ABSTRACT
Current methods for isolating RNA from budding yeast require lengthy and laborious steps such as freezing and heating with phenol, homogenization with glass beads, or enzymatic digestion of the cell wall. Here, extraction with a solution of formamide and EDTA was adapted to isolate RNA from whole yeast cells through a rapid and easily scalable procedure that does not require mechanical cell lysis, phenol, or enzymes. RNA extracted with formamide-EDTA can be directly loaded on gels for electrophoretic analysis without alcohol precipitation. A simplified protocol for downstream DNase treatment and reverse transcription reaction is also included. The formamide-EDTA extraction of yeast RNA is faster, safer, and more economical than conventional methods, outperforms them in terms of total yield, and greatly increases throughput.
Keywords: Formamide, high-throughput, RNA isolation, RT-PCR
Introduction
Isolation of high-quality RNA from cells requires efficient disruption of the cellular structure with simultaneous inhibition of RNases to minimize RNA degradation. Many advanced RNA extraction reagents developed for this purpose, such as concentrated guanidinium salt-based solutions,1 do not work efficiently in baker's yeast because of the presence of the cell wall resistant to their action. Commonly used methods for isolating RNA in yeast require lengthy and labor-intensive steps such as homogenization of cells with glass beads or subjecting the cells to cycles of heating and freezing in the presence of acidic phenol and SDS.2,3 Neither method can be easily adapted for high-throughput analysis. Alternatively, the yeast cell wall can be removed by enzymatic digestion, followed by cell lysis and RNA purification.4,5 The principal disadvantage of this approach is the necessity of prolonged incubation of live cells in buffer with high osmolality, which can lead to significant changes in gene expression.6
In this communication, we present a simple protocol for isolation of RNA from Saccharomyces cerevisiae by heating cells in a mixture of 98% formamide and 10 mM EDTA (FAE solution). A similar use of formamide was described in an RNA isolation method termed RNAsnap™.7 The RNAsnap™ protocol, however, required homogenization of yeast cells with glass beads, thus offering little advantage over other existing yeast methods. Unlike previously described procedures, our protocol does not require mechanical or enzymatic cell lysis, offering easy scalability and the possibility of processing multiple samples in screening applications at a low cost. Our results show that extraction with FAE efficiently recovers different classes of cellular RNA with the yield exceeding that of other commonly used methods. We anticipate that isolation of RNA from yeast using FAE would benefit a broad range of studies in this important model eukaryotic organism.
Results and discussion
Extraction of yeast RNA by a combination of formamide, EDTA and temperature
Formamide is a solvent that denatures nucleic acids and proteins and is commonly used to denature RNA before gel electrophoresis.8 Formamide is also a convenient storage medium for RNA that effectively protects it from degradation by RNases.9 While attempting to use formamide for denaturation of yeast RNPs, we unexpectedly found that incubation of intact yeast cells with formamide released cellular RNA into solution, with the amount of the extracted RNA increasing at elevated temperatures (Fig. 1A). Additional tests indicated that RNA yield could be further increased by addition of chelating reagents such as EDTA or citrate-containing buffer, whereas altering ionic strength, pH or thiol reducing reagents did not have an appreciable effect (Fig. 1A).
Figure 1.
