ABSTRACT
Circular RNA (circRNA) generated by alternative splicing represents a special class of non-coding RNA molecule. CircRNAs are abundant in the eukaryotic cell cytoplasm and have a characteristic organization, timing of action and disease specificity. In contrast to linear RNA, circRNAs are resistant to RNA exonuclease. Consequently, circRNA escapes normal RNA turnover and this improves circRNA stability. CircRNAs can be degraded by microRNA (miRNA) and this results in linearization of the circRNA, which can then act as competitor to endogenous RNA. Through interactions with disease-related miRNA, circRNA can play an important regulatory role in specific diseases. Furthermore, circRNAs have significant potential to become new clinical diagnostic markers.
KEYWORDS: Circular RNA, miRNA sponge, non-coding RNA, Alternative splicing, competing endogenous RNA, microRNA
Introduction
Circular RNA (circRNA) describes a class of annular RNA molecules comprising one or more exons. This type of RNA molecule is abundant in eukaryotic cells, is highly organized, acts in a temporal manner and is disease specific. CircRNA was originally described as a “viroid,” consisting of a covalently closed circular RNA molecule that was pathogenic to particular higher plants.1,2 Subsequently, researchers studying the tumor suppressor gene DCC, human Ets-1 and mouse Sry, found that the corresponding RNAs were processed by precursor mRNA (pre-mRNA) and were annular, which demonstrated that circRNA existed in eukaryotic cells.3-6 Only a few circRNAs were originally reported, most likely as a result of very low expression levels. These molecules were considered to be by-products generated by an error in alternative splicing and were even described as a “genetic accident” or the result of experimental error.6,7
With the rapid advancement of RNA sequencing methodologies and bioinformatics, progressively more circRNAs have been discovered following large scale transcriptome analyses. Danan et al.7 found circRNA were present transcriptome-wide in archaeal cells and suggested that circRNAs had potential biologic function(s). Salzman et al.,8 performed RNA analysis on a variety of human cells and not only detected circRNA but also suggested were a general feature of the human transcriptome. In excess of 25,000 species of circRNA were identified in human fibroblasts cells via a high-throughput RNA sequencing method (RNA-seq).9 CircRNA was considered as a byproduct of RNA splicing which could be degraded by small interfering RNA (siRNA).10 Ye et al.11 revealed that circRNAs are prevalent in plants and have common, characteristic features that distinguish them from circRNAs in animals. Memczak et al.12 isolated RNA from humans, mice and nematodes, and subsequently sequenced and bioinformatically analyzed them. They found that circRNAs were abundant but had restricted expression to certain tissues and to developmental stages and concluded that circRNAs were involved in post-transcriptional control. Hansen et al.13 revealed that circRNA can influence gene regulation and that circRNAs can act as microRNA (miRNA) “sponges” through their action in competing with endogenous RNA. Although the field of circRNA research spans over 30 years, research has been hindered primarily as a result of limitations in the technology to isolate, identify and study circRNAs. Recently, discoveries of circRNAs' biofunctions12-14 have accelerated the research effort. Here, we focus on functions of circRNA and supporting evidence for the role of circRNA in diseases.
The formation of characteristics of circRNA
The biogenesis of circRNA
Generally, eukaryotic RNA processing removes non-coding intron sequences by pre-mRNA splicing, and then links protein-coding exons to form the mature linear RNA molecule. Higher eukaryotes can produce multiple mature RNAs and their respective protein products through alternative splicing. About 90% of human genes are expressed as pre-mRNA splicing isoform alternatives,15 which play important roles in the functional diversity of the genome and proteome. A non-canonical form of alternative splicing produces circRNA, however the detailed mechanism of circRNA biogenesis remains elusive. The most plausible mechanism for RNA circularization is back-splicing,9 where a downstream splice donor (SD) site is ligated to an upstream splice acceptor (SA) site reversely.
