Abstract
Polarization sensitive optical coherence tomography (PSOCT) with serial sectioning has enabled the investigation of 3D structures in mouse and human brain tissue samples. By using intrinsic optical properties of back-scattering and birefringence, PSOCT reliably images cytoarchitecture, myeloarchitecture and fiber orientations. In this study, we developed a fully automatic serial sectioning polarization sensitive optical coherence tomography (as-PSOCT) system to enable volumetric reconstruction of human brain samples with unprecedented sample size and resolution. The 3.5µm in-plane resolution and 50µm through-plane voxel size allow inspection of cortical layers that are a single-cell in width, as well as small crossing fibers. We show the abilities of as-PSOCT in quantifying layer thicknesses of the cerebellar cortex and creating microscopic tractography of intricate fiber networks in the subcortical nuclei and internal capsule regions, all based on volumetric reconstructions. as-PSOCT provides a viable tool for studying quantitative cytoarchitecture and myeloarchitecture and mapping connectivity with microscopic resolution in the human brain.
Introduction
The human brain consists of 80 – 100 billion interconnected neurons. To date, our knowledge of how these neurons organize and connect to enable complex functions is remarkably limited. Directly imaging the anatomical substrate requires a new generation of imaging technologies that afford microscopic resolution and the capability to examine cubic centimeters of tissue. The Big Brain project (Amunts et al., 2013) has made significant contributions in reconstructing a whole human brain with 7404 histological sections. The brain was sectioned with 20µm slices, stained with silver impregnation and registered in 3D for cytoarchitectural investigation and modeling. One challenge with the histological method is the slice-specific distortions that are introduced due to cutting, slice mounting, dehydration and staining (Arsigny et al., 2005; Dauguet et al., 2007; Osechinskiy and Kruggel, 2010). These distortions make 3D registration extremely difficult and prone to errors that prevent accurate reconstruction at the micron scale (Mosaliganti et al., 2006; Yushkevich et al., 2006).
Blockface imaging with serial sectioning overcomes this problem and enables accurate volumetric reconstruction due to the absence of tissue distortion (Odgaard et al., 1990). Blockface imaging techniques employ a microscope that operates in a reflective mode and thus can image the superficial surface of tissue in a thick block. Slicing is conducted after imaging. In this way, 3D reconstruction is easily accomplished by stacking all the blockface images, without requiring sophisticated inter-slice registration algorithms. With advances in 3D microscopy, including two-photon (Tsai et al., 2003), confocal (Sands et al., 2005), structure illumination (Mertz, 2011; Osten and Margrie, 2013), and molecular or genetically encoded labeling techniques, several groups have reported brain-wide reconstruction of cytoarchitecture and connectivity in rodents (Economo et al., 2016; Ragan et al., 2012; Yuan et al., 2015). The advantage of cellular resolution enables the investigation of neuronal architecture, and potentially connectivity mapping across the brain.
Optical coherence tomography (OCT) expands the advantages of blockface imaging by facilitating reconstructions of larger volumes of tissue and by utilizing endogenous sources of contrast. OCT uses optical interferometry to image depth-resolved tissue structures at a resolution of 1–20 µm (Huang et al., 1991). OCT collects back-scattered light from the tissue and enables probing of different structures based on the scattering information. Due to the endogenous contrast, OCT has been utilized in various human applications (see reviews by (Adhi and Duker, 2013; Bezerra et al., 2009; Drexler and Fujimoto, 2008; Gambichler et al., 2015)) including the brain both ex vivo and in vivo (Assayag et al., 2013; Bohringer et al., 2009; Boppart et al., 1998). Previous studies have shown that OCT with 3.5 µm resolution was capable of discriminating the laminar structure of the cerebral cortex in human brain samples (Magnain et al., 2014). At higher resolution of 1.3 µm, optical coherence microscopy (OCM) has the capability of identifying single neurons and fiber tracts in the human cortex (Magnain et al., 2015, 2016). Polarization sensitive optical coherence tomography (PSOCT) is a variation of conventional OCT (de Boer et al., 1997) that uses polarized light to probe birefringence, an optical property of anisotropic structures commonly seen in tissue such as collagen, tendon, muscle and nerve fibers. Due to the birefringence of the myelin sheath that surrounds many axons, PSOCT has demonstrated the ability to distinguish gray matter from white matter, and to measure the orientation of fiber tracts both in mouse (Nakaji et al., 2008; Wang et al., 2016, 2011) and human (Wang et al., 2014b) brains. In combination with manual serial sectioning, we have previously reported 3D reconstruction of brain samples with 15 µm in-plane resolution and slice thickness of 100 – 200 microns, which allows the creation of undistorted maps of fibers, cortical layers and subcortical regions at a mesoscopic resolution (Wang et al., 2014a).
In this study, we advance the PSOCT technique to a fully automated system for human brain imaging, named automatic serial sectioning polarization sensitive optical coherence tomography (as-PSOCT), which dramatically enhances the efficacy for imaging and processing, and provides an appealing solution for big data management. We improve the resolution of PSOCT to 3.5 µm to enable the investigation of single-cell layers and small crossing fibers in the brain. The pipeline allows imaging and reconstruction of cubic centimeter tissue blocks over multiple days without the need for human intervention. Quantification of cerebellar cortical layer thickness shows the significant advantages of 3D cytoarchitectural analysis overcoming the bias based on 2D slice estimations. In addition, microscopic tractography with 30 µm is created to reveal the intricate fiber networks that are beyond the capability of diffusion MRI techniques.
Methods
Materials
Two human brains (mean age 53.5 ± 12.0 y.o., 1 male and 1 female) were obtained from the Massachusetts General Hospital Autopsy Suite. The brains were neurologically normal without previous diagnosis of neurological deficits. The samples were fixed by immersion in 10% formalin for at least two months. The post-mortem interval did not exceed 24 h. The brains were cut into cubic centimeter blocks. Cerebellar and thalamic regions were included in the study.
System description
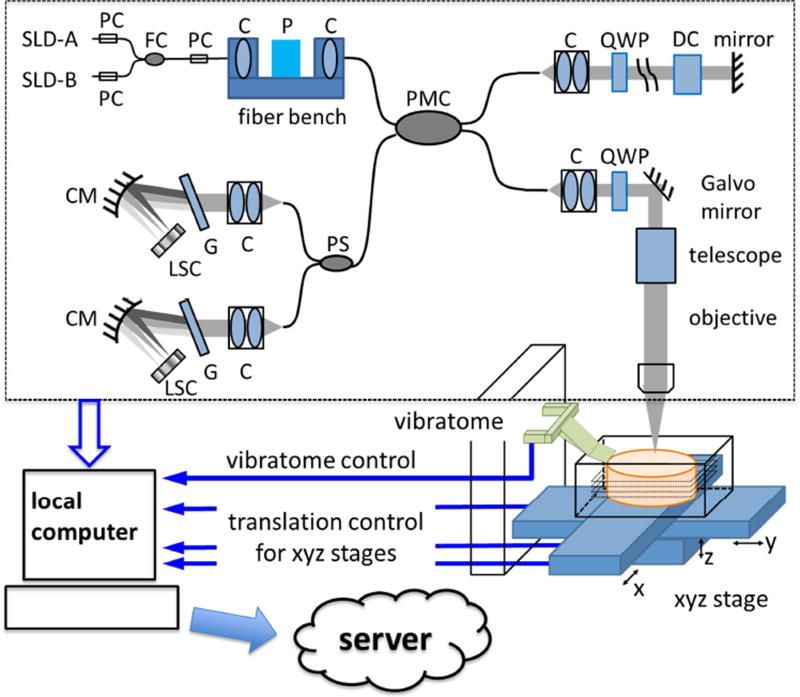
The as-PSOCT system is composed of an in-house built spectral domain PSOCT, motorized xyz translational stages, a vibratome to section the tissue block. A local computer coordinates PSOCT acquisition, xyz stage translation and vibratome slicing for automatic imaging of tissue blocks. The local computer communicates with a computing sever to transfer data and perform imaging reconstruction in parallel to data acquisition (Figure 1). Customized software is optimized for the pipeline of data acquisition, tissue sectioning, and image reconstruction
Fig. 1.
System schematic of as-PSOCT. The dashed block shows the home built spectral domain PSOCT. C, collimator; CM, concave mirror; DC, dispersion compensation block; FC, fiber coupler; G, grating; LSC, line scan camera; P, polarizer; PC, polarization controller; PMC, polarization-maintaining fiber coupler; PS, polarization splitter; QWP, quarter-wave plate; SLD, superluminescent diode.
Polarization sensitive optical coherence tomography
The PSOCT setup was previously introduced in Wang et al., 2016 (black box of Figure 1). Briefly, the light source consists of two broadband super-luminescent diodes (Thorlabs Inc., LSC2000C), with a center wavelength of 1300 nm and a bandwidth of 170 nm. The axial resolution was estimated to be 3.7 µm in tissue (with a refractive index of 1.4). Linearly polarized light was launched into a polarization-maintaining fiber based interferometer. Achromatic quarter-wave plates were placed in the sample and reference arms, respectively, to ensure circular polarization incident on the sample and 45° linear polarization returned from the reference arm. In the sample arm, we used a 4× telescope and a 10× water immersion microscope objective (Zeiss, NA 0.3) to obtain a lateral resolution of 3.5 µm. Light interference from the reference and sample arms was split into orthogonal polarization states by a polarization splitter, and directed to two identical spectrometers. Each spectrometer consisted of a 600 lines/mm grating and a 2048-pixel line scan camera (Sensors Unlimited Inc.) to acquire the spectral interference at an A-line rate of 51 kHz. With a scanning range of 1×1 mm2, one volumetric acquisition composed of 350 A-lines by 350 B-lines takes 3.1 sec.