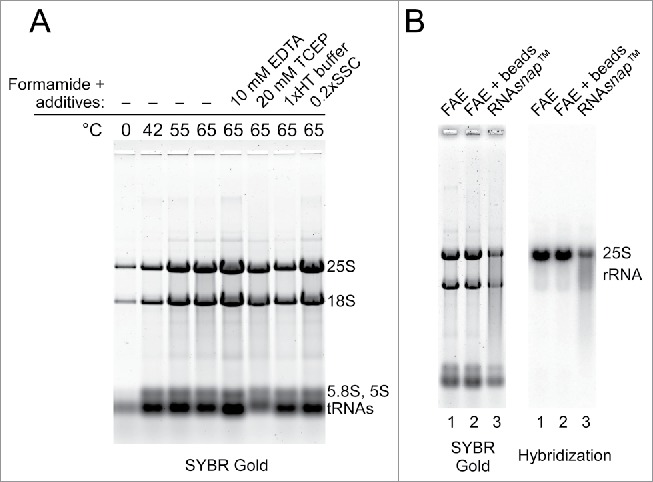
Efficient extraction of yeast RNA by heating yeast cells in the presence of formamide and EDTA. (A) Effects of temperature and additives on the efficiency of RNA extraction by formamide. A total of 2.5 OD600 units of BY4741 cells (culture at OD600 = 5.0) was resuspended in formamide (40 μl per 1.0 OD600 unit of cells), incubated for 5 min at the indicated temperature, vortexed for 10 s and centrifuged for 1 min at 12,500 rpm in a microcentrifuge. Equal volumes of each supernatant were separated on a 1% agarose gel and RNA was stained with SYBR Gold. Incubations at 65°C were either in pure formamide or in formamide with the addition of 10 mM EDTA, 20 mM TCEP, 1 × HT electrophoresis buffer, or 0.2 × SSC. (B) FAE extraction preserves RNA integrity. Electrophoretic analysis of 2.5 μg RNA prepared from the same culture by one-step FAE extraction (lane 1), FAE extraction with additional vortexing of cells for 8 min using 0.5 mm glass beads (lane 2) and RNAsnap™ (lane 3). RNA was transferred to a membrane and analyzed by northern hybridization with a probe that detects 25S rRNA.
Recently, Stead and coworkers developed a method for extraction of RNA from Gram-negative bacteria, in which cells were incubated for 7 min at 95°C in a solution consisting of 18 mM EDTA, 0.025% SDS, 1% 2-mercaptoethanol, and 95% formamide.7 These authors also reported that their method, which they called RNAsnap™, worked in yeast but required cells to be vortexed for 10 min with zirconia beads.7 Our observations (Fig. 1A) were consistent with the ability of formamide-containing solutions to extract RNA from cells, as reported by Stead et al. One important practical difference was that no cell homogenization was used during our extractions, and there were additional differences in temperature and buffer composition. To examine how significant these differences were for the quality and yield of the extracted RNA, we isolated RNA from the same yeast culture using formamide–EDTA at 65°C either with or without mechanical lysis and by using the RNAsnap™ protocol. The extracted RNA was then resolved on a formaldehyde-agarose gel, followed by staining with SYBR Gold and northern hybridization to assess integrity of the large rRNA species (Fig. 1B). This analysis showed that both formamide–EDTA and RNAsnap™ solutions effectively released RNA from yeast cells, but significant degradation of RNA was observed in a sample processed according to the RNAsnap™ protocol (Fig. 1B). It thus appears that the higher incubation temperature and additional components in the RNAsnap™ extraction solution (which was originally designed for bacterial cells) are neither necessary nor optimal for yeast RNA extraction. Interestingly, processing of the yeast cell suspension with glass beads did not improve RNA yield during formamide–EDTA extraction (Fig. 1B, compare lanes 1 and 2), suggesting that the combination of formamide, EDTA and elevated temperature was sufficient to drive RNA release from cells. We also found only minor differences in the amount of RNA released by performing extraction at temperatures between 65° and 75°C, while temperatures >75°C led to increased RNA degradation (not shown). Based on these observations, we used 98% formamide – 10 mM EDTA (FAE) at 70°C throughout the subsequent work.
Comparison of FAE-based and hot phenol extraction
In traditional methods of isolating RNA from yeast, significant hands-on time is required for breaking up the cells, removal of proteins with organic reagents and alcohol precipitation of the RNA. Extraction of RNA with the FAE solution does not involve cumbersome mechanical cell lysis steps or toxic organic reagents, has few pipetting steps and yields RNA ready for gel analysis in less than 20 min (Fig. 2A). In addition to allowing quick processing of multiple samples, the FAE extraction procedure was found to scale up easily while maintaining RNA yield and purity (Table 1).
Figure 2.

