ABSTRACT
Cis-natural antisense transcripts (cis-NATs) are long noncoding RNAs transcribed from the opposite strand and overlapping coding and noncoding genes on the sense strand. cis-NATs are widely present in the human genome and can be involved in multiple mechanisms of gene regulation. Here, we describe the presence of cis-NATs in the 3′ distal region of the c-MYC locus and investigate their impact on transcriptional regulation of this key oncogene in human cancers. We found that cis-NATs are produced as consequence of the activation of cryptic transcription initiation sites in the 3′ distal region downstream of the c-MYC 3′UTR. The process is tightly regulated and leads to the formation of two main transcripts, NAT6531 and NAT6558, which differ in their ability to fold into stem-loop secondary structures. NAT6531 acts as a substrate for DICER and as a source of small RNAs capable of modulating c-MYC transcription. This complex system, based on the interplay between cis-NATs and NAT-derived small RNAs, may represent an important layer of epigenetic regulation of the expression of c-MYC and other genes in human cells.
KEYWORDS: c-MYC, epigenetics, long non coding RNA, natural antisense transcripts, small RNAs, transcriptional regulation
Introduction
Cis-natural antisense transcripts (cis-NATs) are long noncoding RNAs transcribed from the opposite strand totally or partially overlapping annotated protein-coding or non-protein-coding genes.1,2 NATs are widespread in eukaryotic genomes with about 40 to 70% of human and mouse genes having NATs.3,4 NATs have been implicated in multiple regulatory mechanisms, including RNA masking,5,6 alternative splicing,7 genomic imprinting,8 chromatin remodeling9 and RNA editing.10 Cis-NATs have been classified on the basis of their orientation relative to the associated gene as head-to-head (5′ to 5′), tail-to-tail (3′ to 3′) or fully overlapping.11 Sense-antisense transcript pairs are co-expressed or inversely correlated more frequently than expected by chance, indicating interdependence between the two transcriptional units.11 High conservation and coupling of sense-antisense expression are more frequent for the tail-to-tail NAT-gene pairs. The function of many of the identified NATs is still unknown and novel mechanisms underlying the functional interaction between overlapping NAT-gene pairs are emerging. For instance, TALAM1, a recently identified cis-NAT to the human lncRNA MALAT1, interacts with the sense transcript in cis and regulates the 3' cleavage, processing and nuclear accumulation of MALAT1.12
Sense-antisense overlapping transcripts are also processed by DICER or DICER-like proteins to produce small interfering RNAs (siRNAs).13 This phenomenon has been extensively investigated in A. thaliana where cis-NAT-derived siRNAs (nat-siRNAs) are formed in response to specific stimuli and interfere with the sense transcript of the NAT-gene pair.14 A genome-wide analysis of cis-NAT expression in A. thaliana has identified many siRNAs enriched in the overlapping region of NAT-gene pairs and suggests that NAT-siRNAs may be involved in silencing of the gene transcript.14,15 In S. pombe, primary endogenous siRNA (endo-siRNAs) are generated by overlapping sense-antisense transcripts in a DICER-dependent manner.16 The biogenesis of endo-siRNAs is controlled at multiple levels, underlying the importance of these regulatory mechanisms.16 Endo-siRNAs derived from naturally occurring double stranded RNAs are much rarer in somatic mammalian cells. Endo-siRNAs are present in higher amounts in mouse oocytes.17,18 However, only few other examples have been reported, including murine testis, kidney19 and brain.20 Endo-siRNAs have been found in human embryonic kidney cells, but their production in differentiated human cells is still controversial.5,21
Cis-NATs can overlap promoters, introns, exons, enhancers and untranslated regions of the corresponding genes.1,22 Cis-NATs are generally produced at much lower levels than the overlapping gene transcripts and their expression is controlled by structured and regulated transcription initiation sites similar to the gene promoters. Promoters of transcribed cis-NATs present RNA polymerase 2 (RNAPol2) binding and active chromatin marks, such as histone H3 acetylation at K27 and K9 (H3K9Ac and H3K27Ac).23 Conversely, promoters of silent cis-NATs have low H3K9Ac and H3K27Ac and are associated with repressive histone marks. Histone deacetylases (HDACs) remove acetyl groups from histones and deplete activation marks from chromatin.24 Interestingly, HDACs are also found associated with promoters and gene bodies of actively transcribed genes.24 During transcription, HDACs are thought to take part in cycles of deposition and removal of acetyl group on histones and to facilitate transcriptional initiation and elongation.24-26 Paradoxically, treatment with HDACs inhibitors represses transcription of many active genes.24-26 Furthermore, a tight control over cis-NATs transcription is also required to avoid interference with overlapping and surrounding genes on the opposite strand and this might involve histone modifications catalyzed by HDACs and histone acetyltransferases (HATs). In fission yeast the HDAC Clr6 prevents antisense transcription and has a protective function suppressing antisense cryptic initiation sites, suggesting that HDACs might exert similar functions in mammalian cells.27
In this study, we investigated the presence and function of cis-NATs in the c-MYC locus. c-MYC is an oncogene activated in several human cancers.28 We identified cis-NATs overlapping the c-MYC gene and extending downstream the 3′-untraslated region (3′UTR). We show that cis-NATs in the 3′ distal region respond to changes in the chromatin status within the c-MYC locus and are involved in fine tuning the transcription of the gene.
Materials and methods
Cell lines, small interfering RNA transfection and drug treatment
Human cell lines and culturing conditions have been described previously.29,30 The siGL3 Negative Control siRNA29 and DICER siRNA were purchased from Ambion. Cells were transfected with siRNAs using Interferin (Polyplus). Small RNAs generated by in vitro DICER reaction were transfected in PC3 cells using Lipofectamine (Invitrogen). Cells were treated with Trichostatin A (TSA, Alexis Biochemicals), SAHA (suberanilohydroxamic acid, Cayman Chemical) or DMSO (Sigma).
RNA extraction
Total RNA was obtained from cell lines by phenol:chloroform extraction. RNA samples were treated with DNase I (Qiagen) and then precipitated with sodium acetate (Sigma). To examine intracellular distribution of the transcripts cells were lysed in 100 mM Tris HCl (pH 7.4), 100 mM NaCl, 2.5 mM MgCl2, 40 µg/ml digitonin (RSB-100).31 After centrifugation the supernatant containing cytosolic RNA was collected. Pellets were suspended in RSB-100 with 0.5% Triton X (RSB-100T) and after centrifugation the supernatant containing nuclear RNA was recovered. The final pellets were suspended in RSB-100T, sonicated and centrifuged. The supernatant containing chromatin-bound RNA was recovered. RNA was extracted from all the collected fractions using TriReagent (Invitrogen).
Reverse transcriptase polymerase chain reaction (RT-PCR)
RT-PCR was performed using Verso 1 Step kit Thermostart (ThermoScientific). Samples were analyzed by agarose gel electrophoresis followed by staining with GelRed (Biotium) and digital imaging with αImager (Innotech) with indicated primers (Supplementary Table 1). For strand-specific RT-PCR only the forward primers were added to the reverse transcriptase reaction to amplify antisense strand selectively. Experiments were repeated two or more times to ensure reproducibility and representative images are shown. Optimal conditions for each primer set (e.g., amount of starting RNA and PCR amplification cycles) were determined in preliminary experiments. To detect c-MYC and Actin mRNA by RT-PCR total RNA (50 ng) was subjected to 22 and 20 cycles of amplification, respectively. To detect NATs by strand-specific RT-PCR 100 ng of total RNA, following directional RT, were subjected to 30 cycles of PCR amplification. These conditions ensured linear amplification of the target RNAs and therefore a semi-quantitative assessment of their amounts. Negative (i.e., no RNA; no RT step) and positive (i.e., genomic DNA) control reactions were performed to determine the specificity of the produced amplicons and the absence of genomic contaminants.
