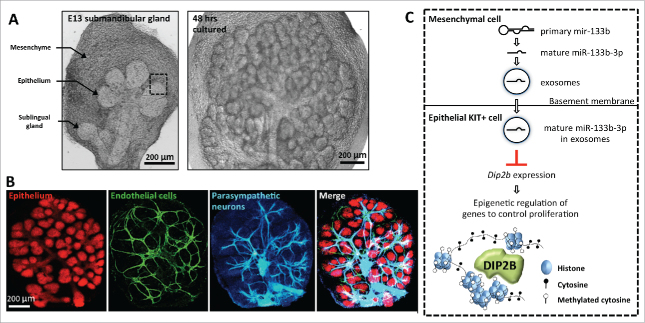
It has been suggested that the miRNAs in exosomes are used as mobile signals for cell-to-cell,19 and even for fetal-maternal communication.28 However, most studies have examined exosomal miRNA transport systemically by injection of exogenous exosomes in vivo or by using cultured cell lines in vitro, which may be different from physiologic conditions. Recently, we demonstrated that endogenous miRNAs are transported via exosomes between tissues during mammalian organogenesis.29 We used ex vivo culture of fetal ICR mouse submandibular glands (SMG) at embryonic day 13 (E13), which has been a well-studied model organ to study epithelial-mesenchymal interactions.30,31 The E13 SMG is easily cultured in minimal media and serum-free conditions ex vivo and undergoes branching morphogenesis, which is robust and consistent (Fig. 1A). The E13 SMG is composed of a condensed neural crest-derived mesenchyme containing an epithelial rudiment, which can be physically separated and recombined in ex vivo culture, to enable us to treat each tissue independently with reagents and recombine them after the treatment. Additionally, the isolated epithelium can be cultured in the absence of mesenchyme and undergoes branching morphogenesis as long extracellular matrix and growth factors are provided.30
Figure 1.
Tissue-tissue interactions in the fetal mouse submandibular gland (SMG) at embryonic day 13 (E13). (A) Ex-vivo culture of an E13 SMG shown after 2 and 48 hrs of culture. The epithelium, mesenchyme and sublingual gland are highlighted with arrows in E13 SMG. Scale bar is 200 µm. (B) E13 SMG cultured for 48 hrs contains multiple tissues that interact and communicate with each other. The epithelium was stained for E-cadherin (anti-ECAD), endothelial cells for PECAM (anti-CD31), and neurons for neuronal tubulin (anti-Tubb3). Scale bar is 200 µm. (C) Model of epithelial-mesenchymal interactions involving exosomal microRNAs (miRNAs) in SMG. A primary miRNA is transcribed from genomic DNA and processed into a mature form in the mesenchyme. The mature miRNA, e.g., miR-133b-3p, is loaded into exosomes as a cargo and transported from mesenchyme to epithelium across the basement membrane. The miR-133b-3p downregulates the expression level of DIP2B, an epigenetic regulator, in the epithelial KIT+ cells to control epithelial progenitor proliferation. The dashed square in (A) and (C) highlights a tissue boundary between epithelium and mesenchyme.
Classic recombination experiments with epithelium and mesenchyme from different organs suggest that the embryonic SMG mesenchyme plays an instructive role for epithelial branching and differentiation.32-34 Thus, it has been accepted that the SMG mesenchyme provides mobile signals to the epithelium leading to a change in epithelial progenitor cell fate, although all the signals of the tissue-tissue interactions have not been defined (Fig. 1B).35-38 We previously profiled the miRNAs expressed in both SMG epithelium and mesenchyme.39,40 Since recent studies suggested that exosomal miRNAs mediate intercellular communication,1,18,19 we therefore investigated whether the E13 SMG in culture releases exosomes containing miRNAs and discovered that they act as mobile genetic signals to mediate epithelial-mesenchymal interactions.
