Abstract
Many bacteria encode biosynthetic proteins that produce a vast array of natural products. These compounds are often synthesized during host invasion as they function as virulence factors. In addition, such secondary metabolites have yielded numerous molecular scaffolds with pharmaceutical and clinical importance. The gene clusters that encode proteins responsible for synthesis of these compounds are typically silenced or “cryptic” under laboratory growth conditions, hampering discovery of novel lead compounds. We report here that MftR is a global repressor of secondary metabolite synthesis in Burkholderia thailandensis and that urate functions as a physiologically relevant inducer of gene expression. Biosynthetic gene clusters under MftR control include those associated with production of the antimicrobial bactobolins, the iron siderophore malleobactin, and the virulence factor malleilactone. MftR also controls additional genes associated with survival in a host environment, such as genes encoding components of the type III secretion system (T3SS) and proteins linked to anaerobic respiration. This observation not only has implications for understanding activation of gene regulatory networks during host invasion, but it also paves the way for isolation of novel therapeutic leads.
Keywords: Natural product, gene regulation, secondary metabolites, MftR, urate, Burkholderia thailandensis
INTRODUCTION
The ecologically diverse genus Burkholderia includes the serious human pathogens B. pseudomallei and B. mallei and the closely related B. cepacia complex opportunistic pathogens, which primarily infect immunocompromised individuals such as persons with cystic fibrosis.1 B. thailandensis is a saprophyte that only causes mammalian infections at high doses. However, it conserves a number of genes encoding factors that are critical for pathogenicity of the more virulent species, such as the type III secretion system and quorum sensing systems that are key to control of virulence gene expression.2
B. thailandensis also shares with the virulent species a number of gene clusters that include predicted polyketide synthase (PKS) or nonribosomal peptide synthetase (NRPS) genes that are associated with synthesis of natural products. Such compounds include iron siderophores and toxins that enable the bacteria to compete with other species and they confer increased fitness and play important roles in establishment and propagation of infection.3 Biosynthetic gene clusters are usually tightly regulated, resulting in silencing or low-level expression under standard growth conditions, and identification of inducers of gene expression in vivo has proven challenging. While the structures of several natural products have been elucidated and their biological function deduced, the products of several such gene clusters remain to be identified. Since many clinically relevant antibiotics and other therapeutics are derived from natural products, triggering the expression of these cryptic gene clusters under laboratory growth conditions is of significant interest.4, 5
Quorum sensing (QS) may contribute to awakening of these silent gene clusters. QS involves the synthesis and secretion of signaling molecules that are detected by other members of the population, thereby allowing the bacteria to synchronize their gene expression programs.6 Such signaling molecules include N-acyl-homoserine lactones (AHLs), which are synthesized by members of the LuxI protein family and detected by LuxR-type regulators. In B. thailandensis, QS has been implicated in controlling expression of ScmR, a LysR-family transcription factor that in turn regulates expression of several biosynthetic gene clusters.7 B. thailandensis and B. pseudomallei each encode three AHL-based QS systems, of which two are conserved in B. mallei.8–10 In addition, two orphan LuxR homologs that lack a cognate LuxI have been identified in B. thailandensis, BtaR4 (also named MalR) and BtaR5.
The QS systems are interconnected and linked to signaling through a separate class of compounds, the 4-hydroxy-2-alkylquinolines (HAQs), which may also contribute to control expression of virulence genes.11 The operon encoding 4-hydroxy-2-heptylquinoline (HHQ) is conserved in B. thailandensis (and B. pseudomallei and B. cenocepacia) and extended by two additional open reading frames to generate hmqABCDEFG, responsible for synthesis of the methylated HHQ derivative 4-hydroxy-3-methyl-2-alkylquinoline (HMAQ).12, 13 It has been suggested that only the methylated derivatives play a role in quorum sensing in Burkholderia spp. since HHQ does not appear to accumulate.
We have previously characterized the transcription factor MftR from B. thailandensis and shown that repression of the mftR gene and a divergent operon (Fig. 1A) is relieved by urate.14, 15 Since MftR binds urate in vitro, MftR was therefore predicted to repress the divergent genes, with urate acting as the inducer. Urate is the product of xanthine dehydrogenase (XDH), which is also a key enzyme in purine salvage (Fig. 1C). In mammalian cells, bacterial infection may lead to conversion of XDH to xanthine oxidase, which produces reactive oxygen species (ROS) in addition to urate as an initial defense against the infection.16, 17 Bacterial infections have been reported to result in urate levels in excess of 200 μM and to be worsened by enhanced xanthine oxidase activity.18 This suggests that bacterial pathogens may encounter urate due to activation of host xanthine oxidase and that urate may function as a signaling molecule to elicit virulence gene expression. The affinity of MftR for urate is ~6 μM,15 suggesting that it responds to physiologically relevant concentrations of urate.
Figure 1.
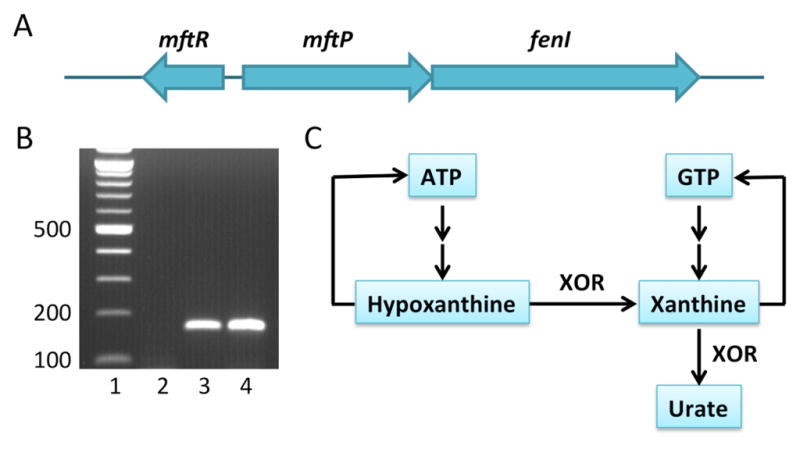
Locus encoding MftR and relevant steps in purine metabolism. A. The gene encoding MftR (BTH_I2391; new locus tag BTH_RS24400) is divergently oriented to an operon encoding the major facilitator superfamily transporter MftP and a glycosyl hydrolase, FenI. B. Verification of mftP-fenI operon by PCR amplification of fragment representing part of the mftP open reading frame. Lane 1, Mw marker; lane 2, negative control using RNA as template; lane 3, positive control using genomic DNA as template; lane 4, test sample with cDNA synthesized using fenI-specific primer as template; detection of an amplicon with mftP-specific primers reflects that cDNA synthesized with fenI–specific primer generated cDNA representing mftP. C. Abridged version of purine metabolism. Xanthine oxidoreductase (XOR) catalyzes conversion of hypoxanthine to xanthine to favor synthesis of GTP. Conversion of xanthine to urate diverts purines away from the salvage pathway. Hypoxanthine may be converted to ATP, while xanthine may be converted to GTP, both in several steps. The xanthine dehydrogenase (XDH) version of XOR primarily transfers substrate-derived electrons to NAD+ to generate NADH, whereas xanthine oxidase has decreased affinity for NAD+ and transfers electrons to molecular oxygen to produce reactive oxygen species.