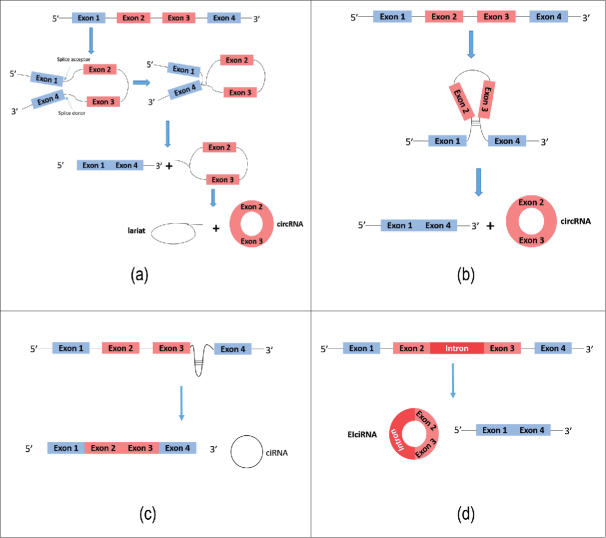
CircRNA is generated by special alternative splicing of pre-mRNA primarily by two means, exon circularization and intron circularization. With regard to exon circularization, Jeck et al.9 proposed two models of circRNA formation; intron-pairing-driven circularization and lariat-driven circularization. In the first model, within the spliced lariat containing exon skipping, a downstream SD is joined to an upstream SA and undergoes internal splicing to form a circRNA (Fig. 1a). In another model, two introns first form a cyclic structure via complementary base pairing and then excision of introns is executed to form the circRNA (Fig. 1b). Jeck et al.9 suggested that the presence of long flanking introns and the existence of increased numbers of complementary ALU repeat sequences about circularized exons was consistent with both of the models described above. Recently, Wang et al.16 found that general splicing factors and cis-elements regulate back-splicing by non-canonical splicing regulatory rules. Zhang et al.17 discovered almost 10 thousand circRNAs in human embryonic stem cell line H9 and showed that exon circulation depends on the presence of flanking intronic complementary sequences. Furthermore, they found that the efficiency of exon circularization is related to RNA pairing across flanking introns or to competition within individual introns.17 Moreover, they showed that single gene loci produce many exon circularization events, which expands our understanding of how genes are regulated. For those genes lacking noticeable flanking intronic secondary structure, Barrett and his coworkers18 discovered that circRNA can be spliced from an exon-containing lariat precursor, and they suggested this could explain why circRNA was considered to be a by-product of exon skipping. Besides non-canonical ‘back-splicing’, Starke, et al.19 demonstrated a circular configuration of circRNAs HIPK3 and LPAR1, and found that RNA circularization requires canonical splice sites without specific circRNA exon sequences. But circularization efficiency can be modulated by potential flanking intron structures, indicating that circRNA biogenesis requires the canonical spliceosomal machinery. Recently, Zhang et al.20 found that back-splicing is far less favorable than canonical splicing and back-splicing circularization is associated with fast PoI II elongation rate. Evidence also showed that shorter exons were less efficient than longer exons in circularising, and the intronic repeats cooperate with the exonic sequences for circularization to occur.21 Previously, Zhang et al. reported that circular intronic RNA (ciRNA) represented yet another type of circular RNA molecule derived from intron circularization22 (Fig. 1c). Li et al.23 identified exons that are circularized with introns retained between them, in a type of circRNA associated with RNA polymerase II in human cells and these were termed exon-intron circRNAs or ElciRNAs (Fig. 1d). EIciRNAs were found exclusively in the cell nucleus. Evidence showed that RNA-editing enzyme ADAR1 antagonizes circRNA biogenesis.24 Computational identification methods allowed Liu et al.25 to postulate that thermodynamic and conformational properties of flanking introns are associated with circRNA formation.
Figure 1.
Formation of circRNAs. (a) Exon-skipping leads to a lariat, which undergoes internal splicing to form a circRNA. (b) Complementary base pairing is required, which brings non-sequential donor-acceptor pairs into opposition, allowing circularization to occur. (c) Circular intronic RNA (ciRNA) represents another type of circular RNA molecule derived from intron circularization. (d) Exons of EIciRNA are circularized with introns ‘retained’ between exons.
