Abstract
The ribosome is one of life’s most ancient molecular machines that has historically been viewed as a backstage participant in gene regulation, translating the genetic code across all kingdoms of life in a rote-like fashion. However, recent studies suggest that intrinsic components of the ribosome can be regulated and diversified as a means to intricately control the expression of the cellular proteome. In this review, we discuss advances in the characterization of ribosome post-translational modifications (PTMs) from past to present. We specifically focus on emerging examples of ribosome phosphorylation and ubiquitylation, which are beginning to showcase that PTMs of the ribosome are versatile, may have functional consequences for translational control, and are intimately linked to human disease. We further highlight the key questions that remain to be addressed to gain a more complete picture of the array of ribosome PTMs and the upstream enzymes that control them, which may endow ribosomes with greater regulatory potential in gene regulation and control of cellular homeostasis.
Within the central dogma of molecular biology, that describes the linear flow of information from DNA to RNA to protein, translational control is considered the final step. The ribosome, a multi-component assembly of RNA and protein, is responsible for accurately decoding messenger RNAs (mRNAs) and converting their sequences into proteins. Since the first electron microscopy observation of the ribosome as “a particular component of small dimensions and high density” in 1955 [1], decades of research have led to the discovery of multiple mechanisms that regulate translation. However, most of these investigations have mainly focused on additional translation factors (e.g. translation initiation and elongation factors) rather than the ribosome itself [2,3]. In all organisms, Ribosomal Proteins (RPs) and ribosomal RNAs (rRNAs) make up the small and large ribosomal subunits. The space between the two subunits harbors the catalytic activity of the ribosome. Although the majority of the RNA and protein components, including the peptidyl transferase center that catalyzes peptide bond formation, are well-conserved throughout all kingdoms of life from bacteria to humans, ribosomes have undergone structural and compositional changes throughout evolution that reflect differences in the translational mechanisms utilized and the complexity of the proteomes synthesized [4,5].
Observations that ribosomes within a single species may differ in composition have been reported since the 1970s, primarily from the bacterial response to antibiotics or changing growth conditions [6,7]. Despite these intriguing observations, ribosomes have traditionally been considered housekeeping structures, invariable in composition within the same species. Only recently has this perception been challenged by studies demonstrating that the absence or presence of specific RPs or rRNA segments actually has functional significance and can tune the ribosome to preferentially translate specific mRNAs. Under stress, bacteria express a site-specific endonuclease that cleaves both mRNAs, which renders them leaderless, as well as rRNAs, to form specialized ribosomes that can only translate these cleaved mRNA species [8]. In mammalian cells, a key example of ribosome heterogeneity is Rpl38/eL38, a core RP, which is differentially expressed in specific regions of the developing mouse embryo, such as the somites that give rise to the mammalian body axis. Reduction of Rpl38/eL38 in a mouse model revealed that it does not affect global translational regulation but is selectively required for the translation of specific Hox genes that control patterning of the mammalian body plan through cis-regulatory elements residing in their 5′ untranslated regions (UTRs) [9,10•]. Emerging studies that highlight the functional significance of different RPs serving specific roles are reviewed elsewhere [11,12] and they suggest that the ribosome is not merely a static machine, but rather a dynamic one whose intrinsic components can be regulated and diversified to promote intricate regulation of translation.
It is tempting to hypothesize that an additional layer of dynamic regulation to ribosome activity may be achieved by RP PTMs such as phosphorylation and ubiquitylation. PTMs are covalent modifications that can change the stability, subcellular localization, and/or interaction partners of the modified substrate and can regulate such properties within developmental and physiological timescales. Initial proteomic studies of ribosomes crudely isolated through centrifugation methods attempted to identify PTMs via database searches of either intact RPs or digested RP-peptides [13–15]. However, PTM assignment based on mass difference, such as acetylation or methylation, is not comprehensive and is prone to misinterpretation as specific masses could result from a combination of multiple PTMs or could be due to artifacts arising from sample preparation methods. Recent advances in PTM enrichment methods from digested peptides, combined with high-resolution quantitative mass spectrometry (MS), permit systems-level PTM mapping for phosphorylation and ubiquitylation, whereby the conjugation sites of the modification can also be identified with higher confidence [16,17]. Interestingly, many of these near-comprehensive static PTM maps or dynamic PTM snapshots in response to different perturbations contain RPs as candidates [18–22,23••]. Although the presence of diverse PTMs increases the combinatorial diversity of ribosomes, analogously to the histone PTMs that make up the histone code, our understanding of the mechanisms that underlie ribosomal PTMs and their potential functional roles in regulating translational output is in its infancy. In this review, we will summarize recent findings, concentrating on examples of phosphorylation and ubiquitylation of RPs identified by both focused, case-by-case studies and by unbiased, large-scale proteomic studies that enrich for a specific PTM.