Comparison of the one-step FAE and hot phenol extraction methods. (A) Outline of a small-scale RNA isolation procedure using FAE extraction. For routine RNA analysis from yeast cultures, a convenient microcentrifuge tube format illustrated here is to start with 1 OD600 unit of cells and perform lysis in 30–40 μL of FAE. The typical yield we obtain is ∼22–30 μg total RNA when using mid-log phase BY4741 cells. (B) Spectrophotometric measurements of the yield and purity of RNA isolated from a mid-log phase culture. FAE-extracted sample was analyzed without additional purification. (C) Gel analysis of 3.5 μg RNA prepared using hot phenol (lane 1), one-step FAE extraction (lane 2), and FAE extraction followed by additional purification with RNazol RT (lane 3). Individual RNA species were detected by northern hybridizations. (D) Recovery of different RNAs using FAE and hot phenol extraction. Total RNA was extracted by the 2 methods from the same log-phase BY4741 culture, separated by gel electrophoresis, and transferred to a membrane. The membrane was stained with methylene blue and hybridized with radiolabeled probes to detect the indicated mRNAs, mitochondrial rRNAs, and tRNAs. (E) Large rRNA precursors can be efficiently extracted with FAE. RNA was isolated from cells in log-phase at 30°C or cells that were heat shocked at 39°C for 15 min to suppress productive ribosome biogenesis.12 2 μg total RNA (as estimated by A260 absorbance) was loaded in each lane. The membrane was hybridized with oligonucleotide probe 00612 to detect pre-rRNAs.
Table 1.
Total RNA yield obtained in the small- and large-scale FAE procedures.
| Culture volume, ml | Total OD600 | Volume of FAE, μl | Estimated RNA yield, μg per OD600 of culture | A260/280 |
|---|---|---|---|---|
| 1 | 0.83 | 40 | 24.6 ± 4.5 | 2.14 ± 0.01 |
| 25 | 17.5 | 700 | 27.6 ± 0.6 | 2.13 ± 0.02 |
RNA was isolated using FAE extraction from a mid-log culture of BY4741 cells. Data indicate mean values ± range in 3 (1 ml) or 2 (25 ml) replicate preparations.
To better characterize RNA isolated with FAE, we compared it with RNA prepared by the widely used hot phenol method of yeast RNA isolation.2 Starting with a BY4741 yeast culture in log phase, we obtained an approximately 3-fold higher yield of material absorbing at 260 nm with FAE as compared with the standard hot phenol protocol (Fig. 2B). Gel analysis of samples prepared by one-step FAE extraction showed a slight shift in the migration of RNA bands compared with the RNA isolated with hot phenol (Fig. 2C, D). This change in electrophoretic mobility is likely due to compounds coextracted from cells because it disappears when samples are additionally purified (Fig. 2C, compare lanes 2 and 3). The mobility shift does not preclude good electrophoretic separation of RNA, with sharp bands observed on a gel for a wide range of RNA species (Fig. 2C-E). FAE- and hot phenol-extracted samples loaded on a gel based on their absorbance at 260 nm showed comparable amounts of different RNA types including small RNAs (tRNA, snoRNAs), mRNAs, and rRNAs from the cytoplasm and mitochondria (Fig. 2C, D). FAE more efficiently recovered mitochondrial rRNAs than hot phenol, while RPL3 mRNA was more efficiently extracted with hot phenol. We also observed comparable recovery of very large (6–7 kb) 35S and 32S pre-rRNA species in the FAE and hot phenol procedures, although a somewhat better yield was obtained using hot phenol for another large precursor, 27S pre-rRNA (Fig. 2E). Thus, FAE extracts more RNA from cells in total than hot phenol, but some RNA-specific variations in extraction efficiencies can be observed in both methods.
Use of FAE-extracted RNA in downstream enzymatic applications
To use RNA in enzymatic reactions and other situations when highly pure RNA is required, FAE-extracted samples can be diluted with water followed by conventional purification approaches such as alcohol precipitation, phenol/chloroform extraction or purification with commercial reagents such as RNAzol RT (see Materials and Methods). We were also interested in whether FAE-extracted samples can be used directly without additional purification in RT-PCR. In the past, we successfully used purified RNA dissolved in formamide for RT-PCR analysis by diluting formamide to concentrations tolerated by the enzymes. However, one concern was that crude FAE-extracted samples might be contaminated with RNases potentially coextracted with RNA from cells. Such nucleases may be inactive in formamide,9 but become reactivated in aqueous solutions. RNA might be vulnerable, for example, when it is incubated at 37°C in buffers containing Mg2+ during DNase treatment before cDNA synthesis, a typical first step in RT-PCR. To test RNA stability under these conditions, we compared an unpurified FAE-extracted RNA sample with a sample that was thoroughly purified using phenol/chloroform to remove potential RNase contaminants (Fig. 3A). The results of the gel and hybridization analysis showed no visible difference between RNA in these 2 samples, indicating lack of significant nuclease activities present in the FAE extract.
