Abstract
Objective:
To evaluate whether the anti-LINGO-1 antibody has immunomodulatory effects.
Methods:
Human peripheral blood mononuclear cells (hPBMCs), rat splenocytes, and rat CD4+ T cells were assessed to determine whether LINGO-1 was expressed and was inducible. Anti-LINGO-1 Li81 (0.1–30 μg/mL) effect on proliferation/cytokine production was assessed in purified rat CD4+ T cells and hPBMCs stimulated with antibodies to CD3 +/– CD28. In humans, the effect of 2 opicinumab (anti-LINGO-1/BIIB033; 30, 60, and 100 mg/kg) or placebo IV administrations was evaluated in RNA from blood and CSF samples taken before and after administration in phase 1 clinical trials; paired samples were assessed for differentially expressed genes by microarray. RNA from human CSF cell pellets was analyzed by quantitative real-time PCR for changes in transcripts representative of cell types, activation markers, and soluble proteins of the adaptive/innate immune systems. ELISA quantitated the levels of CXCL13 protein in human CSF supernatants.
Results:
LINGO-1 is not expressed in hPBMCs, rat splenocytes, or rat CD4+ T cells; LINGO-1 blockade with Li81 did not affect T-cell proliferation or cytokine production from purified rat CD4+ T cells or hPBMCs. LINGO-1 blockade with opicinumab resulted in neither significant changes in immune system gene expression in blood and CSF, nor changes in CXCL13 CSF protein levels (clinical studies).
Conclusions:
These data support the hypothesis that LINGO-1 blockade does not affect immune function.
Classification of evidence:
This study provides Class II evidence that in patients with MS, opicinumab does not have immunomodulatory effects detected by changes in immune gene transcript expression.
MS is characterized by multifocal inflammation in the CNS, resulting in axonal demyelination and neurodegeneration. Although the relapse rate decreases with immunomodulatory treatments, brain atrophy progresses leading to disability.1 Therefore, CNS repair in MS lesions, including remyelination and axonal regeneration, by blocking inhibitor signaling, including LINGO-1 (leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1), could be a helpful therapeutic approach to slow or halt disease progression. Opicinumab (anti-LINGO-1; BIIB033) is a human immunoglobulin G1 (IgG1) aglycosyl monoclonal antibody and was tested as a possible neuroreparative agent in people with acute optic neuritis2 and relapsing forms of MS (RRMS). It targets LINGO-1, which is selectively expressed in the CNS, and plays a pivotal role in axonal myelination and regeneration regulation.3
Opicinumab is not expected to have immunomodulatory effects given that it targets a CNS-specific protein. To evaluate further whether anti-LINGO-1 has any effect on immune function, we analyzed LINGO-1 expression in immune cells and the effect of LINGO-1 blockade using cells from in vitro preclinical and phase 1 studies. First, we assessed LINGO-1 expression in rat and human lymphocytes in vitro and examined effects of an anti-LINGO-1 antibody on T-cell proliferation and cytokine production. Second, we assessed whether administration of opicinumab to patients with MS resulted in any significant changes in immune gene transcript expression in blood and CSF relative to placebo. In addition, we assessed the levels of inflammation associated with chemokine (C-X-C motif) ligand 13 (CXCL13) in CSF.
METHODS
Primary research question and classification of evidence.
Does opicinumab have any immunomodulatory effects? The investigations described provide Class II evidence that in patients with MS, opicinumab does not have immunomodulatory effects detected by changes in immune gene transcript expression or the levels of the chemokine CXCL13 protein in CSF (e-Methods, http://links.lww.com/NXI/A22).
Preclinical studies.
Lymphocyte isolation and stimulation.
Human peripheral blood mononuclear cells (hPBMCs) were isolated from healthy human peripheral blood using Ficoll-Paque PLUS density centrifugation (GE Healthcare Bio-Sciences, Pittsburgh, PA); 1 × 106 hPBMCs in 0.1 mL of culture medium (RPMI 1640 supplemented with 2 mM l-glutamine, 25 mM HEPES, 100 U/mL penicillin, 100 U/mL streptomycin, 55 µM 2-ME, and 10% heat-inactivated fetal calf serum; Invitrogen [Thermo Fisher Scientific Inc., Waltham, MA]) were seeded in 96-well plates. Rat CD4+ T cells were isolated from the spleen using anti-CD4–labeled magnetic beads (Miltenyi Biotec Inc., San Diego, CA) and magnetic separation. Cell purity (90%–95%) was determined using fluorescein isothiocyanate–labeled anti-CD4 (BD Biosciences, San Jose, CA), followed by flow cytometry analysis. Isolated cells were stimulated with 20 ng/mL phorbol myristate-13-acetate (Cell Signaling Technology, Inc., Danvers, MA) and 1 µg/mL ionomycin (Sigma-Aldrich Corp., St. Louis, MO; P/I), 100 ng/mL lipopolysaccharide (Sigma-Aldrich Corp.), 5 µg/mL phytohemagglutinin (Sigma-Aldrich Corp.), or anti-CD3 antibody (human [OKT3] or rat [G4.18]; Thermo Fisher Scientific Inc.) as indicated for 2–3 days.
Western blotting.
Western blotting methods are described elsewhere.4 Briefly, cells were lysed in RIPA buffer with a protease inhibitor cocktail, separated by SDS-PAGE, transferred to nitrocellulose membrane, and incubated with 2 µg/mL of 3C11 (a murine monoclonal anti-LINGO-1 antibody; Biogen, Cambridge, MA). Lysates extracted from cultured neocortical neurons isolated from E18 rats were used as a positive control.