Phosphorylated ribosomes in sickness and in health
The first inducible PTM identified at the ribosome was the phosphorylation of mammalian Rps6/eS6, which was discovered almost 40 years ago by analyzing ribosomes from regenerating livers, utilizing radioactive phosphate incorporation and two dimensional (2D) gel electrophoresis [24]. Phosphorylation events on Rps6/eS6 are mapped to five sites that can all be catalyzed by S6K1/S6K2 kinases. RSK, PKA, and CK1 kinases can also phosphorylate distinct sites of Rps6/eS6, whereas all the phosphorylation can be reversed by phosphatase PP1 (Figure 1a) [25]. Numerous studies have shown that Rps6/eS6 phosphorylation occurs downstream of multiple external stimuli (e.g. growth factors), which are transduced via the PI3K and mTOR pathways. As a result, Rps6/eS6 phosphorylation is frequently used as a readout for mTORC1 activation [25]. Moreover, in mice physiological or pharmacological stimuli that activate neurons, also lead to an increase in Rps6/eS6 phosphorylation [26,27]. This observation led to the development of phosphorylated Rps6/eS6 as a marker of activated neurons and antibodies that specifically recognize this modification enabled the enrichment and isolation of ribosomes and associated mRNAs translated in response to neuronal activation [27]. A knock-in mouse model in which all five Rps6/eS6 phosphorylation sites were mutated, along with mouse models deficient in S6K1/S6K2 kinases, paved the way for a better understanding of the role of Rps6/eS6 phosphorylation. Surprisingly, Rps6/eS6 phosphorylation-deficient mice are viable and only display subtle, tissue-specific phenotypes, for example, smaller pancreatic beta cells accompanied by impaired glucose homeostasis [28], and smaller myoblasts with decreased muscle mass [29]. Moreover, studies using Rps6/eS6 phosphorylation-deficient mice in different cancer mouse models suggested a role of Rps6/eS6 phosphorylation in the initiation of pancreatic cancer, but found it was dispensable for AKT-mediated thymic lymphomas [30–32]. Therefore, although the regulatory inputs leading to Rps6/eS6 phosphorylation have been extensively studied and Rps6/eS6 phosphorylation has been successfully used as a readout of these inputs, the physiological roles of this modification Rps6/eS6 are not yet entirely known.
Figure 1. Ribosomes are remodeled: examples of RP phosphorylation and ubiquitylation events.

(a) Phosphorylation (represented by −P) of Rps6 has been observed downstream of multiple growth signals and pharmacological stimuli. The functional role of Rps6 phosphorylation has been elucidated in mouse models lacking Rps6 phosphorylation sites. Phosphorylation of Rpl13a occurs as part of the innate immune response and leads to the separation of this RP from assembled ribosomes to carry out a transcript-specific translation program as part of the GAIT complex. Rpl13a phosphorylation has been identified in human monocytic cells. Phosphorylation of Rps15 is critical for the neurotoxicity phenotype seen in Parkinson’s disease models in human neurons and Drosophila. Pencil symbol denotes the enzymes responsible for attaching the PTM to the RPs, whereas scissors symbol denotes the enzymes reversing the PTM. Pencil or scissors symbols in green indicates that the specific enzymes are known for this particular PTM; whereas pencil or scissors in white indicates the enzymes remain unknown.
(b) Ubiquitin modifications (represented by magenta circles) at the ribosome can lead to diverse outcomes: non-degradative K63 linked ubiquitin chains or mono-ubiquitin signals can modify RPs in response to stress conditions. For example, in response to oxidative stress in yeast, RPs are modified by K63 linkages. Upon ER stress, cytoplasmic ribosomes, but not ER-ribosomes are decorated with mono-ubiquitin on multiple different RPs as shown in a tissue-culture based system. Ubiquitin signaling at the ribosome as part of the mRNA-protein quality control pathway is versatile and can modify nascent chains with degradative K48-linked ubiquitin chains while also modifying RPs with mono-ubiquitin signals. Ubiquitin signaling can also mark ribosome biogenesis factors and may lead to ribosome functional diversity during neural crest cell differentiation.
A second prominent example of RP phosphorylation comes from human monocytic cells and the interferon (IFN)-γ-mediated innate immune response (Figure 1a). Upon IFN-γ incubation, the DAPK1-ZIPK kinase-signaling cascade results in the phosphorylation of Rpl13a/uL13 at stoichiometric levels at a single serine. This process results in the release of Rpl13a/uL13 from the assembled large ribosomal subunit [33]. Released phosphorylated Rpl13a/uL13 has an extra-ribosomal function as an essential component of the interferon-γ-activated inhibitor of translation (GAIT) complex, which binds to a defined element in the 3′ UTRs of a select group of inflammation-related mRNAs to inhibit their translation [34]. Phosphorylation of Rpl13a/uL13 is the rate-limiting step in GAIT complex-mediated translational repression. After the complex is recruited to target mRNAs, phosphorylated Rpl13a/uL13 interacts specifically with the initiation factor eIF4G and suppresses translation by blocking the recruitment of the small ribosomal subunit [33,35]. Interestingly, DAPK1-ZIPK kinases themselves are translational targets of phosphorylated Rpl13a/uL13, thereby forming a negative feedback loop [33].