We show here that MftR functions as a global repressor of numerous biosynthetic gene clusters and quorum sensing systems and as a regulator of other genes linked to survival in a host environment. This observation not only has implications for understanding activation of gene regulation networks during the environmental stress encountered during host invasion, but it also paves the way for isolation of novel therapeutic scaffolds by upregulating cryptic natural product gene clusters.
RESULTS AND DISCUSSION
While several bacterial species encode transcription factors that share homology with MftR, including the ability to bind urate,19–21 MftR is mainly encoded by Burkholderia species (Fig. S1). Members of the B. pseudomallei group that includes B. thailandensis conserve the mftP-fenI operon encoding a predicted efflux pump and a FenI family enzyme with predicted glycosyl hydrolase activity (Fig. 1A), whereas B. cepacia complex pathogens lack fenI. In contrast, phytopathogens and environmental species such as B. xenovorans and B. glumae encode an MftR homolog in a different genomic context. These distinctions closely match the recent phylogenetic classification of Burkholderia spp.1 To confirm the predicted mftP-fenI operon, cDNA was prepared using a fenI-specific primer, followed by PCR amplification using mftP-specific primers. A product of the expected size was obtained, confirming the annotated operon (Fig. 1B).
The MftR regulon
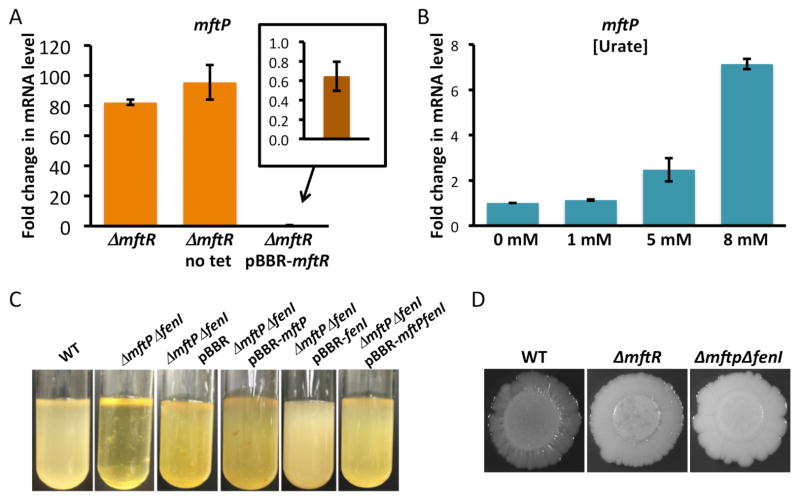
As noted above, MftR was predicted to repress mftR and mftP-fenI by direct binding to the intergenic region, with urate acting as an inducer.14, 15 To confirm this prediction, repression of the mftP-fenI operon by MftR was verified by targeted deletion of mftR followed by determination of transcript levels. As shown in Fig. 2A, expression of mftP was increased 80–100-fold in ΔmftR, while complementation with plasmid-encoded MftR completely restored repression. Consistent with observed repression, confirmed binding sites for MftR overlap with the predicted promoter elements for the divergent genes (Fig. S2A–B).
Figure 2.
Phenotypes of ΔmftR and ΔmftPΔfenI strains. A. Change in mftP transcript levels relative to WT determined by qRT-PCR for ΔmftR (grown with tetracycline or without antibiotic (no tet)) and ΔmftR-pBBR-mftR. Inset shows relative mftP transcript level in ΔmftR-pBBR-mftR. Transcript levels were calculated using 2−ΔΔCT relative to WT using reference gene GAPDH. Error bars represent standard deviation of three replicates. B. Gradual increase in mftP transcript levels with increasing concentrations of urate added to WT cells (1, 5, and 8 mM) calculated using 2−ΔΔCT relative to WT grown without urate. Error bars represent standard deviation of three replicates. C. Clumping in liquid culture resulting from deletion of fenI. Identified cultures were agitated just prior to capturing images, which are representative of three separate cultures. D. Colony morphology of mutant strains. Images were captured after incubation for ~5 days at 37 °C.
For a clue to physiological roles of MftP and Fen1, we examined a disruptant strain containing a transposon insertion in mftP (expected to be polar and to disrupt expression of fenI as well). Disruption of mftP-fenI resulted in very marked clumping during growth as well as a smooth colony morphology (Fig. 2C–D). Complementation with mftP did not prevent clumping, rather it resulted in the clumps attaining an orange tint, and expression of mftP-fenI was required to restore the wild-type growth phenotype. This indicates that absence of FenI is associated with clumping and conversely, that the increased expression of fenI that occurs in presence of the inducing MftR ligand (urate) would be expected to promote dispersal of cells. Burkholderia species produce several exopolysaccharides (EPS), but mainly cepacian, a major virulence determinant and participant in surface adhesion and biofilm maturation.22 A possible function of the glycosyl hydrolase activity predicted for FenI could therefore be cleavage of glycosidic linkages between constituent sugars in EPS, an activity predicted to lead to reduced clumping.
To define the MftR regulon, we isolated total RNA from wild-type B. thailandensis E264 and the corresponding ΔmftR strain after culturing each strain in absence or presence of 5 mM urate. The cDNA reads were mapped to the corresponding genes on the two B. thailandensis chromosomes. Comparison of genes exhibiting a difference in expression level of at least two-fold (q < 0.01) in wild-type and ΔmftR strains yielded 331 genes upregulated in absence of MftR (Table S1) and 70 genes downregulated (Table S2), indicating that MftR negatively regulates most genes in its regulon, either directly or indirectly. A comparison of genes differentially expressed on addition of urate to the wild-type strain yielded 321 genes upregulated (Table S1) and 45 genes downregulated at least two-fold in presence of urate (Table S2).