5′. Rapid amplification of cDNA ends (5′RACE)
5′ RACE was performed with gene-specific primers for antisense transcripts (Supplementary Table 1) using 5′ RACE System (Invitrogen) and RNA from PC3 cells treated with SAHA (2.5 and 10 µM) or DMSO. cDNA was purified, tailed with dCTP and amplified consecutively with gene specific primers and either Abridged Anchor primer or Abridged Universal Amplification primer provided in the 5′RACE system kit. Final PCR products were cloned into pGEM-T Easy vector (Promega) and sequenced.
Immunoblotting
Cells were lysed in 0.5% SDS, 0.5% NP40, 140 mM NaCl and 10 mM Tris-HCl, pH 7.5. Gel electrophoresis and immunoblotting were done as already described.29 Immunoblots were developed using antibodies directed to c-MYC (BD Biosciences), β-tubulin (Santa Cruz), acetylated histone H3 (H3Ac) (Millipore).
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with formaldehyde and processed as described.29 Antibodies toward acetyl-Histone H3 and RNA polymerase 2 (RNAPol2) (Millipore)29 were used for immunoprecipitation. Quantitative real time PCR (qPCR) was performed using SYBR Green FAST qPCR (KAPA Biosystem) on an ABI Step One Plus (Applied Biosystems). The amount of input and immunoprecipitated DNA was calculated in reference to standard curves. Data are presented as fraction of immunoprecipitated DNA relative to input DNA.
In vitro transcription
Templates for in vitro transcription were prepared from 200 ng of genomic DNA amplified by Pwo SuperYield DNA Polymerase (Roche) with primers Myc +5866 Fw and T7prom-Myc +6558 Rev, containing also T7 promoter sequence, for NAT 6558; with primers Myc +5906 Fw and T7 prom-Myc +6531 Rev, for NAT6531 (Supplementary Table 1). PCR products were then purified and in vitro transcribed by T7 RNA Polymerase from Escherichia coli BL 21/pAR 1219 (Roche) for 15 min at 37°C. DNA was digested by DNAse I at 37°C for 15 min and RNA cleaned by LiCl Precipitation Solution (7.5 M) (Thermo Scientific). Production of the correct transcripts was verified by denaturating polyacrylamide gel electrophoresis.
DICER cleavage assay
In vitro transcribed NAT6531 or NAT6558 (3 µg) were heat-denatured at 95°C for 1 min and immediately chilled on ice for 5 min. Transcripts were folded in 25% glycerol, 0.05% Triton-X, 1 mM MgCl2, 50 mM NaCl, 30 mM TrisHCl (pH 6.8) for 15 min at 25°C. An aliquot of the reaction was suspended in denaturing loading 2X buffer (TBE 2X, 61.6% formamide, 2.4 M urea) and kept as denatured RNA control size. Folded RNA (1 µg) was digested for 2 h at 37°C with Turbo DICER (AMS Biotechnology) according to the manufacturer's instructions. A second aliquot of folded RNA was incubated in parallel in absence of the enzyme, as control for nonspecific fragmentation. Denatured, folded and diced NAT6531 and NAT6558 were loaded on native 2% agarose TAE gel and run 10V/cm. Small and long RNA fractions (cut off 200 nt) from DICER cleavage reaction were also isolated using mirVana™ miRNA Isolation Kit (Ambion), following the manufacturer's instructions, and used for small RNAs cloning, Northern blot analysis and cell transfection.
Isolation and cloning of small RNAs derived from DICER cleaved NAT6531
Small RNAs derived from DICER digestion of NAT6531 were purified by mirVana™ miRNA Isolation Kit (Ambion) and cloned using miRCAT Small RNA Cloning Kit (Integrated DNA Technologies) according to the manufacturer's protocol. A total of 18 clones were sequenced.
Northern blotting
DICER digestion products of in vitro transcribed NAT6531 and NAT6558 (50 ng) were suspended in denaturing loading 2X buffer, loaded on 5% or 12% denaturing Urea PAGE, transferred to nylon membranes, positively charged (Roche) and cross-linked by EDC for 2 h at 60°C.32 Northern blotting to detect long transcripts or small RNAs was performed using Digoxigenin NHS Ester (DIG) LNA probe 6333 or 6083 (Exiqon), respectively.33,34 Signal detection was obtained by DIG Nucleic Acid Detection Kit (Roche). For specific detection of endogenous antisense transcripts, DIG-labeled RNA probes covering the full-length NATs and internally labeled with digoxigenin-UTP were synthesized from PCR templates by T7 RNA polymerase according to the DIG RNA labeling kit protocol (Roche). Briefly, 10–12 µg of RNA samples extracted from cells transfected for 72 h with siGL3 or siDICER (100 nM) and then treated with SAHA or DMSO for additional 6 h were diluted in denaturing loading 2X buffer. RNA was separated on 12% or 15% denaturing Urea-PAGE, transferred to nylon membranes and cross-linked by UV. Membranes were incubated overnight with DIG labeled RNA probes.
GRO-Seq and RNA-Seq data analysis
Raw data from global nuclear run-on sequencing (GRO-Seq) performed in various cell lines were downloaded. Sources of the data are reported in Supplementary Table 2. For each data set, raw reads in fastq files were extracted, quality controlled and aligned with bowtie2 (version 2.2.1)35 allowing up to 1 mismatch and accepting uniquely mapping reads on the reference genome GRCh37.p13 provided by Genecode. Samtools (version 0.1.19)36 was used to convert sam to bam files and sort them. Using HOMER (Hypergeometric Optimization of Motif EnRichment) suite of tools for Motif Discovery and next-generation sequencing analysis,37 we predicted genome-wide all the transcription events in each data set (“findPeaks.pl”) allowing the minimum size of 50 bp for transcript body detection. Only the transcripts that were not classified as isoforms of annotated genes were selected. Alignments of paired-ends, stranded RNA-Seq reads data were obtained from the platform http://genome.crg.es/encode_RNA_dashboard/hg19 for Hela and K562 cells. Samples with polyA-enriched cytosolic RNA and polyA-enriched and polyA-depleted nuclear RNA were considered for the analysis. Regions of interest were selected and indexed with samtools. To visualize GRO-seq and RNA-seq data Integrative Genomics Viewer (IGV) was used (version 2.3.32). Alignment of the RNA-seq raw reads was obtained for each panel inspecting mapped reads in the format of sorted, indexed bam files directly uploaded into the IGV visualization tool. This allows to display windows of read alignments in bam files and to show the distribution of mapped reads within the region of interest (each window) in the individual samples. The analysis of reads distribution does not take into account the corresponding library size or the total number of mapped reads as normalization of raw reads is not required for intra-sample comparison.
RNA secondary structure prediction and sequence analysis software
Mfold, a RNA structure prediction tool based on the energy minimization model,38 was used to examine the folding and secondary structural elements in NAT6531 and NAT6558. Sequence search was performed using BLAST (Basic Local Alignment Search Tool, https://blast.ncbi.nlm.nih.gov/Blast.cgi). To search for potential microRNA-like binding sites in the c-MYC locus we used StarMiR,39 a web server tool within the Sfold suite that identifies sites based on feature similarities with experimentally defined miRNA binding events. The entire coding sequence (CDS) of c-MYC and 2 kb in the 5′ (promoter-5′UTR) and 3′ (3′UTR) region were examined.