A more recent study showed how phosphorylation of an RP can be critical for the etiology of the neurodegenerative Parkinson’s disease (PD) (Figure 1a) [36]. Rps11/uS17, Rps15/uS19, and Rps27/eS27 were found to be the major interactors of LRRK2, a kinase frequently mutated in familial and sporadic PD. Intriguingly, 19 of 67 RPs tested can be phosphorylated directly by LRRK2 [37••]. For Rps15/uS19, the mutation of a single phosphorylation site rescues the neurotoxicity caused by the LRRK2 mutation in Drosophila, and phospho-mimetic Rps15/uS19 is neurotoxic, demonstrating that Rps15/uS19 is a critical pathogenic LRRK2 substrate. Consistently, Rps 15/uS19 is found to be hyperphosphorylated in postmortem brain lysates from PD patients that inherited the LRRK2 mutation. LRRK2 kinase has been suggested to have other substrates pertinent for endocytosis, cytoskeleton remodeling, and autophagy [38]. LRRK2 has also been previously suggested to enhance translation by phosphorylating 4E-BP1, which in return promotes the interaction of 4E-BP1 and AGO2 and therefore disrupts microRNA-dependent translational repression of certain a few mRNAs [39]. However, this finding did not pinpoint whether 4E-BP1 phosphorylation is causative for neurodegeneration. To gain mechanistic insights into the neurotoxicity caused by phosphorylated Rps15/uS19, future studies should aim to understand whether different subcellular pools of Rps15/uS19 are phosphorylated and whether this modification results in alterations of ribosome biogenesis, since Rps15/uS19 is known to be important for nuclear export of the pre-40S complex [40]. Moreover, the phospho-deficient form of Rps15/uS19 appears to attenuate the global translational increase observed with LRRK2 mutations as measured by radioactive methionine incorporation, although the molecular mechanisms leading to this remain to be determined [37••]. It is plausible that the phosphorylated Rps15/uS19 may be important for the differential translation of specific mRNAs, which when altered may underlie neurodegeneration. Because such information is not available by analyzing global protein synthesis, applying techniques such as ribosome profiling, which measures ribosome occupancy and translation efficiency of each transcript genome-wide [41], to phospho-mutant Rps15/uS19 cells could provide a high-resolution view of the translational impact of this PTM.
While the very first study that identified the Rps6/eS6 phosphorylation in 1974 concluded that “only a single RP is phosphorylated” in mouse livers [24], this is likely due to the sub-stoichiometric levels of the phosphorylated forms of other RPs which were below the detection limit of 2D gels. Recent technological advancement in phosphoproteomic methods that combine strong cation exchange chromatography with phospho-peptide enrichment methods has increased the dynamic range and sensitivity for the identification of phosphoproteins [16]. Using these techniques, many groups have investigated phosphorylation sites proteome-wide, demonstrating changes during stem cell differentiation, across different mouse tissues, and during difference stages of the mammalian cell cycle [18–20]. Although these studies did not concentrate on ribosomes particularly, a close examination of these data suggests that phosphorylation of RPs may change between different physiological conditions. For instance, RPs are phosphorylated throughout the mammalian cell cycle and contain the consensus kinase recognition site for CDK1 [19,42], a kinase that orchestrates essential steps of mitosis [42,43]. Additional evidence lies in work exploring CDK1 activity, which was found to phosphorylate Rps3/uS3 in vitro [44]. Interestingly, ribosome profiling across cell cycle stages revealed that specific transcripts are preferentially translated during different stages of the cell cycle, even though translation is globally repressed [45,46]. Another PTM, O-GlcNAcylation, can compete with phosphorylation for modifying substrate serines and threonines, and the balance of these two marks was suggested to work in concert to control cell division [47]. While O-GlcNAc profiling experiments found multiple RPs to be enriched [48], it remains to be formally investigated as to whether O-GlcNAcylation and phosphorylation of RPs can cross-talk and may, either individually or in combination, confer specificity to the ribosome in the translation of cell cycle-regulated mRNAs.
Ubiquitin comes in various sizes and shapes at the ribosome
A second prevalent PTM at the ribosome is ubiquitylation, whereby a ubiquitin peptide consisting of 76 amino acids is covalently attached to a lysine residue in a substrate protein. Ubiquitin itself contains seven lysines, which, in addition to its N-terminus, can serve as sites for further ubiquitin chain assembly in vivo [49]. The most abundant ubiquitin chain is formed via K48 linkages that result in the proteasome-dependent degradation of substrate proteins. K63 linked chains, on the other hand, do not trigger degradation but rather allow for the formation of signaling complexes [50]. A 2D gel approach using wild type (WT) or K63 linkage mutant strains revealed that non-degradative K63 linkages modify Rpl28/uL15 in yeast and that ubiquitylation of Rpl28/uL15 was low during G1 and higher during the S-G2 phases of the cell cycle [51]. Although ubiquitylation of Rpl28/uL15 is conserved in mammalian cells, the ubiquitin enzymes responsible for the modification or the direct functional roles of Rpl28/uL15 ubiquitylation remain unknown.