The characteristics of circRNA
The current literature attributes important characteristics to circRNA. CircRNA is produced by special alternative splicing and circRNA is abundant in eukaryotic cells,6 and recently Li et al. reported that circRNA is enriched and stable in exosomes.26 CircRNA is highly abundant in human cells, with the quantities of circular molecules exceeding those of associated linear mRNA by more than 10-fold in some cases.8,9 RNase R, a 3′ to 5′ exoribonuclease, can degrade linear RNA whereas circRNA is RNase R resistant, which makes circRNA more stable than linear RNA.9,27,28 CircRNA is highly conserved,8,9 and some circRNAs show evidence of rapid evolutionary changes.17,22 Most circRNAs derive from exons, while a small number are formed by direct intronic circularization. CircRNA molecules are rich in the miRNA response element (MRE), and can act as competing endogenous RNA (ceRNA) by interacting with miRNA and subsequently releasing target genes from miRNA and thus play a miRNA sponge role in cells.13 Most circRNA are non-coding RNA.8 Finally, one of the conserved features of circRNA biogenesis is presence of reverse complementary matches (RCMs) in introns, which can be used to predict and experimentally validate circRNA.24 Recently, evidence showed that back-splicing largely occurs post-transcriptionally.20
The main function of circRNA
CircRNA is involved in post-transcriptional regulation
An antisense human circRNA derived from the cerebellar degeneration-related protein 1 transcript (CDR1as), contains at least 60 binding sites for microRNA-7 (miR-7) that are conserved, which allows this circRNA to suppress miR-7 activity and leads to an increase in miR-7 target gene expression.12,13 CDR1as and miR-7 co-expression is found at high levels in parts of the developing midbrain.12 In zebrafish embryos, injection of circular CDR1as resulted in reduced midbrain size, while injecting the miR-7 precursor could only partially rescue this effect, suggesting that the effect of CDR1as was the result of an interaction with miR-7, at least in part.12 That CDR1as can behave in vivo as an miR-7 sponge,29 is consistent with such an interaction. Moreover, by predicting microRNA-138 (miR-138) target sites in the testis-specific Sry RNA, Hansen et al.13 identified 16 putative miR-138 target sites, and they also determined that Sry circRNA could be captured by biotin-labeled miR-138, indicating again that Sry circRNA can behave as a miR-138 sponge. Yan Li et al.26 made the interesting observation that when mimics of miR-7 were introduced into cells, the levels of CDR1as circRNA within exosomes were significantly decreased, but were increased in cells, suggesting that circRNAs sorting into exosomes was controlled by changing levels of associated miRNA in producer cells, at least in part. Furthermore, they found that in recipient cells, CDR1as-containing exosomes inhibited miR-7-induced growth.26 Recently, Li et al.30 reported that circRNA cir-ITCH, controlled the expression of itchy E3 ubiquitin protein ligase (ITCH) by acting as a sponge for miR-214, miR-17 and miR-7 in esophageal squamous cell carcinoma (ESCC). Moreover, circHIPK3, which is a derivative of Exon2 from the HIPK3 gene, acts as a sponge for 9 miRNAs (miR-654, miR-584, miR-379, miR-338, miR-193a, miR-152, miR-124, miR-29a and miR-29b) with 18 potential binding sites,31 and researchers have reported previously that all these nine miRNAs could serve as growth-suppressive miRNAs.32-38
CircRNAs can act as miRNA sponges, competitively inhibiting transcriptional regulation of miRNA, and in this respect they represent a new type of ceRNA regulator (Fig. 2a). Compared with other ceRNAs, circRNAs are abundant, highly stable in cells and contain more binding sites for miRNA, which allows them to bind or release miRNA more rapidly and stably. However, the specific mechanism of the interaction between circRNA and miRNA requires further study.
Figure 2.
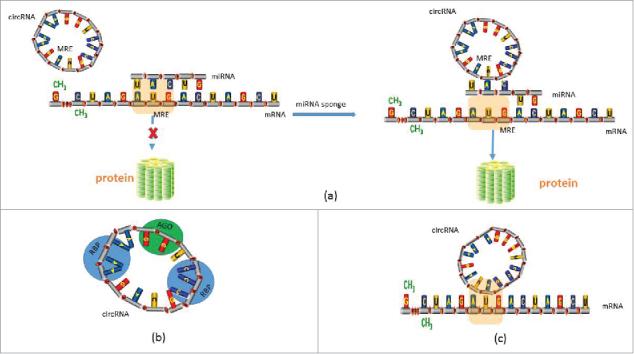
The main function of circRNA. (a) CircRNA acts as miRNA sponge. (b) CircRNA binds with proteins. (c) circRNA interacts with RNA.