Mixed lymphocyte reaction and cytokine production.
Human PBMCs were isolated from 1 healthy individual each as stimulators (irradiated with sublethal dose radiation, 137Cs) and responders. Stimulators/responders were resuspended to 1 × 106 cells/mL in the culture medium (see earlier); 100 µL of each was added to 96-well plates containing concentrations of 0.1, 1.0, 10, or 30 μg/mL anti-LINGO-1 antibody (Li81; earlier version of opicinumab, with identical amino acid sequence, but produced in a different Chinese hamster ovary cell line; Biogen), LFA3TIP (IgG1 fusion protein that can block mitogen-induced T-cell proliferation [alefacept; Biogen]; positive control),5 or isotype control antibody (human IgG1; Biogen). Cells were incubated for 5 days, and 1 µCi [methyl-3H] thymidine (GE Healthcare Bio-Sciences) was added to each well during the last 18 hours of culture. Proliferation was measured by thymidine incorporation using a FilterMate Harvester and 1450 MicroBeta liquid scintillation counter (PerkinElmer, Waltham, MA). Similar triplicate cultures were used to measure the level of cytokine secretion in the presence of 10 µg/mL Li81. Supernatants were collected after 3 days of culture and assayed for cytokine production using the SearchLight multiplex ELISA system (Aushon Biosystems, Billerica, MA). Recombinant human tumor necrosis factor α (20 ng/mL) (PeproTech, Rocky Hill, NJ) was added to the cell cultures as a positive control.
T-cell proliferation and cytokine production.
Purified rat CD4+ T cells or hPBMCs were cultured in triplicate in 96-well plates in the presence of anti-CD3 (0.2 µg/mL) or anti-CD3 plus anti-CD28 antibodies (0.2 µg/mL; Thermo Fisher Scientific Inc.) with 0.1, 1.0, 10, and 30 μg/mL Li81 (as used by this group previously in experimental autoimmune encephalomyelitis [EAE] studies6). For proliferation assays, cells were cultured for 3 days, and 1 µCi [methyl-3H] thymidine was added to each well during the last 18 hours and measured as described above. For cytokine production, rat CD4+ T cells were incubated under the same conditions for 2 days. Supernatants were collected, and cytokine production was measured as described above.
Clinical studies.
Standard protocol approvals, registrations, and patient consents.
Samples were collected in studies approved by institutional review boards, and written informed consent was obtained from all participants.3,7
Clinical study design and participants.
A phase 1, randomized (with allocation concealment) multiple ascending dose (MAD) study was performed in participants with RRMS or secondary progressive MS (SPMS; ClinicalTrials.gov identifier NCT01244139) to test the safety/tolerability of opicinumab; IV doses were administered on days 0 and 14.3 Blood and CSF samples from participants in the higher dose groups (30, 60, 100 mg/kg opicinumab, or placebo) were used in these analyses (figure e-1 at http://links.lww.com/NXI/A22). CSF cell pellet samples from healthy controls were provided through a previous study with Dr. Keith Edwards.7
Sample collection, RNA isolation, and quantification.
Peripheral blood samples and CSF, via lumbar puncture, were collected from study participants at days 0 and 28, 2 weeks after the second opicinumab dose.
Peripheral blood samples were collected using the PAXgene Blood RNA system (PreAnalytiX GmbH, Hombrechtikon, Switzerland) and maintained frozen at −70°C. RNA was extracted from PAXgene-preserved blood with the Agencourt RNAdvance Blood kit according to the manufacturer's specifications (Beckman Coulter, Inc., Indianapolis, IN). RNA integrity was assessed using the HT RNA Reagent Kit (Caliper Life Sciences, Hopkinton, MA) using a LabChip GX (PerkinElmer). RNA samples with an RNA Quality Score of >8.0 were considered to be of sufficient quality for microarray analysis.
For CSF samples, 10 mL aliquots were centrifuged immediately after collection at 10,000g to pellet the cells, which were immediately resuspended in RLT lysis buffer (Qiagen, Hilden, Germany) and frozen at −70°C. RNA was extracted from the resuspended cell pellets using the RNeasy Plus Micro Kit (Qiagen). RNA integrity was measured using a Bioanalyzer system (RNA 6000 Pico Kit; Agilent Technologies, Santa Clara, CA). CSF supernatant was maintained frozen.
RNA transcript analysis.
RNA from peripheral blood and CSF cell pellets was analyzed using the Affymetrix high-throughput microarray expression profiling methodology (Thermo Fisher Scientific Inc.).8 For blood RNA samples, automated sample amplification and biotin labeling were performed using the Ovation RNA Amplification System V2, Ovation Whole Blood Reagent, and Encore Biotin Module (NuGEN Technologies, Inc., San Carlos, CA) according to the manufacturer's recommendations. For CSF RNA samples, total RNA (1–5 ng) was reverse transcribed and amplified to complementary DNA (cDNA) using the Ovation Pico WTA System V2 (NuGEN Technologies Inc.); the cDNA samples were then treated as per the blood RNA.
For blood and CSF samples, 2-µg biotin-labeled cDNA probe was hybridized to Affymetrix GeneChip HT HG-U133+ PM array plates (Thermo Fisher Scientific Inc.), using modified conditions.8 Washing/staining was completed as per the GeneChip Expression Analysis Technical Manual for HT Array Plates Using the Genechip Array Station (Affymetrix; Thermo Fisher Scientific Inc.) with modifications.8 Processed GeneChip array plates were scanned using a GeneTitan scanner (Affymetrix; Thermo Fisher Scientific Inc.).