Exosomal miRNAs are transported from mesenchyme to epithelium in fetal SMG
We cultured E13 SMG without serum on top of floating filters with a 100 nm pore size for 48 hrs and collected the conditioned medium. The filter pore size may help to prevent transfer of large extracellular vesicles secreted to the conditioned medium. Sequential centrifugations were performed to remove cell debris and larger EV such as apoptotic bodies, and microvesicles. Finally, an exosome-containing pellet was acquired by ultracentrifugation and analyzed by transmission electron microscopy to observe the size of the exosomes, western blot to confirm the presence of exosome markers, and electrophoresis for RNA detection. Together, our results confirmed that E13 SMG produce exosomes carrying small RNAs.
To examine if exosomes are transported between epithelium and mesenchyme, we recombined tissues labeled with fluorescent BODIPY-ceramide, which labels cell membranes and exosomes. We observed ceramide-signals in the non-labeled epithelium after 12 hrs of culture with a ceramide-labeled mesenchyme. Furthermore, we treated the ceramide-labeled mesenchyme with brefeldin A,41 which inhibits exosome release resulting in reduced BODIPY-ceramide vesicles in the non-labeled epithelium. Conversely, treatment of mesenchyme with monensin,42 a chemical to enhance exosome release, increased BODIPY-ceramide vesicles in the epithelium. These results suggest that exosomes are transported from mesenchyme to the epithelium in E13 SMGs. We also recombined ceramide-labeled epithelium with non-labeled mesenchyme, but the signal was barely detectable in the non-labeled mesenchyme after 12 hrs of culture.
Next, to examine whether the small RNAs in exosomes contain miRNAs, we used qPCR arrays containing 377 miRNAs. We profiled miRNAs in the intact SMG and the exosome fraction from the media. The exosome fraction contained 81 miRNAs and miR-133a-3p, miR-133b-3p, and miR-409-3p were significantly enriched in this fraction compared to the gland and, not surprisingly, these three miRNAs have the sequence motif important for exosome loading (CCCU).43 Interestingly, the mature form of miR-133b-3p was detected in the SMG epithelium but the primary transcript of the miRNA was not, but was in the mesenchyme, suggesting that the mature miR-133b-3p is processed and transported from the mesenchyme to the epithelium. Furthermore, tissue recombination experiments with brefeldin A treatment to suppress exosome release, reduced mature miR-133b-3p levels in the epithelium, further supporting that transmission of mature miR-133b-3p occurs from mesenchyme via exosomes across the tissue boundary. The other 80 exosomal miRNAs could be also transported to the epithelium, and analysis of all their primary and mature forms remains to be investigated. Importantly, miRNA transport was observed and measured without using any exogenous or transgenic overexpressed miRNAs, suggesting that the miRNA transport occurs under physiologic condition during SMG organogenesis (Fig. 1C).
We showed that miRNA mobile signals from the mesenchyme interact with the acceptor tissue, e.g., the SMG epithelium, although we cannot exclude the possibility that miRNA transport occurs in the other direction, from epithelium to mesenchyme. Recent reports showed that EVs contain other non-coding small RNAs such as piRNA, snRNA, snoRNA, and tRNAs as well as miRNAs.19 Furthermore it has been shown that there are various ways to transport RNAs between cells such as nanotube tunnels, gap junctions, and intercellular bridges formed by incomplete cytokinesis, suggesting that cells within complex tissues also communicate with each other with potentially multiple ways to tranport RNAs.1
Regulation and functional roles of exosomal miRNAs during SMG development
Given that SMG epithelium is composed of heterogeneous cells and multiple cell layers, some open and important questions remain. Which epithelial cells can accept the mesenchymal exosomes? Our data suggested uptake of exosomal miR-133b-3p from mesenchyme by epithelial cells expressing KIT that is a progenitor marker of end buds.29 This was demonstrated by knockdown of miR-133b-3p in epithelium, resulted in both reduction of end bud morphogenesis and an increase of DIP2B, a target of miR-133b-3p, in KIT+ cells. However, KIT+ cells in an endbud are heterogeneous, i.e., KIT+K5+ cells, KIT+K14+ cells, and KIT+K19+ cells.44 Additionally, KIT+ cells are distributed throughout an end bud, indicating that some KIT+ cells are located in more peripheral regions while others are in the center of the end bud. Therefore, which KIT+ cell populations accept the exosomal miR-133b-3p in vivo remains unclear.