The data acquisition computer was a high performance local computer with an eight-core processor, 128GB memory, and a 12TB hard disk. The customized acquisition software was written in C++. The software implemented a multithread approach to perform data reconstruction in real time, including mapping from wavelength space to k-space, dispersion compensation (Cense et al., 2004), Fourier transform for depth-profile creation, and contrast generation. The PSOCT produces three types of contrast in a single measurement: reflectivity represents the back scattering of light from the tissue; retardance results from the birefringent components in the tissue; and optic axis orientation represents the in-plane orientation of the fiber axis (Wang et al., 2010). The software also included algorithms for finding the tissue surface using probabilistic filtering (Geman and Geman, 1984), and saving only 2D e-face images by obtaining the average intensity projection (AIP), maximum intensity projection (MIP), average retardance, and dominant orientation within a desired depth range (Magnain et al., 2014; Wang et al., 2014a). Both of AIP and MIP are produced based on the reflectivity contrast. AIP indicates the average light attenuation along beam propagation, whereas MIP manifests small structures at certain depth that may have high back-scattering or refractive index compared to the surrounding medium.
Automated tiling and serial sectioning
The surface areas of the brain sample blocks we investigated were centimeter squares that were over one hundred times greater than that of the field-of-view of a single image tile. As a result, image tiles from multiple scans were stitched together to form a full section. Motorized xyz stages (xy stages: M-415.PD and M-405.DG, Physik Instrumente; z stage: customized with Parker Hannifin Co.) were incorporated to translate the sample under the PSOCT between image tiles (Figure 1). The maximal travel distance for x-, y-, and z- axis was 150 mm, 75 mm, and 25 mm, with corresponding positioning accuracy of 10 µm, 1 µm, and 5 µm, respectively. A customized vibratome (Ragan et al., 2012) was mounted adjacent to the PSOCT to cut off a superficial slice of the tissue upon completion of the full area scan. The blade was an unbacked stainless steel single edge blade (Razor Blade Co.). The cutting speed was 0.5 mm/s. Customized software written in C++ controlled the xyz translation and vibratome sectioning. The control software communicated with the acquisition software to coordinate data acquisition and tissue positioning.
Data transfer and volumetric reconstruction
At the beginning of the serial scan, a data transmission pipeline was established between the local acquisition computer and a computational server at the Martinos Center. Upon completion of one scan, the data was immediately transferred to the server, while the next block of acquisition threads proceeded at the same time. As a consequence, the reconstruction process could be executed in parallel with the data acquisition. In post-processing, automatic scripts were implemented in two steps. First, the individual tiles of en-face images were stitched to create an entire slice using rigid registration to compute the overlap between tiles and linear blending for image fusion in the Fiji software (Preibisch et al., 2009). Then the slices were stacked to form a volume. The in-plane resolution of the volumetric reconstruction is 3.5 µm resulting from the transverse resolution of PSOCT, while the voxel size in z-direction is determined by the slice thickness, typically 50–60 µm.
Experimental procedures
We investigated two blocks of cerebellum, located in the cerebellar cortex and deep nuclei, respectively, and also the subcortical regions of the thalamus and internal capsule. Each volume consisted of 36 – 90 slices with a thickness of 50 – 60 microns. Each image tile covered a volume of 1 × 1 × 2.6 mm3 (xyz). However, the effective imaging depth in fixed human brain tissue was only 200 – 300 µm due to light attenuation. The original voxel size of PSOCT was 2.9 × 2.9 × 2.6 µm3 (xyz). For large-scale volumetric reconstructions, we represented each image tile by en-face images, which were obtained by integrating the volumetric image over a defined depth range set to the thickness of the tissue slice. As a consequence, the voxel size in the z-direction was the slice thickness. We used a 50% overlap between image tiles in the xy-plane to permit image averaging to reduce speckle noise through angular compounding (Desjardins et al., 2007) and to ensure a smooth fusion of the image tiles.
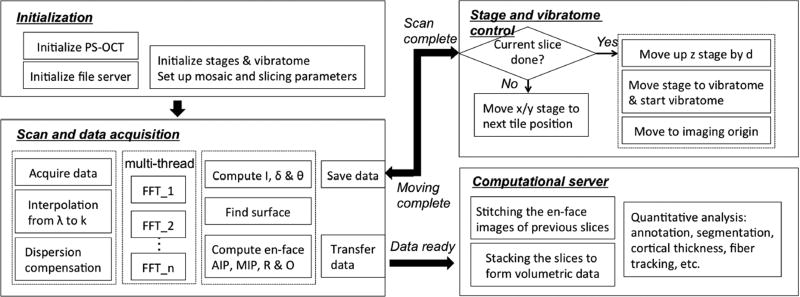
During the experiment, the tissue block was mounted in a water bath and surrounded by agarose blocks to protect against shear forces during cutting. The bath was positioned under the microscope objective and mounted on the xyz stages. Tiling and sectioning parameters were pre-defined before the acquisition, including the origin of the first tile, the translational distance between tiles, the number of tiles in x-, y-, z-directions for one slice, the number of slices, and the slice thickness. Upon completion of each tile, the sample was translated to the position of the next tile and the acquisition was triggered. The vibratome sectioned a slice with predefined thickness and allowed deeper regions to be imaged. The cutting accuracy is typically within a few microns for a 50µm thick slice. The scanning and sectioning procedures were repeated until the entire tissue block was complete. The workflow pipeline is illustrated in the flow chart of Fig. 2.
Fig. 2.
The workflow pipeline of as-PSOCT.
Quantitative data analysis
Data analyses including segmentation, cortical thickness estimation, 3D orientation estimation and tractography were all performed based on the volumetric reconstruction.
Segmentation
Segmentation of the cerebellar cortex was manually performed in Freeview software (part of the FreeSurfer software package, Fischl, 2012). The boundaries of the molecular layer, the granular layers, and white matter were drawn based on the volumetric retardance image, and labels were created for each of the structures. The segmentation results were then used to compute the layer thickness for cerebellum as described in the following section.
Thickness estimation
A voxel-based algorithm was applied to estimate the thickness of the molecular and the granular layers for the cerebellar cortex (Aganj et al., 2009). For every voxel in the volumetric data, the algorithm drew lines in different 3D directions inside the segmented area, computed the length of the intersection of each line with the layer, and selected the shortest length as the estimated thickness.
As a comparison, the thickness of the molecular and the granular layers were also estimated based on 2D slice, using the same algorithm. The slices were selected on xy-, xz- or yz-planes from the volumetric data. The algorithm drew lines in different directions parallel to the selected plane and estimated the thickness accordingly.
2D tractography
Tractography was applied on the xy-plane using the conventional method for diffusion tensor imaging in Diffusion Toolkit (Wang et al., 2007). The optic axis orientation was used to track the fibers, while the orientation for the z-direction was set to 0. The fiber tracking algorithm is based on the spherical harmonic basis method (Hess et al., 2006). Tracts were created with a maximum angular threshold of 45° for tracking, and masked by the retardance image to include the white matter only.
3D orientation estimation and tractography
The axis of fibers running in the brain is denoted by a vector v⃗ = [vx, vy, vz] (1) in Cartesian coordinates, which can also be represented by the in-plane angle θ and the through-plane angle α as v⃗ = [cos α · cos θ, cos α ∙ sin θ, sin α] (2). θ was directly obtained by the optic axis orientation. Here we derived α computationally based on the volumetric image of retardance. As the image features such as edges and spacing between fiber bundles indicated the directionality of fiber axis, we used a gradient-based method named “structure tensor” to estimate the fiber orientation (Bigun, 1987). The voxel size of the retardance image was downsampled to 30 µm isotropic with the purpose of reducing the speckle noise and enhancing the anisotropic appearance of the fiber tracts. The structure tensor was applied to consecutive xz-sections and yz-sections of the volumetric data, respectively. The generated orientation map described the angular representation of fiber axis projected onto xz- and yz-planes, respectively (tan−1 (vx/vz) and tan−1 (vy/vz)). By connecting (1) and (2), we obtained the α angles separately from the orthogonal planes. For each voxel in the volumetric data, we averaged the two α angles to obtain the through-plane orientation.
3D Tractography was performed in Diffusion Toolkit based on the estimated orientation. The same tracking algorithm was used as for 2D tractography. Tracts were created with a 45° angular threshold, and masked by the retardance image to contain the white matter only.
Ex-vivo structural MRI and diffusion-weighted MRI
Before cutting a brain hemisphere into smaller blocks and imaging with as-PSOCT, we first acquired MRI data on one sample, including 750µm diffusion-weighted images at 3T and 120µm structural images at 7T.
Diffusion-weighted images were collected using a 3D steady state free precession (SSFP) sequence on a 3T TIM Trio whole body scanner (Siemens Medical Solutions, Erlanger, Germany) with a Siemens 32 channel head coil, TR = 30.19 ms, α = 60°, TE = 25.10 ms, at 750µm isotropic resolution. Diffusion weighting was applied along 60 directions distributed over the unit sphere (effective b-value = 2434 s/mm2) (Miller et al., 2012) with eight b0 images. Q-Ball reconstruction was performed using Diffusion Toolkit. The fiber tracking algorithm is based on the Spherical Harmonic Basis method. Tracts were created using a 60° angular threshold, and masked so they are only contained within the approximate brain volume.
MRI data was acquired using a 3D FLASH sequence on a 7T human scanner from Siemens (Siemens Medical Systems, Erlangen, Germany) with a custom-built 7-channel RF receive coil, TR=60ms, α=10°, 20°, 45°, TE=30ms at 120µm isotropic resolution. We estimated the underlying tissue parameters that are the source of image contrast in standard gradient echo sequences(Fischl et al., 2004).
Registration of dMRI to as-PSOCT
Registration between the dMRI and the as-PSOCT data was manually completed in two steps, using the Freeview software. We first registered the 750-µm resolution b0 image to the 120-µm resolution anatomical MR image (FLASH), and the FLASH image to the as-PSOCT retardance image, independently. Then we concatenated the two registrations to align the dMRI data to the as-PSOCT space. The dMRI tractography was transformed accordingly, with a volume crop that matched the sample size of as-PSOCT imaging. Inclusion of the MRI data at 120-µm resolution as an intermediate registration step was necessary, because the dramatic resolution gap between dMRI and as-PSOCT made the feature matching less accurate and reliable.