A more recent study in yeast, leveraging quantitative MS techniques, performed similar comparative experiments between WT and K63 linkage mutant strains in response to hydrogen peroxide stress (Figure 1b). Interestingly, nearly 25% of these targets (~30 proteins) were RPs and translation elongation factors [52••]. In this system, by testing candidate enzymes that are known to form K63 linkages, the authors identified RAD6/BRE1 as the enzyme that conjugates K63 linkages. UBP2 is the major deubiquitylating enzyme (DUB) that reverses this modification. Moreover, it was discovered that UBP2, rather than RAD6/BRE1, acted as a potential redox sensor and was regulated upon hydrogen peroxide stress. Because the relative stoichiometry of ubiquitylated proteins depends on the interplay between ubiquitylating enzymes and the reverse reaction catalyzed by DUBs, this study demonstrated that specific enzymes that reverse RP PTMs likely exist and remain to be identified. Multiple RPs were found to be modified by K63 linkages, and when these RPs are mapped to the ribosome structure, they are positioned throughout the ribosome without any localization pattern. Furthermore, in the K63 linkage mutant strain changes in global protein abundance upon oxidative stress were observed using quantitative MS [52••]. It will be important to determine whether these changes in protein synthesis are due to indirect alterations to protein degradation dynamics, or linked to differences in the modification of specific RPs and possible subsequent changes in translational control. With respect to these questions, a ribosome profiling study that monitored changes upon hydrogen peroxide stress in yeast suggested that short upstream ORFs or non-AUG ORFs are preferentially translated [53]. Therefore, it remains to be investigated whether the oxidative stress-induced K63 polyubiquitylation of ribosomes can specifically influence the stringency of start codon recognition. High-resolution cryo-electron microscopy (cryo-EM) structures of human ribosomes demonstrate that certain mammalian RPs have greater solvent-exposed domains when compared to their yeast homologs, and may act as possible platforms for additional PTMs [5]. It will be important to determine whether such RP modifications in response to oxidative stress are also conserved in mammalian cells, for example in neuronal cells that are especially vulnerable to oxidative stress [54].
Evidence for a functional role of mammalian RP-ubiquitylation has started to accumulate from a recent, unbiased study that examined ubiquitylation dynamics in response to drugs that induce ER unfolded protein response (UPR) (Figure 1b) [23••]. UPR is a well-studied response to proteotoxic stress and facilitates the protein-folding capacity of the cell by allowing selective translation of specific mRNAs while repressing the translation of most mRNAs to maintain ER protein homeostasis [55,56]. The proteome-wide ubiquitylation targets upon ER stress were determined by employing an antibody that recognizes the di-glycyl remnant on the ubiquitylated lysine of the substrate protein following trypsin digestion [23••,57]. Specifically, trypsin digestion cleaves off the rest of the ubiquitin leaving two C-terminal glycine residues of ubiquitin attached to the substrate lysine, thereby allowing the identification of the modified lysines. Analysis of the proteome-wide data revealed that ER stress results in dynamic levels of ubiquitylation on RPs. Small ribosomal subunit RPs, Rps2/uS5 and Rps3/uS3, were shown to be mono-ubiquitylated at sub-stoichiometric levels of 5–15% in response to drugs that induce ER folding stress as well as to drugs that inhibit translation elongation [23••]. Since ribosomes exist in multiple subcellular locations, it is critical to distinguish whether ribosome PTMs are present on the assembled ribosomes or instead demarcate RPs, as in the case of Rpl13a/uL13, for their extra-ribosomal roles. Interestingly, the authors showed that the mono-ubiquitylated small subunit RPs are predominantly in the cytoplasmic ribosome pool as opposed to ER-associated pool, and are present in assembled 80S ribosomes but absent from translating ribosomes. Mono-ubiquitylation typically affects protein–protein interactions rather than substrate turnover [49], thereby suggesting that these modifications on the ribosome may act as nucleation sites for interactions with additional, yet uncharacterized proteins. One feature of the UPR is the ability to decrease global translation at the ER, a function that when blocked leads to cell death [58]. Mutating the specific mono-ubiquitylation sites on the small subunit RPs (Rps2/uS5 and Rps20/uS10) renders the cell more susceptible to cell death upon UPR induction, suggesting a critical function for these ubiquitylation events [23••].
In order to define the upstream molecular mechanisms leading to RP mono-ubiquitylation upon stress conditions, it is critical to identify the ubiquitylating and DUB enzyme networks that can perform and reverse these modifications. Since the human genome encodes for an estimated number of 1000 enzymes that can recognize and attach ubiquitin to specific substrates [49], testing each of these enzymes for RP ubiquitylation may be challenging. Therefore, the ubiquitin-related enzymes that can interact with the mammalian ribosomes should be investigated by following a more focused, selective ribosome enrichment strategy with the potential to connect the ribosome with yet-unidentified enzymatic activities. It is also possible that known ribosome-interacting E3 enzymes that ubiquitylate nascent chains as part of a multilayered mRNA-protein quality control mechanism [51] could also be potential candidates for enzymes that directly modify the ribosome. LTN1 (mammalian homolog Listerin) is one example of mRNA-quality control related ubiquitin enzymes and is necessary and sufficient for ubiquitylation of truncated polypeptides at stalled ribosomes [59]. LTN1 binds to the large ribosomal subunit and is absent from assembled or translating ribosomes [60,61]. In addition, genetic screens in yeast using reporter constructs designed to induce stalled translation identified another ubiquitin ligase, HEL2, and implicated it in the initial response to stalled translation [32,62]. Moreover, the ATPase UPF1 [63,64], which contains a ubiquitin ligase domain [65], and NOT4 ubiquitin ligase [66] are additional candidates since they are involved in mRNA-protein quality control pathways and co-sediment with ribosomes in fractionation experiments.