Only for CSF cell pellet RNA samples, quantitative real-time PCR (qPCR) analysis was performed using the 96.96 Dynamic Arrays for the microfluidic BioMark system (Fluidigm Corporation, San Francisco, CA). A custom qPCR panel was developed to evaluate gene transcripts representative of specific cell types, activation markers, and soluble proteins of the adaptive and innate immune systems (table e-1, http://links.lww.com/NXI/A22). All qPCR probe sets were designed using Primer Express v3.0 with TaqMan (Applied Biosystems; Thermo Fisher Scientific Inc.) minor groove binder quantification default settings.
CXCL13 assay.
The CXCL13-Imperacer assay (Chimera Biotec GmbH, Dortmund, Germany) was developed to quantify CXCL13 protein in human CSF. Cell-free CSF supernatants collected at days 0 and 28 were used. Human CXCL13-binding capture antibody (MAb801 clone 53610) was diluted in coating buffer, immobilized onto 8-well Imperacer microplate modules, and incubated overnight. The coated plates were then washed and blocked with chimera direct block solution immediately before use.
A calibration curve and samples were prepared according to the standard protocol, including a 1:2 dilution step for both samples and calibration curve in buffer SDB2100 for antigen stabilization and minimization of matrix effects. The sample dilution buffer additionally contained a 1:300 dilution of the CXCL13-specific Imperacer antibody-DNA detection conjugate CHI-CXCL13.
Samples/calibration curve with detection conjugate were incubated overnight at 4°C. After a final washing step, PCR mix was applied to the wells and the sealed plate was placed into the Imperacer reader instrument for real-time qPCR signal generation. Signals in real-time qPCR were recorded as fluorescence increase and documented as Rn (fluorescence) and Ct (threshold cycle) values.
Data analyses.
Affymetrix microarray data analyses.
Affymetrix scans were subject to quality control tests, including visual inspection of the distribution of raw signal intensities and assessment of RNA degradation, relative log expression, and normalized unscaled standard error. CEL files that passed quality control were normalized using the GC-content–based Robust Multiarray Average method.9,10
Significant differentially expressed genes associated with administration of opicinumab were studied as follows. Paired participant samples (days 0 and 28) in each dose group were subjected to a paired t test. Genes that exhibited an absolute fold change >1.5, average normalized intensity >4, and Benjamini-Hochberg corrected p value <0.05 were considered differentially expressed; each dose group was analyzed independently. All calculations and analyses were performed using R and Bioconductor computational tools.11
qPCR data analyses.
For normalization, the Ct geometric mean for the housekeeping genes GAPDH, YWHAZ, and ACTB was calculated for each sample (see supplementary material).12,13 For each participant, the fold change in expression of each gene transcript was calculated relative to baseline (day 0) and compared with the placebo group using a t test.
CXCL13 data analyses.
Ct values were converted to delta Ct values by subtracting Ct from Ctmax to obtain signal values directly proportional to the antigen concentration. Quantitative data analysis was performed by 4-parameter sigmoidal fit (logistic model) of a spiked calibration curve (target recombinant antigen CXCL13 in artificial CSF), which was included in each experiment (see supplementary material). Calculated concentrations are provided as picograms per milliliter.
RESULTS
LINGO-1 is not detectable in hPBMCs, rat splenocytes, and rat CD4+ T cells in vitro.
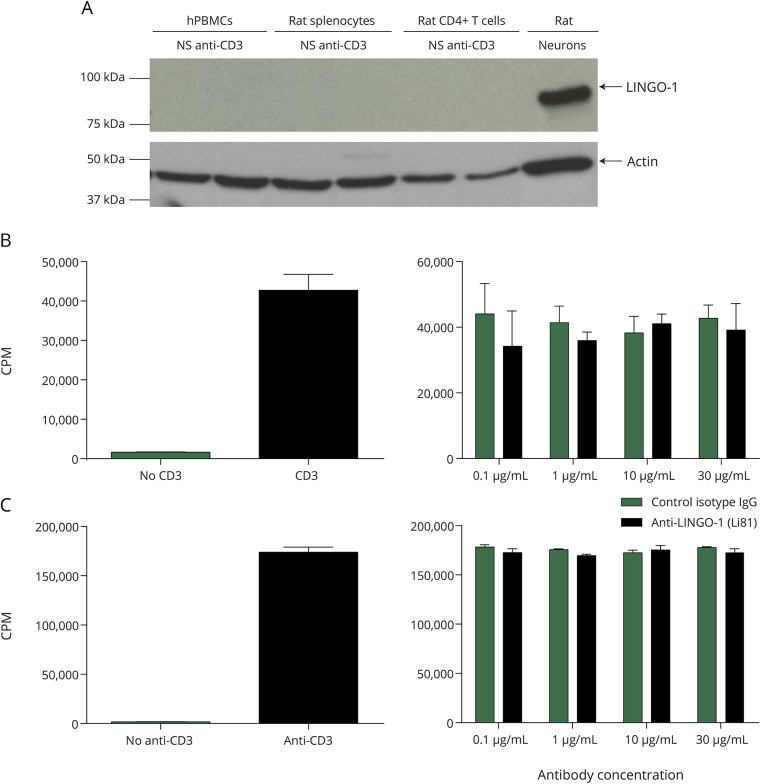
To determine whether LINGO-1 was expressed and was inducible in hPBMCs, rat splenocytes, and rat CD4+ T cells, Western blots for LINGO-1 protein were performed from cultured cells stimulated with anti-CD3 antibodies. Unlike cultured neocortical neurons of the rat (positive control), LINGO-1 expression was not detectable in any of these cell types (figure 1A). In separate experiments, hPBMCs were stimulated with phorbol myristate-13-acetate/ionomycin, phytohemagglutinin, or lipopolysaccharide to assess whether LINGO-1 might be induced via alternative signaling pathways. Again, LINGO-1 protein was not detected in those cells (figure e-2, http://links.lww.com/NXI/A22).