Histology
Selected slices from the serial scanning and reconstruction were processed with Nissl stain (Amunts et al., 1999; Augustinack et al., 2005) for cell body identification and Gallyas stain (Miller et al., 2012; Pistorio et al., 2006) for myelinated fiber identification. The stained slices were digitized with a camera mounted on an 80i Nikon Microscope (Microvideo Instruments, Avon, Massachusetts) with a 4× microscope objective, yielding a pixel size of 1.9 µm. We used the image series workflow (“SRS Image workflow”) provided by Stereo Investigator (MBF Bioscience, Burlington, Vermont) to automatically mosaic the entire slice, and the tiles were then stitched with the same Fiji software as used for the PSOCT images. The histological stains were used to validate the as-PSOCT images for neuronal architecture and fiber identification.
Results
Quantitative cytoarchitecture and myeloarchitecture
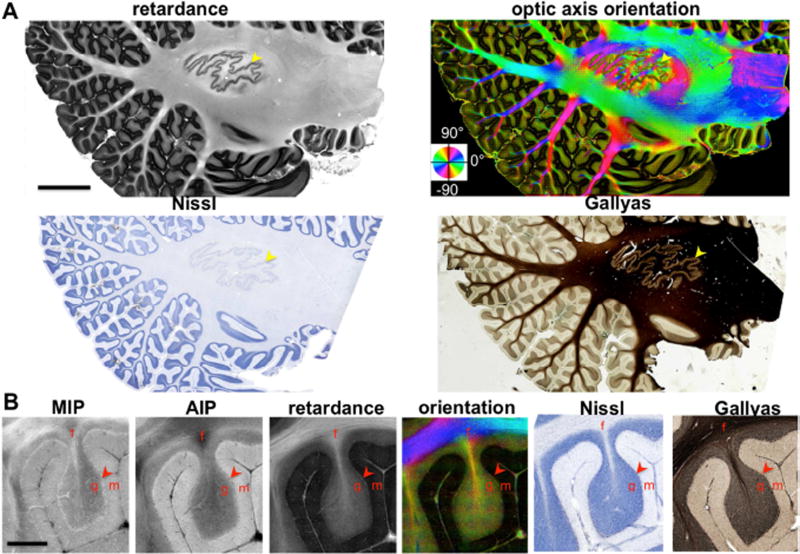
The as-PSOCT provides prominent contrasts delineating the cytoarchitecture and myeloarchitecture of a parasagittal cerebellar section (Fig. 3), spanning an area of 5.5 × 2.7 cm2 and composed of 9179 individual image tiles. The retardance contrast identifies the laminar structure of cerebellar cortex including the molecular layer and the granular layer, as well as the subfolial fissure, which refers to the white matter in the folia. The high resolution of as-PSOCT allows clear discrimination of the layer boundaries. The architectural organization revealed by as-PSOCT is highly correlated with the Nissl stain and Gallyas stain. On magnified images (B), the Purkinje cell line is clearly visible as a band of relatively high retardance (arrowhead) compared to the surrounding molecular and granular layers, possibly due to the birefringence from the regular membrane alignment. One advantage of PSOCT over conventional OCT is that the retardance reliably detects the fiber tracts where the AIP and MIP images may miss due to the dependence of through-plane fiber orientation. The brightness variations in the retardance may imply extensive axon crossing or fibers running nearly perpendicular to the imaging plane.
Fig. 3.
as-PSOCT imaging of a 15 cm2 parasagittal section of the human cerebellum showing cytoarchitecture and in-plane fiber orientations by retardance, optic axis orientation, respectively, and validated by Nissl stain and Gallyas stain. A) The yellow arrowheads indicate the location of the dentate nucleus which forms a thin layer. The orientation map was color coded according to the color wheel, with the intensity weighted by retardance. B) The cortical layers and the fiber tracts are shown in a single folium by MIP, AIP, retardance, optic axis orientation, Nissl stain and Gallyas stain. The red arrowheads indicate the Purkinje cell layer. Labels: f, fiber tracts; g, granular layer; m, molecular layer. Scale bars: A, 8 mm; B, 1 mm.
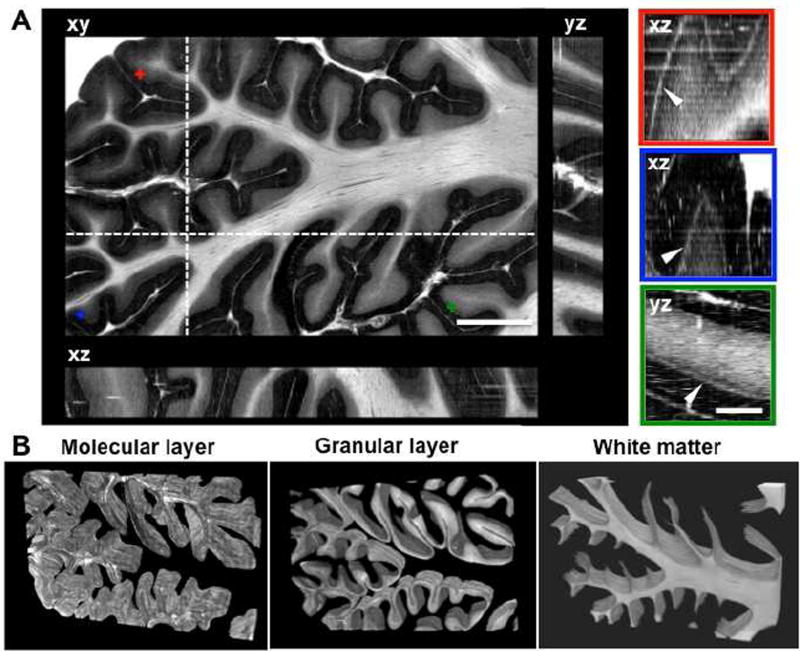
To investigate cerebellar cytoarchitecture in 3D, a block of a cerebellar lobule with a total size of 16 × 10 × 1.8 mm3 was imaged with as-PSOCT. The volumetric reconstruction included 36 slices of 50 µm thick, each composed of 1000 individual tiles. Figure 4A shows the retardance images on orthogonal viewing planes. The xy-plane represents the en-face image, while the yz-plane and xz-plane represent cross sections of the stacked slices. Although all cortical layers of the cerebellum are visible on the 2D en-face images, the folding patterns of the cortex are only unveiled in 3D space. One way to examine the accuracy of volumetric reconstruction is to follow the trajectory of a continuous microscopic structure. As the arrowheads point out on the right panels of Fig. 4A, the Purkinje cells forming a regularly aligned layer of 20 – 50 µm thickness exhibit smooth transitions on xz- and yz-planes. Here we are interested in the alignment of the Purkinje cell layer instead of single cells, because the en-face image creation reduces the resolution in z-axis to 50 µm, leading the profile of single cell unresolvable. The intensity variation across slices perceived as alternating brightness (right panels of Fig. 4A) was caused by an imaging focus shift where the wrapped cerebellar folia were unevenly sectioned, or a residual piece of slice overlaid on top of the tissue block, obscuring the imaging field of view.
Fig. 4.
Retardance images of a block of cerebellar lobule (16 × 10 × 1.8 mm3). A) The folded cerebellar cortex is shown on orthogonal viewing planes (xy, coronal; xz, axial; yz, sagittal). The locations of the axial and sagittal planes are indicated by the dashed lines on xy-plane. The panels on the right demonstrate the Purkinje cell layer (white band indicated by the arrowhead), viewed on cross-sections of the stacked slices. The locations of the Purkinje cells are indicated by the crosses with corresponding colors on the xy-plane. Scale bars: left, 2.5 mm; right, 0.6 mm. B) Volume rendering of segmented molecular layer, granular layer and white matter for the cerebellar lobule.
The prominent retardance contrast in the cerebellar cortex provides information that enables laminar structure segmentation and thickness estimation. Fig. 4B shows the segmented molecular layer, granular layer and white matter in volume rendering. Within the 1.8 mm thick block, wrapping of the folia was clearly revealed on both molecular and granular layers, while the patterns are not fully appreciated on 2D planes. The white matter fibers form a sheet like appearance following the long axis of the cerebellar folia (medial-lateral direction). Branching of fiber bundles are observed as well. The current segmentation does not include the Purkinje cell layer, but treats it as a boundary between the molecular and granular layers. Having a total volume of 225 mm3 for the cerebellar lobule, the volume fraction of the molecular layer, granular layer and white matter is roughly evenly distributed, which takes 37.7%. 31.8%, and 30.5%, respectively.
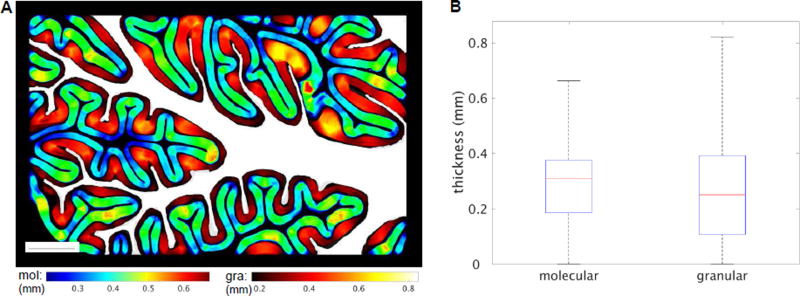
Based on the laminar segmentation, the thickness of the molecular and granular layers is estimated using a voxel-based algorithm and visualized in a selective slice (Fig. 5A). The value in every voxel represents a thickness of the corresponding layer calculated through the voxel. Different color-coding is used for molecular (jet) and granular (hot) layers to reveal the spatial relationship for the estimation. The granular layer exhibits more variations, with the greatest thickness seen in the convex curvatures of the cortex. In contrast, the thickness of the molecular layer is more uniform across gyri and sulci. The statistical graph (Fig. 5B) elaborates that the range of thickness in the granular layer is wider and the maximum thickness is much greater compared to the molecular layer. A skeleton for the two layers is drawn correspondingly and the mean thickness is estimated on the skeleton. The thicknesses of the molecular and granular layer are 313.0±96.7 µm and 322.1±158.7 µm (mean ± std), respectively.