Another remaining question; how can the exosomes target KIT+ cells or potentially subset of KIT + cells? There may be two possibilities for uptake of the exosomes in KIT + cells. First, it was shown that basement membrane (BM) in E13 SMG was perforated by numerous microscopic holes at tips of the expanding end buds (KIT+ cells).45 The holes (average 1.6 µm2 in diameter) at the end buds are potentially large for exosomes to pass through. Importantly, the microperforations were absent in duct areas.45 Therefore, if the mesenchymal exosomes are mainly transported through the holes to epithelium in E13 SMG, holes at tips in endbuds may allow KIT+ cells to accept mesenchymal exosomes. Interestingly, BM in E11 lung and kidney was also perforated by holes,45 suggesting that epithelial-mesenchymal interaction with exosomes could also occur in these organs.
Second, adhesion molecules in exosomes might mediate exosome uptake in KIT+ epithelial cells. It was shown that human tumor cells secrete exosomes with integrins that are selectively packaged and target to extracellular matrix-enriched cellular areas in distant organs, suggesting that unique exosomal integrins may selectively adhere and establish a niche for sites of future metastasis.46 In fetal mouse SMG, various integrins are expressed in epithelium and mesenchyme.30 Although it is not examined whether the mesenchymal exosomes in E13 SMG contain integrins as a cargo,29 these adhesion molecules in the exosomes may mediate exosome uptake in KIT+ epithelial cells that express integrins. For example, Itga4 expression levels are quite different between KIT+K5+ cells and KIT+ cells at E15 SMG (sgmap.nidcr.nih.gov), suggesting that subset of KIT+ cells express distinct integrins. Taken together, both microperforation at end buds in BM or/and differential integrin expressions in KIT+ cells could affect where mesenchymal exosomes are delivered.
What is the role of exosomal miRNAs on differentiation during organ development? A major function of the fetal mesenchyme that has been proposed is to instruct the epithelium.32,33 Exosomal miRNAs from the mesenchyme may regulate epithelial differentiation as well as morphology, and thus play a major instructive role in early epithelial development. Functional differentiation with the occurrence of proacinar cells and secretory products begins in vivo at E15 and continues to birth.30 Interestingly, microperforations in the BM were associated with early developmental stages and were not detected at E15.45 Thus, exosome transport may be reduced at E15, which could potentially be one of triggers for epithelium to begin differentiation, which raises the possibility that miRNAs are an effective way to repress differentiation of the epithelium during branching morphogenesis. Given that gene expression during development is regulated by multiple growth factors, the transcriptome of the immature epithelium must respond and integrate multiple growth, maintenance and differentiation cues simultaneously, using cassettes of genes in gene regulatory networks (GRNs). As reported, exosomal miR-133b-3p targets Disco interacting protein 2 (DIP2B), which is associated with methylated genomic DNA in heterochromatin (Fig. 1C),29 may influence GRNs at the epigenetic level, rather than by downregulating the expression of an individual gene within a GRN. Thus, exosomal miRNAs downregulate expression of the epithelial epigenetic machinery to allow expansion of undifferentiated epithelial progenitor cells at early stages of development during growth and branching until the epithelium is ready for further cell differentiation.