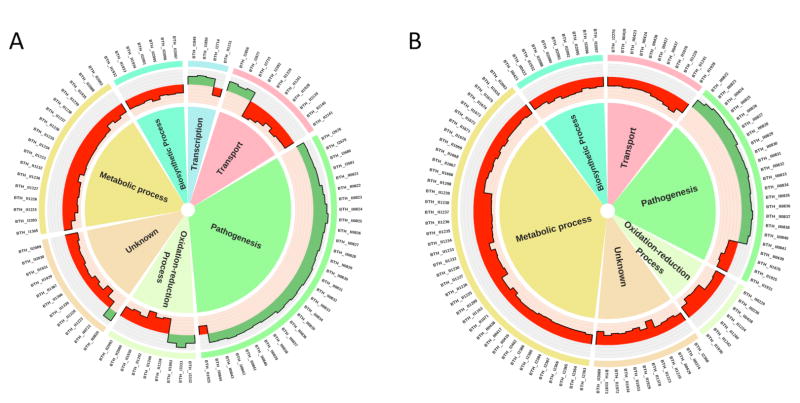
Examination of functional categories of genes that are differentially expressed on deletion of MftR or on addition of urate to WT cells revealed significant overlap (Tables S1 and S2). In general, genes associated with pathogenesis are downregulated in ΔmftR or in presence of urate, whereas genes linked to metabolic or biosynthetic processes or transport are upregulated (Fig. 3). Among all genes downregulated either in ΔmftR or in presence of urate (Table S2), 76% are downregulated to some extent in both conditions, with 41% downregulated two-fold or more in both conditions. For genes that exhibit increased expression in either condition (Table S1), 88% are expressed at a higher level in both conditions, with 34% upregulated two-fold or greater in both conditions.
Figure 3.
Circos plots showing differential gene expression. A. Higher levels of expression in wild-type compared to ΔmftR in green (genes activated by MftR) and reduced levels in red (genes repressed by MftR). Functional categories of genes identified in the central ring. B. Differential expression of genes in wild-type cells grown without or with 5 mM urate; genes repressed in presence of urate in green and genes induced by urate in red. Functional categories of genes identified in the central ring. The height of the bars corresponds to transcript level. Genes for which log2-fold change >|2| and P<0.05 are included.
As expected, genes encoding MftR, MftP, and FenI were significantly upregulated (10–30-fold) in ΔmftR. Notably, urate did not result in upregulation of these genes, which were previously seen to be upregulated ~14-fold when cultures were grown with 10 mM urate.15 The affinity of MftR for each of the two DNA sites in the mftR-mftP intergenic DNA is ~0.7 nM and its affinity for urate is ~6 μM.15 The Cheng-Prusoff equation23 relates the IC50 (the concentration of inhibitor – urate in this case – required for half-maximal inhibition) and affinity for the ligand that is being competed off (DNA). When the affinity for DNA is very high, the Cheng-Prusoff equation predicts that IC50 will be greater than the inhibition constant Ki (which corresponds to the ~6 μM affinity for urate), whereas the IC50 would be closer to the Ki when the affinity for DNA is low. We therefore considered if induction of mftP expression would be dependent on the concentration of urate; mftP expression was not induced at 1 mM urate and it increased with urate concentration, with ~7-fold induction with 8 mM urate (Fig. 2B). Urate, which is dissolved in NaOH, precipitates on addition to the culture medium due to the decrease in pH, confounding an estimate of actual concentrations in the culture; in addition, cellular content will depend on uptake. The exact cellular concentration of urate notwithstanding, the observation that mftP expression gradually increases with the amount of urate added to the culture medium leads to the inference that higher urate levels are required to relieve MftR-mediated repression of mftR and mftP-fenI. This observation also leads to the prediction that gene promoters to which MftR binds with lower affinity may be more sensitive to urate. The physiological implication is that some genes are induced at a urate concentration at which others remain repressed, perhaps effecting a temporal pattern of gene induction as the MftR ligand accumulates.
Differential expression of genes associated with anaerobic growth
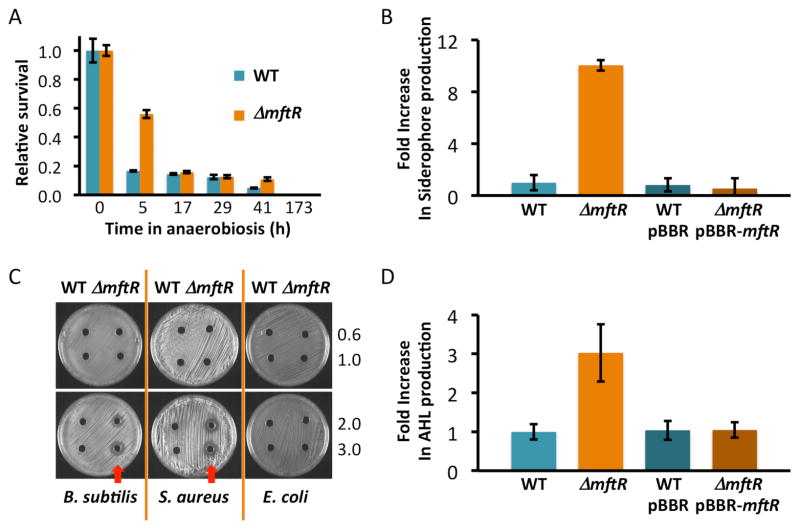
Deletion of mftR or addition of urate to WT cells led to 5–10-fold upregulation of genes encoding proteins that participate in the arginine deiminase pathway (arcDABC; BTH_I2383-2386; Table S1). This pathway has been implicated in anaerobic growth on rich medium, with the final step generating ATP.24, 25 Also upregulated were divergent operons encoding proteins required for synthesis of poly-hydroxybutyrate (PHB; BTH_II0416-0418) and ATP synthase (BTH_II0419-0427), respectively. ATP synthase utilizes the proton gradient for ATP synthesis, but it also functions to hydrolyze ATP to increase proton-motive force, a function that has been linked to survival under anaerobic conditions in P. aeruginosa.24 Upregulation of atpA-2 encoding F1F0 ATP synthase α subunit on deletion of MftR or on addition of 5 mM urate to WT cells was verified by qRT-PCR (Fig. S3A). In oxygen-deprived B. pseudomallei, several classes of genes were upregulated, including arcDABC, ATP synthase genes, and genes encoding PHB.26 Evidently, deletion of MftR resulted in upregulation of several genes that are also induced under anaerobiosis. Consistent with elevated expression of such genes, ΔmftR cells exhibited greater survival when maintained under low oxygen conditions for ~5 h, following which survival of ΔmftR cells approached that of WT cells (Fig. 4A).
Figure 4.
Phenotypes resulting from deletion of MftR. A. Survival under anaerobic conditions. Cells were transferred to airtight Hungate tubes at t = 0 h and grown in LB without antibiotics; survival was determined by plating at the indicated times. Data are normalized to viability at t = 0 h. Teal, WT; orange, ΔmftR. Error bars represent standard deviation of three replicates and are determined by standard propagation of errors. B. Siderophore production determined spectrophotometrically; data are represented as change in absorbance relative to control and normalized to absorbance change recorded for WT cells. Teal, WT; orange, ΔmftR; dark teal, WT-pBBR (empty vector); brown, ΔmftRpBBR- mftR. Error bars represent standard deviation of three replicates. C. Detection of antibacterial compounds in cell-free supernatants (from cells grown in LB without antibiotics). Supernatants were applied to holes in agar plates on which the indicated bacterial strains were spread. Culture supernatants were collected at OD600 of 0.6, 1.0, 2.0, and 3.0, as indicated at the right. For each set of plates, supernatant from WT was applied to the left and ΔmftR to the right. Clearing zones represent growth inhibition (arrows). D. AHL production determined spectrophotometrically; data are represented as change in absorbance relative to control and normalized to absorbance change recorded for WT cells. Color code as in (B). Error bars represent standard deviation of three replicates.