Fig. 5.
Thickness estimation for molecular and granular layers of cerebellar cortex. A) Voxel-based thickness map shows on a slice located in the middle of the block. The molecular layer (mol) is color coded in jet, and the granular layer (gra) in hot. The white matter is shown in white as an anatomical reference. Scale bar: 2.5 mm. B) Comparison of thickness distribution between molecular layer and granular layer in boxplot.
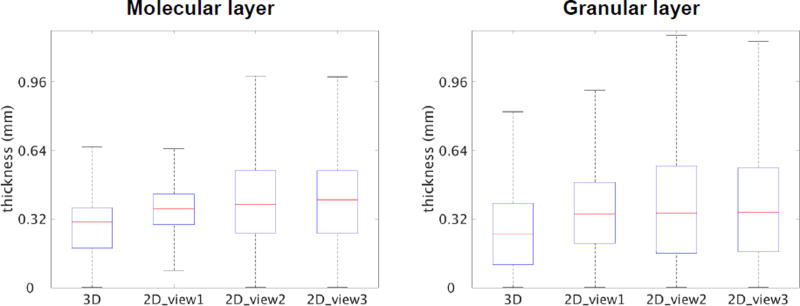
To compare the 3D estimation with the conventional 2D assessment, the thicknesses for the molecular and granular layers were also calculated based on individual slices. The slices were obtained from the volumetric data either parallel or orthogonal to the physical sectioning plane. The 2D analysis results in a consistent overestimation in the thickness for both molecular and granular layers (Fig. 6).
Fig. 6.
Comparison of 3D and 2D thickness estimation for the molecular layer and granular layer of the cerebellar cortex. 3D estimation was conducted on the volumetric data, while 2D estimation was performed on slices either parallel (2D_view1) or orthogonal (2D_view2 and 2D_view3) to the physical sectioning plane.
Orientation mapping and microscopic tractography
The optic axis orientation describes in-plane fiber orientation, which represents the projection of the 3D fiber orientation onto the en-face plane. This contrast elucidates the diversity of fiber tracts and facilitates fiber bundle grouping and tracking. Fig. 3 and 7 demonstrates the in-plane orientation map in the cerebellar lobules, the cerebellar deep nuclei region and the cerebellar peduncles on the parasagittal sections. The fissure in the folia exhibits a uniform orientation, which complies with the geometry of the fiber tracts. However, as the fibers merge into the deep white matter, a separation of multiple traces was observed (Fig. 7C). In regions where anatomical features of fiber alignments cannot be discriminated, the orientation information provides evidence for clustering of fiber bundles and tracking their directions. For example, the peduncles connecting the cerebellum with the rest of the brain are differentiated by their orientation (green, blue and magenta on Fig. 3). The boundary of the deep nuclei with respect to the middle peduncle manifests by the orientation alterations as fibers in the deep nuclei run upward connecting the superior peduncle (Granziera et al., 2009). The use of orientation information to discriminate fiber bundles in human cerebellum has been reported before with a high-angular-resolution diffusion tractography (Takahashi et al., 2013).
Fig. 7.
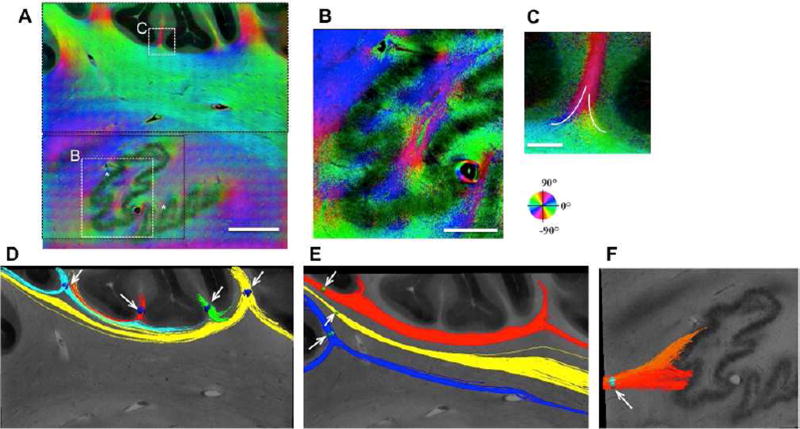
Optic axis orientation map (A-C) and 2D tractography (D-F) in the cerebellum. The orientation is encoded according to the color wheel, and the brightness is weighted by retardance. The label of * indicates the deep nucleus where retardance is low. The differed colors of the orientation indicate fiber splitting and merging (B and white traces drawn in C), which are clearly revealed by tractography (D-F). D) Tracts showing inter-folium connectivity patterns. E) Split paths within a fiber bundle. F) The pathways to the cerebellar deep nuclei. The white arrows in D-F indicate the position of seeds through which fiber tracts are created. Scale bars: A, 2 mm; B, 800 µm; C, 300 µm.
2D tractography is applied based on optic axis orientation assuming that fiber axis is parallel to the imaging plane. This is appropriate for most of the fiber bundles in cerebellar lobules viewed in the parasagittal plane, where through-plane angles are small (Takahashi et al., 2013). The fibers created by placing seeds in adjacent subfolial fissures reveal the split trajectories of fissure and the merging into adjacent bundles, providing evidence for inter-folia connections (Fig. 7D). The split of the fiber bundle extending from a cerebellar lobule into multiple traces are obvious on the tractography map as well (Fig. 7E).
Unlike cerebellar lobules, fiber orientations around the deep nuclei are more complex where fibers from all the cerebellar lobules converge and project to and from the spinal cord, brainstem and cerebrum. The transition of the orientation colors indicates the turning of fibers and their spatial interactions with the thin layer of the nucleus (Fig. 7B), which is clearly shown on the tractography (Fig. 7F).
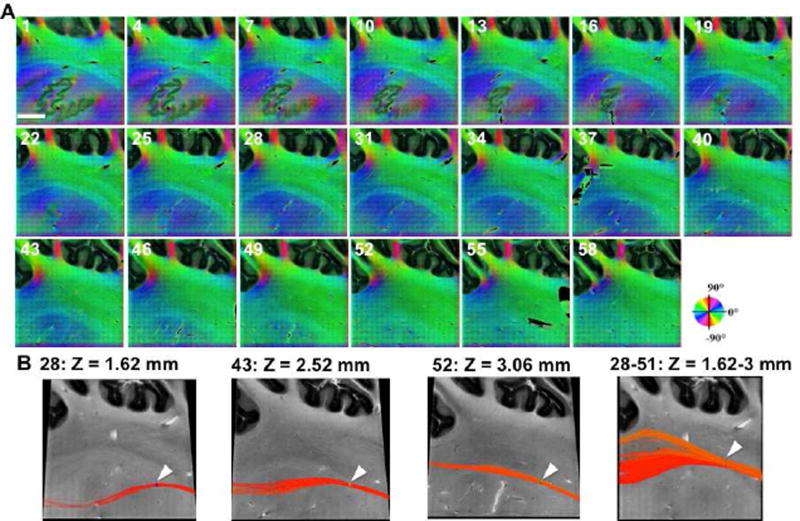
To map fiber orientation in the vicinity of the deep nuclei, a sample block of 10 × 10 × 3.6 mm3 was imaged in parasagittal sections. The entire volume contains 60 slices, each having a thickness of 60 µm and 400 individual tiles. Figure 8A shows the consecutive orientation maps, from medial to lateral direction at an interval of 180 µm (every three slices), where fibers in the dentate nucleus connecting to the superior peduncle (slice 1–46) and the transition to middle peduncle (slice 49–58) are included. The white matter in the deep nuclei region exhibits a major orientation of 40–60 degrees (slices 1–21 Fig. 8A), which runs oblique through the plane extending to the superior peduncle. As the dentate nucleus disappears towards the lateral cerebellum, the central fiber group exhibits a reduced orientation angle (slice 31–46), and horizontal fiber tracts become apparent (slice 49–58). The trajectory alterations of the fiber bundles are elucidated by the 2D tractography in Fig. 8B.
Fig. 8.
Consecutive slices of fiber orientation maps in a cerebellar block (10 × 10 × 3.6 mm3). A) The maps shown on parasagittal sections are separated by 180 µm and labeled with slice numbers, from medial to lateral. Each slice represents a 60 µm thickness. The orientation is encoded according to the color wheel, and the brightness is weighted by retardance. Scale bar: 2.5 mm. B) 2D tractography reveals directional changes of fiber trajectory across slices. The origin of Z = 0 is the first slice. The white arrows indicate the location of seeds through which fiber tracts are created.
The superior resolution of as-PSOCT allows exploitation of intricate fiber organizations in 3D space and orientation mapping with unprecedented details. An example is shown in subcortical regions including the thalamus, the Reticular nucleus and the internal capsule (ic), where small fiber tracts with diameters of 20–50 µm are embedded in the nuclei and extensive crossing is present (Fig. 9). The sample block has a volume of 13 × 10 × 3.1 mm3, with 62 slices each of which is 50 µm thick. The volumetric reconstruction allows an exploration of the spatial structures and connections (A). We select 2 sections in the external medullary lamina (eml) (1) and internal capsule (2) and inspect the organizations on sagittal planes (Mai et al., 2015). The sagittal view illustrates the cross sections of eml bundles inter-mingled together. The dominantly superior-anterior orientation of the internal capsule is revealed in both coronal and sagittal planes.
Fig. 9.