The exosomal miRNAs from the mesenchyme may also play a role in specifying epithelial cell fate. This idea is suggested by tissue recombination experiments of either embryonic mammary or pituitary epithelium with embryonic SMG mesenchyme (C3H/HeM mouse), showing that the mesenchyme was instructive.33,34 The mammary epithelium at E16 was recombined with SMG mesenchyme at E14, and the mammary epithelium resembled SMG in morphology, although functional differentiation of the epithelium was similar to mammary gland.34 The exact mesenchymal signals that induce a change of mammary epithelial morphology are still unknown.47 Likewise, in the pituitary epithelium at E9-E10 (C3H/HeM mouse), the E14 SMG mesenchyme induced α-amylase and adrenocorticotropic hormone expression, indicating that the SMG mesenchyme induced both salivary- and pituitary-specific differentiation.33 Distinct tissue-specific transcriptomes and/or GRNs could be responsive to common epigenetic regulators that specify or direct epithelial cell fate. Thus, embryonic SMG mesenchyme-derived exosomal miRNAs could direct a tissue-specific transcriptome toward SMG-like transcriptome, which might result in SMG morphogenesis or/and differentiation.48
Communication with exosomal miRNAs is also associated with salivary gland dysfunction. Sjögren's syndrome (SS) is a systemic autoimmune disease of the exocrine glands, such as salivary glands and lacrimal glands. It has been reported that miRNA expression profiles in minor salivary glands were different between SS patients and healthy donors,49 and saliva from SS patients contained distinct exosomal-miRNAs.50 Interestingly, ebv-miR-BART13-3p, a miRNA expressed by the Epstein Barr Virus (EBV), was detected at high level in salivary glands of SS patient despite the EBV usually infects B cells.49 A following study revealed that the ebv-miR-BART13-3p was transported from the infected B cells to salivary epithelial cells via exosomes, and downregulated expressions of a protein involved in Ca2+ signaling, stromal interacting molecule 1 (STIM1), suggesting a functional correlation between the ebv-miR-BART13-3p and impairment of saliva production in the SS.51
Exosomal miRNA-based diagnostics and therapies
Since EVs, including exosomes, are circulating in body fluids and reflect physiologic states of donor cells,19 they have been a focus of active research for use in diagnostics, understanding pathogenesis and for therapeutics (Fig. 2A). Cancer cells secrete exosomes, which act as messengers to communicate with themselves as well as other normal cells.52 In fact, melanoma cells secrete exosomes, microvesicles, and apoptotic bodies during culture.53 The EV subsets in the conditioned medium of melanoma cells were isolated by differential centrifugation and then small RNA sequencing detected 252 miRNAs in all samples of which 23 mRNAs were unique to exosomes. Another 125 miRNAs in exosomes were also detected in both/either microvesicles or apoptotic bodies, suggesting that many identical miRNAs were loaded in all EVs secreted by cancer cells.53 These data highlight that in the discovery and characterization phases of defining miRNA signatures of a particular cancer or cell type, standardization of exosome isolation methods will be required to avoid contamination of other EVs that may lead to erroneous interpretation.54
Figure 2.

Model of exosomal microRNA (miRNA) applications from development to therapy. (A) Clinical applications with exosomal miRNAs. Exosomal miRNAs in body fluids can be useful as non-invasive biomarkers for monitoring the physiologic and disease conditions of the human body. Potential uses of exosomes as miRNA carriers have also been proposed for therapeutics. The representative organs shown require epithelial-mesenchymal interactions during development. (B) Exosomal miRNAs derived from SMG mesenchyme guide the development of therapeutics. Treatment with mesenchymal exosomes could deliver endogenous or artificial miRNAs packaged in the exosomes to cells in vitro or to organs ex vivo. These miRNAs could regulate gene expression and influence the transcriptome to direct proliferation and/or differentiation of the recipient organs, which could have potential to repair or regenerate damaged tissues and restore function.
It will be important to determine whether exosomes could enhance regenerative therapy. Tissue regeneration using either gene or cell-based therapy has been proposed as therapy to restore damaged-organ function including SMG (Fig. 2A).55Exosomes could protect miRNA cargo from endogenous RNases, and allow the miRNAs to reach their target cells.1 In addition, exosomes usually contain CD47, an integrin-associated transmembrane protein, which plays a functional role to protect cells from phagocytosis as in a “don't eat me” signal.56-61 It was reported that the half-life of exosomes overexpressing CD47 in circulation was longer than exosomes without CD47, which facilitated therapeutic effects in pancreatic cancer.61 Therefore, either exogenous exosomes engineered with CD47 and artificial miRNAs or endogenous exosomes carrying miRNAs from an organ tissue could be useful for therapeutic applications during organ regeneration.
