ABSTRACT
Reinitiation after translation of short upstream ORFs (uORFs) represents one of the means of regulation of gene expression on the mRNA-specific level in response to changing environmental conditions. Over the years it has been shown-mainly in budding yeast-that its efficiency depends on cis-acting features occurring in sequences flanking reinitiation-permissive uORFs, the nature of their coding sequences, as well as protein factors acting in trans. We earlier demonstrated that the first two uORFs from the reinitiation-regulated yeast GCN4 mRNA leader carry specific structural elements in their 5′ sequences that interact with the translation initiation factor eIF3 to prevent full ribosomal recycling post their translation. Actually, this interaction turned out to be instrumental in stabilizing the mRNA·40S post-termination complex, which is thus capable to eventually resume scanning and reinitiate on the next AUG start site downstream. Recently, we also provided important in vivo evidence strongly supporting the long-standing idea that to stimulate reinitiation, eIF3 has to remain bound to ribosomes elongating these uORFs until their stop codon has been reached. Here we examined the importance of eIF3 and sequences flanking uORF1 of the human functional homolog of yeast GCN4, ATF4, in stimulation of efficient reinitiation. We revealed that the molecular basis of the reinitiation mechanism is conserved between yeasts and humans.
KEYWORDS: ATF4, eIF3, GCN4, mRNA, ribosome, reinitiation, translational control
Introduction
Translation of mRNA has four stages: initiation, elongation, termination, and ribosome recycling. During recycling, the post-termination 80S ribosome is first split into the small 40S and large 60S subunits by the energy-depended action of ABCE1.1 However, mRNA and deacylated tRNA remain bound to the small subunit and must be removed in the second step by a joint action of either canonical initiation factors eIF1, eIF1A and eIF3, or by eIF2D (also known as Ligatin) or by the heterodimer MCT1-DENR.2-5 As such, ribosome recycling can be considered as the link between translation termination and initiation because termination, recycling and initiation use several factors in common, like for example eIF3.6-8 Even though ribosome recycling naturally captures the translational cycle, there are specific exceptions where the completion of the full recycling step is undesirable or even detrimental, and the termination reaction is followed by reinitiation (REI) on the same mRNA molecule at a site downstream of the stop codon (reviewed in6,9,10). Translation reinitiation is a gene-specific regulatory mechanism where upon translation of the so-called REI-permissive short upstream ORF (uORF) only the large 60S subunit and deacylated tRNA are recycled, whereas the mRNA is retained on the post-termination 40S subunit to allow REI downstream. It has been well established that most of relatively widespread uORFs across all eukaryotic genomes in principle inhibits expression of the main ORF by preventing the fully recycled ribosome to reach its start site. Hence, existence of REI-permissive uORFs, which are often part of intricate regulatory circuits together with REI-non-permissive uORFs, is very critical as it enables-upon various stimuli-efficient expression of a main ORF. Importantly, various oncogenes, proteins involved in differentiation, development, cell cycle, stress response, learning and memory forming can be found on the list of REI-regulated mRNAs (see for example11-13).
Practically since the onset of this scientific direction, the textbook example of an mRNA regulated via REI has been the yeast GCN4 gene encoding a very potent transcriptional activator.14 The GCN4 mRNA contains four short uORFs in its 5′ leader, out of which the first two (uORF1 and uORF2) are highly REI-permissive, while the remaining two (uORF3 and uORF4) allow only negligible levels of REI.15,16 Their specific effects in combination with stress-induced changes in the level of one of the key initiating complexes composed of Met-tRNAiMet and eIF2·GTP (the so-called ternary complex-TC) create a fail-safe mechanism that allows GCN4 translation only under specific stresses.15,17 The trick is that the distance between the REI-permissive vs. non-permissive uORFs is long enough that under non-stress conditions (characteristic of high TC levels) most of the post-termination 40S ribosomes scanning downstream from uORF1 or uORF2 stop codons will reacquire the TC before AUG of uORF3 or uORF4 has been reached-as a result the GCN4 protein cannot be made. At the same time, it is short enough to ensure that under specific stress conditions (characteristic of low TC levels, when longer time is