Figure 3.
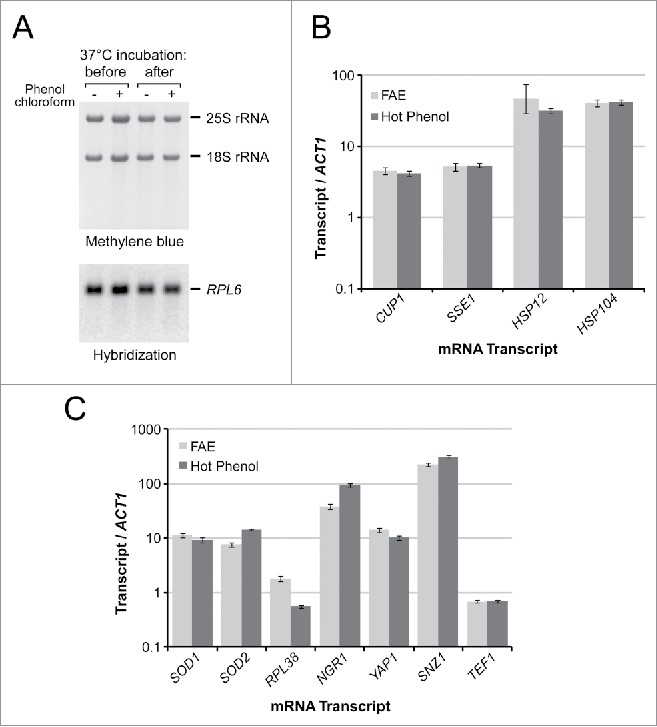
FAE-extracted RNA in downstream enzymatic applications. (A) Lack of degradation of FAE-extracted RNA after a 15-min incubation in DNase buffer at 37°C. RNA was prepared using one-step FAE extraction or additionally purified with phenol-chloroform. After the incubation, RNA was precipitated, separated on an agarose gel, and transferred to a membrane for hybridization. (B) Quantitative RT-PCR analysis of induction of heat shock genes relative to normal growth conditions (30°C log phase) using RNA isolated by the FAE and hot phenol extraction. Data represent mean and SD values in technical triplicates. (C) Quantitative RT-PCR analysis of gene expression in stationary phase relative to log phase cultures. Samples were analyzed in the same way as in panel B.
As a test case for RT-PCR-based gene expression analysis, we exposed log phase BY4741 cells to 39°C for 15 min to induce a heat shock response. RNA from these cells and control cells maintained at 30°C was extracted using single-step FAE extraction and conventional hot phenol purification. The relative increase in transcripts associated with the heat shock response was assessed using ACT1 as an internal reference transcript. The quantitative RT-PCR data (Fig. 3B) demonstrated highly concordant increases in levels of the heat shock-inducible CUP1, SSE1, HSP104 and HSP12 mRNAs in samples prepared by the FAE and hot phenol methods. For a second test, we used stationary phase cells, which have a thickened cell wall and high polysaccharide content, making cell lysis less efficient as well as lowering the purity of cell extracts. To compare the efficiency of FAE and hot phenol extraction in stationary cultures, we grew BY4741 cells for 7 d and examined changes in expression of several target genes relative to exponentially growing cultures. The RT-PCR results obtained with both extraction techniques demonstrated comparable increases in relative levels of the SOD1, SOD2, NGR1, YAP1 and SNZ1 transcripts, known to be induced in stationary phase cells, while genes involved in translation (RPL38, TEF1) did not show a similar increase relative to ACT1 mRNA (Fig. 3C). Based on these results, the direct RT-PCR protocol using unpurified FAE-extracted RNA appears to yield results comparable with the standard RNA purification workflow, while reducing time and labor involved in sample preparation.