Figure 1. Anti-LINGO-1 (Li81) has no effect on activated T-cell proliferation.
(A) Western blot analysis with anti-LINGO-1 (3C11) shows that it was readily detected in neurons but not in activated human peripheral blood lymphocytes, rat splenocytes, or rat CD4+ T cells. (B and C) Stimulation with anti-CD3 antibody activated 3H thymidine incorporation into rat CD4+ T cells (B, left panel) and hPBMC (C, left panel), and in both cases, this was unaffected by the addition of anti-LINGO-1 (Li81) antibody at concentrations up to 30 μg/mL (B and C, right panels). CPM = counts per minute; hPBMC = human peripheral blood mononuclear cell; IgG = immunoglobulin G; NS = no stimulation.
Anti-LINGO-1 (Li81) has no effect on activated T-cell proliferation in vitro.
hPBMCs were activated with anti-CD3 antibody, and T-cell proliferation was determined by 3H thymidine incorporation after 72 hours of incubation (figure 1B, left panel as a positive control). Anti-LINGO-1 (Li81) did not show any effect on anti-CD3–activated T-cell proliferation, as evidenced by comparable uptake of 3H thymidine in the presence of either anti-LINGO-1 or isotype control IgG (figure 1B, right panel). Anti-LINGO-1 did not affect proliferation in the human mixed lymphocyte reaction (MLR) compared with the positive control, an anti-LFA3TIP monoclonal antibody (1E6), which showed significant inhibition for the MLR (figure e-3A, http://links.lww.com/NXI/A22). Similarly, anti-LINGO-1 did not show any effect on the proliferation of rat CD4+ T cells activated with anti-CD3+/– anti-CD28 antibodies (figure 1C and figure e-3B, http://links.lww.com/NXI/A22).
Anti-LINGO-1 (Li81) does not affect production of cytokines by activated T cells in vitro.
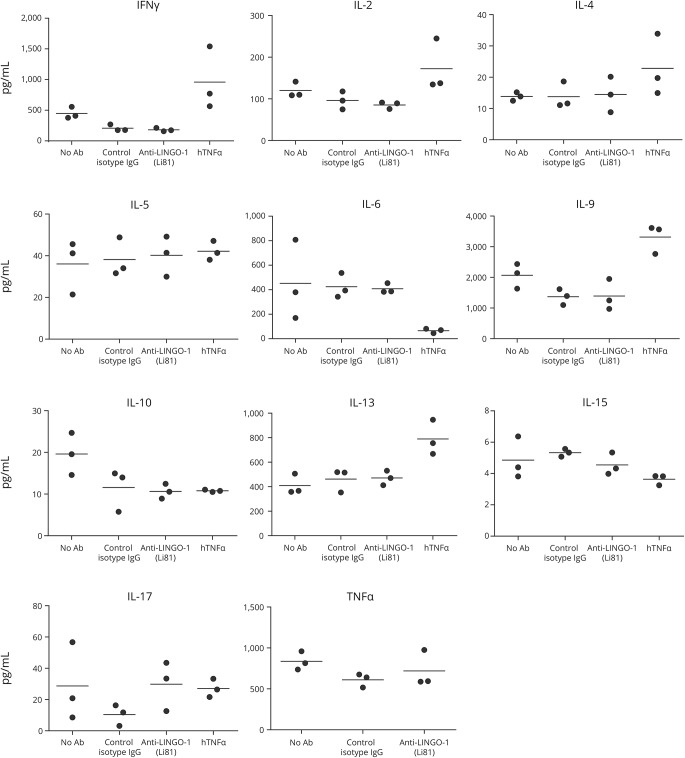
Anti-LINGO-1 (Li81) did not show any effect on cytokine production in the human MLR. By contrast, in the presence of human recombinant tumor necrosis factor α, the levels of several cytokines increased significantly (figure 2). Similarly, no change in cytokine production was observed in the presence of different concentrations of anti-LINGO-1 in anti-CD3 antibody–activated rat CD4+ T cells (figure e-4, http://links.lww.com/NXI/A22). Similar results were observed when the assay was conducted with rat CD4+ T cells activated by anti-CD3 plus anti-CD28 antibodies (data not shown).
Figure 2. Anti-LINGO-1 (Li81) does not affect cytokine production.
Anti-LINGO-1 (Li81) does not affect cytokine production in the hPBMC, as determined by ELISA. Ab = antibody; hPBMC = human peripheral blood mononuclear cell; IFN = interferon; IgG = immunoglobulin G; IL = interleukin; TNF = tumor necrosis factor.
Demographics of study participants.