Results
Antisense transcription in the c-MYC locus
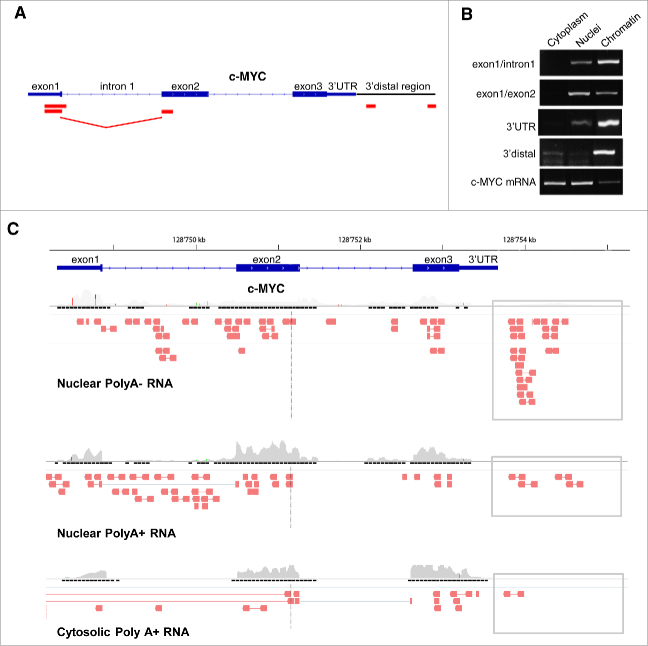
We searched for evidence of antisense transcription overlapping the c-MYC gene using strand-specific RT-PCR (ssRT-PCR) with primer sets spanning different segments of the locus from the 5′UTR to the region downstream the 3′UTR (Fig. 1A). RNA was isolated from PC3 cells, a prostate cancer cell line with high c-MYC expression. Antisense transcripts were detected in the region overlapping the 5′UTR, exon1, exon 2, 3′UTR and up to ∼2 kb downstream the 3′UTR (3′distal region). The antisense transcripts were present preferentially in the nuclear and chromatin fraction with the 3′UTR and 3′distal transcripts almost exclusively associated to the chromatin (Fig. 1B). As control of the fractionation procedure, mature c-MYC mRNA was found in the cytoplasm and nuclei. We observed also an antisense transcript comprising sequences of both exon1 and exon2, due to the splicing of the intervening intron 1 (Fig. 1A). The spliced exon1/exon2 antisense transcript was more abundant in the soluble nuclear than in the chromatin-bound fraction, suggesting that the transcript was released from chromatin after splicing (Fig. 1B).
Figure 1.
Antisense transcription in the c-MYC locus. (A) Map of the c-MYC locus and position of the detected antisense transcripts (red boxes). (B) Expression and intracellular distribution of antisense transcripts overlapping and downstream the c-MYC gene in PC3 cells determined by strand-specific RT-PCR. (C) Distribution of antisense reads overlapping the c-MYC gene detected by RNA-Seq in polyA-depleted (top) and polyA-enriched (middle) nuclear RNA and polyA-enriched cytosolic RNA (bottom) in HeLa cells. Gray boxes, 3′distal NATs.
To support our findings, we interrogated public RNA-Seq data sets in the Encode performed in various cell types (http://genome.crg.es/encode/). We inspected stranded RNA-Seq data from polyadenylated (polyA)-enriched and polyA-depleted RNA extracted from the cytosolic and nuclear fractions of HeLa cells (Fig. 1C) and K562 cells (Supplementary Fig. 1A). PolyA-depleted nuclear RNA is enriched of unstable transcripts that are retained in the nucleus or on the chromatin and do not undergo maturation and export to the cytoplasm.40 In line with our data, we found evidence of antisense transcription overlapping the c-MYC locus. Specifically, antisense transcripts were seen in the 3′distal region downstream the 3′UTR in the polyA-depleted nuclear fraction (Fig. 1C, top panel). Antisense transcripts covering the exon1-exon2 region were present both in the polyA-depleted and polyA-enriched nuclear fraction (Fig. 1C, top and middle panel). Conversely, antisense transcripts in the 3′distal region of the c-MYC locus were relatively less in the polyA-enriched cytosolic fraction (Fig. 1C, lower panel), in line with limited maturation and export to the cytoplasm of these transcripts. These data suggested that the presence of cis-NATs in the c-MYC locus is a common phenomenon. To further support this point, we examined additional prostate cancer cell lines for evidence of antisense transcription in the 3′ distal region of the gene using ssRT-PCR (Supplementary Fig. 1B). Transcripts in antisense orientation were detected, although with different levels, in all the 5 human cell lines examined.
Antisense transcripts in the c-MYC locus are controlled by the local histone H3 acetylation level
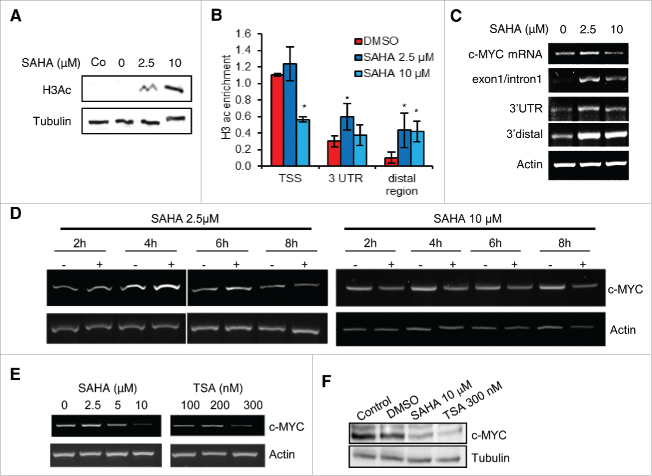
Antisense transcription may be controlled by local histone acetylation and deacetylation.27 Accordingly, we examined the changes in distribution of acetylated histone H3 (H3Ac), a mark of transcriptionally active chromatin, within the c-MYC locus after treatment with the pan-HDAC inhibitor suberanilohydroxamic acid (SAHA). A 6 h treatment with SAHA increased global H3Ac in a dose-dependent manner, indicating effective inhibition of histone deacetylation (Fig. 2A). At the level of the c-MYC locus, H3Ac was high at the TSS while lower or barely detectable at the 3′UTR and the distal downstream region of the gene in control cells (Fig. 2B). SAHA (10 μM) significantly reduced H3Ac at the c-MYC TSS, despite the dose-dependent increase in global H3Ac. Notably, SAHA selectively increased H3Ac at the 3′UTR and even more at the 3′ distal downstream region (4-fold increase). This redistribution of chromatin marks was also reflected in changes in RNAPol2 binding at the c-MYC TSS and in 3′ distal region (Supplementary Fig. 2A). Interestingly, RNAPol2 occupancy increased at the low dose of SAHA (2.5 μM) both at the TSS and at the 3′distal region.
Figure 2.
Histone H3 acetylation and antisense transcription in the c-MYC locus. (A) Global histone H3 acetylation (H3Ac) determined by Western blotting in PC3 cells untreated (control, Co) or treated with vehicle only (0) or SAHA (2.5–10 μM) for 6 h. Tubulin was used as loading control. (B) Acetylated histone H3 in the c-MYC locus determined by chromatin immunoprecipitation in PC3 cells treated with vehicle (DMSO) or SAHA (2.5–10 μM) for 6 h. Data are normalized to input. (C) c-MYC mRNA and antisense transcript expression determined by strand-specific RT-PCR in PC3 cells treated with vehicle (DMSO) or SAHA (2.5–10 μM) for 6 h. Actin mRNA is used as control gene. (D) c-MYC mRNA level determined by RT-PCR in PC3 cells treated with 2.5 (left) and 10 (right) μM SAHA for 2–8 h. (E) c-MYC mRNA expression determined by RT-PCR in PC3 cells treated with vehicle (DMSO), SAHA or TSA for 24 h. (F) c-MYC protein level determined by Western blotting in PC3 cells untreated (Control) or treated with vehicle (DMSO), SAHA (10µM) or TSA (300 nM) for 24 h.
To assess how the changes in H3Ac impacted locally on the transcriptional activity across the c-MYC locus, we examined c-MYC mRNA and antisense transcripts after treatment with SAHA. c-MYC mRNA and the antisense transcripts overlapping exon1/intron1 and the 3′UTR increased slightly at the lower dose (2.5 μM), whereas they decreased at the higher dose (10 μM), indicating a dose-dependent response to SAHA (Fig. 2C). Conversely, the 3′ distal antisense transcripts increased considerably with both doses of SAHA, reflecting the local increase of H3Ac. The concomitant changes in histone marks, RNAPol2 binding and c-MYC mRNA suggest that SAHA likely affects c-MYC transcription via activation or repression of transcriptional initiation and elongation.