Insights into which ubiquitin ligases may modify RPs came from two recent studies that sought to determine whether ZNF598, the mammalian homolog of HEL2 also has a role in the response to stall-inducing sequences on reporter constructs (Figure 1b) [67••,68••]. These studies suggested that ZNF598 may achieve such a role by mono-ubiquitylating primarily Rps10/eS10. By using a mutant Rps10/eS10 that cannot be ubiquitylated, this modification was shown to be important for resolving ribosomes stalled at the poly(A) sequences of the reporter constructs. These findings highlight the possibility that the sequence context of the mRNA may influence a ubiquitin ligase interacting with the ribosome to modify a core RP for proper mRNA-protein quality control. Additional work will be required to fully elucidate the detailed mechanisms and the importance of ZNF598-dependent Rps10 modification for physiologically stalled ribosomes. Interestingly, ZNF598 may affect the ER stress-dependent ubiquitylation of some but not all of the RPs to different levels, suggesting that other yet-unidentified ubiquitin ligases exist and may also have specificity for certain RPs [23••,67••,68••].
Ubiquitin is a versatile signaling molecule, which has also been directly linked to controlling the stoichiometry of RPs in the cell. In eukaryotes, ribosome biogenesis starts with precursor rRNA synthesis in the nucleolus. RPs that are translated in the cytoplasm are imported into the nucleus where rRNA and RPs are assembled via multi-step processes and then are exported to the cytoplasm with the help of many trans-acting factors [69]. It has been shown that in both mammalian and yeast cells, RPs are translated in excess and are degraded in a proteasome-dependent manner in the nucleus [70••,71,72]. A recent study in yeast shows that when RPs do not incorporate into mature cytoplasmic ribosomes, they are ubiquitylated by TOM1 and subsequently undergo proteasome-dependent degradation. This pathway is conserved in mammalian cells, and although the exact mechanism of recognition remains to be identified, it was shown that TOM1 modifies lysines of RPs that become available only if they do not interact with rRNA [70••].
In humans, there are ~70 additional factors that can affect rRNA processing without known yeast homologs, suggesting that mammalian ribosomes may be assembled by adopting unique strategies and/or by different trans-acting factors [73]. A recent study showed that ubiquitin signaling can modify a ribosome biogenesis factor, TCOF1, during human neural crest differentiation (Figure 1b) [74••]. The role of TCOF1 in ribosome biogenesis has been extensively studied and mutations in Tcof1 cause Treacher Collins syndrome, a craniofacial disorder characterized by loss of cranial neural crest cells [75,76]. Tcof1 knockdown or Tcof1 haploinsufficiency in mice results in pre-rRNA synthesis defects [77]. In a recent study aimed at identifying neuronal-lineage specific substrate adaptors of CUL3 ubiquitin ligases, both TCOF1 and another rRNA processing factor, NOLC1, were found to be mono-ubiquitylated by the KBTBD8 CUL3-adaptor [74••]. Moreover, at specific time points in neural crest differentiation, knockdown of the Kbtbd8 adaptor or even Tcof1 itself did not affect ribosome biogenesis or general protein synthesis. Instead mono-ubiquitylated TCOF1 was shown to act as a protein-interaction scaffold. Mono-ubiquitylated TCOF1 preferentially interacted with NOLC1, RNA polymerase I, H/ACA complex that results in rRNA pseudouridylation, and SSU proteins that control small ribosomal subunit maturation [78]. Ribosome profiling studies upon knockdown of Kbtbd8 at specific differentiation time points suggested that certain transcripts were differentially translated. Interestingly, it has previously been shown that rRNA pseudouridylation affects the translation of specific mRNAs [79,80]. This finding leads to the hypothesis that mono-ubiquitylated TCOF1, by recruiting rRNA modification and SSU enzymes, may result in the formation of ribosomes that favor the translation of select transcripts during neural crest differentiation. However, characterization and identification of potentially distinct cytoplasmic ribosomes due to TCOF1 mono-ubiquitylation remains to be demonstrated. It is also of particular interest to determine whether TCOF1 is modified in the nucleolus or in the cytoplasm, since most of TCOF1 is present in the nucleolus [77]. Furthermore, since knockdown of Kbtbd8 may affect translation independently of its role in TCOF1 mono-ubiquitylation, it will be critical to determine the functional consequences of mutating TCOF1 such that it can no longer be ubiquitylated.
In summary, emerging studies, albeit few in number, suggest that the ribosome is at the nexus of diverse PTMs. By coupling phosphorylation and ubiquitylation enrichment methods with stringent ribosome enrichment strategies, it will be increasingly feasible to determine comprehensively how many RPs are modified by PTMs. A major challenge that remains is to define the functional consequences of the RP PTMs. Examples highlighted here at the level of RP phosphorylation show that these modifications serve as markers of mTORC1 activity and neuronal activation, can control the extra-ribosomal pool of an RP upon specific stimuli to regulate translation of certain transcripts, and are critical for the pathogenesis of PD. Ubiquitin signaling has proven to be a versatile means of modifying RPs and highlights how diverse linkages and modifications of the ribosome have different functional outcomes. Notable examples of RP ubiquitylation occur when specific transcripts must be preferentially translated despite global translational repression, such as during the cell cycle or upon ER folding stress. The ubiquitin signal can also modify nascent polypeptide chains to integrate mRNA and protein quality control pathways, to monitor ribosome stoichiometry, and to modify ribosome biogenesis factors. Currently, in addition to the antibody-based enrichment of ubiquitin di-glycyl remnants from digested peptides, there are additional tools to study specific ubiquitin linkages such as linkage-specific antibodies [81–83]. Combining these tools with stringent ribosome enrichment strategies will lead to a more complete understanding of ubiquitin signaling at the ribosome.