In summary, hot formamide extraction of yeast RNA with the FAE solution is simple, uses low-cost reagents, does not require mechanical lysis of cells, and efficiently extracts a diverse range of RNAs. Using this method can facilitate RNA analysis in many types of yeast studies. Our observations also support the broad applicability of cellular RNA extraction with formamide-based reagents, as initially proposed by Stead et al,7 and suggest that exploring this technology may lead to effective RNA isolation approaches in other species.
Materials and methods
Reagents
The FAE solution consists of 98% formamide and 10 mM EDTA, prepared by mixing high-quality formamide (e.g., Sigma F9037) and RNA-grade 0.5M EDTA, pH 8.0. The FAE solution can be prepared in advance and stored in aliquots at −80°C or −20°C for several months.
Harvesting cells
BY4741 cells were grown in liquid YPD or synthetic media at 30°C. Before collecting cells, OD600 of cultures was measured with an Eppendorf Biophotometer. OD600 units of cell biomass, used to determine the amount of the FAE solution during cell lysis, were defined as OD600 × volume in ml (for example, precipitating 0.5 ml of culture with OD600 = 2.0 makes 1 OD600 unit). Cells were harvested by centrifugation for 30 s at 15,000 rpm in 1.5-ml microcentrifuge tubes using a tabletop microcentrifuge. After the cells were pelleted, medium was removed by pipetting or aspiration. Attention should be paid at this step to ensure complete removal of medium before FAE addition. This can be done by quick recentrifugation of the pelleted cells and careful aspiration of any residual medium from the top of the pellet. For larger culture volumes (5–25 ml), we pelleted cells first in 50-ml centrifuge tubes, resuspended pellets in 1 ml water, transferred the cells into 1.5-ml microfuge tubes and centrifuged them twice to remove liquid completely as described above. An additional wash of cell pellets with water may also be needed in experiments that involve treatments with high-salt media or any substances that can alter electrophoretic mobility of RNA if samples are loaded directly on gels without purification.
Using frozen cells
Cell pellets can be frozen and kept at −80°C until lysis. To minimize RNA degradation, freezing should be done quickly (e.g., in racks prechilled at −80°C, using alcohol-dry ice bath, or in liquid nitrogen). For the same reason, frozen cells should not be allowed to thaw before being mixed with FAE.
FAE extraction of RNA
Cell pellets in 1.5-ml microcentrifuge tubes prepared as described above (fresh or frozen) are resuspended in FAE by pipetting or brief vortexing. For optimal results, a minimum of 30–40 μl FAE per 1.0 OD600 unit of cell culture is recommended. The cell suspension in FAE is heated at 70°C for 10 min to extract RNA, briefly vortexed, and centrifuged at 12,500–15,000 rpm (14,000–21,000 × g) in a tabletop microcentrifuge at room temperature for 1–2 min. The FAE supernatant containing RNA is then transferred to a fresh tube. To avoid carrying over cells, we typically leave 5–10 μl supernatant on top of the cell pellet. The RNA in FAE can be used directly for gel analysis or optionally purified if desired. RNA in FAE can be stored at −20°C or −80°C for at least several months.
Purification of RNA from FAE extracts
RNA can be precipitated from FAE extracts by first diluting them 5–10-fold with RNase-free water, followed by optional phenol-chloroform extraction and then adding 1/10 volume 3M sodium acetate (pH 5) and one volume of isopropanol. To obtain high-purity RNA using RNAzol RT (Molecular Research Center, Inc.), we mixed FAE extracts with RNAzol RT in a 1:3 to 1:7 ratio, added 0.4 volume of water relative to the combined volume of FAE and RNAzol RT and followed the manufacturer's protocol thereafter. The adjustment in the amount of water compared with the original protocol was necessary to ensure proper phase separation when the RNAzol reagent is mixed directly with formamide.