During anaerobic growth, nitrate is the favored electron acceptor, and respiratory nitrate reduction is much more efficient compared to the arginine deiminase pathway. However, concurrent signals of low oxygen and presence of nitrate (or a metabolite thereof) is required for expression of respiratory nitrate reductase genes. These genes are activated by NarL, which is part of the NarXL two-component system that responds to nitrate. In P. aeruginosa, NarL was also reported to repress the arcDABC operon.27 Notably, B. thailandensis narXL genes (BTH_I1849-1850) were downregulated ~6-fold in ΔmftR. It is therefore conceivable that reduced expression of B. thailandensis NarL is responsible for the observed increase in arcDABC expression.
The expression of nitrate reductase genes (BTH_I1851-1856) was also reduced in ΔmftR, consistent with reduced expression of the activator NarL. While narXL genes were also downregulated on addition of urate, nitrate reductase genes were upregulated (Table S1), suggesting a urate-mediated regulation that does not depend on MftR. One of the end-products of urate metabolism is ammonia (B. thailandensis is predicted to encode all enzymes required for degradation of urate (Fig. S4)). Considering that several Pseudomonas species, which are closely related to Burkholderia, have been reported to exhibit heterotrophic nitrification activity,28 one possibility is that B. thailandensis is likewise capable of converting ammonia to nitrite and nitrate; accumulation of these compounds would function with NarXL to upregulate cognate genes. Alternatively, the antioxidant properties of urate may produce a condition that emulates hypoxia, as evidenced by the observation that growth of anaerobes is enhanced when media is supplemented with antioxidants (including urate).29 That narXL expression is reduced in presence of urate may serve to balance expression of the respiratory nitrate reduction pathway with that of arginine fermentation.
Repression of genes encoding T3SS components in mftRΔ
The type III secretion system (T3SS), which functions to deliver effector toxins into host cells is critical for virulence.30 The B. pseudomallei T3SS is required for escape from the phagosome into the host cytosol, and it is downregulated during growth in macrophages.31, 32 A regulatory cascade has been reported in which BsaN is required for activation of genes encoding T3SS effector and translocon components.33 In B. thailandensis, expression of genes encoding the T3SS was reduced in mftRΔ and on addition of urate (that would be produced by cytosolic xanthine oxidase), perhaps reflecting its role only during early stages of infection. The B. thailandensis bsaN gene is among those downregulated (~6-fold), which could explain the similarly reduced expression of other T3SS genes. The downregulation of bsaN in mftRΔ or on addition of urate to WT cells was verified by qRT-PCR (Fig. S3B).
Regulation of biosynthetic gene clusters and quorum sensing
For enumeration of differentially expressed biosynthetic gene clusters, we applied a more stringent cutoff of log2 = 1.5 (an ~3-fold change in gene expression), but included all genes in operons in which at least one gene fits this description when comparing wild-type and ΔmftR or wild-type cells grown in absence or presence of urate (Table 1); neighboring genes encoding potential (or confirmed) transcriptional regulators were also noted. In general, the fold-change in expression level was greater for early gene(s) in each operon. Increased expression of select genes in ΔmftR and in WT cells grown with 5 mM urate was validated by qRT-PCR (Fig. S5).
Table 1.
MftR-controlled gene clusters encoding proteins with roles in quorum sensing and synthesis of secondary metabolites
| Locus tag BTH_ | Gene product/Secondary metabolite | log2 range mftRΔ/WT | log2 range WT(U)/WT(N) |
|---|---|---|---|
|
| |||
| I1952-1954 (+) | NRPS (unknown product) | 0.8 to 1.0 | 1.0 to 1.2 |
| I1955 (+) | 0.7 | 1.2 | |
| I1956-1967 (+) | 0.5 to 1.3 | 1.2 to 1.9 | |
| I1968-1971 (+) | 0.6 to 0.9 | 1.0 to 1.5 | |
|
| |||
| I2357-2358 (−) | 0.6 to 0.7 | 0.8 to 1.3 | |
| I2359-2367 (−) | Burkholdac | 0.8 to 1.8 | 1.8 to 2.6 |
| I2368 (−) | 1.8 | 2.4 | |
| I2369 (+) | AraC | 0.9 | 0.7 |
|
| |||
| I2414 (−) | 1.0 | 0.7 | |
| 2415-2418 (−) | Malleobactin | 1.0 to 1.4 | 0.6 to 0.7 |
| I2419 (+) | 1.4 | 0.4 | |
| I2420 (+) | 0.9 | 0.4 | |
| I2421-2423 (−) | 1.0 to 1.3 | 0.2 to 0.4 | |
| I2424-2427 (−) | ECF sigma factor | 1.5 to 2.2 | -0.2 to 0.1 |
|
| |||
| I2437a (+) | Capistruin | 1.5 | 1.3 |
| I2438-2440 (+) | 0.9 to 1.2 | 0.4 to 0.9 | |
|
| |||
| II0229-0234 (+) | Pyoverdine | 0.3 to 0.9 | 0.6 to 2.1 |
|
| |||
| II0803 (−) | Oxidoreductase | 1.6 | 0.9 |
| II0804 (−) | BtaI3 | 1.9 | 1.2 |
| II0805 (−) | BtaR3 | 1.9 | 1.4 |
|
| |||
| II1209-1219 (+) | PKS/NRPS | 0.3 to 1.0 | 0.8 to 2.3 |
| II1220-1221 (−) | 0.8 to 1.1 | 0.4 to 0.5 | |
| II1222 (+) | Bactobolin | 3.6 | 3.3 |
| II1223 (−) | 4.4 | 4.3 | |
| II1224 (−) | 4.6 | 4.4 | |
| II1225 (−) | 4.6 | 4.4 | |
| II1226-1228 (−) | BtaI2 | 2.7 to 4.8 | 1.4 to 4.0 |
| II1229-1230 (+) | 3.4 to 4.3 | 2.8 to 3.3 | |
| II1231 (−) | BtaR2 | 2.6 | 1.7 |
| I1232 (−) | 3.3 | 2.7 | |
| II1233-1241 (+) | 3.8 to 4.6 | 3.7 to 4.7 | |
| II1242 (−) | 5.1 | 4.4 | |
|
| |||
| II1662-1672 (−) | Thailandamide | 0.3 to 1.2 | 0.6 to 2.8 |
| II1673-1674 (−) | 0.5 | 2.2 to 2.4 | |
| II1675-1676 (−) | 0.6 to 0.8 | 2.3 to 2.4 | |
| II1680-1681 (−) | BtaR5 | 0.8 to 0.9 | 0.8 |
|
| |||
| II1823 (−) | Pyochelin | 1.6 | 0.5 |
| II1824-1828 (−) | 0.7 to 1.3 | 0.2 to 0.7 | |
| II1829 (−) | 1.5 | -0.3 | |
| II1830-1833 (+) | 0.8 to 1.2 | 0.4 to 0.7 | |
|
| |||
| II1923-1924 (+) | Nitroreductase | 1.7 to 2.1 | 0.1 to 0.3 |
| II1925 (+) | Chitin-binding | 2.2 | 2.9 |
| II1926-1927 (+) | Reductase | 0.9 to 1.3 | 0.8 to 1.6 |
| II1928 (−) | Membrane protein | 2.5 | 2.6 |
| II1929-1935 (−) | HMAQ | 2.6 to 2.9 | 2.5 to 3.3 |
|
| |||
| II2084-2086 (−) | 0.3 to 1.2 | 0.4 to 0.8 | |
| II2087 (−) | MalR (BtaR4) | 1.8 | 1.2 |
| II2088 (+) | 3.1 | 2.3 | |
| II2089 (+) | 3.7 | 3.4 | |
| II2090-2099 (+) | Malleilactone | 1.6 to 3.3 | 1.6 to 3.0 |
Predicted operons according the Burkholderia Genome Database are indicated by a range of locus tags, with the coding strand identified by (+) and (−). Identified natural products encoded by the corresponding gene clusters are highlighted in boldface. Log2-fold changes show that MftR functions as a repressor in all instances and that urate induces expression. For annotation of each gene product, see Supplementary Table S1.