needed to reacquire the TC), majority of these ribosomes will rebind the TC after bypassing the REI-non-permissive uORFs-as a result these will be skipped and the GCN4 translation eventually initiated. Over the years it has been demonstrated that the REI potential of uORF1 and uORF2 is determined by: (i) the presence of the AU-rich motif in the 3′ sequence; (ii) their defined length and coding triplets composition; (iii) specific REI-promoting elements (RPEs) situated in their 5′ sequences; and (iv.) the functional interaction of some of the RPEs (namely RPE i. and iv. of uORF1 and RPE v. of uORF2) with the N-terminal domain (NTD) of the a/TIF32 subunit of the translation initiation factor 3 (eIF3) within the context of the post-termination mRNA·40S complex.15,16,18-24 The favorable location of the a/TIF32-NTD on the 40S subunit next to the mRNA exit channel25-27 led to an idea that while the eIF3-bound 40S ribosome scans through the region upstream of uORF1 (or uORF2) and translates it as the fully assembled 80S ribosome still bound by eIF3, the RPEs progressively fold into a specific secondary structure. Upon termination, eIF3 interacts with these RPEs to specifically stabilize only the small ribosomal subunit on the uORF1 (or uORF2) stop codon. Thanks to this incomplete ribosomal recycling, the post-termination 40S subunit can, upon acquisition of other essential eIFs, subsequently resume scanning for REI downstream.21 Actually, continued presence of some eIFs on early elongating ribosomes as a prerequisite for efficient REI had been a long standing hypothesis18 that was strongly supported by our most recent yeast work.28 With the help of a newly developed in vivo RNA-protein Ni+2 pull down (Rap-Nip) assay we have clearly demonstrated that eIF3 does travel with early elongating ribosomes and interacts with RPEs in vivo, and this eIF3s ability is critical for stimulation of efficient reinitiation downstream of REI-promoting uORFs. Besides eIF3, the mRNA-delivery eIF4F complex, and particularly the central one-third fragment of eIF4G interacting with eIF3 and eIF4A, was also suggested to remain bound to early elongation ribosomes and promote efficient REI, at least in an in vitro reconstituted mammalian system.29 However, firm experimental evidence is lacking in this case.
Here we set out to examine whether the just described molecular mechanism of REI relying on cis-acting features of REI-permissive uORFs and eIF3 is conserved between yeasts and humans. We used an extensively studied mRNA encoding transcriptional activator ATF4 (the mammalian functional homolog of yeast GCN4) that contains two uORFs as a reporter that we mutagenized. We also knocked down several eIF3 subunits, in particular eIF3a (implicated in REI in yeasts21) and eIF3h (shown to stimulate REI in plants30,31), and checked their effects on REI efficiency in human cells. Our analysis revealed that the ATF4s uORF1 is in analogy to uORF1 of GCN4 also surrounded by cis-acting features, with those occurring in its 5′ leader specifically structured, that ensure its permissiveness for REI. Furthermore, we also show that human eIF3h (like its plant counterpart) enhances efficiency of REI.
Novel insights into the molecular mechanism of reinitiation in human cells
Sequences flanking uORF1 of ATF4 substantially increase its reinitiation potential
As mentioned above, mammalian ATF4 mRNA contains only two uORFs in its leader in contrast to four uORFs of yeast GCN4 (Fig. 1A). However, only the first uORF1 of the two fulfills the requirements of a typical short uORF with a REI-potential because it is composed of only three sense codons and the distance between its stop codon and AUG of uORF2 is in most species 87 nucleotides or close to it. (Based on Kozak 1987,32 the optimal distance ensuring efficient REI in mammals is 80 nt and more.) This could enable a similar mode of regulation under stress vs. non-stress conditions like in the case of GCN4 despite the fact that ATF4s uORF2 is markedly different from GCN4s uORFs 3 and 4. It is too long to be even considered as an uORF with some REI potential (59 amino acids residues) and, most importantly, its sequence partially overlaps the ATF4 ORF in a different reading frame. Therefore, according to the current model, all ribosomes that reinitiate on uORF2 will under normal conditions terminate past the ATF4 AUG and thus prevent its translation.33,34 Nonetheless, taking into account the striking similarity between the GCN4s and ATF4s uORF1 with respect to their arrangement and proposed function, we were curious to examine what else they have in common. In other words, we asked whether ATF4s uORF1 utilizes an identical molecular strategy to that of GCN4s uORF1.
Figure 1.