A time-course experiment confirmed the biphasic response of c-MYC mRNA to the low dose of SAHA (Fig. 2D). A slight increase of c-MYC mRNA was seen at early times (4–6 h) followed by a decrease at later times (8 h) in cells treated with 2.5 μM SAHA. Conversely, treatment with 10 μM SAHA consistently decreased c-MYC mRNA within 2–8 h. Furthermore, c-MYC mRNA (Fig. 2E) and protein (Fig. 2F) were consistently reduced after 24 h of treatment with the HDAC inhibitors SAHA (10 μM) and trichostatin (TSA, 300 nM). Thus, inhibition of HDACs represses c-MYC transcription concomitantly with a change in antisense transcription and histone H3 acetylation in the 3′ distal region of the gene, leading to the hypothesis that antisense transcripts generated in this region might intervene in c-MYC transcriptional repression.
Cryptic antisense initiation sites are activated by HDAC inhibition in the 3′distal region
We performed 5′RACE to identify the 5′ends and possible transcription initiation sites of the antisense transcripts in the 3′UTR and the 3′distal region. RNA was extracted from cells untreated or treated with 2.5 and 10 µM SAHA. After the 5′RACE the products were separated by gel electrophoresis, purified and sequenced. In untreated cells a single transcript was detected with a putative initiation site at position +6531 with respect to the c-MYC TSS corresponding to about 1.3 kb downstream the 3′UTR (Fig. 3A, lane 1). Surprisingly, rather than increasing the level of this transcript, SAHA induced the appearance of alternative transcripts. In cells incubated with 2.5 and 10 μM SAHA we found 2 antisense transcripts with putative initiation sites at positions +6558 and +7281, respectively (Fig. 3A, lane 2 and 3).
Figure 3.
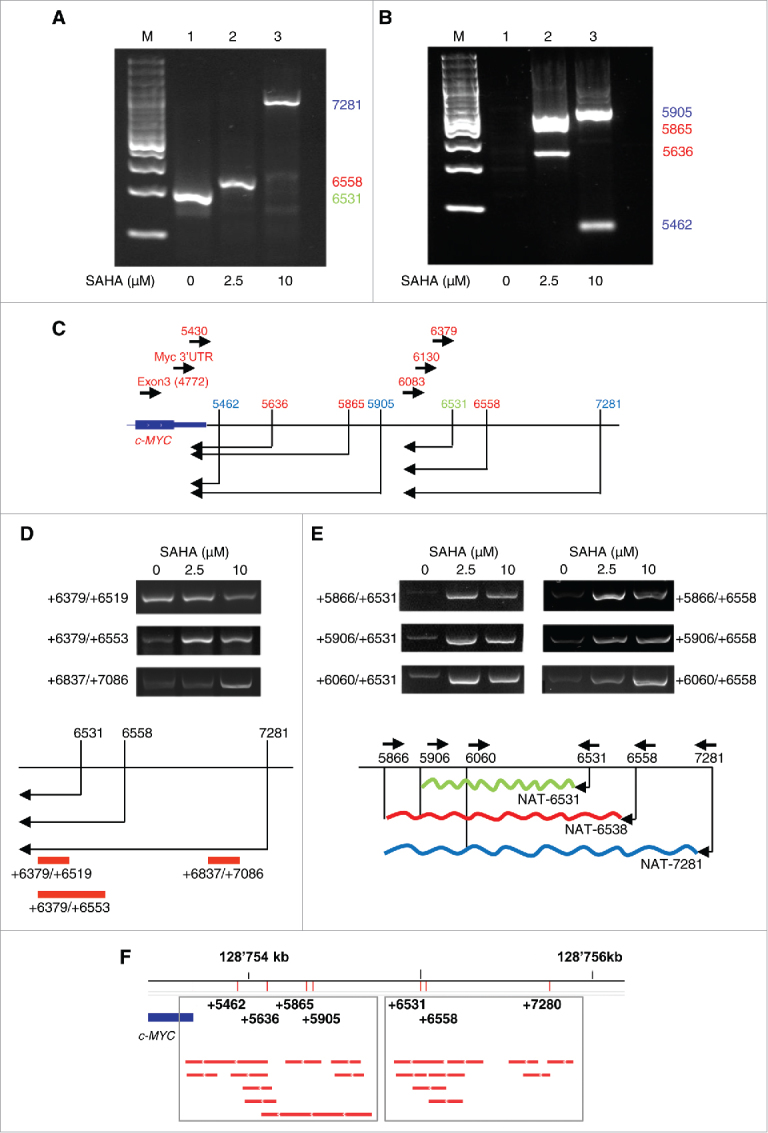
Mapping antisense transcripts in the 3′ distal region of the c-MYC gene. (A-B) Analysis of products generated by 5′RACE starting from position +6083 (A) and position +4772 (exon 3) (B). RNA was isolated from control (1) and SAHA treated PC3 cells (2, 3). M, marker (C) Position of the antisense transcripts and of primers used for 5′RACE experiments in A and B. (D-E) Expression and extension of antisense transcripts by strand-specific RT-PCR (top panels) and schematic representation of transcript initiation sites and PCR primer positions (bottom panels). (F) Predictions of antisense transcripts in the 3′ distal region based on GRO-Seq data from multiple cell lines. Numbers indicate the 5′ends of the antisense transcripts found by 5′RACE in PC3 cells.
We observed a similar phenomenon with the appearance of diverse antisense initiation sites activated upon SAHA treatment in the region overlapping the c-MYC 3′UTR (Fig. 3B). In untreated cells we did not detect any antisense transcript in this region (Fig. 3B, lane 1). However, after treatment with 2.5 µM SAHA 2 main 5′RACE products, corresponding to antisense transcripts putatively starting at the positions +5636 and +5865, respectively, were found (Fig. 3B, lane 2). At 10 µM of SAHA a different pattern was observed with a first antisense transcript starting at position +5462 and a second one at position +5905 (Fig. 3B, lane 3). These results indicated that chromatin remodeling consequent to gradual HDAC inhibition results in broad and complex changes in antisense transcription across the c-MYC locus with induction of different transcripts at the low and high dose of SAHA. Fig. 3C shows a summary of the putative initiation sites and antisense transcripts identified by 5′RACE in the region overlapping and downstream the c-MYC 3′UTR. Using a primer (+6083) that excluded the most abundant transcripts seen by 5′RACE in this region, we detected low level transcripts extending only few hundred bases further downstream from the 3′UTR (Supplementary Fig. 3). Interestingly, no amplification products were obtained by 5′RACE using a more distal primer (+6379). These findings indicate that the antisense transcripts detected in the 3′UTR and in the more 3′ distal region derive from distinct transcriptional units, whose boundary resides between position +6083 and +6379.
To estimate the extension of the antisense transcripts initiated at each of the sites found downstream the 3′UTR, we performed directional ssRT-PCR with primer sets that partially discriminated between the various transcripts (Fig. 3D). In line with the 5′RACE results, we detected the transcript starting at +6531 (primer set +6379/+6519) in untreated cells (Fig. 3D). In cells treated with SAHA the alternative initiation sites at position +6558 (2.5 μM) and +7281 (10 μM) generated longer transcripts detected with the more distal primer sets (+6379/+6553 and +6837/+7086) and overlapping the transcript initiated at position +6531 (NAT6531) in control cells.
To estimate also the extension of the transcripts in the 3′direction, we performed ssRT-PCR with primers (+5866, +5906 and +6060) positioned at increasing distance from the putative initiation sites at +6531 and +6558 (Fig. 3E). In cells treated with SAHA the detected transcripts that extended up to the position the most downstream primer, +5866 (Fig. 3E). Therefore, the NATs induced in presence of by SAHA (NAT6538 and NAT7281) were longer and extended further toward the c-MYC 3′UTR than in untreated cells.
To have additional evidence of the antisense transcripts identified in the 3′ region of the c-MYC locus we interrogated global nuclear run-on sequencing (GRO-Seq) data sets. GRO-Seq has the advantage of detecting nascent transcripts independently of stability, polyadenylation and intra-cellular localization and therefore has greater probability to identify low abundance unstable transcripts.41 Consulting the in-house built compendium of GRO-Seq data sets from multiple cell types we found antisense transcripts in the 3′UTR and 3′ distal region overlapping those experimentally identified by ssRT-PCR and 5′RACE in PC3 cells, thus confirming and extending our observations (Fig. 3F).