Several well-characterized gene clusters for which the corresponding metabolite(s) have been structurally and functionally identified were repressed by MftR and induced in presence of urate. A gene cluster annotated as two adjacent operons, BTH_I2357-2358 and BTH_I2359-2367, was previously shown to encode proteins responsible for synthesis of burkholdacs, which function as histone deacetylase inhibitors. These compounds were reported to accumulate on induction of a construct expressing the adjacent AraC family transcription factor encoded by BTH_I2369, suggesting that this transcription factor functions as an activator.34 This otherwise poorly expressed gene cluster was also reported to be induced in presence of trimethoprim.5 Genes encoding proteins involved in synthesis of burkholdacs, including AraC, are markedly induced on deletion of ScmR, a LysR-family transcription factor recently reported to control expression of several biosynthetic gene clusters.7 The scmR expression is increased 2–3-fold in ΔmftR or on addition of urate, whereas mftR expression is unaffected by deletion of ScmR, suggesting that MftR acts upstream of ScmR (Table S1).
Malleobactins, encoded by BTH_I2414-2426, function as iron siderophores and the corresponding genes encoding NRPSs and iron siderophore transport proteins in B. pseudomallei were shown to be upregulated under iron-limiting conditions.35 BTH_I2427 encodes a predicted extracytoplasmic function (ECF) sigma factor, which in B. pseudomallei is involved in regulation of the malleobactin gene cluster. While the gene encoding the ECF sigma factor is upregulated ~5-fold in absence of MftR, no effect of urate was observed; expression of other genes in the malleobactin cluster was also less affected by urate addition than by absence of MftR, consistent with a role for ECF in gene regulation. We speculate that the inability of urate to induce expression may be linked to the affinity of MftR for the ecf gene promoter.
The antibacterial compound capistruin is a ribosomally synthesized 19-amino acid peptide that is post-translationally modified.36 In B. thailandensis, the capistruin peptide precursor is encoded by BTH_I2437a and the modifying enzymes by BTH_I2438-2440. These genes are also repressed by ScmR.7
Most of the biosynthetic gene clusters under MftR control are encoded on chromosome II. A predicted pyoverdine biosynthetic cluster encoded by BTH_II0229-00234 was upregulated ~2-fold in mftRΔ and on addition of urate. Pyoverdines are the primary siderophores in P. aeruginosa, where their production is under control of both an ECF sigma factor (the ortholog of BTH_I2427) and quorum sensing.37 The more significant upregulation on addition of urate compared to deletion of mftR suggests that urate causes upregulation by an additional mechanism, possibly by chelating iron and inducing an ironlimiting condition.
The siderophore pyochelin has lower affinity for iron compared to other compounds, including pyoverdine.38 In the iron-limited environment of a host, siderophores are important virulence factors, facilitating iron uptake by means of TonB-dependent receptors.39 The B. thailandensis pyochelin gene cluster (BTH_II1823-1833) is upregulated in ΔmftR, along with genes encoding TonB and the associated ExbB/ExbD transporter (BTH_II2024-2026) and a TonB-dependent receptor encoded in an operon that also includes hemin transport proteins (BTH_II2139-2141). This suggests that MftR controls expression of multiple iron-acquisition systems. To determine if ΔmftR cells indeed produce elevated levels of siderophores, we used a universal siderophore assay in which chromeazurol S and hexadecyltrimethylammonium bromide complexes with ferric iron to produce a blue color; in presence of a siderophore, the color will change from blue to orange. As shown in Fig. 4B, ΔmftR cells produced markedly higher levels of siderophores, whereas siderophore production returned to basal levels on complementation with plasmid-borne mftR.
Phenazines such as pyoverdine are redox active and may act as alternate electron acceptors, and their synthesis has been associated with reduced wrinkling during colony growth.40 While wild-type B. thailandensis forms wrinkled colonies to maximize oxygen uptake through a larger surface area, ΔmftR cells formed smooth colonies, consistent with increased production of alternate electron acceptors (Fig. 2D). The smooth colony morphology of ΔmftPΔfen1 is reminiscent of the delayed wrinkling reported when P. aeruginosa is deleted for MexGHI-OpmD, the efflux system through which phenazines are exported;41 a possible explanation for the smooth colony phenotype characteristic of ΔmftPΔfen1 is therefore that MftP functions to efflux these compounds.
Some gene clusters include both PKS- and NRPS-encoding genes; one such gene cluster encodes proteins involved in synthesis of bactobolins, some of which are potent antibiotics.42, 43 Many genes in the bactobolin gene cluster are upregulated >20-fold in absence of MftR (Table 1 and Fig. S5). Expression of genes encoding bactobolin biosynthetic proteins is regulated by BtaR2, a LuxR-type regulator encoded by BTH_II1231.10 The cognate AHL is synthesized by BtaI2 (BTH_II1227), which is upregulated >15-fold in ΔmftR or on addition of urate. Global analysis of gene expression in a strain deleted for BtaR2 indicated that BtaR2 is dedicated to control of the bactobolin gene cluster.44 Consistent with previous reports that bactobolins were particularly active against Gram-positive bacteria,45 we observed a greater zone of inhibition in an agar well diffusion assay using cell-free culture supernatant of ΔmftR cells compared to WT (Fig. 4C).