RT-PCR using FAE-extracted RNA
A total of 1.5 μg of RNA was treated with 2 units of DNase I (Thermo Fisher) for 15 min at 37°C in a 100 μl reaction mixture supplemented with the manufacturer's buffer. The final formamide concentration in this reaction coming from FAE should not exceed 5%. RNA was purified by phenol/chloroform extraction and precipitated with isopropanol. RNA was next annealed with 0.4 μM oligo(dT)18 at 65°C for 5 min and reverse transcribed using the RevertAid enzyme (Thermo Fisher) for 1.5 h at 42°C. Reaction products were diluted with water and 1/30 was amplified by PCR with the SYBR FAST Universal qPCR Master Mix (Kapa Biosystems). All samples were analyzed in triplicate, using ACT1 as an internal control.
Other methods
Hot phenol RNA extraction was done according to ref. 2 For RNAsnap™, we followed the protocol described in ref. 7 Electrophoretic analysis of RNA was performed using 1% agarose–0.4M formaldehyde gels containing HEPES-triethanolamine (HT) buffer.10 Gels were stained with SYBR Gold (Thermo Fisher) and scanned using a Typhoon 9410 laser imager in a fluorescence mode. Northern blotting analysis was done using radiolabeled oligonucleotide probes.11 qPCR primers were designed using Primer Express v3.0 (Thermo Fisher). Sequences of primers and probes are available upon request.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We wish to thank members of the Pestov and Shcherbik laboratories for their input.
Funding
This work was supported by the NIH grants GM114308 to N.S. and GM074091 to D.G.P.
References
- 1.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat Protoc 2006; 1:581-5; PMID:17406285; https://doi.org/ 10.1038/nprot.2006.83 [DOI] [PubMed] [Google Scholar]
- 2.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 1990; 18:3091-2; PMID:2190191; https://doi.org/ 10.1093/nar/18.10.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collart MA, Oliviero S. Preparation of yeast RNA. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, . Current protocols in molecular biology. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2001; PMID:18265096; https://doi.org/ 10.1002/0471142727.mb1312s23 [DOI] [PubMed] [Google Scholar]
- 4.Achilles J, Stahl F, Harms H, Müller S. Isolation of intact RNA from cytometrically sorted Saccharomyces cerevisiae for the analysis of intrapopulation diversity of gene expression. Nat Protoc 2007; 2:2203-11; PMID:17853877; https://doi.org/ 10.1038/nprot.2007.322 [DOI] [PubMed] [Google Scholar]
- 5.Klassen R, Fricke J, Pfeiffer A, Meinhardt F. A modified DNA isolation protocol for obtaining pure RT-PCR grade RNA. Biotechnol Lett 2008; 30:1041-4; PMID:18246302; https://doi.org/ 10.1007/s10529-008-9648-y [DOI] [PubMed] [Google Scholar]
- 6.Suzuki T, Iwahashi Y. RNA preparation of Saccharomyces cerevisiae using the digestion method may give misleading results. Appl Biochem Biotechnol 2013; 169:1620-32; PMID:23325148; https://doi.org/ 10.1007/s12010-012-0051-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stead MB, Agrawal A, Bowden KE, Nasir R, Mohanty BK, Meagher RB, Kushner SR. RNAsnap: A rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res 2012; 40:e156; PMID:22821568; https://doi.org/ 10.1093/nar/gks680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehrach H, Diamond D, Wozney JM, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry 1977; 16:4743-51; PMID:911786; https://doi.org/ 10.1021/bi00640a033 [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P. Solubilization in formamide protects RNA from degradation. Nucleic Acids Res 1992; 20:3791-2; PMID:1379361; https://doi.org/ 10.1093/nar/20.14.3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansour FH, Pestov DG. Separation of long RNA by agarose-formaldehyde gel electrophoresis. Anal Biochem 2013; 441:18-20; PMID:23800830; https://doi.org/ 10.1016/j.ab.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, Pestov DG. Quantitative northern blot analysis of mammalian rRNA processing. Methods Mol Biol 2016; 1455:147-57; PMID:27576717; https://doi.org/ 10.1007/978-1-4939-3792-9_12 [DOI] [PubMed] [Google Scholar]
- 12.Kos-Braun IC, Jung I, Koš M. Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. PLoS Biol 2017; 15:e2000245; PMID:28282370; https://doi.org/ 10.1371/journal.pbio.2000245 [DOI] [PMC free article] [PubMed] [Google Scholar]