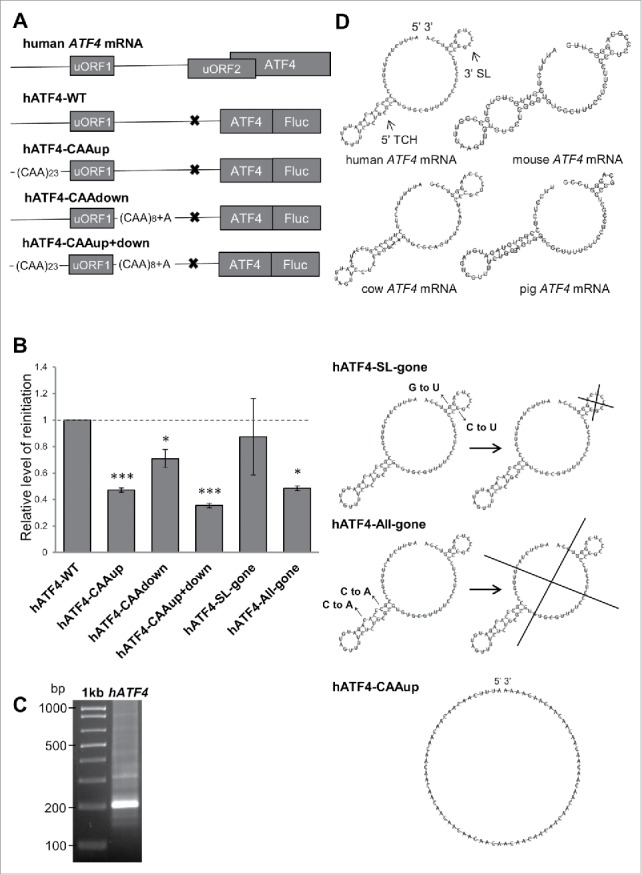
Flanking sequences of human ATF4s uORF1 individually contribute to ensure its high propensity for reinitiation. A) Schematics of the human ATF4 mRNA and mutagenized constructs used in this study. Please note that the inhibitory effect of uORF2 was neutralized by mutating its AUG to AGG to simplify the analysis, because thus modified constructs could be analyzed without using a stress inducer. In “CAAup,” a major part of the original sequence upstream of uORF1 was replaced by 23 CAA triplets (the transcriptional start site and 9 nts immediately preceding AUG of uORF1 that are buried in the mRNA binding channel of the ribosome terminating on uORF1 were left intact, as in case of GCN4s uORF121); in “CAAdown,” the original sequence encompassing 25 nts immediately following the stop codon of uORF1 was replaced by 7 CAA triplets followed by one CAAA tetranucleotide; in “CAAup+down,” both of these substitutions were combined. B) All constructs shown in A) were transfected into HEK293T cells and subjected to Dual luciferase assay normalized to mRNA levels (Fluc/Rluc) as described in Materials and Methods. The statistical analysis was performed using One sample t test; statistical significance is indicated by stars (one star means P ≤ 0.05, 3 stars P ≤ 0.001). C) 5´RLM-RACE of human ATF4 cDNA prepared from total RNA derived from HEK293T cells. DNA sample was separated by gel electrophoresis in a 2% agarose gel, cut out and processed to be sequenced (the obtained sequence indeed corresponded to the 5′ UTR of human ATF4 mRNA - NM_182810 in NCBI). Size markers in base pairs are indicated in the left. D) Secondary structure predictions of the entire 5′ sequence of ATF4s uORF1 in indicated mammals as determined by the RNA Vienna package software.35 The bottom panels depict the “CAAup” mutation and 2 double-point substitutions engineered to disrupt either the individual structures or the 5′ UTR fold as whole. Please see the main text for further details.
To answer this question, we first isolated total RNA from human HEK293T cells and using the 5′ RLM-RACE system from Ambion, generated cDNA carrying full-length 5′ UTR of human ATF4 and precisely mapped its transcriptional start site (Fig. 1C). We then replaced the 5′ and 3′ sequences (either individually or in combination) of human ATF4s uORF1, which might hypothetically correspond to the 5′ RPEs and 3′ AU-rich motif of GCN4s uORF1, with stretches of supposedly linear (CAA)n triplets (Fig. 1A). In detail, we replaced 69 nts upstream of uORF1 (in “CAAup”) and 25 nts downstream of uORF1 (in “CAAdown”); in addition we combined these mutations in a single construct “CAAup+down.” The resulting mutant variants were introduced into the uORF1-only ATF4-Luc construct containing solitary uORF1 kindly provided by the Wek's laboratory33 and the luciferase activity, as an indicator of the REI efficiency, was measured in HEK293T cells and normalized to mRNA levels of individual constructs. Please note that the inhibitory effect of uORF2 was neutralized by mutating its AUG to AGG implying that these constructs could be analyzed without using a stress inducer. As shown in Fig. 1B, both sequences flanking uORF1 are-in a striking analogy to the GCN4s uORF1-required for efficient REI. Replacement of the 5′ sequence (“CAAup”) decreased the REI efficiency to a greater extent (down to ∼47%) than the replacement of the 3′ sequence (“CAAdown;” down to ∼71%), suggesting that its contribution is significantly greater. Interestingly, the opposite is true in case of the GCN4s uORF121. The combination of both mutations (“CAA up+down”) produced a fully additive effect-downregulation to ∼35% (Fig. 1B). Together these findings strongly indicate that both upstream and downstream sequences of uORF1 independently contribute to its overall REI potential by more than 60%, suggesting that this important translational control mechanism is evolutionary conserved.