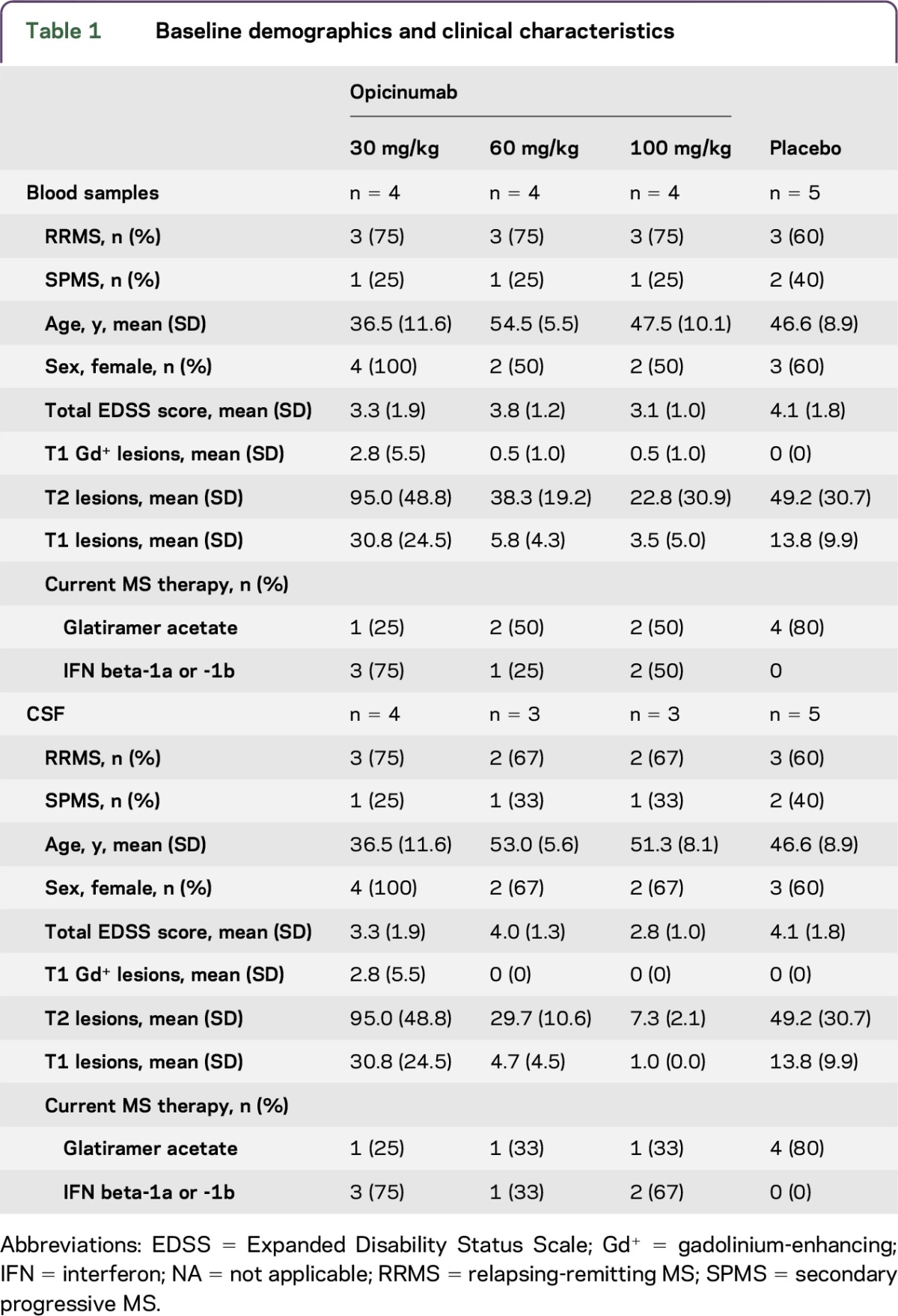
Paired blood and CSF RNA samples from 17 (RRMS, n = 12; SPMS, n = 5) and 15 (RRMS, n = 10; SPMS, n = 5) participants in the MAD study, respectively, were successfully isolated and analyzed (figure e-1 at http://links.lww.com/NXI/A22, and table 1).3 Samples from 4 to 5 participants per dose group were included in the blood analysis, and samples from 3 to 5 in the CSF analysis (table 1). The placebo and 3 treatment groups were well balanced with respect to the type of MS, baseline age, sex, and Expanded Disability Status Scale score. The majority were middle-aged women with mild-to-moderate disability. However, there were some differences in their concomitant anti-inflammatory disease-modifying treatments for MS, with more participants randomized to placebo taking glatiramer acetate, whereas participants randomized to opicinumab were more often treated with interferon beta.
Table 1.
Baseline demographics and clinical characteristics
Treatment with 2 doses of opicinumab does not alter gene expression in the blood or CSF of participants with MS.
A paired t test was performed on the gene expression array data for each dose group across all participants to understand how gene expression at day 28 compared with the pretreatment day 0 samples. Results using a Benjamini-Hochberg p value correction indicated that there were no differentially expressed genes identified in the peripheral blood or CSF following treatment with opicinumab relative to the placebo control group (table e-2, http://links.lww.com/NXI/A22).