Volumetric retardance image (A) and optic axis orientation maps (B, C) delineate the fiber networks in the thalamus and internal capsule. A) Volumetric reconstruction revealing 3D organization of fiber tracts in thalamus and internal capsule regions. Sections 1 and 2 show the fiber tracts of the external medullary lamina (eml) and the internal capsule (ic), respectively, on the yz-plane representing cross sections of stacked slices. B) The orientation is encoded according to the color wheel, and the brightness is weighted by AIP. C) Zoom-in of the optic axis orientation in consecutive slices revealing fiber-crossing patterns. The arrowheads indicate the locations of crossing. The origin of Z = 0 is at the first slice. The map uses the same color and intensity coding as in (B). The location of those panels is indicated by the white ROI on B. Scale bars: A, 2 mm; B, 2.5 mm; C, 1 mm. S: superior; I: inferior; L: left; R: right; A: anterior; P: posterior.
Figure 9B elucidates the colored orientation maps of fibers in the thalamus and the internal capsule on a coronal section, weighted by the intensity of AIP (see Supplemental Video 1 as well). The AIP contrast is selected as the weight map because it facilitates the visualization of crossing-fiber patterns, by highlighting the in-plane fibers while suppressing the visualization of those running through the plane. The orientation map reveals three major groups in the internal capsule: those running in superior-inferior direction (red), extending from the thalamic nuclei to the white matter (blue and magenta), and crossing horizontally (green). Crossing of fibers in the internal capsule have been previously identified on the overlaid images of multiple tracer injections in primate brains (Schmahmann and Pandya, 2006). as-PSOCT is able to directly visualize microscopic fiber crossing in a single case and reveal the trajectory due to its micron-scale resolution. Fig. 9C shows a 3.3 × 3.3 mm2 panel from two groups of consecutive slices, where crossing fibers are observed in all directions. By following the orientation and location of the same fibers across slices, we can recover the individual trajectories (indicated by the arrowheads). It is noted that each of these panels is about the size of 2×2 voxels in in-vivo diffusion MRI, and thus resolving those crossing patterns would be extremely difficult.
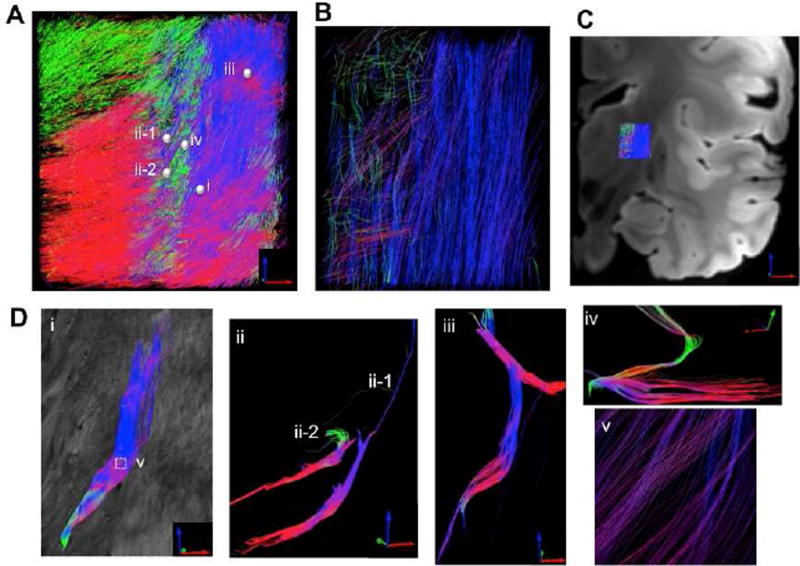
To recover the full axis of fibers running in 3D space, we computationally derived the through-plane orientation based on the volumetric image of retardance and generated 3D tractography maps by combining both in-plane angle and through-plane angles in the thalamus and internal capsule regions. Fig. 10 presents as-PSOCT tractography at a resolution of 30 µm (A, D). The tracts are shown with a 99% skip (i.e. displaying 1 out of every 100 streamlines) in order to make individual tracts visible (see Supplemental Video 2 and 3 as well). The details of trajectory and crossing patterns are investigated in small groups of fiber tracts (D). Five seeds with a radius of 85–90 µm were selected (small spheres on A), located in the thalamus (ii-1 and ii-2), reticular nucleus (iv), and internal capsule (i and iii). Fibers passing through a small seed could diverge and experience substantial crossing along the path (i, v). The two thalamic fiber tracts exhibit different turning directions and distinct pathways as they joined the white matter (ii-1 and ii-2). In the reticular nucleus and internal capsule, various fiber patterns are exposed at microscopic scale (iii, iv).
Fig. 10.
as-PSOCT 3D tractography with 30 µm resolution (A) compared with registered dMRI tractography at 750 µm resolution (B). C) The location where tractography is applied in the brain is indicated on the anatomical MRI image. D) i – iv demonstrate various fiber trajectories revealed by as-PSOCT that are not resolvable with dMRI. The tracts are created by the seeds (spheres i, ii-1, ii-2, iii, and iv) selected on (A). v shows the zoom-in view of fiber crossing on ROI-i.
To compare the microscopic fiber maps with conventional tractography, Fig. 10B shows an ex-vivo dMRI result at 750 µm resolution. The dMRI was obtained on the same sample and registered to the as-PSOCT data. The tractography was displayed with no skip. It is clear that the dMRI tracts are much more sparse and lack detailed delineations of turning and crossing patterns as revealed by as-PSOCT (Fig. 10D). The 750µm resolution dMRI illustrates that fibers in the internal capsule are dominantly in the superior-inferior direction, and the paths from the thalamus to the internal capsule are not well defined. In addition, the tracts in the reticular nucleus are not detected due to their compact size.
Discussion
OCT and PSOCT have been used to image cytoarchitecture, myeloarchitecture and fiber orientations in ex-vivo human brain. Owing to its utilization of endogenous contrast mechanisms, this imaging approach is applicable to the entire human brain across subject populations. By incorporating a vibratome into the imaging system, volumetric reconstruction of macroscopic brain samples was accomplished. Comparing to existing brain-wide imaging techniques, as-PSOCT requires minimal sample preparations therefore favoring broad applicability to the postmortem human brain. Most brain-wide techniques using labeling are restricted to small animal studies that require either injection of contrast agent while the animals are alive or using transgenetic models. In addition, existing whole-brain labeling techniques combined with tissue clearing are currently only possible for very small blocks of human brain tissue, due to the high density of myelin in the white matter, the problem of tissue integrity caused by large variations of post-mortem interval, and the complications of tissue fixation.
Previous investigations of OCT/PSOCT in the human brain were either 2D or with relatively low resolution. In Wang et al., 2014a, a medulla sample was reconstructed with an in-plane resolution of 15 µm and slice thickness of 150 µm. The 1.4 cm3 sample was completed entirely manually in 40 hours. The post-processing, apart from acquisition, took much longer considering the amount of data for network transmission, reconstruction, stitching and stacking. In this study, we developed an automatic serial sectioning PSOCT system that enabled automatic 3D imaging and reconstruction of human brain samples at unprecedented resolution and scales.
There are several significant contributions in the as-PSOCT. 1) The fully automated system integrating an imaging, sample translation and slicing pipeline enables continuous blockface imaging in a few days without the need for human interventions. Compared to previous studies (Wang et al., 2014a, 2014b) all relying on manual operations, as-PSOCT demonstrates a 100 times speed-up in 3D imaging and reconstruction. 2) The PSOCT yields an optical resolution of 3.5 µm both laterally and axially. At a pixel size of 2.9 µm and slice thickness of 50 µm, we were able to achieve volumetric reconstruction of a 0.6-cm3 sample in a total of 48,000 image tiles. 3) The optimized software pipeline achieving parallel data acquisition, real-time processing, data transferring, and post-processing promotes the 3D reconstruction in a computationally efficient approach. As a result, 3D reconstruction can be achieved almost at the same time as the completion of block imaging.
The ability to save en-face maps in as-PSOCT offered us a viable solution to manage the huge amount of data. The data size continuously generated from one experiment, if saving as raw data, could be as large as 50 TB, which would impose an overwhelming demand for data transmission, storage and processing. The rate of data generation is 0.4 GB/s with two 16-bit 2048-pixel cameras operating at 51 kHz, close to the limit of the writing speed for solid-state drives and far beyond the capability of conventional hard disks. In as-PSOCT, we saved the reconstructed 2D maps for each slice, which provides a 1000 times data reduction compared to directly saving the volumetric data in one scan.
The as-PSOCT system significantly advances the scalability of microscopic human brain imaging, which makes the technique realistically applicable to populations of human brain samples instead of just one or two cases. This allows the as-PSOCT to address critical questions that have been very difficult for traditional histology in a single study, such as inter-subject variability, group difference between normal and pathological conditions, as well as relationship among various brain regions.
Undistorted 3D reconstruction
3D reconstruction of histological images have the significant drawback of irregular distortions that are specific for every slice, including nonlinear deformation, tearing, overlapping and damage (Arsigny et al., 2005; Song et al., 2013). The convoluted nature of the human brain could lead to registration errors much greater than the size of anatomical structures, with no way to verify whether the distortions have been accurately removed. as-PSOCT technology overcomes the problem by adopting a strategy that performs the scan before slicing. As imaging is always conducted in the intact tissue block, 3D reconstruction is realized by stacking the sections without the need for any non-rigid inter-slice registrations. This procedure yields volumetric reconstruction with micrometer-scale accuracy. We have shown that as-PSOCT allows us to visualize the regular alignment of the Purkinje cells in 3D that are 20–50 microns and follow tracts in complex networks such as the internal capsule, indicating that the junctions between slices reveal no local distortions. The quality of the reconstruction mainly depends on the vibratome slicing that should vary within a few microns for a 50–60 µm thick slice. In brain regions such as cortex where structures are highly convoluted with empty space in between, slicing may result in uneven surfaces or residual tissue pieces on top of the tissue that alter the light intensity and focus during imaging and thus introduce artifact in volumetric reconstruction. Surface unevenness or tissue damage caused by physical slicing may also lead to geometric distortions or discontinuity of small-scale structures in 3D reconstructions. However, given that we are interested in resolving laminar structures and nuclei instead of single cells and fiber tracts instead of single axons, the distortion is not visible in volumetric reconstructions. It is worthwhile mentioning that one prominent advantage of OCT/PSOCT is the 3D capability which resolves the structure in depth with a resolution of 2–20 µm (Huang et al., 1991), up to a few hundreds of microns deep in the human brain. This imaging penetration is much greater than the physical slice thickness, sufficient to preserve overlapping regions between two consecutive sections for inter-slice registration. As a result, local distortions can be corrected during post-processing. Embedding the tissue in gelatin or agarose with matched stiffness may improve the quality of slicing (Magnain et al., 2014).