Given the fact that the 5′ sequence of the GCN4s uORF1, as well as the 5′ sequence of a single uORF of another yeast transcriptional activator YAP1, contain specific structural and sequence-specific REI-promoting cis-acting features, the RPEs,21 we next investigated whether the 5′ sequence of ATF4s uORF1 also adopt some specific structure, and if so, whether it is also important for efficient REI. Therefore, we subjected the entire region preceding the ATF4 uORF1 to in silico modeling by the RNA Vienna package software.35 As in case of GCN4, our prediction was based on the fact that the 5′ sequence is not a standalone molecule with a rigid structure but its fold forms and changes dynamically as the sequence emerges from the ribosomal mRNA exit pore.21 Hence, we divided the 5′ UTR of uORF1 into two consecutive segments and first folded the extreme 5′ segment, which formed a stable triple-circle hairpin (“5′ TCH”) (Fig. 1D). After that we added the other segment and continued with modeling of the entire 5′ sequence as it emerged from the mRNA exit pore with the initially identified 5′ triple-circle hairpin structure “pre-folded.” As a result, a stem-loop formed proximal to the 3′ end (“3′ SL”), in addition to the 5′ triple-circle hairpin. These structures and their spacing not only resemble similar structures representing GCN4s RPEs ii. and iv.,21 they also seem to be very well conserved at least among other mammalian species (Fig. 1D).
To examine the prospective physiologic importance of these structures, we further used in silico modeling to design and test minimal mutations disrupting one or the other structure using the same reporter system as described above. The first mutation, “SL-gone” (where C64 and G73 were both mutated to Us), was designed to disrupt the 3′ stem-loop; however, its effect on luciferase activity was very mild (∼13% reduction) indicating that this stem-loop contributes to REI only negligibly (Fig. 1B). We did not find any computational prediction that would disrupt selectively only the 5′ triple-circle hairpin, hence as the second mutation we chose “All-gone” (where C17 and C21 were both mutated to As), which disrupts both structures. Strikingly, the effect of this mutation showed the same dramatic drop in the luciferase activity as the “CAAup” construct (down to ∼49%) strongly suggesting that the 5′ triple-circle hairpin highly likely is what lies behind the REI-promoting effect of 5′ sequences of ATF4 uORF1 (Fig. 1B). However, at present we cannot tell whether it is the entire specific structure or only some sequential motif within this structure, like for example the apical circle that is required for its function in promoting REI.
eIF3h promotes translation reinitiation in human cells
As mentioned above, RPE i. and iv. of the GCN4s uORF1 specifically interact with the N-terminal domain of the a/TIF32 subunit of eIF3 and this interaction is instrumental for stabilizing the 40S·mRNA post-termination complex. Hence the next obvious question we asked was whether human eIF3 also contributes to efficient REI on the ATF4 mRNA. To our knowledge, the prospective role of eIF3a in REI has never been tested in the past; however, there are a few reports implicating the eIF3h subunit in REI in plants.30,31 To address this question, we individually reduced the eIF3a and eIF3h expression by knocking them down with On-target plus siRNA system (Dharmacon) as described before,36,37 and measured the luciferase activity in thus treated HeLa cells transfected with human ATF4-Luc constructs bearing either uORF1 alone (“uORF1-only”) or none of the uORFs (“d-all”) (Fig. 2A). The latter construct was used for normalization purposes. As a control we used cells treated with non-targeting siRNA, as well as cells knocked down for eIF3k. The knock down efficiency for all 3 eIF3 subunits was as observed before37-expression was reduced by ∼70–80% (Fig. 2B). Please note that we used HeLa instead of HEK293T cells owing to the fact that the efficiency of downregulation of all eIF3 subunits is significantly greater in HeLa cells.36 Also note that the eIF3k knock down results in the loss of only two non-essential subunits (eIF3k by itself and its interacting partner eIF3l) from the rest of human 12-subunit eIF3, the eIF3h knock down eliminates itself plus both eIF3kandl, whereas the eIF3a knock down pretty much destroys the entire eIF3 complex leaving intact only the “Yeast-Like-Core” assembly composed of the eIF3b–eIF3i–eIF3g subunits (Fig. 2B).36,37 As shown in Fig. 2C, the eIF3k knock down displayed practically no impact on the efficiency of REI; similarly the eIF3a knock down produced only an insignificantly modest reduction (by ∼11%). However, the eIF3h knock down led to a statistically significant reduction by ∼34%. Importantly, the eIF3a knock down as the only knock down downregulated general translation initiation rates as judged from our measurements of the “d-all” construct; this is expected given the detrimental consequences of the eIF3a knock down on the overall integrity of the entire eIF3 complex and its function in general initiation.36,37 Hence, we cannot conclude anything specific regarding its involvement in REI in mammals. However, the fact that the eIF3h knock down (co-downregulating also the expression of the eIF3kandl dimer) clearly impacted the efficiency of REI, whereas the eIF3k knock down (co-downregulating only the eIF3kandl dimer) showed no impact whatsoever, suggests that eIF3h does enhance efficiency of REI also in humans.