We performed also an analysis of the effects of SAHA on transcription in the c-MYC locus in immortalized normal prostate epithelial cells.30 Normal prostate epithelial cells have lower basal level of c-MYC mRNA compared with prostate cancer PC3 cells.29 SAHA, both at 2.5 and 10 μM, reduced c-MYC level (Supplementary Fig. 4). This was associated with reduced production of antisense transcripts in the 3′ distal region in SAHA-treated epithelial cells, unlike the induction seen in PC3 cells. Thus, the dynamic relation between mRNA and antisense transcription in the c-MYC locus may differ substantially between cancer and normal epithelial cells, although further studies are clearly needed.
NAT6531 is processed by DICER into small RNAs
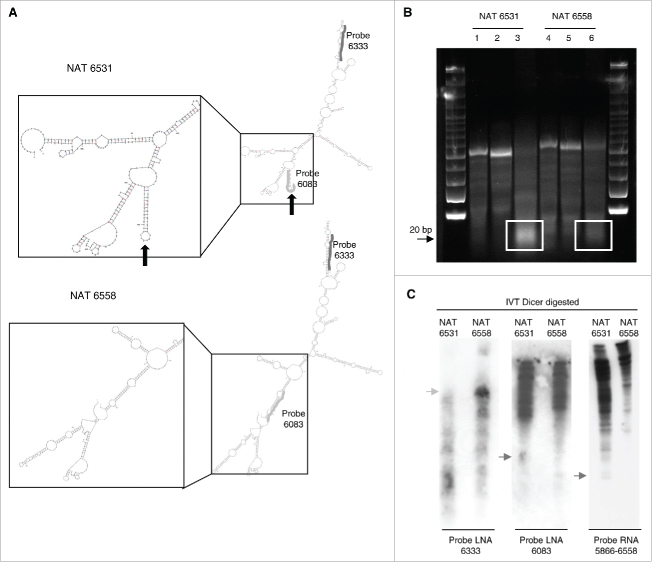
Our data indicate the presence of two close antisense transcripts, NAT6531 and NAT6558, along with a more distal one, NAT7281, in control and SAHA treated PC3 cells. NAT6531 and NAT6558 differ only for few extra bases at 5′ and 3′ ends in the longer transcript. Using the RNA structure prediction algorithm Mfold38 we examined the possibility that this difference in the sequence and length could translate into differential folding and formation of distinct secondary structures of the NATs. RNA secondary structure is an important determinant of the function of noncoding RNAs defining recognition sites for RNA binding proteins, microRNA binding, RNA modifications and processing.42-44 We found overall a high similarity in the predicted folding of NAT6531 and NAT6558 (Fig. 4A). However, there were small local differences, particularly evident at the level of a short stem-loop present in NAT6531 and absent in NAT6558.
Figure 4.
Differential folding and cleavage by DICER of 3′ distal antisense transcripts in the c-MYC locus. (A) Predicted secondary structure of NAT6531 (top) and NAT6558 (bottom) using Mfold. Left, close-up of the stem-loop structure present in NAT6531 and missing in NAT6558. Position of short LNA probes 6083 (light gray) and 6333 (dark gray) relative to the folded NATs is shown. (B) Cleavage of in vitro transcribed NAT6531 (lanes 1–3) and NAT6558 (lanes 4–6) by DICER. Samples contained NATs either denaturated (lanes 1 and 4), refolded (lanes 2 and 5) or refolded and incubated with DICER (lane 3 and 6). White boxes, region of small RNA DICER products. (C) Northern blot analysis of NAT6531 and NAT6558 in vitro transcribed and incubated with DICER. Electrophoresis was run on 5% (left panel) or 12% (middle and right panels) polyacrylamide gels. Blots were hybridized with probes LNA 6333, LNA 6083 and RNA 5866–6558. Light gray arrow, full length transcripts; dark gray arrows, DICER-processed small RNAs.
Short stem-loops can be recognized by RNA processing enzymes such as DICER and may represent sites for generation of small RNAs.45-47 We hypothesized that the short stem-loop in NAT6531 could serve as substrate for DICER cleavage and consequently be the precursor of small RNAs. To test this hypothesis, we transcribed NAT6531 and NAT6558 in vitro and, after folding, incubated them with human recombinant DICER. The products of the reactions were examined by agarose gel electrophoresis (Fig. 4B). In line with our hypothesis, NAT6531 was processed by DICER in vitro to produce a distinct population of short RNAs. After incubation with DICER we observed a decrease of the full length transcript and the appearance of RNA products of the size expected for DICER-processed small RNAs (∼20 nt in length) (Fig. 4B, lane 3). Notably, incubation of NAT6558 with DICER did not lead to accumulation of similar small RNA products (Fig. 4B, lane 6).
To confirm detection of the small RNAs generated from NAT6531 by DICER and determine the origin, DICER reaction products were run on denaturating polyacrylamide gels, blotted and hybridized with probes corresponding to different regions of NAT6531 and NAT6558. The LNA probe LNA6333 hybridizes to a portion common to both NAT6531 and NAT6558, while the LNA probe LNA6083 recognizes the sequence within the predicted stem-loop in NAT6531 (Fig. 4A). The third probe (RNA 5866–6558) corresponds to the full length NAT6558 sequence. Northern blots performed after in vitro DICER digestion showed the decrease of full length NAT6531 detected by LNA6333 (Fig. 4C). Small RNA products were recognized in NAT6531 containing reactions by LNA6083 and the full length (5866–6558) RNA probe, confirming that NAT6531 was processed by DICER. On the other hand, Northern blots with all three probes showed that most of NAT6558 was not cleaved and processed by DICER. Furthermore, hybridization with LNA6083 indicated that most of the DICER-produced small RNAs derived from the stem-loop in NAT6531 (Fig. 4C, middle panel).
Cloning of small RNA generated from NAT6531
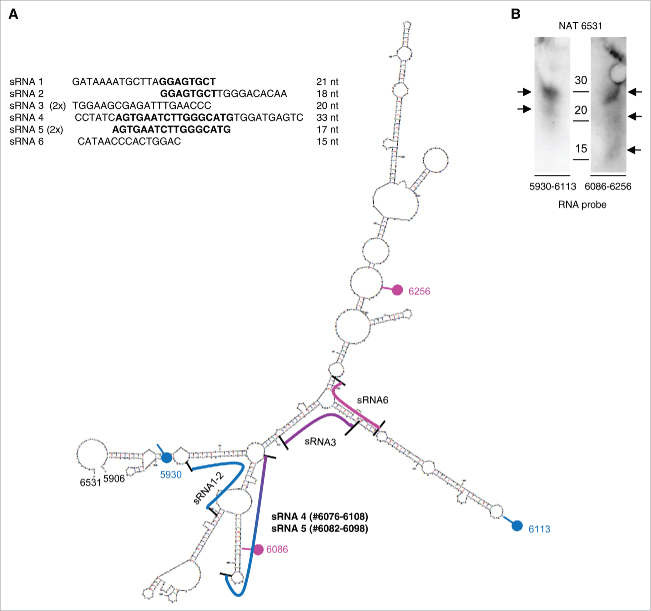
To identify the small RNAs derived from NAT6531 through DICER cleavage, we sequenced the products generated in vitro by DICER using the NAT as substrate. The sequenced small RNAs clustered in two distinct regions of the NAT (Fig. 5A). A first sequence (sRNA5, 17 nt in length) corresponded to the stem-loop region identified by in silico analysis and Northern blotting with LNA6083. We found also a longer sequence (sRNA4, 33 nt in length) overlapping sRNA5 and close to the region considered as the potential source of sRNAs. Considering the location and abundance of the two sRNAs, it is likely that sRNA4 and sRNA5 represent an intermediate cleavage product and the major small RNA generated by DICER, respectively. Northern blots performed with RNA probes (5930–6113 and 6086–6256) designed to recognize distinct regions of NAT6531 detected various small RNA products (Fig. 5B). The two bands detected by both probes could correspond in size to the sRNA4 (33-nt) and the shorter sRNA5 (17-nt). The shorter small RNA product seen only with RNA probe 6086–6256 could correspond to the sRNA6 (15-nt) derived from a secondary region of processing of NAT6531.