The studies summarized in this review provide crucial insights and pave the way for more directed investigations. For example, it will be important to define the mechanisms for how PTMs affect ribosome activity, such as whether they can play structural roles on the ribosome, control ribosome subunit assembly, stabilize mature ribosome complexes, or influence specificity in translational control. Stoichiometry of the RP modification should be considered as well as the localization of modified RPs on the ribosome structure. For example, if RPs near the mRNA entry or exit channels are modified, one can envision that these PTMs may affect the interaction of the ribosome with specific mRNA classes and thus may alter the rate of elongation or specificity of termination. From recent cryo-EM studies, we also know that ribosome structures are dynamic during translation [84•] and that binding of trans-acting factors can result in conformational changes in the ribosome at sites distinct from their initial binding site [85]. Therefore, it is also tempting to hypothesize that modifications of an RP distant to conventional mRNA binding surfaces may still be critical, and could lead to the recruitment of additional RNA binding proteins to impart translational specificity.
The histone code describes the interplay of modifications at histones whose hierarchical assembly and disassembly have been functionally dissected [86]. In addition to the examples of RP phosphorylation and ubiquitylation discussed above, the spectrum of PTMs is rapidly expanding and RPs may potentially be modified with other types of PTMs. It remains to be determined whether RP PTMs follow underlying mechanistic rules that exclude or enable combinations of modifications to cross-talk and thereby increase the complexity and dynamics of the translation machinery (Figure 2). We are now poised to understand the consequences of RP PTMs towards translational control in more detail. For example, one can perform ribosome profiling with site-specific RP mutants that can no longer be modified. Also, interactions of kinases or ubiquitylating enzymes on the ribosome can be mechanistically studied using cryo-EM techniques. Moreover, systematic approaches to identify the writers, erasers, and downstream trans-factors that could read ribosome PTMs are an area of research that requires extensive study. Finally, establishing antibodies that can recognize site-specific PTMs will help to understand the distribution of these RP PTMs within different tissues, during different physiological states, or during the course of embryonic development. It has been recently shown that translation of the mammalian genome is intricately controlled in space and time [87]. The approaches outlined here to identify RP PTMs and to characterize their functional consequences are likely to reveal the potential dynamics of how ribosome modifications may directly influence protein expression within cells and developing organisms.
Figure 2. Is there a ribosome code?
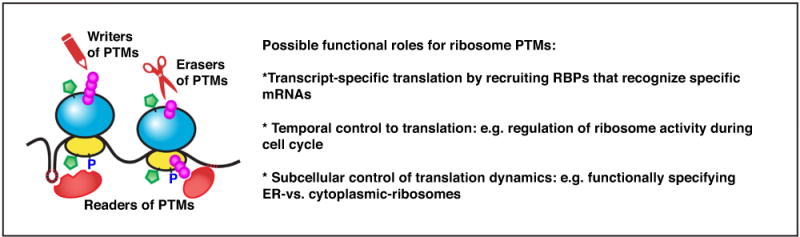
Diverse PTMs observed in RPs include O-GlcNAcylation (represented by green pentagons), phosphorylation, and ubiquitylation. The enzymes that can catalyze (writers) or reverse (erasers) the PTMs may interact with and modify different sub-pools of ribosomes, leading to increased dynamics of ribosome heterogeneity. Analogous to the ‘histone code’, the possibility of downstream trans-factors that can recognize combinations of these PTMs (readers) remains to be investigated. It is possible to envision that PTMs can recruit RNA binding proteins (RBPs) to the ribosome that can specifically recognize certain structures or sequence motifs within selective groups of mRNAs.
Acknowledgments
We apologize to those whose work we were unable to cite due to size limitations. This work was supported by the New York Stem Cell Foundation Award (M.B.), NIH R21HD086730 (M.B), Mallinckrodt Foundation Award (M.B.), and Pew Scholars Award (M.B.). D.S. is a Philip O’Bryan Montgomery, Jr., MD fellow of the Damon Runyon Cancer Research Foundation and postdoctoral fellow of the American Heart Association. M.B. is a New York Stem Cell Foundation Robertson Investigator.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• Of special interest
•• of outstanding interest
- 1.Palade GE. A small particulate component of the cytoplasm. J Biophys Biochem Cytol. 1955;1:59–68. doi: 10.1083/jcb.1.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 3.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yusupova G, Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu Rev Biochem. 2014;83:467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- 5.Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 6.Byrgazov K, Vesper O, Moll I. Ribosome heterogeneity: another level of complexity in bacterial translation regulation. Curr Opin Microbiol. 2013;16:133–139. doi: 10.1016/j.mib.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deusser E, Wittmann HG. Ribosomal proteins: variation of the protein composition in Escherichia coli ribosomes as function of growth rate. Nature. 1972;238:269–270. doi: 10.1038/238269a0. [DOI] [PubMed] [Google Scholar]
- 8.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. This study showed that subsets of Hox mRNAs contain a structural element in their 5′UTRs that allows specialized ribosomes containing the ribosomal protein Rpl38 to selectively translate them. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinman JD. Pathways to specialized ribosomes: the Brussels lecture. J Mol Biol. 2016;428:2186–2194. doi: 10.1016/j.jmb.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SW, Berger SJ, Martinovic S, Pasa-Tolic L, Anderson GA, Shen Y, Zhao R, Smith RD. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc Natl Acad Sci U S A. 2002;99:5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odintsova TI, Muller EC, Ivanov AV, Egorov TA, Bienert R, Vladimirov SN, Kostka S, Otto A, Wittmann-Liebold B, Karpova GG. Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J Protein Chem. 2003;22:249–258. doi: 10.1023/a:1025068419698. [DOI] [PubMed] [Google Scholar]