Other biofunctions of circRNA
Enuka, et al.39 found that in mammary cells, miRNA seed matches in circRNAs didn't significantly enrich, suggesting that circRNAs may have other biofunctions. Indeed, in addition to acting as a miRNA sponge, circRNA can bind with RNA-binding protein (RBP) (Fig. 2b), or couple with other RNA via complementary base pairing40,41 (Fig. 2c). Zhang et al.22 found that knockdown of ciRNAs could reduce their parent genes' expression, and such ciRNAs are abundant in the nucleus. One such RNA, ci-ankrd52, is a positive controller of PoI II transcription. In addition, circRNA can bind with Argonaute (AGO), influencing the transcription and translation of mRNA12,13 (Fig. 2B). Furthermore, Du et al.42 demonstrated that circRNA circ-Foxo3 could bind to cell cycle proteins CDK2 and p21, which results in the formation of a ternary complex, thereby arresting CDK2 function and blocking progression of the cell cycle. Furthermore, in rabbit reticulocyte lysate, circRNAs are translated into long proteins without involvement of any specific elements such as a cap structure, or poly-A tail for internal ribosome entry via a rolling circle amplification mechanism. This transition system facilitates protein synthesis by cyclisation, using a much simpler RNA as a template.43
The role of circRNA in diseases
CircRNAs play important regulatory roles in diseases through interactions with disease-related miRNAs. Hansen et al.44 found that miR-7 and CDR1as are co-expressed in the murine brain, and could influence midbrain development.12 Ghosal et al.45 found that CDR1as may be associated with Parkinson disease. Moreover, a deficiency in the ciRS-7 (also called CDR1as) sponging effect could up-regulate miR-7 expression,46 which probably results in down-regulating Alzheimer disease-related targets including ubiquitin protein ligase A (UBE2A).47,48 miR-7 is closely coupled to CDR1as in placental mammals, and modifying the miR-7/miR-671/CDR1as axis may play an important role in diseases like cancer.49 Our own work50 provides evidence that a circRNA microarray can be used to screen for differential circRNA expression in cases of HPH (hypoxia-induced pulmonary hypertension) compared with controls, and that circRNAs may be involved in the regulatory mechanisms underlying HPH. Xu et al.51 identified a novel target of the CDR1as/miR-7 pathway, Myrip, which regulates insulin granule secretion, and identified an additional target, pax6, which promotes insulin transcription. These results suggest that the CDR1as/miR-7 pair interact and strongly influence insulin secretion, and they may becoming a new target for therapies aimed at improvement of β cell function in diabetes.
Deep RNA profiling in various mouse tissues was used by You et al.52 to show that among these various tissues, circRNAs were markedly enriched in the brain and that these circRNAs originated from gene loci that encode synapse-related functional proteins. A circRNA (circHomer1_a) which was derived from a linear transcript of the Homer1 gene, could compete with the biogenesis of Homer1b/c mRNA, because its head-to-tail junction uses the splicing donor of intron 5, which is essential for splicing Homer1b/c transcripts, and this competition could maintain the homeostatic synaptic downscaling.52 Filippenkov et al. also showed that circRNAs containing sequences of the gene's 5′ untranslated region (5′UTR) of the human sphingomyelin synthase 1 gene (SGMS1) were found predominantly in different parts of the brain, and regulated the function of the SGMS1 gene in the brain,53 while the SGNS1 gene encodes a transmembrane protein participating in sphingomyelin synthesis, intracellular molecular transport regulation, cholesterol metabolism, cell proliferation, and apoptosis.54-56 CircRNAs are very abundant in the mammalian brain, as well as through neuronal differentiation, especially in synapses, and with their high stability, neuronal termini might use them and they might perform as synaptic tags to maintain a molecular memory.57 Using their own algorithm, Szabo et al.58 revealed a noticeable induction of general, as well as tissue specific circRNAs. They found developmentally induced circRNAs, which are processed by the minor spliceosome, and these minor spliceosome donors spliced into circRNA at un-annotated, instead of annotated exons, suggesting a possibly important developmental role for circRNA in humans.
Cir-ITCH was expressed at low levels in ESCC compared with peritumoral tissue, and may inhibit ESCC by regulating the Wnt pathway.30 Recently, Li et al.26 identified 257 new circRNAs in exosomes from the serum of cancer patients compared with healthy individuals. This shows the significant translational potential of exo-circRNAs (exosome circRNAs) as circulating biomarkers for the diagnosis of cancer. Other researchers also reported that circRNAs has_circ_002059,59 Rev3I, IGSF11, MAML2 and LPP60 may be biomarkers for carcinoma diagnosis or for carcinogenesis. Qu et al.61 explored the expression profile of circRNAs in 4 pancreatic ductal adenocarcinoma (PDAC) samples and paired adjacent normal tissues, indicating that circRNAs may contribute to PDAC initiation and progression.