Figure 5.
Identification of NAT6531 derived small RNAs. (A) Sequences of small RNAs cloned from DICER-processed NAT6531 (left) and their position in the predicted secondary structure of NAT6531 (right). Nucleotide position of sRNA4 and sRNA5 are reported. Blue and pink dots indicate the boundaries of the RNA probes 5930–6113 and 6086–6256, respectively, used in Northern blots. (B) Northern blot analysis of small RNAs generated in vitro by DICER using NAT6531 as substrate and hybridized with RNA probe 5930–6113 (left) and RNA probe 6086–6256 (right). Arrows indicate the position of small RNA products.
Sequence analysis revealed a unique site matching the full sequence of sRNA5, the main NAT6531-derived DICER product, in the human genome and corresponding to the site in the 3′distal region of the c-MYC locus. No other sequence with full length identity or complementarity was found. Despite the absence of fully complementary sites, we hypothesize that sRNA5 could act as a miRNA-like molecule interacting with partial complementarity to the target sequences. To explore this possibility we use StarMiR39 from the Sfold package to interrogate the c-MYC locus for potential binding sites of sRNA5. StarMiR searches for miRNA binding sequences using CLIP-based predictions.39 Using this approach we found multiple sites (n = 73; 53 sites in promoter/5′UTR and 20 sites in CDS) predicted to be compatible with miRNA-like binding (Supplementary Table 3). Seventeen sites had the best fit with canonical miRNA binding features. Best examples are presented in Supplementary Fig. 6. Interestingly, 14 of the best 17 sites were in the c-MYC promoter and 3 in intron 1. No sites were found in the 3′UTR. Together, these results support the hypothesis that NAT6531-derived small RNAs could regulate transcription by acting locally on transcripts generated in the promoter or in part in the gene body. However, more work is needed to determine the functionality of the identified sites and their impact on c-MYC transcription.
DICER processes NAT6531 and generates endogenous small RNAs in cells
To determine whether DICER is involved in processing the endogenous cis-NATs in the c-MYC locus, we knocked down DICER in PC3 cells using siRNA (Figure Supplementary 6A). RNA from untreated and SAHA treated cells was examined by electrophoresis on denaturating polyacrylamide gels followed by Northern blotting with an RNA probe (probe 5865–6558) complementary to a region common to all distal cis-NATs (Fig. 6A, top panel). Low abundance of the antisense transcripts makes their detection by Northern blot challenging. Nevertheless, probing the long RNA region of the gel we detected transcripts corresponding to the c-MYC cis-NATs in all samples (Fig. 6A, top panel, and Supplementary Fig. 6B-C). Importantly, the size of the transcripts increased in SAHA treated cells compared with control cells, reflecting the increase of the longer more distal NATs expected on the basis of the 5′RACE and RT-PCR. Interestingly, cis-NATs detected in control cells and cells treated with 2.5 μM SAHA after depletion of DICER had similar size, indicating that cleavage of the cis-NATs detected in control cells required DICER and was abolished in its absence.
Figure 6.
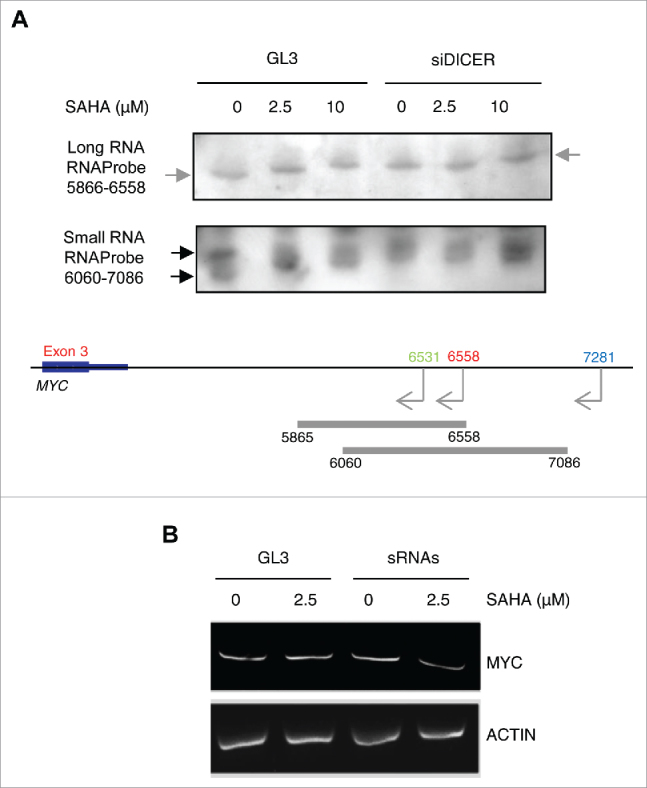
DICER cleavage of 3′ distal NATs and production of endogenous small RNA in cells. (A) Northern blot analysis of 3′ distal NATs (top panel) and small RNAs (bottom panel) in PC3 cells untreated or treated with SAHA for 6 h after transfection with control (siGL3) or siDICER for 72 h. Gray and black arrows indicate the position of NATs and small RNAs, respectively. Bottom, schematic representation of the NATs in the c-MYC locus and position of RNA probes used for Northern blotting. (B) c-MYC mRNA level in PC3 cells transfected for 72 h with in vitro NAT6531-derived small RNA (5 nM) or control siGL3 and then treated with SAHA for additional 6 h. β-actin mRNA was used as control.
To detect small RNAs derived by c-MYC cis-NATs in cells, we probed the small RNA region of the gel with probe 6060–7086. Small RNAs were detected in untreated PC3 cells matching in size the sRNA4 and sRNA5 produced in vitro by DICER cleavage of NAT6531 (Fig. 6A, bottom panel, and Supplementary Fig. 6B). In SAHA treated cells longer products were detected. Similarly, after DICER depletion only longer bands were seen both in control and SAHA treated cells, suggesting that production of the mature small RNAs was inhibited by both SAHA and DICER knockdown. These data indicate that DICER is involved in the cleavage of the full length NAT6531 and the production of NAT-derived small RNAs in cells.
Our data suggest that the NAT6531-derived small RNAs could be involved in regulation of c-MYC transcription in response to HDAC inhibition. To determine whether this was the case, we purified the in vitro DICER-produced small RNAs and transfected them in untreated and SAHA treated PC3 cells. SAHA (2.5 μM) slightly increased c-MYC mRNA (Fig. 6B). Transfection of NAT6531-derived small RNAs prevented the increase of c-MYC mRNA in SAHA treated cells, indicating that the small RNAs had a repressive effect on c-MYC transcription. These results suggest that the switch between NAT6531 and NAT6558 induced by local changes in histone H3 acetylation in the 3′ distal region can contribute to the regulation of transcription of c-MYC by permitting or preventing the synthesis of NAT6531-derived small RNAs, which might locally control transcription within the c-MYC locus.
Because DICER is involved in chromatin modifications and small RNA processing we also examined how DICER affected c-MYC mRNA levels in presence or absence of SAHA. We found that DICER knockdown reduced, although slightly, c-Myc mRNA in control cells (Supplementary Fig. 7). In presence of SAHA, c-MYC mRNA was further reduced by DICER knockdown. Indeed, DICER depletion abolished the induction of c-MYC expression by low dose of SAHA and enhanced c-MYC repression at the highest dose. Although this is apparently in contrast with the effects of DICER on NAT6531-derived small RNAs, it should be noted that DICER knockdown increased the level of the 3′distal NAT6558 and specially NAT7281 (Fig. 6A). The longer NAT has a repressive function and likely overrides any change in small RNA processing resulting from DICER depletion. This is particularly evident at the highest dose of SAHA at which NAT7281 becomes prominent and may in turn block c-MYC transcription.