- 15.Yu Y, Ji H, Doudna JA, Leary JA. Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci. 2005;14:1438–1446. doi: 10.1110/ps.041293005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Stechow L, Francavilla C, Olsen JV. Recent findings and technological advances in phosphoproteomics for cells and tissues. Expert Rev Proteomics. 2015;12:469–487. doi: 10.1586/14789450.2015.1078730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu G, Jaffrey SR. The new landscape of protein ubiquitination. Nat Biotechnol. 2011;29:1098–1100. doi: 10.1038/nbt.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 19.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiskin E, Bionda T, Dikic I, Behrends C. Global analysis of host and bacterial ubiquitinome in response to Salmonella typhimurium infection. Mol Cell. 2016;62:967–981. doi: 10.1016/j.molcel.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Higgins R, Gendron JM, Rising L, Mak R, Webb K, Kaiser SE, Zuzow N, Riviere P, Yang B, Fenech E, et al. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol Cell. 2015;59:35–49. doi: 10.1016/j.molcel.2015.04.026. This study showed that in mammalian cells under ER folding stress, cytoplasmic but not ER-associated ribosomal proteins are modified with mono-ubiquitin. Mutations within these ribosomal proteins that abrogate mono-ubiquitylation show that this modification is involved in cell death upon ER folding stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gressner AM, Wool IG. The phosphorylation of liver ribosomal proteins in vivo: evidence that only a single small subunit protein (S6) is phosphorylated. J Biol Chem. 1974;249:6917–6925. [PubMed] [Google Scholar]
- 25.Meyuhas O. Ribosomal protein S6 phosphorylation: four decades of research. Int Rev Cell Mol Biol. 2015;320:41–73. doi: 10.1016/bs.ircmb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 27.Knight ZA, Tan K, Birsoy K, Schmidt S, Garrison JL, Wysocki RW, Emiliano A, Ekstrand MI, Friedman JM. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell. 2012;151:1126–1137. doi: 10.1016/j.cell.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruvinsky I, Katz M, Dreazen A, Gielchinsky Y, Saada A, Freedman N, Mishani E, Zimmerman G, Kasir J, Meyuhas O. Mice deficient in ribosomal protein S6 phosphorylation suffer from muscle weakness that reflects a growth defect and energy deficit. PLoS One. 2009;4:e5618. doi: 10.1371/journal.pone.0005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alliouachene S, Tuttle RL, Boumard S, Lapointe T, Berissi S, Germain S, Jaubert F, Tosh D, Birnbaum MJ, Pende M. Constitutively active Akt1 expression in mouse pancreas requires S6 kinase 1 for insulinoma formation. J Clin Invest. 2008;118:3629–3638. doi: 10.1172/JCI35237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalaileh A, Dreazen A, Khatib A, Apel R, Swisa A, Kidess-Bassir N, Maitra A, Meyuhas O, Dor Y, Zamir G. Phosphorylation of ribosomal protein S6 attenuates DNA damage and tumor suppression during development of pancreatic cancer. Cancer Res. 2013;73:1811–1820. doi: 10.1158/0008-5472.CAN-12-2014. [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay R, Ray PS, Arif A, Brady AK, Kinter M, Fox PL. DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Mol Cell. 2008;32:371–382. doi: 10.1016/j.molcel.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- 35.Kapasi P, Chaudhuri S, Vyas K, Baus D, Komar AA, Fox PL, Merrick WC, Mazumder B. L13a blocks 48S assembly: role of a general initiation factor in mRNA-specific translational control. Mol Cell. 2007;25:113–126. doi: 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson’s disease. Annu Rev Genomics Hum Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37••.Martin I, Kim JW, Lee BD, Kang HC, Xu JC, Jia H, Stankowski J, Kim MS, Zhong J, Kumar M, et al. Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson’s disease. Cell. 2014;157:472–485. doi: 10.1016/j.cell.2014.01.064. This study showed that a ribosomal protein is the main substrate of a kinase mutated in Parkinson’s disease and that ribosomal protein phosphorylation is critical for the neurodegenerative phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taymans JM, Nkiliza A, Chartier-Harlin MC. Deregulation of protein translation control, a potential game-changing hypothesis for Parkinson’s disease pathogenesis. Trends Mol Med. 2015;21:466–472. doi: 10.1016/j.molmed.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Errico A, Deshmukh K, Tanaka Y, Pozniakovsky A, Hunt T. Identification of substrates for cyclin dependent kinases. Adv Enzyme Regul. 2010;50:375–399. doi: 10.1016/j.advenzreg.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 44.Yoon IS, Chung JH, Hahm SH, Park MJ, Lee YR, Ko SI, Kang LW, Kim TS, Kim J, Han YS. Ribosomal protein S3 is phosphorylated by Cdk1/cdc2 during G2/M phase. BMB Rep. 2011;44:529–534. doi: 10.5483/bmbrep.2011.44.8.529. [DOI] [PubMed] [Google Scholar]