Wang et al.62 reported that circular RNA100783 could regulate phosphoprotein expression during CD28 related CD8(+) T cell aging, indicating that circular RNA100783 might be a novel biomarker for global immunosenescence. Recent evidence showed that a circRNA, heart-related circRNA (HRCR), can prevent cardiac hypertrophy and heart failure by acting as an endogenous sponge for miR-223.63 Other studies revealed that circRNAs can be biomarkers for the diagnosis and progression of diseases like laryngeal squamous cell cancer,64 and pre-eclampsia,65 which highlights the importance of circRNAs in human diseases.
Methods for the identification of circRNA
Molecular methods
CircRNAs have no 3′ end compared with linear exonic RNAs, leading to slower migration in a gel than the same length linear RNAs, and this difference in migration can be augmented by increasing the degree of gel cross-linking.66 However, circRNAs have shorter total nucleotide sequences which make them migrate further in a gel with low cross-linking. Based on these characteristics, a standard northern blot can be used to identify circRNAs.9,67 CircRNAs can be linearized into predictable, single products following RNase H degradation or by single nicking using hydrolysis.5,68 This allows the identification of circRNAs by 2-dimensional gel or gel trap electrophoresis.44,66,69 Enzymatic methods provide good supporting evidence for circRNA studies. RNase R exonuclease,27 terminator exonuclease13 and tobacco acid phosphatase13 can degrade most linear RNAs, but not circRNAs. Thus, it is possible to enrich circular transcripts following quantitative analysis of data gathered before and after treatment with these 3 enzymes.
Like circRNAs, lariat RNA is also exonuclease insensitive,27 and migrates more slowly than linear RNA on a gel.66 However, circRNA has a backsplice sequence, which allows circRNA to be easily distinguished from lariat RNA. Moreover, lariat RNA can be preferentially depleted by treatment with a debranching enzyme before exonuclease digestion. Thus the two forms of RNA can be easily distinguished from each other.
Genomic methods
Genomic studies use two general approaches to identify exonic circRNAs. One method is to use existing transcript models and select the candidate junctions,8,70 while the other is to identify the joins through matching the reading frames to the genomic sequence.71 circRNAs were first successfully identified in a genomic study as the result of analyzing independent mapping of paired-end reads sequence from opposite ends of a single cDNA fragment.8 The other strategy identified circRNAs following analysis of rRNA-depleted RNA-seq data from nematode and mammalian cells.12 Danan et al.7 and Jeck et al.9 used high-throughput sequencing methods to analyze RNase R insensitive RNAs. Such “CircleSeq” technology allows for more extensive analysis of circRNAs, but requires more RNA input and is highly sensitive to contamination by exonuclease enzyme.
Other methods
Recently, Glažar et al.72 built an online database called “circBase,” which contains data sets of circRNA that were merged and unified. This database provides a platform for circRNA research and can be used to identify known and new circRNAs. CircNet (http://circnet.mbc.nctu.edu.tw/) is another useful database that provides information about new circRNAs, integrated miRNA-target networks, circRNA isoform expression profiles, genomic annotations of circRNA isoforms, and sequences of circRNA isoforms.73 Gao et al.74 presented a novel method called “CircRNA identifier” (CIRI) for the identification as well as the annotation of circRNA from RAN-seq reads. They used this method to identify and validate the intronic /intergenic circRNAs in the human transcriptome. Song et al.75 developed a computational pipeline called “UROBORUS” to indentify circRNAs within total RNA-seq data, which can be accessed freely at http://uroborus.openbioinformatics.org/. The UROBORUS tool can detect lowly expressed circRNAs in total RNA-seq, without RNase R treatment. Zheng et al.76 developed their deepBase v1.0 to deepBase v2.0 (http://biocenter.sysu.edu.cn/deepBase/), providing more comprehensive expression and evolution profiles of long non-coding RNAs (lncRNAs), circRNAs and small RNAs.
Conclusion
The nomenclature for circRNAs is not standardized yet. The same circRNA, has been called different names depending on the perspectives of particular research groups, which leads to inconsistency in the naming of circRNA. For example, “ciRS-7” indicates that the circRNA binds to miR-7, while “CDR1as” indicates that the same circRNA is related to CDR1. Kosik77 recommended that this circRNA should be called circR-1. Thus, the standardization of nomenclature for circRNAs is an important and pressing issue.