Discussion
This study demonstrates the presence of multiple cis-NATs overlapping the gene and the 3′distal region of the c-MYC locus and highlights the complex relationship between cis-NAT production and transcriptional regulation. We show that the production of distinct cis-NATs in the c-MYC locus is controlled epigenetically by the level of histone H3 acetylation in the 3′UTR and 3′ distal region (Fig. 7). We hypothesize that fine tuning of the cis-NATs in the 3′distal region modulates the c-MYC transcriptional output in part by controlling the production of regulatory small RNAs. c-MYC is a key oncogene activated in several human cancers.28 Understanding the mechanisms controlling c-MYC expression and underlying its deregulation in cancer may provide insights in the pathogenesis of human cancer and assist in the development of new therapeutic strategies.
Figure 7.
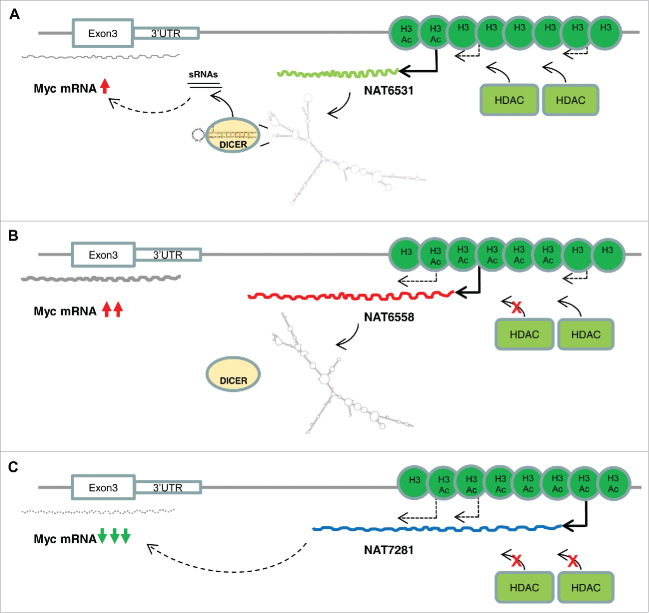
Regulation of transcriptional activity in the c-MYC locus by antisense transcription and histone H3 acetylation. (A) Histone deacetylases (HDACs) prevent activation of the cryptic initiation sites of the distal NATs. In basal condition NAT6531 is transcribed and processed by DICER to produce small RNAs, which attenuate c-MYC mRNA transcription likely acting in miRNA-like mode. (B) Mild inhibition of HDACs by low dose of SAHA increases histone H3 acetylation, activates NAT6558 transcription and block production of NAT6531 and NAT6531-derived small RNAs relieving the block on c-MYC transcription. (C) Strong inhibition of HDACs by high dose of SAHA induces hyper-acetylation in the 3′distal region and activates transcription of the more distal NAT7281 that overrides production of both NAT6531 and NAT6558. In this condition c-MYC mRNA transcription is strongly repressed.
We show that treatment of cancer cells with HDAC inhibitors, such as SAHA, alters the pattern of cis-NATs in the c-MYC locus and affects c-MYC transcription. We observed a complex non-linear relationship between HDAC inhibition, NAT production and c-MYC expression in terms of dose and time dependence of the effects. The relationship between gene-NAT pairs has been shown in many cases to be complex and dose-dependent. For instance, the clock genes Frequency in Neurospora and Period in mammals are both associated with NATs that are tightly regulated to establish repressive heterochromatin through DNA methylation.48 Interestingly, before the induction of heterochromatin, expression of the NATs creates a transcriptionally permissive status that promotes sense transcript expression.48 The response to HDAC inhibition in terms of transcription of NATs and c-MYC mRNA also is non-linear. The low dose of SAHA apparently induced a permissive status that results in transiently increased transcription of c-MYC mRNA. The high dose of SAHA conversely represses c-MYC transcription. The effects on the c-MYC transcription are reflected in changes of cis-NATs in the 3′distal region of the gene, suggesting that the antisense transcripts might be implicated in controlling the status of the gene.
c-MYC and other highly expressed genes are known to be repressed upon HDAC inhibition.25 Gene repression has been proposed to be due to a block of transcriptional elongation and not to the activation of indirect repressive factors.25,26 HDACs have been implicated in transcriptional elongation by limiting histone acetylation in gene bodies and intergenic regions and promoting the recruitment of positive elongation factors.26 HDAC inhibitors alter the distribution of acetylated histones with consequent repositioning of negative elongation factors, such as NELF.26 However, how these changes in global histone acetylation are translated locally in activation or repression of specific genes is still unclear. Here we show that inhibition of HDACs results in localized chromatin changes in the 3′distal region of the c-MYC locus and consequent activation of cryptic initiation sites of antisense transcripts. These findings, therefore, uncover an additional element, cis-NATs, in the complex mechanisms regulating transcription in response to changes in the chromatin status. We found that the production of cis-NATs in the c-MYC locus depends on the degree of local histone H3 acetylation whose gradual increase induces different patterns of antisense transcription. Our data further link changes in the transcriptional status of the c-MYC gene to the activation of distinct cis-NATs in the 3′ distal region in response to modulation of HDAC activity. We believe that these observations highlight an important layer of transcriptional control involving local chromatin modifications and production of cis-NATs, which represent the key elements linking HDAC activity and transcriptional elongation.
We have investigated the mechanism by which cis-NATs might influence c-MYC transcription. Treatment with HDAC inhibitors shift the balance between NAT6531 and NAT6558. We hypothesized that the difference in length and sequence between the two transcripts affects their function. In silico predictions point to a critical difference in the folding and secondary structure of the two NATs. NAT6531is capable of forming a stem-loop that is missing in NAT6558. As a consequence of this, NAT6531 acts as an effective substrate of DICER and a source of small RNAs, which might affect c-MYC transcription.
DICER is known to cleave long double stranded RNAs generated by transposable elements or convergent transcriptional units.49 We found that NAT6531 is efficiently cleaved by DICER in vitro. DICER is sufficient to cleave NAT6531 in vitro and produce small RNAs of discrete sizes using the intramolecular stem-loop and independently of intermolecular double-stranded RNA. Processing by DICER into small RNAs did not occur with the highly similar NAT6558, suggesting that the intramolecular stem-loop dictates the recognition and cleavage of NAT6531. In A. thaliana DICER-like 1 (DCL-1) mediates processing of stem-loop structures within long noncoding RNAs as shown by small RNA sequencing and structure analysis.45 Generation of small RNAs by DICER from hairpins formed at transcription start sites also has been documented in human cells.50,51 Small RNAs derived from tRNA are produced by non-canonical DICER activity.52,53 snoRNAs have been shown to act as non-canonical substrates of DICER in a DROSHA-independent manner.46,47 Sequencing of DICER binding sites revealed many non-canonical substrates for dicing in humans and C. elegans.54 In addition, a DICER isoform, whose expression is driven by a retrotransposon promoter in mouse oocytes, is able to generate efficiently endo-siRNAs.55 This DICER isoform is able to process intramolecular RNA stems, in addition to intermolecular double-stranded RNA to produce small RNAs. Interestingly, this event depends on higher cleavage activity of the isoform and substrate availability, indicating that non-canonical DICER activity may be driven by conditions enhancing substrate-enzyme interaction.55
In summary, we propose that the transcription of NAT6531 and NAT6558 is regulated by histone H3 acetylation and is modulated by HDAC activity. The interplay between NAT6531 and NAT6558 changes the ability of DICER to recognize a stem-loop element in NAT6531 and cleave it to produce small RNAs. In this context, cis-NATs and NAT-derived small RNAs act as rheostats to integrate and fine tune the activity of multiple competing inputs in a highly complex regulatory circuit. The NAT6531-derived small RNAs per se exert a repressive function on c-MYC mRNA transcription, possibly by modulating transcription initiation and elongation. The precise mechanism by which the NAT-derived small RNAs exert their regulatory function has to be further investigated. Sequence and motif analyses predict that NAT-derived small RNAs may act as miRNA-like molecules targeting sequences in the c-MYC promoter and intron 1. cis-NATs and NAT-derived small RNAs might integrate locally inputs derived from histone modifications, chromatin binding proteins and RNA processing activities to modulate the gene transcriptional output. It is interesting that at higher doses of HDAC inhibitors a longer antisense transcript, NAT7281, becomes prevalent. The presence of NAT7281 is associated with strong repression of c-MYC transcription and shutdown of NAT6531, NAT6558 and NAT6531-derived small RNAs, suggesting that an additional more potent repressive mechanism likely overrides the fine tuning system provided by the interplay between the shorter cis-NATs. DICER's role in this process is also complex and multifaceted. DICER is required for processing of NAT6531 and production of NAT-derived small RNAs. DICER, however, influences also the overall level of cis-NATs, including NAT7281. Therefore, knocking down DICER alters this equilibrium in favor of the more distal NAT7281 resulting in downregulation of c-MYC transcription.