The advantage of using miRNA is that multiple genes or critical genes within a GRN may be targeted, and thus have a broader effect on a specific biologic process rather than on a single gene/signaling pathway (Fig. 2B).62 Also, miRNAs could be used to redirect the transcriptome in acceptor cells,48 tissues in vitro and organs in vivo, which could result in functional regeneration. However, a single dose of exosomes may have only transient therapeutic effect.63 Therefore, the identification of exosome-secreting cells may be useful, to either isolate exosomes from culture media, or to use the cells directly, in combination with a stem/progenitor cell for cell-based therapy (Fig. 2A). Indeed, mesenchymal stem cell (MSC)-based therapies are at the forefront of regenerative medicine, and accumulating evidence suggests that the regenerative effects of MSCs are at least in part due to extracellular vesicles that may contain miRNA.63,64
Conclusions
We have mainly discussed how the exosomal miRNAs as developmental signals participate in epithelial-mesenchymal interactions during SMG organogenesis. Identifying miRNAs that regulate the organogenesis provides potential therapeutic targets for tissue repair or regeneration of damaged adult organs. Future challenges include determining how many copies of particular miRNAs are in exosomes and how this is controlled,65 how the exosomal receptors target miRNA delivery to recipient epithelial cells, what biologic significance individual miRNA signals play in vivo organogenesis, whether functional redundancy occurs, whether other developing organs also communicate with exosomal miRNAs between tissues, and whether there is tissue-specificity of miRNAs transported via exosomes in other developing organs that orchestrate GRNs required for epithelial-mesenchymal interactions.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research at the National Institutes of Health, USA and by JSPS KAKENHI grants JP15K11060, JP15K20370 and JP17K08520, and by a grant for Basic Science Research Projects from The Sumitomo Foundation, Japan.
References
- 1.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13:328–35. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011;30:3553–63. doi: 10.1038/emboj.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voinnet O, Baulcombe DC. Systemic signalling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- 4.Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–45. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Benkovics AH, Timmermans MC. Developmental patterning by gradients of mobile small RNAs. Curr Opin Genet Dev. 2014;27:83–91. doi: 10.1016/j.gde.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Pyott DE, Molnar A. Going mobile: non-cell-autonomous small RNAs shape the genetic landscape of plants. Plant Biotechnol J. 2015;13:306–18. doi: 10.1111/pbi.12353 [DOI] [PubMed] [Google Scholar]
- 8.Jose AM. Movement of regulatory RNA between animal cells. Genesis. 2015;53:395–416. doi: 10.1002/dvg.22871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–5. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 10.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–9. doi: 10.1126/science.1068836 [DOI] [PubMed] [Google Scholar]
- 11.Shih JD, Hunter CP. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA. 2011;17:1057–65. doi: 10.1261/rna.2596511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marre J, Traver EC, Jose AM. Extracellular RNA is transported from one generation to the next in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2016;113:12496–501. doi: 10.1073/pnas.1608959113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grainger RM, Wessells NK. Does RNA pass from mesenchyme to epithelium during an embryonic tissue interaction? Proc Natl Acad Sci U S A. 1974;71:4747–51. doi: 10.1073/pnas.71.12.4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feuermann Y, Robinson GW, Zhu BM, Kang K, Raviv N, Yamaji D, Hennighausen L. The miR-17/92 cluster is targeted by STAT5 but dispensable for mammary development. Genesis. 2012;50:665–71. doi: 10.1002/dvg.22023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 17.Leonardo TR, Schultheisz HL, Loring JF, Laurent LC. The functions of microRNAs in pluripotency and reprogramming. Nat Cell Biol. 2012;14:1114–21. doi: 10.1038/ncb2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 19.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, et al.. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maas SL, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017;27:172–88. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hristov M, Erl W, Linder S, Weber PC. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood. 2004;104:2761–6. doi: 10.1182/blood-2003-10-3614. [DOI] [PubMed] [Google Scholar]