Figure 2.
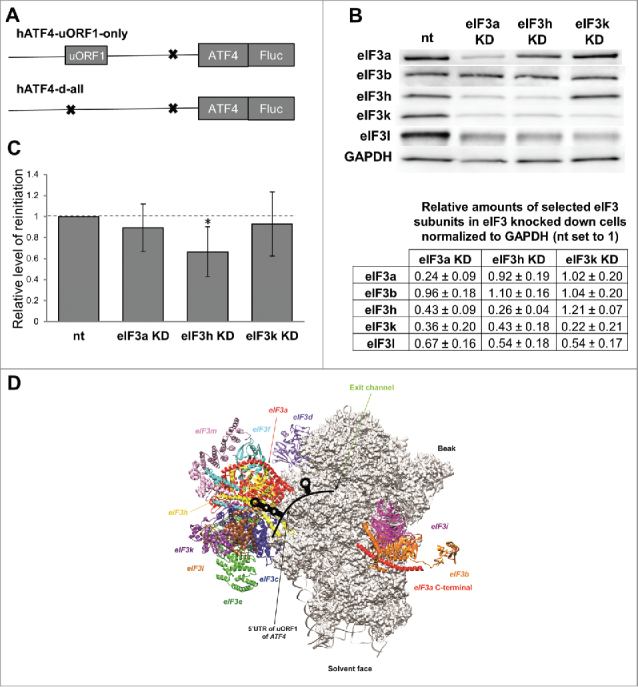
The eIF3h subunit of human eIF3 enhances efficiency of resumption of scanning from ATF4s uORF1 for reinitiation downstream. A) Schematics of hATF4-Fluc constructs used in C). B) Efficiency of siRNA-mediated downregulation and co-downregulation of protein levels of selected eIF3 subunits normalized to house-keeping GAPDH and Non-targeted (nt) control cells estimated by Western blotting. C) Relative Firefly luciferase signals obtained from HeLa cells knocked down for indicated eIF3 subunits transfected with either the “uORF1-only” or “d-all” constructs (the latter was used for normalization purposes), expressed as relative percentages of Fluc signals obtained from Non-targeted control (nt) cells. The Firefly luciferase signals were individually normalized to mRNA levels of each reporter, which were beforehand normalized to the spike RNA added before the RNA extraction. Statistical analysis was performed using One sample t test; statistical significance is indicated by stars (one star means P ≤ 0.05). D) Graphical illustration of the proposed arrangement of the post-termination complex on ATF4s uORF1 with its secondary structures interacting with the eIF3h subunit of eIF3 to promote resumption of scanning for REI on the ATF4 mRNA. Depicted is the exit channel view of the 48S PIC (adopted from47) illustrating 12 color-coded eIF3 subunits with eIF3a and eIF3h indicated by an arrow. The 5′ UTR of the ATF4s uORF1 highlighting its secondary structures is shown in black.
Concluding remarks
Two questions we asked in this article were: (1) Is there any mechanistic resemblance in the modus operandi between REI-permissive uORFs from mRNA leaders of functional homologs from two rather diverse eukaryotic organisms like yeasts (GCN4) and humans (ATF4)?; and (2) Does eIF3a and/or eIF3h promote reinitiation in mammals? The answer is yes to both questions. Flanking sequences of ATF4s uORF1 independently contribute to significantly boost the basic level of REI that this uORF allows. In addition, its 5′ sequence contains two well conserved structural features-the 5′ triple-circle hairpin and the 3′ stem-loop-that resemble the structural features of GCN4s uORF1 and the former of which seems to be fully responsible for the observed effect. Finally, whether or not eIF3a promotes REI as in budding yeast cannot be judged from our analysis; however, eIF3h does seem to be involved like in plants. In fact, it is interesting to note that human eIF3h seems to adopt a similar position on the ribosome to the REI-promoting N-terminal domain of yeast eIF3a/TIF32; i.e., right next to the mRNA exit channel (Fig. 2D), where it could interact with the 5′ triple-circle hairpin post uORF1 translation. This further supports the idea that in the 12-subunit eIF3 complex, eIF3h has a direct role in stimulating reinitiation.