The anti-proliferative polyunsaturated polyketide thailandamide is produced by proteins encoded by BTH_II1662-1676, and the genes are under control of the orphan LuxR homolog BtaR5 encoded by BTH_II1681.46 Expression of genes encoding proteins involved in malleilactone biosynthesis (BTH_II2084-2099) is also under control of an orphan LuxR (BtaR4) that is encoded by BTH_II2087 and renamed MalR.47 Malleilactone promotes virulence, as evidenced by prolonged survival of Caenorhabditis elegans fed B. thailandensis lacking an essential malleilactone biosynthesis gene.3 The malleilactone gene cluster is normally silent, but is among the several gene clusters induced by trimethoprim.5, 47
The BtaI3-BtaR3 quorum sensing system (BTH_II0804-0805) is upregulated ~3-fold in ΔmftR (Table 1 and Fig. S5). We examined production of AHL using the Agrobacterium tumefaciens NTL4/pZLR4 reporter strain, which harbors a plasmid that confers AHL-sensitive production of β-galactosidase. Cell-free B. thailandensis supernatants were mixed with A. tumefaciens NTL4/pZLR4 in presence of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal), and the absorbance at 635 nm was recorded. As shown in Fig. 4D and S6, deletion of MftR resulted in increased AHL production whereas AHL production in cells complemented with pBBR-mftR was indistinguishable from that in WT cells.
Numerous HMAQs are produced by proteins encoded within the hmqABCDEFG operon (BTH_II1929-1935).12 All genes within this cluster were upregulated at least 6-fold in ΔmftR or on addition of urate (and verified by qRT-PCR; Fig. S5). In B. ambifaria, inactivation of genes within this cluster resulted in increased AHL production, while the opposite was reported on inactivation of a structural gene in B. pseudomallei, reflecting a complex QS network.12, 13 ScmR was reported to activate expression of genes encoding HMAQs;7 the increased expression of the hmaq gene cluster in mftRΔ may therefore be a consequence of increased scmR expression.
Induction by trimethoprim
As noted above, trimethoprim was identified as an inducer of certain biosynthetic gene clusters.5, 47 A recent analysis of the B. thailandensis metabolome demonstrated that a large number of compounds accumulate when cells are exposed to trimethoprim,4 suggesting that trimethoprim is a global inducer of secondary metabolite biosynthesis, much like urate is a global inducer of biosynthetic gene clusters that are repressed by MftR. In addition to bactobolins, capistruin, HMAQ, burkholdacs, malleilactone, and thailandamide, trimethoprim was shown to elicit production of terphenyl.4 Terphenyl is produced by proteins encoded by BTH_II0204-0206, a gene cluster that was modestly upregulated in absence of MftR.
While the biosynthetic gene clusters that are upregulated in presence of urate correspond to the compounds that accumulate in cultures grown with trimethoprim, trimethoprim is unlikely to be direct inducer of the MftR regulon, as DNA binding by MftR is unaffected by the addition of trimethoprim (Fig. S7A). Trimethoprim is a folate biosynthesis inhibitor, and it inhibits enzymatic reactions that require tetrahydrofolate derivatives, including steps in purine biosynthesis Under conditions of tetrahydrofolate deficiency, the biosynthetic intermediate 5-aminoimidazole-4-carboxamide-ribonucleotide (AICAR) was reported to accumulate and increased synthesis of both xanthine and adenine transporters was observed.48 Since AICAR did not inhibit DNA binding by MftR either (Fig. S7B), we speculate that other intermediates may accumulate or that an increased expression of purine transporters may lead to increased levels of inducing ligands for MftR.
Model for gene regulation by MftR
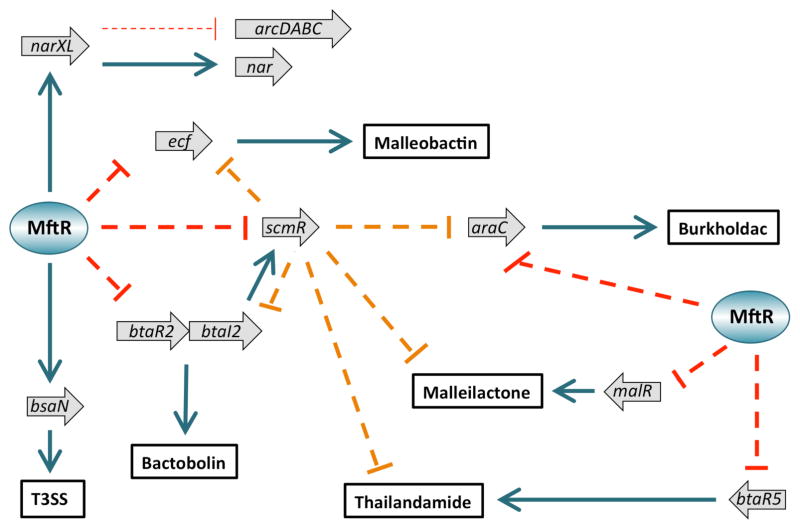
Successful host invasion is associated with activation of gene regulatory networks that operate together to control production of virulence factors and enhance fitness. Our data suggest that MftR functions at the top of a hierarchy of regulators that controls expression of genes linked to anaerobic respiration, T3SS, quorum sensing, and production of secondary metabolites. For example, MftR directly or indirectly activates expression of bsaN, and BsaN in turn activates expression of T3SS genes; MftR also activates narXL, and NarL has been implicated in repression of the arcDABC gene cluster and activation of nitrate reductase genes (nar) (Fig. 5).
Figure 5.
Model for gene regulation by MftR. Representative genes that are under MftR control are represented by grey block arrows. Activation is indicated by solid arrows (teal) and repression by dashed lines (red). End products are identified in bold frame. Thin line represents inferred regulation based on data from P. aeruginosa. All identified secondary metabolites accumulated after addition of trimethoprim.4
MftR also represses the gene encoding ScmR, which was recently reported to control biosynthetic gene clusters in response to quorum sensing signals.7 Both MftR and ScmR repress btaRI2, however, the increased ScmR production expected in mftRΔ cannot overcome the derepression of btaRI2 due to deletion of MftR. BtaRI2 in turn activates expression of the bactobolin biosynthetic gene cluster. Since AHL produced by BtaI2 functions to activate scmR, we cannot rule out that MftR-mediated repression of scmR is indirect and due to repression of BtaRI2 (Fig. 5). A similar pattern of gene regulation by MftR and ScmR pertains for ecf; both transcription factors repress ecf, yet the phenotype of the mftR deletion is ecf derepression, despite presumably increased ScmR levels in ΔmftR. ECF has been implicated in control of the malleobactin biosynthetic gene cluster. Likewise, increased repression of the araC gene linked to activation of the burkholdac biosynthetic gene cluster would be expected in ΔmftR if araC were regulated by ScmR only; however, the opposite is observed, with increased araC expression in ΔmftR, indicating separate control of araC by the two transcription factors (Fig. 5). The malleilactone and thailandamide biosynthetic gene clusters were also reported to be under ScmR control, however, the genes encoding the cognate transcription factors, MalR and BtaR5, were not.7 By contrast, both malR and btaR5 are repressed by MftR, clearly indicating distinct regulatory pathways for MftR and ScmR in these cases.