Quantitative cytoarchitecture and myeloarchitecture of human brain
The cytoarchitectural investigations of the brain include cell types and populations, and at a more moderate resolution, laminar structure and architectonic boundaries between cortical regions. The back-scattering contrast of OCT has shown the ability to identify individual neurons and quantify laminar structure comparable with the results of Nissl staining (Magnain et al., 2015, 2014). In this study, we exploit the value of retardance contrast for cytoarchitectural identification. Retardance is attributed to tissue birefringence and introduced while polarized light propagates through anisotropic molecules or alignments along a single axis at sub-wavelength scale. In the human cerebellum, retardance is advantageous as it enables the discriminations of fiber tracts as well as all the cortical layers. By integrating the 3D imaging, as-PSOCT elaborates the advantages to comprehensively unveil the morphology and spatial organizations of microscopic structures. We were able to segment the molecular and granular layers of cerebellar cortex and inspect the convoluted laminar structures in their intrinsic 3D space, which is beyond the capability of MRI or 2D histology.
The volumetric reconstruction also enables a series of quantitative analyses that may be biased on 2D slices. For the first time, we evaluated the thickness of the molecular and granular layer in a volumetric cerebellar lobule. The mean thickness estimation is consistent with an early report by Braitenberg and Atwood (1958) based on morphological observations. The authors found that the thickness of the molecular layer was between 250 µm and 380 µm and the thickness of the granular layer held more variations between 100 µm and 500 µm with greater values towards the convex curvatures. However,Andersen et al. (2003) conducted a stereological study reporting a greater thickness of 615 µm for the molecular layer and 443 µm for the granular layer. The age distribution (19 samples, ages 19–84 years, used in Andersen et al. 2003) may contribute to the differing results. However, the discrepancy could have resulted from a bias inherent in 2D analyses. Our study shows that the thickness prediction on 2D slices bears a consistent overestimation that can be more than three times larger compared to the estimate from the 3D volume. This is because the 2D estimate only searches for a minimal distance in a specific plane, which is a subset of the 3D space. Depending on the orientation of the selective plane with respect to cortical wrapping, the estimation on 2D can be much greater than that obtained on 3D. A recent 7T MRI study with an in-plane resolution of 120 µm reported an average layer thickness of 240 µm in the cerebellar cortex (Marques et al., 2010), which is in general agreement with our current study. However, an underestimation may be implied in the MRI due to the limitation of a partial volume effect. Given its advantages over existing methods, as-PSOCT may serve as an objective tool to resolve these discrepancies in the sparse literature and potentially be useful in new discoveries of normal and pathological morphology.
Literature on quantitative pathological analysis of cerebellar diseases is lacking. It is still unclear whether selective regions or layers of cerebellar cortex are altered in brain diseases. MRI studies have shown global atrophy in neurodegenerative cerebellar diseases such as multiple system atrophy (Brooks et al., 2009). However, the resolution limit of MRI does not allow exploration of individual lobules or cerebellar cortical layers. Histological studies have been sparse and limited to small sample size (Halliday, 2015; Lin et al., 2015; Wakabayashi and Takahashi, 2006). Therefore, quantitative characterization of the pathological impact in the cerebellum is inconclusive. It is imperative for high-resolution 3D imaging tools such as as-PSOCT to be developed to address the questions of pathological importance.
Fiber tracking and microscopic connectivity
The polarization contrasts especially target the fiber tracts in the brain due to the birefringence of myelin. The volumetric images created by as-PSOCT enhances the power of fiber tracking over long distance, especially in regions where current dMRI cannot resolve and 2D histology is difficult to follow, such as small fiber bundles intersecting to form intricate networks and projecting to different regions of the brain. Optic axis orientation is a characteristic measure with light polarization. Unlike other microscopy technologies that extract image features to calculate the orientation information (Budde et al., 2011; Budde and Frank, 2012; Choe et al., 2012; Schilling et al., 2016; Schmitt et al., 2004), the optic axis directly accesses the axis of structural anisotropy that gives rise to the fiber orientation projected in the plane. This orientation information has been validated in several human brain studies involving polarized light, such as polarized light imaging (PLI) (Axer et al., 2001, 2011a, 2011b) and PSOCT (Wang et al., 2014b). Although it is capable of imaging a full section of human brain, PLI only works on 2D histological slices and bears an intrinsic limitation in the axial resolution given by the slice thickness. In contrast, as-PSOCT is advantageous in creating volumetric orientation maps with extremely high resolution. The orientation mapping enables us to examine micro connectivity in the human brain from several perspectives.
Distinctive fiber tracts can be identified and clustered in the white matter by the orientation information in regions where anatomical features are not visible. We elucidated fiber groups of the sobfolial fissure, cerebellar peduncles, and fibers extending from the cerebellar deep nuclei intercrossing the middle peduncles (Fig. 3, 7 and 8). Although previous dMRI studies have reconstructed the primary paths of superior, middle, and inferior peduncles in human cerebellum (Dell’Acqua et al., 2013; Diedrichsen et al., 2009), detailed trajectories connecting the deep nuclei and cerebellar folia are missing due to the resolution limit. In this study, as-PSOCT shows the evidence of the inter-folial connection, splitting of paths from a cerebellar lobule, and the diverging projections to the dentate nucleus. We acknowledge that optic axis orientation is an in-plane measurement and hence 2D tractography may be biased without considering the through-plane angle. With 3D measurement now available, comprehensive connectivity maps can be established for the human cerebellum.
The high resolution of as-PSOCT allows us to explore fiber crossing at multiple scales, from millimeter (cerebellar peduncles) and sub-millimeter (subfolial fissure) down to a few microns (tracts in internal capsule). In the current study, we obtained the 3D orientation computationally by combining in-plane axis orientation and the volumetric anatomical features in the retardance images. The 3D tractography created by as-PSOCT at 30µm resolution demonstrates significant advances over 750µm resolution dMRI. The power of revealing fiber trajectories in compact structures such as the reticular nucleus and extensive crossing patterns in the internal capsule elevates the potential of as-PSOCT to chart the microscopic circuitry in the human brain. In parallel, a 3D axis measurement has been proposed in PSOCT and validated in birefringent samples of tendons and muscles (Liu et al., 2016; Ortega-Quijano et al., 2014; Ugryumova et al., 2009; Wallenburg et al., 2010).
It is noticeable that the orientation map shows a background in some of the gray matter region, such as a green color in the granular layer of the cerebellum (Fig. 3 and Fig. 8). The background color represents a constant offset in the optic axis orientation measurement (Wang et al., 2010), which has been removed for deriving the absolute fiber orientation. However, the optic axis orientation with PSOCT is only meaningful in anisotropic structure where birefringence is above a certain threshold, primarily in fiber tracts. Therefore, the orientation in the granular layer does not indicate directionality.
Outlook: towards whole brain imaging
Fundamentally, as-PSOCT is scalable to the entire human brain. Practical obstacles reside in acquisition time and data management. Multi-MHz OCT systems have been reported with ultrafast swept source and advanced GPU processing (Fechtig et al., 2015; Wieser et al., 2014, 2010; Xu et al., 2014), which has the potential to accelerate the acquisition by 20–200 times compared to the current A-line rate in this study. The spatial resolution and sampling rate are other factors that affect the scan time. The resolution of 3.5 µm used in this study is able to resolve single cell layers and small fiber tracts (but not most individual axons). Nevertheless, the resolution requirements for brain-wide cytoarchitectural boundaries and structural connectivity remain to be investigated. We have shown that with 15 µm lateral resolution, PSOCT was successful in describing cortical layers, subcortical nuclei and brain-wide fiber tracts in rodent (Wang et al., 2014a), and fiber orientations in human medulla (Wang et al., 2014b). Other ways to cut down the total acquisition time include minimizing the overlap between adjacent tiles and increasing the slice thickness to reduce redundant volumes between scans. In addition to imaging hardware and software, sectioning the entire human brain under as-PSOCT is not a trivial issue, due to the limitation of the maximal sample width that a vibratome can slice. The use of a microtome needs to be investigated with a frozen brain (Axer et al., 2011a) or a brain embedded in solid materials. Overall, the as-PSOCT technology provides intriguing opportunities for mapping the human brain in 3D with unprecedented resolution and accuracy. With continuous technical improvement in speed and scalability, the cumulative sample populations will provide a statistically meaningful database for scientists to understand the cytoarchitecture, connectivity and their functional implications in the human brain.