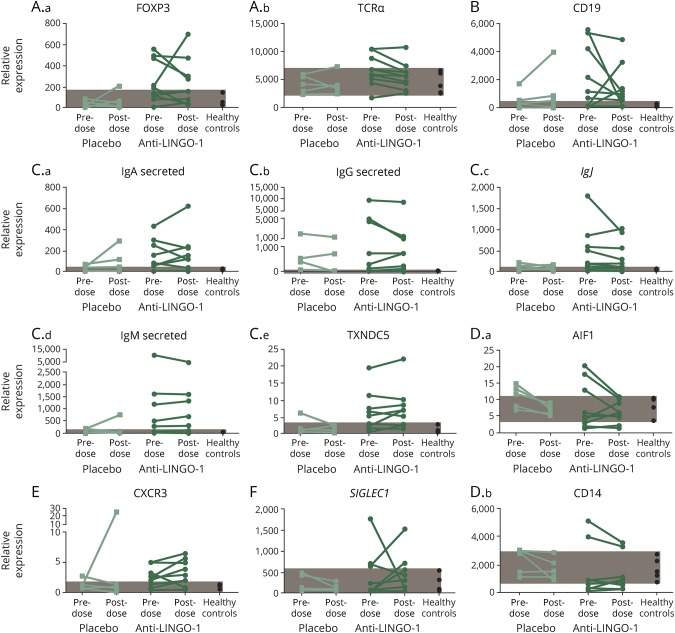
Further analysis of the CSF cell pellets was performed using a targeted immune response qPCR panel (table e-3, http://links.lww.com/NXI/A22). These results indicated no statistically significant changes in gene expression that appeared to be opicinumab dose dependent; the larger differences in fold change values were driven by shifts in only a few participants (table e-3, http://links.lww.com/NXI/A22). Elevated levels of the various RNA transcripts remained largely unchanged in most participants after short-term opicinumab treatment (figure 3).
Figure 3. Gene expression in CSF cell pellets.
Gene expression in CSF cell pellets reflecting (A.a–A.b) T cells, (B) B cells, (C.a–C.e) plasma cells, (D.a–D.b) myeloid cells, (E) T-cell activation, and (F) type I IFN response before and after opicinumab or placebo treatment. Each line connects the 2 time points of an individual patient. Patients from the 30-, 60-, and 100-mg/kg groups have been combined. Healthy range (shaded area) was determined by the minimum and maximum values of 5 healthy control patients evaluated. Ig = immunoglobulin; TCRα = T-cell receptor alpha.
At the molecular level, differences were observed at baseline between the group of participants randomized to placebo and those in the opicinumab groups. Participants assigned to the opicinumab groups pretreatment had higher relative expression levels of some key transcripts representing immune cell types (T, B, and plasma cells), with a concomitant reduction in myeloid markers. In some cases, differences reached statistical significance (p < 0.05; t test, TCR-δ, FOXP3, CXCR5, MX1, and G1P2; figure 3, not shown).
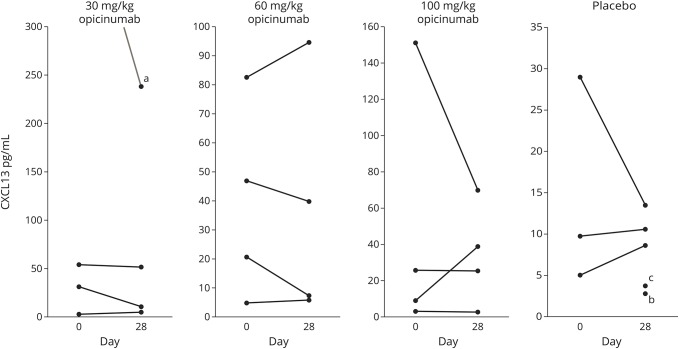
Treatment with opicinumab does not affect CXCL13 levels in CSF.
CSF levels of CXCL13 have been demonstrated to reflect intrathecal inflammation.7,14 In this study, we found that CXCL13 levels were stable in all groups, across the 2 time points evaluated (figure 4), regardless of opicinumab dose. As anticipated, CXCL13 levels highly correlated with the presence of B-cell transcripts in CSF (data not shown).
Figure 4. Opicinumab does not affect the level of CXCL13 in CSF.
Opicinumab does not affect the level of CXCL13 protein in CSF as measured by the CXCL13-Imperacer assay. aCorresponding day 0 (before dose) sample above the limit of quantification. bCorresponding day 0 sample not done. cDay 0 sample below the limit of quantification.
DISCUSSION
In this combined preclinical and clinical study, biochemical and genetic analyses indicated that LINGO-1 expression was not detectable in activated hPBMCs, rat splenocytes, or CD4+ T cells, and blocking LINGO-1 had no effect on in vitro cytokine production or proliferation of hPBMCs or rat T cells. There were no significant changes in the peripheral blood or the CSF in participants with MS exposed to 2 opicinumab doses up to 100 mg/kg. These data add further support to the view that LINGO-1 expression is limited to the CNS15,16 and that the effects of antagonizing LINGO-1 function even at the very high doses tested, with exposures above EC50 lasting up to 4 months,3 do not involve immunomodulatory activity.