According to recent reports, uORFs occur at a much higher frequency in mammalian (∼45%) mRNAs than in yeast (∼13%).11,38-40 Yeast studies on GCN4 and YAP1 mRNA leaders,16,21,41 as well as an early report examining REI efficiency of randomly generated uORFs42 strongly suggest that majority of yeast uORFs are severely REI-non-permissive. Even though uORFs are prevalent translational repressors also in mammals,43,44 there is a prevailing notion that uORFs in mammalian mRNAs (including randomly laboratory-designed uORFs) are in general less repressive for REI than in yeast, usually reducing protein expression by 30 to 80% (i.e., they allow at least some resumption of scanning and reinitiation),11 which suggests that there might be a smaller requirement for specific sequences.9 Our results from both yeast and humans seem to agree with this theory, because whereas eliminating all cis-acting sequences flanking uORF1 or uORF2 of yeast GCN4 fully abolished their REI potential,16 substituting the similar sequences flanking ATF4s uORF1 with unstructured stretches of CAA repeats reduced the efficiency of REI “only” by ∼65% (Fig. 1B; hATF4-CAAupanddown). Hence it does seem likely that most of mammalian uORFs are less inhibitory than in yeast, and only if they form an integral part of some sophisticated stress-related regulatory system often containing more than one uORF, nature equipped some of them with specific cis-acing features that render them highly permissive. What lies behind this difference between budding yeast and humans (mammals)?
It could very well be the nature of the two initiation factors that have been implicated in stimulating REI in one and/or the other organism; i.e., eIF3 and to a lesser extent also eIF4G,20,21,29 and that differ most dramatically between yeast and vertebrates in several aspects. (1) Human eIF3 has practically twice as many subunits than its yeast counterpart; (2) human eIF4G is markedly longer and has more direct interacting partners; and 3), perhaps the most important difference is that mammalian eIF3 does directly interact with eIF4G; however, in yeasts this contact is supposedly only bridged by eIF5 and eIF1 (reviewed in [6,9]).
If we assume that eIF4G and eIF3 are indeed capable to persist throughout uORF translation on the mammalian 80S ribosome to stabilize the post-termination mRNA·40S complex, their direct contact could substantially empower this stabilization process. This would set the basal level of permissiveness for REI in mammals higher than it is set in the budding yeast, where these two factors do not directly interact, which may weaken the eIF4G interaction with elongating ribosomes. In the light of the recent findings, the fact that the recycling factor ABCE1 interacts with the intersubunit face of the 40S subunit even after ribosomal recycling and most likely also promotes the initiation phase in close co-operation with eIF345,46 further suggests that eIF4G could, via its direct connection with eIF3, modulate ribosomal recycling in a way that would favor dissociation of only the 60S subunit and deacylated tRNA, which would stimulate REI. Since reinitiation-as a molecular phenomenon-is also rather interesting from the medical point of view (there is a rapidly growing number of articles reporting contributions of defective uORF functions to various human diseases10), more work is certainly needed to fully understand the mechanistic aspects of this intriguing difference, as well as the reinitiation mechanism as a whole.
Material and methods
Dual luciferase reporter assays
HEK293T cells were grown at 37°C and 5% CO2 in 6-well plates in DMEM (Sigma, cat # D6429) supplemented with 10% FBS (Sigma, cat # F7524). The cells were lysed directly on plate with 1x Glo Lysis Buffer (Promega, cat # E266A) exactly 24 hours after the Firefly and Renilla reporter plasmids transfection with TurboFect (Thermo Scientific, cat # R0531). The lysate was then transferred into a white flat-bottom 96-well plate and part of the lysate was stored for RNA isolation. The Dual-Glo® Luciferase Assay System (Promega, cat # E2940) was used according to the vendor's instructions. The Renilla luciferase signal was used for normalization purposes. Total RNA was isolated using the RNA Blue reagent (Top Bio, cat # R013) according to the manufacturer's instructions. After the Turbo DNase digestion (Ambion, cat # AM2238), cDNA was synthesized using the High-capacity cDNA reverse transcription kit (Applied Biosystems, # 4368813). qPCR was performed using 5 × HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne # 08–25–00020). The mRNA levels of Firefly luciferase were normalized to Renilla luciferase mRNA levels. The obtained qPCR data were used for normalization of measured luciferase activities. qPCR primers are listed in Supplementary Table S1.
siRNA treatment, whole cell extract preparation and Western blotting
HeLa cells were grown at 37°C and 5% CO2 in 6-well plates in DMEM (Sigma, cat # D6429) supplemented with 10% FBS (Sigma, cat # F7524). 24 hours after seeding, cells were transfected with the ON-TARGETplus siRNA cocktail system from Dharmacon at a final concentration of 5 nM (human eIF3a cat # L-019534–00, eIF3h cat # L-003883–00, eIF3k cat # L-020216–02 and Non-Targeting siRNA cat # D-001810–10). INTERFERin (Polyplus, cat # 409) was used as a transfection reagent and transfection was performed according to the vendor's instructions.