Supplementary Material
Acknowledgments
We thank Vivina Siless and Anastasia Yendiki for helpful discussions about tractography analysis and comparison with dMRI. Support for this research was provided in part by the National Institute for Biomedical Imaging and Bioengineering (P41EB015896, R01EB006758, R21EB018907, R01EB019956), the National Institute on Aging (5R01AG008122, R01AG016495), the National Institute for Neurological Disorders and Stroke (R01NS0525851, R21NS072652, R01NS070963, R01NS083534, 5U01NS086625), and was made possible by the resources provided by Shared Instrumentation Grants 1S10RR023401, 1S10RR019307, and 1S10RR023043. Additional support was provided by the NIH Blueprint for Neuroscience Research (5U01-MH093765), part of the multi-institutional Human Connectome Project. IA was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (K01DK101631) and the BrightFocus Foundation (A2016172S). In addition, BF has a financial interest in CorticoMetrics, a company whose medical pursuits focus on brain imaging and measurement technologies. BF's interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adhi M, Duker JS. Optical coherence tomography – current and future applications. Curr. Opin. Ophthalmol. 2013;24:213–221. doi: 10.1097/ICU.0b013e32835f8bf8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aganj I, Sapiro G, Parikshak N, Madsen SK, Thompson PM. Measurement of cortical thickness from MRI by minimum line integrals on soft-classified tissue. Hum. Brain Mapp. 2009;30:3188–3199. doi: 10.1002/hbm.20740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts K, Lepage C, Borgeat L, Mohlberg H, Dickscheid T, Rousseau M-É, Bludau S, Bazin P-L, Lewis LB, Oros-Peusquens A-M, Shah NJ, Lippert T, Zilles K, Evans AC. BigBrain: An Ultrahigh-Resolution 3D Human Brain Model. Science. 2013;340:1472–1475. doi: 10.1126/science.1235381. [DOI] [PubMed] [Google Scholar]
- Amunts K, Schleicher A, Bürgel U, Mohlberg H, Uylings HB, Zilles K. Broca’s region revisited: cytoarchitecture and intersubject variability. J. Comp. Neurol. 1999;412:319–341. doi: 10.1002/(sici)1096-9861(19990920)412:2<319::aid-cne10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Andersen BB, Gundersen HJG, Pakkenberg B. Aging of the human cerebellum: a stereological study. J. Comp. Neurol. 2003;466:356–365. doi: 10.1002/cne.10884. [DOI] [PubMed] [Google Scholar]
- Arsigny V, Pennec X, Ayache N. Polyrigid and polyaffine transformations: A novel geometrical tool to deal with non-rigid deformations – Application to the registration of histological slices. Med. Image Anal., ITKOpen science - combining open data and open source software: Medical image analysis with the Insight Toolkit. 2005;9:507–523. doi: 10.1016/j.media.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Assayag O, Grieve K, Devaux B, Harms F, Pallud J, Chretien F, Boccara C, Varlet P. Imaging of non-tumorous and tumorous human brain tissues with full-field optical coherence tomography. NeuroImage Clin. 2013;2:549–557. doi: 10.1016/j.nicl.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustinack JC, van der Kouwe AJW, Blackwell ML, Salat DH, Wiggins CJ, Frosch MP, Wiggins GC, Potthast A, Wald LL, Fischl BR. Detection of Entorhinal Layer II Using Tesla Magnetic Resonance Imaging. Ann. Neurol. 2005;57 doi: 10.1002/ana.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axer H, Axer M, Krings T, Keyserlingk DGv. Quantitative estimation of 3-D fiber course in gross histological sections of the human brain using polarized light. J. Neurosci. Methods. 2001;105:121–131. doi: 10.1016/s0165-0270(00)00349-6. [DOI] [PubMed] [Google Scholar]
- Axer M, Amunts K, Grässel D, Palm C, Dammers J, Axer H, Pietrzyk U, Zilles K. A novel approach to the human connectome: ultra-high resolution mapping of fiber tracts in the brain. Neuroimage. 2011a;54:1091–1101. doi: 10.1016/j.neuroimage.2010.08.075. [DOI] [PubMed] [Google Scholar]
- Axer M, Grässel D, Kleiner M, Dammers J, Dickscheid T, Reckfort J, Hütz T, Eiben B, Pietrzyk U, Zilles K, Amunts K. High-Resolution Fiber Tract Reconstruction in the Human Brain by Means of Three-Dimensional Polarized Light Imaging. Front. Neuroinformatics. 2011b;5 doi: 10.3389/fninf.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra HG, Costa MA, Guagliumi G, Rollins AM, Simon DI. Intracoronary Optical Coherence Tomography: A Comprehensive ReviewClinical and Research Applications. JACC Cardiovasc. Interv. 2009;2:1035–1046. doi: 10.1016/j.jcin.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigun J. Optimal orientation detection of linear symmetry. Linköping University Electronic Press; 1987. [Google Scholar]
- Böhringer HJ, Lankenau E, Stellmacher F, Reusche E, Hüttmann G, Giese A. Imaging of human brain tumor tissue by near-infrared laser coherence tomography. Acta Neurochir. (Wien) 2009;151:507–517. doi: 10.1007/s00701-009-0248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boppart SA, Brezinski ME, Pitris C, Fujimoto JG. Optical coherence tomography for neurosurgical imaging of human intracortical melanoma. Neurosurgery. 1998;43:834–841. doi: 10.1097/00006123-199810000-00068. [DOI] [PubMed] [Google Scholar]
- Braitenberg V, Atwood RP. Morphological observations on the cerebellar cortex. J. Comp. Neurol. 1958;109:1–33. doi: 10.1002/cne.901090102. [DOI] [PubMed] [Google Scholar]
- Brooks DJ, Seppi K for the Neuroimaging Working Group on MSA. Proposed neuroimaging criteria for the diagnosis of multiple system atrophy. Mov. Disord. 2009;24:949–964. doi: 10.1002/mds.22413. [DOI] [PubMed] [Google Scholar]
- Budde MD, Frank JA. Examining brain microstructure using structure tensor analysis of histological sections. NeuroImage. 2012;63:1–10. doi: 10.1016/j.neuroimage.2012.06.042. [DOI] [PubMed] [Google Scholar]
- Budde MD, Janes L, Gold E, Turtzo LC, Frank JA. The contribution of gliosis to diffusion tensor anisotropy and tractography following traumatic brain injury: validation in the rat using Fourier analysis of stained tissue sections. Brain. 2011;134:2248–2260. doi: 10.1093/brain/awr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Cense B, Chen TC, Park BH, Pierce MC, de Boer JF. In vivo depth-resolved birefringence measurements of the human retinal nerve fiber layer by polarization-sensitive optical coherence tomography. Opt. Lett. 2002;27:1610. doi: 10.1364/OL.27.001610. [DOI] [PubMed] [Google Scholar]
- Cense B, Nassif NA, Chen TC, Pierce MC, Yun S-H, Park BH, Bouma BE, Tearney GJ, de Boer JF. Ultrahigh-resolution high-speed retinal imaging using spectral-domain optical coherence tomography. Opt. Express. 2004;12:2435. doi: 10.1364/OPEX.12.002435. [DOI] [PubMed] [Google Scholar]
- Choe AS, Stepniewska I, Colvin DC, Ding Z, Anderson AW. Validation of diffusion tensor MRI in the central nervous system using light microscopy: quantitative comparison of fiber properties. NMR Biomed. 2012;25:900–908. doi: 10.1002/nbm.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauguet J, Delzescaux T, Condé F, Mangin J-F, Ayache N, Hantraye P, Frouin V. Three-dimensional reconstruction of stained histological slices and 3D non-linear registration with in-vivo MRI for whole baboon brain. J. Neurosci. Methods. 2007;164:191–204. doi: 10.1016/j.jneumeth.2007.04.017. [DOI] [PubMed] [Google Scholar]
- De Boer JF, Milner TE, van Gemert MJ, Nelson JS. Two-dimensional birefringence imaging in biological tissue by polarization-sensitive optical coherence tomography. Opt. Lett. 1997;22:934–936. doi: 10.1364/ol.22.000934. [DOI] [PubMed] [Google Scholar]
- Dell’Acqua F, Bodi I, Slater D, Catani M, Modo M. MR Diffusion Histology and Micro-Tractography Reveal Mesoscale Features of the Human Cerebellum. Cerebellum Lond. Engl. 2013;12:923–931. doi: 10.1007/s12311-013-0503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins AE, Vakoc BJ, Oh WY, Motaghiannezam SMR, Tearney GJ, Bouma BE. Angle-resolved Optical Coherence Tomography with sequential angular selectivity for speckle reduction. Opt. Express. 2007;15:6200–6209. doi: 10.1364/OE.15.006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diedrichsen J, Balsters JH, Flavell J, Cussans E, Ramnani N. A probabilistic MR atlas of the human cerebellum. NeuroImage. 2009;46:39–46. doi: 10.1016/j.neuroimage.2009.01.045. [DOI] [PubMed] [Google Scholar]
- Drexler W, Fujimoto JG. Optical Coherence Tomography: Technology and Applications. Springer Science & Business Media; 2008. [Google Scholar]
- Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, Myers EW, Chandrashekar J. A platform for brain-wide imaging and reconstruction of individual neurons. eLife. 2016;5:e10566. doi: 10.7554/eLife.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Yao G. Mapping local optical axis in birefringent samples using polarization-sensitive optical coherence tomography. J. Biomed. Opt. 2012a;17:110501–110501. doi: 10.1117/1.JBO.17.11.110501. [DOI] [PubMed] [Google Scholar]
- Fan C, Yao G. Mapping local retardance in birefringent samples using polarization sensitive optical coherence tomography. Opt. Lett. 2012b;37:1415. doi: 10.1364/OL.37.001415. [DOI] [PubMed] [Google Scholar]
- Fechtig DJ, Grajciar B, Schmoll T, Blatter C, Werkmeister RM, Drexler W, Leitgeb RA. Line-field parallel swept source MHz OCT for structural and functional retinal imaging. Biomed. Opt. Express. 2015;6:716–735. doi: 10.1364/BOE.6.000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. NeuroImage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Ségonne F, Quinn BT, Dale AM. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004;23(Suppl 1):S69–84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Gambichler T, Pljakic A, Schmitz L. Recent advances in clinical application of optical coherence tomography of human skin. Clin. Cosmet. Investig. Dermatol. 2015;8:345–354. doi: 10.2147/CCID.S69119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geman S, Geman D. Stochastic Relaxation, Gibbs Distributions, and the Bayesian Restoration of Images. IEEE Trans. Pattern Anal. Mach. Intell. PAMI. 1984;6:721–741. doi: 10.1109/TPAMI.1984.4767596. [DOI] [PubMed] [Google Scholar]