LINGO-1 is a CNS-specific membrane-associated protein expressed by neurons and oligodendrocyte progenitor cells, which negatively regulates oligodendrocyte differentiation, myelination, and axonal regeneration.17–19 Inhibition of myelination by LINGO-1 is mediated via mechanisms involving RhoA GTPase19 and by nerve growth factor/TrkA receptor,20 and inhibition of oligodendrocyte differentiation and maturation by blocking the ErbB2 signaling pathway.21 LINGO-1 is expressed only in the CNS,15 and there is no previous evidence that it plays any role in immune biology. In animal models of EAE, antagonism of LINGO-1 by gene knockout or administration of a LINGO-1 antagonistic monoclonal antibody ameliorated disease symptoms and led to remyelination.17,22 However, there was no effect on infiltration of immune cells into the spinal cord. Furthermore, T cells from LINGO-1 knockout mice demonstrated normal proliferation and cytokine production, and encephalitogenic T cells from LINGO-1 knockout EAE animals were capable of inducing EAE in wild-type mice, similar to T cells from wild-type mice.17
Although the blood-brain barrier limits CNS penetration of systemically administered monoclonal antibodies such as opicinumab,6 the restricted expression of LINGO-1 within the CNS allows for safe administration of high systemic doses of opicinumab such that potentially efficacious concentrations in the CNS may be achieved.3 In preclinical studies, the CSF concentration of anti-LINGO-1 associated with EC50 and EC90 was determined to be 30 ng/mL and 100 ng/mL, respectively, corresponding to the serum levels of 30 μg/mL (3 mg/kg) and 100 μg/mL (10 mg/kg), assuming 0.1% blood-brain barrier penetration.6 In humans, doses of 3 mg/kg resulted in mean serum concentrations >EC50, and 10 mg/kg opicinumab achieved EC90. Furthermore, the CSF/serum ratio (0.1%) is similar to that reported for other IgG monoclonal antibody therapies. The doses used in the clinical studies were ≥30 mg/kg and should therefore have been high enough to stimulate an immune effect if one does exist.3,6
There are some limitations to this study because the sample size in the phase 1 clinical trial was small. In the absence of an exhaustive analysis of every type of immune cell, we cannot exclude the possibility of effects in cell types or under conditions that were not tested. Nevertheless, given the extent of interactions of immune cells, it would have been reasonable to expect any such effect to exert indirect effects in the experiments that were performed. In addition, the lack of safety signals (other than hypersensitivity reactions) in the phase 13 and 2a (RENEW)2 studies support a lack of any detectible immunomodulatory effects of opicinumab. Furthermore, the statistical analyses performed on the gene expression data help to protect the scientific and statistical rigor of the experiments and should increase confidence in the negative claim. Another limitation is that only 2 doses of opicinumab were administered. Future studies need to confirm that no changes are observed with a longer treatment duration.
Baseline differences in expression levels of key transcripts between participants in the clinical study were observed and expected owing to variability in MS and the small size of the cohorts randomized at each dose level in the MAD study.3 At baseline, opicinumab-treated participants had higher relative expression levels of IFN-response genes as well as transcripts for T, B, and plasma cell markers, with a concomitant reduction in myeloid markers. Levels of the various markers tended to remain stable after dosing. Differences in gene expression observed at baseline may be a result of difference in concomitant medications or disease heterogeneity. In fact, elevated basal expression of the interferon-inducible transcripts MX1 and G1P2 in the opicinumab-treated patients likely reflects the use of interferons in the treated but not in the placebo participants. The study protocol did not differentiate background MS treatments across classes (interferons vs glatiramer acetate; in the MAD study, natalizumab and fingolimod were excluded per protocol).
Evidence of CNS remyelination was provided by the recently published RENEW study that investigated the efficacy and safety of 100 mg/kg opicinumab in participants experiencing a first acute optic neuritis episode vs placebo; opicinumab-treated participants showed improved latency recovery (measured by full-field visual evoked potential), indicative of remyelination.2 Furthermore, the efficacy/safety of opicinumab (4 doses and placebo, administered concurrently with interferon beta-1a) was evaluated in a recently completed phase 2b trial in active relapsing MS (SYNERGY, ClinicalTrials.gov identifier NCT01864148).23 In SYNERGY, opicinumab missed the primary end point, a linear trend test on a multicomponent measure of improvement in physical function, cognitive function, and disability. Instead, an inverted U-shaped dose-response was seen, suggesting a lack of effect with the lowest/highest opicinumab doses and a potential clinical effect with doses in between.24
Taken together, these novel, combined preclinical and clinical data sets lend further support to the view that LINGO-1 antagonism exerts activity via remyelination and/or neuroaxonal protection rather than immune modulation.19,20 Further analysis of SYNERGY and future clinical data will provide additional evidence on the current assumption, supported by this new data set, that there is little if any potential for any direct interaction of LINGO-1 blockade with immune system function peripherally or intrathecally.
ACKNOWLEDGMENT
The authors thank Dr. Keith Edwards (Multiple Sclerosis Center of Northeastern New York, Latham, NY) for permission to use the CSF cell pellet samples from healthy controls provided through a previous study. They also thank Yinghui Hu (Biogen employee at the time) for performing some of the in vitro work described. Rebecca Gardner, PhD, from Excel Scientific Solutions wrote the first draft of the manuscript based on input from authors, and Elizabeth Cassell from Excel Scientific Solutions copyedited and styled the manuscript as per journal requirements. Biogen reviewed and provided feedback on the article. The authors had full editorial control of the article and provided their final approval of all content.
GLOSSARY
- EAE
experimental autoimmune encephalomyelitis
- EDSS
Expanded Disability Status Scale
- Gd+
gadolinium-enhancing
- hPBMC
human peripheral blood mononuclear cell
- IFN
interferon
- IgG
immunoglobulin G
- MAD
multiple ascending dose
- MLR
mixed lymphocyte reaction
- NA
not applicable
- RRMS
relapsingremitting MS
- SPMS
secondary progressive MS
Footnotes
Supplemental data at http://links.lww.com/NXI/A22
AUTHOR CONTRIBUTIONS
A.R., S.R., S.S., A.D., N.A., and S.M. performed the experiments reported. D.C. was involved in study design, analysis of the clinical data, and writing of the manuscript. R.M. was the local principal investigator for the phase 1 study in participants with MS. S.S. performed the statistical analyses on the clinical samples and S.M. analyzed the preclinical data. R.G. wrote the first draft of the manuscript. All authors contributed to data interpretation and were involved in the review and editing of all drafts of the manuscript.
STUDY FUNDING
These studies were funded by Biogen (Cambridge, MA). Biogen provided funding for medical writing support in the development of this article.