This work focuses on the regulation of c-MYC gene by cis-NATs and NAT-derived small RNAs. However, these findings may have broader implications and open toward new lines of investigation of interdependent cis-acting long and small noncoding RNAs that might represent an important and wide-spread layer of transcriptional tuning and regulation of gene expression.
Supplementary Material
Funding
This work was funded by grants from the Swiss National Science Foundation (Grant no. 310030–153099), Ticino Foundation for Cancer Research, Virginia Boeger Foundation, Fidinam and Ceresio Foundation to CVC. SN and GP were supported by fellowships from the Novartis Foundation and Ticino Foundation for Cancer Research (Mario Luvini fellowship), respectively.
References
- 1.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet. 2013;14:880–93. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 2.Wight M, Werner A. The functions of natural antisense transcripts. Essays Biochem. 2013;54:91–101. doi: 10.1042/bse0540091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al.. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 4.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–7. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faghihi MA ZM, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–8. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 7.Morrissy AS GM, Marra MA. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 2011. 21:1203–12. doi: 10.1101/gr.113431.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berteaux N AN, Cathala G, Genton C, Coll J, Daccache A, Spruyt N, Hondermarck H, Dugimont T, Curgy JJ, Forné T, Adriaenssens E. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. 2008;28:6731–45. doi: 10.1128/MCB.02103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotake Y NT, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011. 30:1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandal AK1 PR, Jha V, Mukerji M. Transcriptome-wide expansion of non-coding regulatory switches: evidence from co-occurrence of Alu exonization, antisense and editing. Nucleic Acids Res. 2013;41:2121–37. doi: 10.1093/nar/gks1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 2005;21:326–9. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Zong X, Nakagawa S, Freier SM, Fei J, Ha T, Prasanth SG, Prasanth KV. Natural antisense RNA promotes 3' end processing and maturation of MALAT1 lncRNA. Nucleic Acids Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Xia J, Lii YE, Barrera-Figueroa BE, Zhou X, Gao S, Lu L, Niu D, Chen Z, Leung C, et al.. Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 2012;13:R20. doi: 10.1186/gb-2012-13-3-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin H, Vacic V, Girke T, Lonardi S, Zhu JK. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol. 2008;9:6. doi: 10.1186/1471-2199-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan C, Wang J, Harrison AP, Meng X, Chen D, Chen M. Genome-wide view of natural antisense transcripts in Arabidopsis thaliana. DNA Res. 2015;22:233–43. doi: 10.1093/dnares/dsv008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu R, Jih G, Iglesias N, Moazed D. Determinants of heterochromatic siRNA biogenesis and function. Mol Cell. 2014;53:262–76. doi: 10.1016/j.molcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al.. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 18.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al.. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlile M, Swan D, Jackson K, Preston-Fayers K, Ballester B, Flicek P, Werner A. Strand selective generation of endo-siRNAs from the Na/phosphate transporter gene Slc34a1 in murine tissues. Nucleic Acids Res. 2009;37:2274–82. doi: 10.1093/nar/gkp088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling KH, Brautigan PJ, Moore S, Fraser R, Cheah PS, Raison JM, Babic M, Lee YK, Daish T, Mattiske DM, et al.. Derivation of an endogenous small RNA from double-stranded Sox4 sense and natural antisense transcripts in the mouse brain. Genomics. 2016;107:88–99. doi: 10.1016/j.ygeno.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Werner A, Cockell S, Falconer J, Carlile M, Alnumeir S, Robinson J. Contribution of natural antisense transcription to an endogenous siRNA signature in human cells. BMC Genomics. 2014;15:19. doi: 10.1186/1471-2164-15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li K, Ramchandran R. Natural antisense transcript: a concomitant engagement with protein-coding transcript. Oncotarget. 2010;1:447–52. doi: 10.18632/oncotarget.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conley AB, Jordan IK. Epigenetic regulation of human cis-natural antisense transcripts. Nucleic Acids Res. 2012;40:1438–45. doi: 10.1093/nar/gkr1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YJ, Greer CB, Cecchini KR, Harris LN, Tuck DP, Kim TH. HDAC inhibitors induce transcriptional repression of high copy number genes in breast cancer through elongation blockade. Oncogene. 2013;32:2828–35. doi: 10.1038/onc.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greer CB, Tanaka Y, Kim YJ, Xie P, Zhang MQ, Park IH, Kim TH. Histone Deacetylases Positively Regulate Transcription through the Elongation Machinery. Cell Rep. 2015;13:1444–55. doi: 10.1016/j.celrep.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–80. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 28.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer Discov. 2015;5:1024–39. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–19. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albino D, Longoni N, Curti L, Mello-Grand M, Pinton S, Civenni G, Thalmann G, D'Ambrosio G, Sarti M, Sessa F, et al.. ESE3/EHF controls epithelial cell differentiation and its loss leads to prostate tumors with mesenchymal and stem-like features. Cancer Res. 2012;72:2889–900. doi: 10.1158/0008-5472.CAN-12-0212. [DOI] [PubMed] [Google Scholar]
- 31.Topisirovic I, Siddiqui N, Lapointe VL, Trost M, Thibault P, Bangeranye C, Piñol-Roma S, Borden KL. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009;28:1087–98. doi: 10.1038/emboj.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–84. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 33.Varallyay E, Burgyan J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc. 2008;3:190–6. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- 34.Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rennie W, Liu C, Carmack CS, Wolenc A, Kanoria S, Lu J, Long D, Ding Y. STarMir: a web server for prediction of microRNA binding sites. Nucleic Acids Res. 2014;42:W114−8. doi: 10.1093/nar/gku376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werner MS, Ruthenburg AJ. Nuclear Fractionation Reveals Thousands of Chromatin-Tethered Noncoding RNAs Adjacent to Active Genes. Cell Rep. 2015;12:1089–98. doi: 10.1016/j.celrep.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan K, Arfat Y, Li D, Zhao F, Chen Z, Yin C, Sun Y, Hu L, Yang T, Qian A. Structure Prediction: New Insights into Decrypting Long Noncoding RNAs. International journal of molecular sciences. 2016;17. doi: 10.3390/ijms17010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature structural & molecular biology. 2013;20:300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 44.Pisignano G, Napoli S, Magistri M, Mapelli SN, Pastori C, Di Marco S, Civenni G, Albino D, Enriquez C, Allegrini S, et al.. A promoter-proximal transcript targeted by genetic polymorphism controls E-cadherin silencing in human cancers. Nature Communications. 2017;8:15622. doi: 10.1038/ncomms15622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma X, Shao C, Jin Y, Wang H, Meng Y. Long non-coding RNAs: a novel endogenous source for the generation of Dicer-like 1-dependent small RNAs in Arabidopsis thaliana. RNA Biol. 2014;11:373–90. doi: 10.4161/rna.28725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–28. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 48.Li N, Joska TM, Ruesch CE, Coster SJ, Belden WJ. The frequency natural antisense transcript first promotes, then represses, frequency gene expression via facultative heterochromatin. Proc Natl Acad Sci U S A. 2015;112:4357–62. doi: 10.1073/pnas.1406130112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 50.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014;156:920–34. doi: 10.1016/j.cell.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5:171. doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–9. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rybak-Wolf A, Jens M, Murakawa Y, Herzog M, Landthaler M, Rajewsky N. A variety of dicer substrates in human and C. elegans. Cell. 2014;159:1153–67. doi: 10.1016/j.cell.2014.10.040. [DOI] [PubMed] [Google Scholar]
- 55.Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–16. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.