- 22.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Köppel T, Jahantigh MN, Lutgens E, et al.. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 23.Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D'Souza-Schorey C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol. 2009;19:1875–85. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123:1603–11. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 26.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borràs FE, Breakefield X, Budnik V, et al.. Vesiclepedia (PLos Biol., 2012). PLOS Biology. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGough IJ, Vincent JP. Exosomes in developmental signalling. Development. 2016;143:2482–93. doi: 10.1242/dev.126516. [DOI] [PubMed] [Google Scholar]
- 28.Vilella F, Moreno-Moya JM, Balaguer N, Grasso A, Herrero M, Martinez S, Marcilla A, Simón C. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development. 2015;142:3210–21. doi: 10.1242/dev.124289. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi T, Lombaert IM, Hauser BR, Patel VN, Hoffman MP. Exosomal MicroRNA Transport from Salivary Mesenchyme Regulates Epithelial Progenitor Expansion during Organogenesis. Dev Cell. 2017;40:95–103. doi: 10.1016/j.devcel.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel VN, Rebustini IT, Hoffman MP. Salivary gland branching morphogenesis. Differentiation. 2006;74:349–64. doi: 10.1111/j.1432-0436.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- 31.Tucker AS. Salivary gland development. Semin Cell Dev Biol. 2007;18:237–44. doi: 10.1016/j.semcdb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Grobstein C. Inductive epitheliomesenchymal interaction in cultured organ rudiments of the mouse. Science. 1953;118:52–5. doi: 10.1126/science.118.3054.52. [DOI] [PubMed] [Google Scholar]
- 33.Kusakabe M, Sakakura T, Sano M, Nishizuka Y. A pituitary-salivary mixed gland induced by tissue recombination of embryonic pituitary epithelium and embryonic submandibular gland mesenchyme in mice. Dev Biol. 1985;110:382–91. doi: 10.1016/0012-1606(85)90097-1. [DOI] [PubMed] [Google Scholar]
- 34.Sakakura T, Nishizuka Y, Dawe CJ. Mesenchyme-dependent morphogenesis and epithelium-specific cytodifferentiation in mouse mammary gland. Science. 1976;194:1439–41. doi: 10.1126/science.827022. [DOI] [PubMed] [Google Scholar]
- 35.Knosp WM, Knox SM, Lombaert IM, Haddox CL, Patel VN, Hoffman MP. Submandibular parasympathetic gangliogenesis requires sprouty-dependent Wnt signals from epithelial progenitors. Dev Cell. 2015;32: 667–77. doi: 10.1016/j.devcel.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knox SM, Lombaert IM, Haddox CL, Abrams SR, Cotrim A, Wilson AJ, Hoffman MP. Parasympathetic stimulation improves epithelial organ regeneration. Nat Commun. 2013;4:1494. doi: 10.1038/ncomms2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knox SM, Lombaert IM, Reed X, Vitale-Cross L, Gutkind JS, Hoffman MP. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science. 2010;329:1645–7. doi: 10.1126/science.1192046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwon HR, Nelson DA, DeSantis KA, Morrissey JM, Larsen M. Endothelial cell regulation of salivary gland epithelial patterning. Development. 2017;144:211–20. doi: 10.1242/dev.142497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi T, Koyama N, Azuma Y, Kashimata M. Mesenchymal miR-21 regulates branching morphogenesis in murine submandibular gland in vitro. Dev Biol. 2011;352:299–307. doi: 10.1016/j.ydbio.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 40.Rebustini IT, Hayashi T, Reynolds AD, Dillard ML, Carpenter EM, Hoffman MP. miR-200c regulates FGFR-dependent epithelial proliferation via Vldlr during submandibular gland branching morphogenesis. Development. 2012;139:191–202. doi: 10.1242/dev.070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–90. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 43.