DISCLOSURE
A. Ranger was employed by and held stock in Biogen. S. Ray was employed by and held stock in Biogen and is employed by Vertex Pharmaceuticals. S. Szak is employed by and holds stock in Biogen. A. Dearth was employed by, received research support from, and held stock in Biogen and is employed by Curis, Inc. N. Allaire is employed by and holds stock in Biogen Idec. R. Murray received travel funding and speaker honoraria from Teva, Biogen, and Genzyme and received research support from Teva, Biogen, Genzyme, and Novartis. R. Gardner is an employee of Excel Scientific Solutions. D. Cadavid is employed by Fulcrum Therapeutics, has patents pending related to the use of drugs that block LINGO-1 to treat demyelination in multiple sclerosis, and holds stock from Biogen. S. Mi is employed by and holds stock in Biogen. Go to Neurology.org/nn for full disclosure forms.
REFERENCES
- 1.Harrison DM. Multiple sclerosis. Ann Intern Med 2014;160:ITC4-2–ITC4-18. [DOI] [PubMed] [Google Scholar]
- 2.Cadavid D, Balcer L, Galetta S, et al. ; RENEW Study Investigators. Safety and efficacy of opicinumab in acute optic neuritis (RENEW): a randomised, placebo-controlled, phase 2 trial. Lancet Neurol 2017;16:189–199. [DOI] [PubMed] [Google Scholar]
- 3.Tran JQ, Rana J, Barkhof F, et al. Randomized phase I trials of the safety/tolerability of anti-LINGO-1 monoclonal antibody BIIB033. Neurol Neuroimmunol Neuroinflamm 2014;1:e18. doi: 10.1212/NXI.0000000000000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Y, Lee X, Shao Z, et al. A DR6/p75NTR complex is responsible for β-amyloid-induced cortical neuron death. Cell Death Dis 2013;4:e579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller GT, Hochman PS, Meier W, et al. Specific interaction of lymphocyte function-associated antigen 3 with CD2 can inhibit T cell responses. J Exp Med 1993;178:211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pepinsky RB, Shao Z, Ji B, et al. Exposure levels of anti-LINGO-1 Li81 antibody in the central nervous system and dose-efficacy relationships in rat spinal cord remyelination models after systemic administration. J Pharmacol Exp Ther 2011;339:519–529. [DOI] [PubMed] [Google Scholar]
- 7.Edwards KR, Goyal J, Plavina T, et al. Feasibility of the use of combinatorial chemokine arrays to study blood and CSF in multiple sclerosis. PLoS One 2013;8:e81007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allaire NE, Bushnell SE, Bienkowska J, Brock G, Carulli J. Optimization of a high-throughput whole blood expression profiling methodology and its application to assess the pharmacodynamics of interferon (IFN) beta-1a or polyethylene glycol-conjugated IFN beta-1a in healthy clinical trial subjects. BMC Res Notes 2013;6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003;31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2001;2:RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer Science+Business Media; 2005. 442 p. [Google Scholar]
- 12.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 14.Bielekova B, Komori M, Xu Q, Reich DS, Wu T. Cerebrospinal fluid IL-12p40, CXCL13 and IL-8 as a combinatorial biomarker of active intrathecal inflammation. PLoS One 2012;7:e48370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi S, Pepinsky RB, Cadavid D. Blocking LINGO-1 as a therapy to promote CNS repair: from concept to the clinic. CNS Drugs 2013;27:493–503. [DOI] [PubMed] [Google Scholar]
- 16.Mi S, Sandrock A, Miller RH. LINGO-1 and its role in CNS repair. Int J Biochem Cell Biol 2008;40:1971–1978. [DOI] [PubMed] [Google Scholar]
- 17.Mi S, Hu B, Hahm K, et al. LINGO-1 antagonist promotes spinal cord remyelination and axonal integrity in MOG-induced experimental autoimmune encephalomyelitis. Nat Med 2007;13:1228–1233. [DOI] [PubMed] [Google Scholar]
- 18.Mi S, Lee X, Shao Z, et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci 2004;7:221–228. [DOI] [PubMed] [Google Scholar]
- 19.Mi S, Miller RH, Lee X, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci 2005;8:745–751. [DOI] [PubMed] [Google Scholar]
- 20.Lee X, Yang Z, Shao Z, et al. NGF regulates the expression of axonal LINGO-1 to inhibit oligodendrocyte differentiation and myelination. J Neurosci 2007;27:220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee X, Shao Z, Sheng G, Pepinsky B, Mi S. LINGO-1 regulates oligodendrocyte differentiation by inhibiting ErbB2 translocation and activation in lipid rafts. Mol Cell Neurosci 2014;60:36–42. [DOI] [PubMed] [Google Scholar]
- 22.Mi S, Miller RH, Tang W, et al. Promotion of central nervous system remyelination by induced differentiation of oligodendrocyte precursor cells. Ann Neurol 2009;65:304–315. [DOI] [PubMed] [Google Scholar]
- 23.Cadavid D, Phillips G, Dong-Si T, Tran J, Xu L. Efficacy and safety of anti LINGO-1 for the treatment of relapsing forms of multiple sclerosis: design of the phase 2 SYNERGY trial. Neurology 2014:82;P3.154. [Google Scholar]
- 24.Cadavid D, Edwards KR, Hupperts R, et al. ; SYNERGY Investigators. Efficacy analysis of opicinumab in relapsing multiple sclerosis: the phase 2b SYNERGY trial. Mult Scler 2016;22(suppl 3):P192. [Google Scholar]