With the discovery of increasing numbers of circRNAs and the identification of their biologic characteristics, researchers are displaying increasing interest in circRNAs. CircRNAs are promising new players in the repertoire of regulatory RNAs and are of potential importance second only to miRNAs. Our current knowledge of circRNA is rudimentary, and much effort is needed to unravel the precise mechanism of circRNA biofunction. However, even our initial forays into circRNA research reveal they act as natural miRNA sponges, and this has expanded the horizon of our knowledge with regard to the natural evolution of animals. The canonical concept that RNA acts as an ordinary messenger between DNA and its encoding protein has been undermined by the potential for circRNAs to be drivers of evolution.77 The tissue specificity and stability of circRNAs make them potentially useful biomarkers. In recent years, studies of the structure and characteristics of circRNA show that circRNA are closely associated with certain diseases. CircRNA may play a vital role in diseases through interactions with disease-related miRNAs. Based on these characteristics, circRNAs have great potential to become a new and important addition to our collection of clinical diagnostic markers and are likely to guide new therapeutic strategies for the treatment and prevention of diseases.
Conflict of interest
The authors declare that they have no conflict of interest regarding the publication of this paper.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81460748, Grant No. 31171129, Grant No. 31171129).
References
- 1.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–56. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, et al.. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–87. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–13. doi: 10.1016/0092-8674(91)90244-S. [DOI] [PubMed] [Google Scholar]
- 4.Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–30. doi: 10.1016/0092-8674(93)90279-Y. [DOI] [PubMed] [Google Scholar]
- 6.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–60. [DOI] [PubMed] [Google Scholar]
- 7.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–42. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen YF, Zhang X, Wang Y, Cao FF, Uzan G, Peng B, Zhang DH. Celastrol targets IRAKs to block Toll-like receptor 4-mediated nuclear factor-kappaB activation. J Integr Med. 2016;14:203–8. doi: 10.1016/S2095-4964(16)60257-1. [DOI] [PubMed] [Google Scholar]
- 11.Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- 12.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 13.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 14.Saha SK, Roy S, Khuda-Bukhsh AR. Ultra-highly diluted plant extracts of Hydrastis canadensis and Marsdenia condurango induce epigenetic modifications and alter gene expression profiles in HeLa cells in vitro. J Integr Med. 2015;13:400–11. doi: 10.1016/S2095-4964(15)60201-1. [DOI] [PubMed] [Google Scholar]
- 15.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–9. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–47. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–11. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–24. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 21.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–47. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al.. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–7. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Han J, Lv H, Liu J, Liu R. Computational identification of circular RNAs based on conformational and thermodynamic properties in the flanking introns. Comput Biol Chem. 2016;61:221–5. doi: 10.1016/j.compbiolchem.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–42. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hentze MW, Preiss T. Circular RNAs: Splicing's enigma variations. EMBO J. 2013;32:923–5. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–13. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al.. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, et al.. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–47. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–76. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 34.Tsuruta T, Kozaki K, Uesugi A, Furuta M, Hirasawa A, Imoto I, Susumu N, Aoki D, Inazawa J. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 2011;71:6450–62. doi: 10.1158/0008-5472.CAN-11-0364. [DOI] [PubMed] [Google Scholar]
- 35.Iliopoulos D, Rotem A, Struhl K. Inhibition of miR-193a expression by Max and RXRalpha activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 2011;71:5144–53. doi: 10.1158/0008-5472.CAN-11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueno K, Hirata H, Shahryari V, Chen Y, Zaman MS, Singh K, Tabatabai ZL, Hinoda Y, Dahiya R. Tumour suppressor microRNA-584 directly targets oncogene Rock-1 and decreases invasion ability in human clear cell renal cell carcinoma. Br J Cancer. 2011;104:308–15. doi: 10.1038/sj.bjc.6606028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostling P, Leivonen SK, Aakula A, Kohonen P, Makela R, Hagman Z, Edsjö A, Kangaspeska S, Edgren H, Nicorici D, et al.. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–67. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- 39.Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–83. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilusz JE, Sharp PA. Molecular biology; A circuitous route to noncoding RNA. Science. 2013;340:440–1. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, et al.. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–22. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bingol B, Sheng M. Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron. 2011;69:22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Lonskaya I, Shekoyan AR, Hebron ML, Desforges N, Algarzae NK, Moussa CE. Diminished parkin solubility and co-localization with intraneuronal amyloid-beta are associated with autophagic defects in Alzheimer's disease. J Alzheimers Dis. 2013;33:231–47. doi: 10.3233/JAD-2012-121141. [DOI] [PubMed] [Google Scholar]