The gene encoding ScmR is upregulated in response to increased cell density, and this results in activation of certain stress response genes and in repression of biosynthetic gene clusters. However, when an inducing ligand for MftR (urate) accumulates, derepression of biosynthetic gene clusters occurs; since exposure to purine metabolites such as urate would occur when bacterial infection elicits host xanthine oxidase activity, we suggest that differential expression of genes in the MftR regulon occurs to enhance bacterial fitness in the host environment. The concomitant induction of scmR may serve as an added layer of control to prevent overproduction of secondary metabolites under conditions of high cell density.
Sequencing of bacterial genomes have revealed that Burkholderia spp. harbor numerous biosynthetic gene clusters. These secondary metabolites have important roles as virulence factors, yet signals that trigger their production in vivo have proven difficult to identify. The discovery that the urate-inducible MftR functions as a global repressor of biosynthetic gene clusters and quorum sensing systems has implications for understanding activation of gene regulation networks during host invasion, and it paves the way for isolation of novel therapeutic scaffolds by upregulating cryptic natural product gene clusters. The pathology of infections with Burkholderia spp. includes the existence of environments with limited oxygen supply, whether in the CF lung characterized by accumulation of mucus, in the abscesses that are associated with B. pseudomallei infections, or in the interior of biofilms. Evidently, MftR also controls expression of numerous genes whose products are required under anaerobiosis, suggesting a key role in adaptation to reduced oxygen environments and thereby virulence.
METHODS
Detailed procedures for construction of ΔmftR, verification of ΔmftRΔfenI, and construction of the complemented strains are described in the Supplemental Information, along with procedures for RNA isolation, RNA seq, data analysis, and qRT-PCR. Sequences of primers used are shown in Supplemental Tables S3 and S4.
Siderophore production was measured using chromazurol S (CAS) and acyl homoserine lactone production was measured using the indicator strain Agrobacterium tumefaciens NTL4/pZLR4, which harbors a plasmid that confers AHL-sensitive production of β-galactosidase. Survival under anaerobic conditions was assessed by incubating cultures in air-tight Hungate tubes. Effect of different compounds on DNA binding by MftR was assessed using electrophoretic mobility shift assays. Detailed procedures for all assays are available in Supplemental Information.
Supplementary Material
Acknowledgments
We thank C. Tikhe for assistance with imaging of colony morphology. The authors declare no conflict of interest. This work was supported by the National Institutes of Health (1R15GM107825 to A. Grove) and by the National Science Foundation (MCB-1714219 to A. Grove). G.W. and M.D. were supported by the National Science Foundation (MCB-1616827 to M.D.), and by the Next-Generation BioGreen21 Program of the Rural Development Administration, Republic of Korea (grant no. PJ011379 to M.D.).
Footnotes
Accession codes. RNA-Seq data have been deposited to the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo, under accession no. GSE100659.
Supporting Information Available
Detailed experimental procedures and sequences of primers used. Tables of differentially regulated genes. Figures showing growth of mutant strains, qRT-PCR validation of gene expression, production of AHL, and DNA-binding assays. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Sawana A, Adeolu M, Gupta RS. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet. 2014;5:429. doi: 10.3389/fgene.2014.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect Immun. 2008;76:5402–5411. doi: 10.1128/IAI.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggins JB, Ternei MA, Brady SF. Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J Am Chem Soc. 2012;134:13192–13195. doi: 10.1021/ja3052156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada BK, Wu Y, Mao D, Bushin LB, Seyedsayamdost MR. Mapping the Trimethoprim-Induced Secondary Metabolome of Burkholderia thailandensis. ACS Chem Biol. 2016;11:2124–2130. doi: 10.1021/acschembio.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seyedsayamdost MR. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A. 2014;111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramoni S, Sokol PA. Quorum sensing systems influence Burkholderia cenocepacia virulence. Future Microbiol. 2012;7:1373–1387. doi: 10.2217/fmb.12.118. [DOI] [PubMed] [Google Scholar]
- 7.Mao D, Bushin LB, Moon K, Wu Y, Seyedsayamdost MR. Discovery of ScmR as a global regulator of secondary metabolism and virulence in Burkholderia thailandensis E264. Proc Natl Acad Sci U S A. 2017;114:E2920–E2928. doi: 10.1073/pnas.1619529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulrich RL, Hines HB, Parthasarathy N, Jeddeloh JA. Mutational analysis and biochemical characterization of the Burkholderia thailandensis DW503 quorum-sensing network. J Bacteriol. 2004;186:4350–4360. doi: 10.1128/JB.186.13.4350-4360.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler JR, Duerkop BA, Hinz A, West TE, Herman JP, Churchill ME, Skerrett SJ, Greenberg EP. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J Bacteriol. 2009;191:5901–5909. doi: 10.1128/JB.00591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duerkop BA, Varga J, Chandler JR, Peterson SB, Herman JP, Churchill ME, Parsek MR, Nierman WC, Greenberg EP. Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J Bacteriol. 2009;191:3909–3918. doi: 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, Williams P. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Diggle SP, Lumjiaktase P, Dipilato F, Winzer K, Kunakorn M, Barrett DA, Chhabra SR, Camara M, Williams P. Functional genetic analysis reveals a 2-Alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem Biol. 2006;13:701–710. doi: 10.1016/j.chembiol.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Vial L, Lepine F, Milot S, Groleau MC, Dekimpe V, Woods DE, Deziel E. Burkholderia pseudomallei, B. thailandensis, and B ambifaria produce 4- hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J Bacteriol. 2008;190:5339–5352. doi: 10.1128/JB.00400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grove A. Urate-responsive MarR homologs from Burkholderia. Mol Biosyst. 2010;6:2133–2142. doi: 10.1039/c0mb00086h. [DOI] [PubMed] [Google Scholar]