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lombaert IM, Abrams SR, Li L, Eswarakumar VP, Sethi AJ, Witt RL, Hoffman MP. Combined KIT and FGFR2b signaling regulates epithelial progenitor expansion during organogenesis. Stem Cell Reports. 2013;1:604–19. doi: 10.1016/j.stemcr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harunaga JS, Doyle AD, Yamada KM. Local and global dynamics of the basement membrane during branching morphogenesis require protease activity and actomyosin contractility. Dev Biol. 2014;394:197–205. doi: 10.1016/j.ydbio.2014.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al.. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–35. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakakura T, Suzuki Y, Shiurba R. Mammary stroma in development and carcinogenesis. J Mammary Gland Biol Neoplasia. 2013;18:189–97. doi: 10.1007/s10911-013-9281-9. [DOI] [PubMed] [Google Scholar]
- 48.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 49.Alevizos I, Alexander S, Turner RJ, Illei GG. MicroRNA expression profiles as biomarkers of minor salivary gland inflammation and dysfunction in Sjogren's syndrome. Arthritis Rheum. 2011;63:535–44. doi: 10.1002/art.30131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–8. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallo A, Jang SI, Ong HL, Perez P, Tandon M, Ambudkar I, Illei G, Alevizos I. Targeting the Ca(2+) Sensor STIM1 by Exosomal Transfer of Ebv-miR-BART13-3p is Associated with Sjogren's Syndrome. EBioMedicine. 2016;10:216–26. doi: 10.1016/j.ebiom.2016.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pitt JM, Kroemer G, Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J Clin Invest. 2016;126:1139–43. doi: 10.1172/JCI87316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lunavat TR, Cheng L, Kim DK, Bhadury J, Jang SC, Lasser C, Sharples RA, López MD, Nilsson J, Gho YS, et al.. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells–Evidence of unique microRNA cargos. RNA Biol. 2015;12:810–23. doi: 10.1080/15476286.2015.1056975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzas EI, Buck AH, de Candia P, Chow FW, Das S, Driedonks TA, et al.. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – an ISEV position paper. J Extracell Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel VN, Hoffman MP. Salivary gland development: a template for regeneration. Semin Cell Dev Biol. 2014;25–26:52–60. doi: 10.1016/j.semcdb.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaur S, Singh SP, Elkahloun AG, Wu W, Abu-Asab MS, Roberts DD. CD47-dependent immunomodulatory and angiogenic activities of extracellular vesicles produced by T cells. Matrix Biol. 2014;37:49–59. doi: 10.1016/j.matbio.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kibria G, Ramos EK, Lee KE, Bedoyan S, Huang S, Samaeekia R, Athman JJ, Harding CV, Lötvall J, Harris L, et al.. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci Rep. 2016;6:36502. doi: 10.1038/srep36502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.EricJ.Brown WAF. Integrin-associated protein (CD47) and its ligands. TRENDS in Cell Biology. 2001;11:130–5. doi: 10.1016/S0962-8924(00)01906-1. [DOI] [PubMed] [Google Scholar]
- 59.Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, Weissman IL. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–85. doi: 10.1016/j.cell.2009.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chao MP, Weissman IL, Majeti R. The CD47-SIRPalpha pathway in cancer immune evasion and potential therapeutic implications. Curr Opin Immunol. 2012;24:225–32. doi: 10.1016/j.coi.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hauser BR, Hoffman MP. Regulatory Mechanisms Driving Salivary Gland Organogenesis. Curr Top Dev Biol. 2015;115:111–30. doi: 10.1016/bs.ctdb.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zappulli V, Friis KP, Fitzpatrick Z, Maguire CA, Breakefield XO. Extracellular vesicles and intercellular communication within the nervous system. J Clin Invest. 2016;126:1198–207. doi: 10.1172/JCI81134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rani S, Ryan AE, Griffin MD, Ritter T. Mesenchymal Stem Cell-derived Extracellular Vesicles: Toward Cell-free Therapeutic Applications. Mol Ther. 2015;23:812–23. doi: 10.1038/mt.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, et al.. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888–93. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]