- 49.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–12. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 50.Shijin X, Xiantao T, Bing L, Jingyan S, Hua D. Study of circular RNA expression profilie of lungs in the mice with hypoxia-induced pulmonary hypertension. Chin J Lung Dis (Electronic Edition). 2015;8:40–44. [Google Scholar]
- 51.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik, Wang M, Glock C, Quedenau C, et al.. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–10. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Filippenkov IB, Sudarkina OY, Limborska SA, Dergunova LV. Circular RNA of the human sphingomyelin synthase 1 gene: Multiple splice variants, evolutionary conservatism and expression in different tissues. RNA Biol. 2015;12:1030–42. doi: 10.1080/15476286.2015.1076611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guillen N, Navarro MA, Surra JC, Arnal C, Fernandez-Juan M, Cebrián-Pérez JA, Osada J. Cloning, characterization, expression and comparative analysis of pig Golgi membrane sphingomyelin synthase 1. Gene. 2007;388:117–24. doi: 10.1016/j.gene.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Shakor AB, Taniguchi M, Kitatani K, Hashimoto M, Asano S, Hayashi A, Nomura K, Bielawski J, Bielawska A, Watanabe K, et al.. Sphingomyelin synthase 1 generated sphingomyelin plays an important role in transferrin trafficking and cell proliferation. J Biol Chem. 2011;286:36053–62. doi: 10.1074/jbc.M111.228593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan N, Ding T, Dong J, Li Y, Wu M. Sphingomyelin synthase overexpression increases cholesterol accumulation and decreases cholesterol secretion in liver cells. Lipids Health Dis. 2011;10:46.doi: 10.1186/1476-511X-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al.. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 58.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J, et al.. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–6. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 60.Zhang C, Wu H, Wang Y, Zhao Y, Fang X, Chen C, Chen H. Expression patterns of circular RNAs from primary kinase transcripts in the mammary glands of lactating rats. J Breast Cancer. 2015;18:235–41. doi: 10.4048/jbc.2015.18.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qu S, Song W, Yang X, Wang J, Zhang R, Zhang Z, Zhang H, Li H. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data. 2015;5:385–7. doi: 10.1016/j.gdata.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12:17. doi: 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al.. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–11. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 64.Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T, Wang X, Li Q, Tian L, Liu M, et al.. Circular RNA: A novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8:932–9. [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang YG, Yang HL, Long Y, Li WL. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG. 2016;123:2113–18. doi: 10.1111/1471-0528.13897. [DOI] [PubMed] [Google Scholar]
- 66.Tabak HF, Van der Horst G, Smit J, Winter AJ, Mul Y, Groot Koerkamp MJ. Discrimination between RNA circles, interlocked RNA circles and lariats using two-dimensional polyacrylamide gel electrophoresis. Nucleic Acids Res. 1988;16:6597–605. doi: 10.1093/nar/16.14.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hurowitz EH, Brown PO. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 2003;5:R2. doi: 10.1186/gb-2003-5-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schindler CW, Krolewski JJ, Rush MG. Selective trapping of circular double-stranded DNA molecules in solidifying agarose. Plasmid. 1982;7:263–70. doi: 10.1016/0147-619X(82)90007-5. [DOI] [PubMed] [Google Scholar]
- 70.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fonseca NA, Rung J, Brazma A, Marioni JC. Tools for mapping high-throughput sequencing data. Bioinformatics. 2012;28:3169–77. doi: 10.1093/bioinformatics/bts605. [DOI] [PubMed] [Google Scholar]
- 72.Glazar P, Papavasileiou P, Rajewsky N. circBase: A database for circular RNAs. RNA. 2014;20:1666–70. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al.. CircNet: A database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao Y, Wang J, Zhao F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang K, Lu W. deepBase v2.0: Identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44:D196–202. doi: 10.1093/nar/gkv1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kosik KS. Molecular biology: Circles reshape the RNA world. Nature. 2013;495:322–4. doi: 10.1038/nature11956. [DOI] [PubMed] [Google Scholar]