- 15.Gupta A, Grove A. Ligand-binding pocket bridges DNA-binding and dimerization domains of the urate-responsive MarR homologue MftR from Burkholderia thailandensis. Biochemistry. 2014;53:4368–4380. doi: 10.1021/bi500219t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin HM, Hancock JT, Salisbury V, Harrison R. Role of xanthine oxidoreductase as an antimicrobial agent. Infect Immun. 2004;72:4933–4939. doi: 10.1128/IAI.72.9.4933-4939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segal BH, Sakamoto N, Patel M, Maemura K, Klein AS, Holland SM, Bulkley GB. Xanthine oxidase contributes to host defense against Burkholderia cepacia in the p47(phox−/−) mouse model of chronic granulomatous disease. Infect Immun. 2000;68:2374–2378. doi: 10.1128/iai.68.4.2374-2378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crane JK, Naeher TM, Broome JE, Boedeker EC. Role of host xanthine oxidase in infection due to enteropathogenic and Shiga-toxigenic Escherichia coli. Infect Immun. 2013;81:1129–1139. doi: 10.1128/IAI.01124-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perera IC, Grove A. MarR homologs with urate-binding signature. Protein Sci. 2011;20:621–629. doi: 10.1002/pro.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deochand DK, Meariman JK, Grove A. pH-Dependent DNA Distortion and Repression of Gene Expression by Pectobacterium atrosepticum PecS. ACS Chem Biol. 2016;11:2049–2056. doi: 10.1021/acschembio.6b00168. [DOI] [PubMed] [Google Scholar]
- 21.Deochand DK, Grove A. MarR family transcription factors: Dynamic variations on a common scaffold. Crit Rev Biochem Mol Biol. 2017 doi: 10.1080/10409238.2017.1344612. In press. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira AS, Leitao JH, Silva IN, Pinheiro PF, Sousa SA, Ramos CG, Moreira LM. Distribution of cepacian biosynthesis genes among environmental and clinical Burkholderia strains and role of cepacian exopolysaccharide in resistance to stress conditions. Appl Environ Microbiol. 2010;76:441–450. doi: 10.1128/AEM.01828-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 24.Glasser NR, Kern SE, Newman DK. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol. 2014;92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platt MD, Schurr MJ, Sauer K, Vazquez G, Kukavica-Ibrulj I, Potvin E, Levesque RC, Fedynak A, Brinkman FS, Schurr J, Hwang SH, Lau GW, Limbach PA, Rowe JJ, Lieberman MA, Barraud N, Webb J, Kjelleberg S, Hunt DF, Hassett DJ. Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J Bacteriol. 2008;190:2739–2758. doi: 10.1128/JB.01683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamad MA, Austin CR, Stewart AL, Higgins M, Vazquez-Torres A, Voskuil MI. Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob Agents Chemother. 2011;55:3313–3323. doi: 10.1128/AAC.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benkert B, Quack N, Schreiber K, Jaensch L, Jahn D, Schobert M. Nitrate-responsive NarX-NarL represses arginine-mediated induction of the Pseudomonas aeruginosa arginine fermentation arcDABC operon. Microbiology. 2008;154:3053–3060. doi: 10.1099/mic.0.2008/018929-0. [DOI] [PubMed] [Google Scholar]
- 28.Castignetti D, Hollocher TC. Heterotrophic nitrification among denitrifiers. Appl Environ Microbiol. 1984;47:620–623. doi: 10.1128/aem.47.4.620-623.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dione N, Khelaifia S, La Scola B, Lagier JC, Raoult D. A quasi-universal medium to break the aerobic/anaerobic bacterial culture dichotomy in clinical microbiology. Clin Microbiol Infect. 2016;22:53–58. doi: 10.1016/j.cmi.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 30.Hauser AR. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol. 2009;7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens MP, Wood MW, Taylor LA, Monaghan P, Hawes P, Jones PW, Wallis TS, Galyov EE. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol Microbiol. 2002;46:649–659. doi: 10.1046/j.1365-2958.2002.03190.x. [DOI] [PubMed] [Google Scholar]
- 32.Chieng S, Carreto L, Nathan S. Burkholderia pseudomallei transcriptional adaptation in macrophages. BMC Genomics. 2012;13:328. doi: 10.1186/1471-2164-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Schroder I, French CT, Jaroszewicz A, Yee XJ, Teh BE, Toesca IJ, Miller JF, Gan YH. Characterization and analysis of the Burkholderia pseudomallei BsaN virulence regulon. BMC Microbiol. 2014;14:206. doi: 10.1186/s12866-014-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biggins JB, Gleber CD, Brady SF. Acyldepsipeptide HDAC inhibitor production induced in Burkholderia thailandensis. Org Lett. 2011;13:1536–1539. doi: 10.1021/ol200225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alice AF, Lopez CS, Lowe CA, Ledesma MA, Crosa JH. Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243. J Bacteriol. 2006;188:1551–1566. doi: 10.1128/JB.188.4.1551-1566.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knappe TA, Linne U, Zirah S, Rebuffat S, Xie X, Marahiel MA. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J Am Chem Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 37.Schalk IJ, Guillon L. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ Microbiol. 2013;15:1661–1673. doi: 10.1111/1462-2920.12013. [DOI] [PubMed] [Google Scholar]
- 38.Cornelis P, Dingemans J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol. 2013;3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krewulak KD, Vogel HJ. TonB or not TonB: is that the question? Biochem Cell Biol. 2011;89:87–97. doi: 10.1139/o10-141. [DOI] [PubMed] [Google Scholar]
- 40.Okegbe C, Price-Whelan A, Dietrich LE. Redox-driven regulation of microbial community morphogenesis. Curr Opin Microbiol. 2014;18:39–45. doi: 10.1016/j.mib.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakhtah H, Koyama L, Zhang Y, Morales DK, Fields BL, Price-Whelan A, Hogan DA, Shepard K, Dietrich LE. The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc Natl Acad Sci U S A. 2016;113:E3538–3547. doi: 10.1073/pnas.1600424113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seyedsayamdost MR, Chandler JR, Blodgett JA, Lima PS, Duerkop BA, Oinuma K, Greenberg EP, Clardy J. Quorum-sensing-regulated bactobolin production by Burkholderia thailandensis E264. Org Lett. 2010;12:716–719. doi: 10.1021/ol902751x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carr G, Seyedsayamdost MR, Chandler JR, Greenberg EP, Clardy J. Sources of diversity in bactobolin biosynthesis by Burkholderia thailandensis E264. Org Lett. 2011;13:3048–3051. doi: 10.1021/ol200922s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majerczyk C, Brittnacher M, Jacobs M, Armour CD, Radey M, Schneider E, Phattarasokul S, Bunt R, Greenberg EP. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol. 2014;196:1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandler JR, Truong TT, Silva PM, Seyedsayamdost MR, Carr G, Radey M, Jacobs MA, Sims EH, Clardy J, Greenberg EP. Bactobolin resistance is conferred by mutations in the L2 ribosomal protein. MBio. 2012;3 doi: 10.1128/mBio.00499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishida K, Lincke T, Behnken S, Hertweck C. Induced biosynthesis of cryptic polyketide metabolites in a Burkholderia thailandensis quorum sensing mutant. J Am Chem Soc. 2010;132:13966–13968. doi: 10.1021/ja105003g. [DOI] [PubMed] [Google Scholar]
- 47.Truong TT, Seyedsayamdost M, Greenberg EP, Chandler JR. A Burkholderia thailandensis Acyl-Homoserine Lactone-Independent Orphan LuxR Homolog That Activates Production of the Cytotoxin Malleilactone. J Bacteriol. 2015;197:3456–3462. doi: 10.1128/JB.00425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stepanek JJ, Schakermann S, Wenzel M, Prochnow P, Bandow JE. Purine biosynthesis is the bottleneck in trimethoprim-treated Bacillus subtilis. Proteomics Clin Appl. 2016;10:1036–1048. doi: 10.1002/prca.201600039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.