For Western blotting, cells were harvested 3 d after siRNA transfection in lysis buffer containing 1M Tris-HCl pH6.8, 20% glycerol, 20% SDS, 2% β-merkaptoethanol and 5% bromphenolblue. All samples were resolved using SDS-PAGE followed by Western blotting. All primary antibodies used in this study are listed in Supplementary Table S2. The Western signals were developed using the SuperSignal West Femto Maximum Sensitivity Substrate from Thermo Scientific (cat # 34096) and detected in a G-Box imager from Syngene using a series of varying exposure times. Signals were processed with Quantity One (BioRad). The resulting values were normalized as indicated in the corresponding figure legend.
For siRNA treatments followed by Firefly luciferase reporter assays, transfection of Firefly reporter plasmids was performed 48 hours after the siRNA treatment and cells were harvested 24 hours later as described above. The Firefly luciferase signal was normalized to the reporter's mRNA level, which was beforehand normalized to the spike RNA (particularly yeast RPL41a mRNA) added before the RNA extraction. In detail, HeLa cells in 6-well plates were lysed in 200 µl of 1x Glo Lysis Buffer (Promega, cat # E266A). 70 µl of this lysate was directly used for the luciferase reporter assay, and another 70 µl was mixed with 2 µl of yeast spike RPL41a mRNA to the final amount of approx. 100 ng per sample, and subsequently also with 750 µl of RNA Blue reagent (Top Bio, cat # R013). The total RNA was isolated according to the manufacturer's instructions. After the Turbo DNase digestion (Ambion, cat # AM2238), cDNA was synthesized using the High-capacity cDNA reverse transcription kit (Applied Biosystems, # 4368813). qPCR was performed using 5 × HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne # 08–25–00020). The signal from Firefly luciferase reporter plasmid was normalized to its mRNA levels, which were already normalized to the spike RPL41a mRNA to correct for any loss during the RNA isolation. The obtained values with individual constructs were finally normalized to the nt siRNA and the “d-all” control construct, the latter of which corrects for defects in general translation initiation.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
We are thankful to Ron Wek for the ATF4 reporter plasmids and advise, to Olga Krýdová for technical and administrative assistance, and to the members of the Valášek laboratory for helpful suggestions.
Funding
This research was supported by the Czech Science Foundation Grant GA15–10116S (to L.S.V.) and Charles University in Prague, project GA UK no. 10315 (to V.H.).
References
- 1.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A. 2011;108:E1392-8. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pisarev AV, Hellen CUT, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286-99. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196-210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787-801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950-62. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valášek LS. 'Ribozoomin’-translation initiation from the perspective of the ribosome-bound Eukaryotic Initiation Factors (eIFs). Curr Protein Pept Sci. 2012;13:305-30. doi: 10.2174/138920312801619385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beznosková P, Cuchalová L, Wagner S, Shoemaker CJ, Gunišová S, von der Haar T, Valášek LS. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLoS Genet. 2013;9:e1003962. doi: 10.1371/journal.pgen.1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beznoskova P, Wagner S, Jansen ME, von der Haar T, Valasek LS. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015;43:5099-111. doi: 10.1093/nar/gkv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45-93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 10.Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip Rev RNA. 2014;5:765-78. doi: 10.1002/wrna.1245. [DOI] [PubMed] [Google Scholar]
- 11.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. PNAS. 2009;106:7507-12. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbosa C, Peixeiro I, Romao L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 2015;25:1848-59. doi: 10.1101/gr.195404.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407-50. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 15.Gunisova S, Valasek LS. Fail-safe mechanism of GCN4 translational control-uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4 expression. Nucleic Acids Res. 2014;42:5880-93. doi: 10.1093/nar/gku204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunisova S, Beznoskova P, Mohammad MP, Vlckova V, Valasek LS. In-depth analysis of cis-determinants that either promote or inhibit reinitiation on GCN4 mRNA after translation of its four short uORFs. RNA. 2016;22:542-58. doi: 10.1261/rna.055046.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201-7. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226-32. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luukkonen BG, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol. 1995;69:4086-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szamecz B, Rutkai E, Cuchalová L, Munzarová V, Herrmannová A, Nielsen KH, Burela L, Hinnebusch AG, Valásek L. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 2008;22:2414-25. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munzarová V, Pánek J, Gunišová S, Dányi I, Szamecz B, Valášek LS. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet. 2011;7:e1002137. doi: 10.1371/journal.pgen.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant CM, Hinnebusch AG. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606-18. doi: 10.1128/MCB.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant CM, Miller PF, Hinnebusch AG. Sequences 5′ of the first upstream open reading frame in GCN4 mRNA are required for efficient translational reinitiation. Nuc Acids Res. 1995;23:3980-8. doi: 10.1093/nar/23.19.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller PF, Hinnebusch AG. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes and Development. 1989;3:1217-25. doi: 10.1101/gad.3.8.1217. [DOI] [PubMed] [Google Scholar]
- 25.Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CH, Cimermančič P, Boehringer D, Sali A, Aebersold R, et al.. Molecular architecture of the 40SeIF1eIF3 translation initiation complex. Cell. 2014;158:1123-35. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valášek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. The yeast eIF3 Subunits TIF32/a and NIP1/c and eIF5 make critical connections with the 40S Ribosome in vivo. Genes Dev. 2003;17:786-99. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouba T, Dányi I, Gunišová S, Munzarová V, Vlčková V, Cuchalová L, Neueder A, Milkereit P, Valášek LS. Small ribosomal protein RPS0 stimulates translation initiation by mediating 40S-binding of eIF3 via its direct contact with the eIF3a/TIF32 Subunit. PLoS One. 2012;7:e40464. doi: 10.1371/journal.pone.0040464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammad MP, Munzarova Pondelickova V, Zeman J, Gunisova S, Valasek LS. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res. 2017;45:2658-74. doi: 10.1093/nar/gkx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pöyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62-75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy B, Vaughn JN, Kim BH, Zhou F, Gilchrist MA, Von Arnim AG. The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA. 2010;16:748-61. doi: 10.1261/rna.2056010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013;32:1087-102. doi: 10.1038/emboj.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438-45. doi: 10.1128/MCB.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269-74. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27-33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofacker IL. RNA secondary structure analysis using the Vienna RNA package. Curr Protoc Bioinformatics. 2004;Chapter 12:Unit 12 2. doi: 10.1002/0471250953.bi1202s04. [DOI] [PubMed] [Google Scholar]
- 36.Wagner S, Herrmannova A, Malik R, Peclinovska L, Valasek LS. Functional and biochemical characterization of human eukaryotic translation initiation factor 3 in living cells. Mol Cell Biol. 2014;34:3041-52. doi: 10.1128/MCB.00663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner S, Herrmannova A, Sikrova D, Valasek LS. Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: The yeast-like core and the octamer. Nucleic Acids Res. 2016;44:10772-88. doi: 10.1093/nar/gkw972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Resch AM, Ogurtsov AY, Rogozin IB, Shabalina SA, Koonin EV. Evolution of alternative and constitutive regions of mammalian 5′UTRs. BMC Genomics. 2009;10:162. doi: 10.1186/1471-2164-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218-23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawless C, Pearson RD, Selley JN, Smirnova JB, Grant CM, Ashe MP, Pavitt GD, Hubbard SJ. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics. 2009;10:7. doi: 10.1186/1471-2164-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilela C, Linz B, Rodrigues-Pousada C, McCarthy JE. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 1998;26:1150-9. doi: 10.1093/nar/26.5.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yun DF, Laz TM, Clements JM, Sherman F. mRNA sequences influencing translation and the selection of AUG initiator codons in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1996;19:1225-39. doi: 10.1111/j.1365-2958.1996.tb02468.x. [DOI] [PubMed] [Google Scholar]
- 43.Johnstone TG, Bazzini AA, Giraldez AJ. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 2016;35:706-23. doi: 10.15252/embj.201592759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chew GL, Pauli A, Schier AF. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun. 2016;7:11663. doi: 10.1038/ncomms11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heuer A, Gerovac M, Schmidt C, Trowitzsch S, Preis A, Kötter P, Berninghausen O, Becker T, Beckmann R, Tampé R. Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat Struct Mol Biol. 2017;24:453-60. doi: 10.1038/nsmb.3396. [DOI] [PubMed] [Google Scholar]
- 46.Mancera-Martinez E, Brito Querido J, Valasek LS, Simonetti A, Hashem Y. ABCE1: A special factor that orchestrates translation at the crossroad between recycling and initiation. RNA Biol. 2017; in press; doi: 10.1080/15476286.2016.1269993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491-5. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