- 45.Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. The translational landscape of the mammalian cell cycle. Mol Cell. 2013;52:574–582. doi: 10.1016/j.molcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanenbaum ME, Stern-Ginossar N, Weissman JS, Vale RD. Regulation of mRNA translation during mitosis. Elife. 2015;4 doi: 10.7554/eLife.07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeidan Q, Wang Z, De Maio A, Hart GW. O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol Biol Cell. 2010;21:1922–1936. doi: 10.1091/mbc.E09-11-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 50.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 52••.Silva GM, Finley D, Vogel C. K63 polyubiquitination is a new modulator of the oxidative stress response. Nat Struct Mol Biol. 2015;22:116–123. doi: 10.1038/nsmb.2955. This study showed that in yeast under oxidative stress, multiple ribosomal proteins are modified with non-degradative ubiquitin linkages. The study also identified enzymes responsible for this modification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci U S A. 2012;109:17394–17399. doi: 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci. 2006;7:278–294. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 56.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 57.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat Struct Mol Biol. 2016;23:7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letzring DP, Wolf AS, Brule CE, Grayhack EJ. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA. 2013;19:1208–1217. doi: 10.1261/rna.039446.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Atkin AL, Schenkman LR, Eastham M, Dahlseid JN, Lelivelt MJ, Culbertson MR. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 64.Kuroha K, Tatematsu T, Inada T. Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep. 2009;10:1265–1271. doi: 10.1038/embor.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carpy A, Krug K, Graf S, Koch A, Popic S, Hauf S, Macek B. Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission Yeast) Mol Cell Proteomics. 2014;13:1925–1936. doi: 10.1074/mcp.M113.035824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Juszkiewicz S, Hegde RS. Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol Cell. 2017;65:743–750.e744. doi: 10.1016/j.molcel.2016.11.039. This study showed that Znf598 mono-ubiquitylates Rps10 in vitro and in vivo. By generating mutant Rps10 that cannot be ubiquitylated, the study showed that this modification is important for resolution of ribosome stalling at poly(A) sequences of a reporter construct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68••.Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. ZNF 598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol Cell. 2017;65:751–760.e754. doi: 10.1016/j.molcel.2016.12.026. This study showed that Znf598 mono-ubiquitylates Rps10 in vivo. By generating mutant Rps10 that cannot be ubiquitylated, the study showed that this modification is important for resolution of ribosome stalling at poly(A) sequences of a reporter construct. In addition, this study also suggests that some of the RP ubiqutylation events upon ER stress are dependent on RACK1 protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 70••.Sung MK, Porras-Yakushi TR, Reitsma JM, Huber FM, Sweredoski MJ, Hoelz A, Hess S, Deshaies RJ. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. Elife. 2016;5 doi: 10.7554/eLife.19105. This study identified the ubiquitin enzyme responsible for degrading excess ribosomal proteins within the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sung MK, Reitsma JM, Sweredoski MJ, Hess S, Deshaies RJ. Ribosomal proteins produced in excess are degraded by the ubiquitin-proteasome system. Mol Biol Cell. 2016;27:2642–2652. doi: 10.1091/mbc.E16-05-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 74••.Werner A, Iwasaki S, McGourty CA, Medina-Ruiz S, Teerikorpi N, Fedrigo I, Ingolia NT, Rape M. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature. 2015;525:523–527. doi: 10.1038/nature14978. This study showed that at a specific timepoint during neural crest specification, modification of a ribosome biogenesis factor with mono-ubiquitin results in preferential binding to ribosome modifying enzymes and other factors that have the potential to generate heterogenous ribosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. The Treacher Collins Syndrome Collaborative Group. Nat Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- 76.Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, Dixon MJ, Trainor PA. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci U S A. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci U S A. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Phipps KR, Charette J, Baserga SJ. The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdiscip Rev RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montanaro L, Calienni M, Bertoni S, Rocchi L, Sansone P, Storci G, Santini D, Ceccarelli C, Taffurelli M, Carnicelli D, et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res. 2010;70:4767–4777. doi: 10.1158/0008-5472.CAN-09-4024. [DOI] [PubMed] [Google Scholar]
- 80.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 81.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 82.Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 83.Matsumoto ML, Dong KC, Yu C, Phu L, Gao X, Hannoush RN, Hymowitz SG, Kirkpatrick DS, Dixit VM, Kelley RF. Engineering and structural characterization of a linear polyubiquitin-specific antibody. J Mol Biol. 2012;418:134–144. doi: 10.1016/j.jmb.2011.12.053. [DOI] [PubMed] [Google Scholar]
- 84•.Behrmann E, Loerke J, Budkevich TV, Yamamoto K, Schmidt A, Penczek PA, Vos MR, Burger J, Mielke T, Scheerer P, et al. Structural snapshots of actively translating human ribosomes. Cell. 2015;161:845–857. doi: 10.1016/j.cell.2015.03.052. This study showed the conformational changes that translating ribosomes can undergo by performing cryo-EM of polysome fractions containing actively translated mRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weis F, Giudice E, Churcher M, Jin L, Hilcenko C, Wong CC, Traynor D, Kay RR, Warren AJ. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol. 2015;22:914–919. doi: 10.1038/nsmb.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 87.Fujii K, Shi Z, Zhulyn O, Denans N, Barna M. Pervasive translational regulation of the cell signalling circuitry underlies mammalian development. Nat Commun. 2017;(8):14443. doi: 10.1038/ncomms14443. [DOI] [PMC free article] [PubMed] [Google Scholar]