- Granziera C, Schmahmann JD, Hadjikhani N, Meyer H, Meuli R, Wedeen V, Krueger G. Diffusion Spectrum Imaging Shows the Structural Basis of Functional Cerebellar Circuits in the Human Cerebellum In Vivo. PLOS ONE. 2009;4:e5101. doi: 10.1371/journal.pone.0005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday GM. Re-evaluating the glio-centric view of multiple system atrophy by highlighting the neuronal involvement. Brain. 2015;138:2116–2119. doi: 10.1093/brain/awv151. [DOI] [PubMed] [Google Scholar]
- Helmstaedter M, Mitra PP. Computational methods and challenges for large-scale circuit mapping. Curr. Opin. Neurobiol., Neurotechnology. 2012;22:162–169. doi: 10.1016/j.conb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess CP, Mukherjee P, Han ET, Xu D, Vigneron DB. Q-ball reconstruction of multimodal fiber orientations using the spherical harmonic basis. Magn. Reson. Med. 2006;56:104–117. doi: 10.1002/mrm.20931. [DOI] [PubMed] [Google Scholar]
- Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, Et A. Optical coherence tomography. Science. 1991;254:1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DJ, Hermann KL, Schmahmann JD. The Diagnosis and Natural History of Multiple System Atrophy, Cerebellar Type. The Cerebellum. 2015:1–17. doi: 10.1007/s12311-015-0728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Black AJ, Wang H, Akkin T. Quantifying three-dimensional optic axis using polarization-sensitive optical coherence tomography. J. Biomed. Opt. 2016;21:070501. doi: 10.1117/1.JBO.21.7.070501. [DOI] [PubMed] [Google Scholar]
- Magnain C, Augustinack JC, Konukoglu E, Frosch MP, Sakadžić S, Varjabedian A, Garcia N, Wedeen VJ, Boas DA, Fischl B. Optical coherence tomography visualizes neurons in human entorhinal cortex. Neurophotonics. 2015;2:015004–015004. doi: 10.1117/1.NPh.2.1.015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnain C, Augustinack JC, Reuter M, Wachinger C, Frosch MP, Ragan T, Akkin T, Wedeen VJ, Boas DA, Fischl B. Blockface histology with optical coherence tomography: A comparison with Nissl staining. NeuroImage. 2014;84:524–533. doi: 10.1016/j.neuroimage.2013.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnain C, Wang H, Sakadžić S, Fischl B, Boas DA. En face speckle reduction in optical coherence microscopy by frequency compounding. Opt. Lett. 2016;41:1925–1928. doi: 10.1364/OL.41.001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai JK, Majtanik M, Paxinos G. Atlas of the Human Brain. Academic Press; 2015. [Google Scholar]
- Marques JP, van der Zwaag W, Granziera C, Krueger G, Gruetter R. Cerebellar Cortical Layers: In Vivo Visualization with Structural High-Field-Strength MR Imaging. Radiology. 2010;254:942–948. doi: 10.1148/radiol.09091136. [DOI] [PubMed] [Google Scholar]
- Mertz J. Optical sectioning microscopy with planar or structured illumination. Nat. Methods. 2011;8:811–819. doi: 10.1038/nmeth.1709. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, Fobbs AJ, Sousa AMM, Šestan N, Wildman DE, Lipovich L, Kuzawa CW, Hof PR, Sherwood CC. Prolonged myelination in human neocortical evolution. Proc. Natl. Acad. Sci. 2012;109:16480–16485. doi: 10.1073/pnas.1117943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosaliganti K, Pan T, Sharp R, Ridgway R, Iyengar S, Gulacy A, Wenzel P, de Bruin A, Machiraju R, Huang K, Leone G, Saltz J. Registration and 3D visualization of large microscopy images. 2006 doi: 10.1117/12.653505. 61442V-61442V-12. [DOI] [Google Scholar]
- Nakaji H, Kouyama N, Muragaki Y, Kawakami Y, Iseki H. Localization of nerve fiber bundles by polarization-sensitive optical coherence tomography. J. Neurosci. Methods. 2008;174:82–90. doi: 10.1016/j.jneumeth.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Odgaard A, Andersen K, Melsen F, Gundersen HJG. A direct method for fast three-dimensional serial reconstruction. J. Microsc. 1990;159:335–342. doi: 10.1111/j.1365-2818.1990.tb03038.x. [DOI] [PubMed] [Google Scholar]
- Ortega-Quijano N, Fanjul-Vélez F, Arce-Diego JL. Polarimetric study of birefringent turbid media with three-dimensional optic axis orientation. Biomed. Opt. Express. 2014;5:287–292. doi: 10.1364/BOE.5.000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osechinskiy S, Kruggel F. Slice-to-Volume Nonrigid Registration of Histological Sections to MR Images of the Human Brain. Anat. Res. Int. 2010;2011:e287860. doi: 10.1155/2011/287860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osten P, Margrie TW. Mapping brain circuitry with a light microscope. Nat. Methods. 2013;10:515–523. doi: 10.1038/nmeth.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistorio AL, Hendry SH, Wang X. A modified technique for high-resolution staining of myelin. J. Neurosci. Methods. 2006;153:135–146. doi: 10.1016/j.jneumeth.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proskurin SG, Meglinski IV. Optical coherence tomography imaging depth enhancement by superficial skin optical clearing. Laser Phys. Lett. 2007;4:824–826. doi: 10.1002/lapl.200710056. [DOI] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands GB, Gerneke DA, Hooks DA, Green CR, Smaill BH, Legrice IJ. Automated imaging of extended tissue volumes using confocal microscopy. Microsc. Res. Tech. 2005;67:227–239. doi: 10.1002/jemt.20200. [DOI] [PubMed] [Google Scholar]
- Schilling K, Janve V, Gao Y, Stepniewska I, Landman BA, Anderson AW. Comparison of 3D orientation distribution functions measured with confocal microscopy and diffusion MRI. NeuroImage. 2016;129:185–197. doi: 10.1016/j.neuroimage.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN. Fiber Pathways of the Brain. Oxford University Press; New York: 2006. [Google Scholar]
- Schmitt O, Pakura M, Aach T, Hömke L, Böhme M, Bock S, Preusse S. Analysis of nerve fibers and their distribution in histologic sections of the human brain. Microsc. Res. Tech. 2004;63:220–243. doi: 10.1002/jemt.20033. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Suppl 1):S208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smithson KG, MacVicar BA, Hatton GI. Polyethylene glycol embedding: a technique compatible with immunocytochemistry, enzyme histochemistry, histofluorescence and intracellular staining. J. Neurosci. Methods. 1983;7:27–41. doi: 10.1016/0165-0270(83)90016-x. [DOI] [PubMed] [Google Scholar]
- Song Y, Treanor D, Bulpitt A, Magee D. 3D reconstruction of multiple stained histology images. J. Pathol. Inform. 2013;4:7. doi: 10.4103/2153-3539.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Song JW, Folkerth RD, Grant PE, Schmahmann JD. Detection of Postmortem Human Cerebellar Cortex and White Matter Pathways Using High Angular Resolution Diffusion Tractography: A feasibility study. NeuroImage. 2013a;68:105–111. doi: 10.1016/j.neuroimage.2012.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Song JW, Folkerth RD, Grant PE, Schmahmann JD. Detection of postmortem human cerebellar cortex and white matter pathways using high angular resolution diffusion tractography: A feasibility study. NeuroImage. 2013b;68:105–111. doi: 10.1016/j.neuroimage.2012.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PS, Friedman B, Ifarraguerri AI, Thompson BD, Lev-Ram V, Schaffer CB, Xiong Q, Tsien RY, Squier JA, Kleinfeld D. All-Optical Histology Using Ultrashort Laser Pulses. Neuron. 2003;39:27–41. doi: 10.1016/S0896-6273(03)00370-2. [DOI] [PubMed] [Google Scholar]
- Ugryumova N, Jacobs J, Bonesi M, Matcher SJ. Novel optical imaging technique to determine the 3-D orientation of collagen fibers in cartilage: variable-incidence angle polarization-sensitive optical coherence tomography. Osteoarthritis Cartilage. 2009;17:33–42. doi: 10.1016/j.joca.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Vermeer KA, Mo J, Weda JJA, Lemij HG, de Boer JF. Depth-resolved model-based reconstruction of attenuation coefficients in optical coherence tomography. Biomed. Opt. Express. 2013;5:322–337. doi: 10.1364/BOE.5.000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Takahashi H. Cellular pathology in multiple system atrophy. Neuropathology. 2006;26:338–345. doi: 10.1111/j.1440-1789.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- Wallenburg MA, Wood MF, Ghosh N, Vitkin IA. Polarimetry-based method to extract geometry-independent metrics of tissue anisotropy. Opt. Lett. 2010;35:2570–2572. doi: 10.1364/OL.35.002570. [DOI] [PubMed] [Google Scholar]
- Wang H, Akkin T, Magnain C, Wang R, Dubb J, Kostis WJ, Yaseen MA, Cramer A, Sakadžić S, Boas D. Polarization sensitive optical coherence microscopy for brain imaging. Opt. Lett. 2016;41:2213–2216. doi: 10.1364/OL.41.002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Al-Qaisi MK, Akkin T. Polarization-maintaining fiber based polarization-sensitive optical coherence tomography in spectral domain. Opt. Lett. 2010;35:154. doi: 10.1364/OL.35.000154. [DOI] [PubMed] [Google Scholar]
- Wang H, Black AJ, Zhu J, Stigen TW, Al-Qaisi MK, Netoff TI, Abosch A, Akkin T. Reconstructing micrometer-scale fiber pathways in the brain: Multi-contrast optical coherence tomography based tractography. NeuroImage. 2011;58:984–992. doi: 10.1016/j.neuroimage.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhu J, Akkin T. Serial optical coherence scanner for large-scale brain imaging at microscopic resolution. NeuroImage. 2014a;84:1007–1017. doi: 10.1016/j.neuroimage.2013.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhu J, Reuter M, Vinke LN, Yendiki A, Boas DA, Fischl B, Akkin T. Cross-validation of serial optical coherence scanning and diffusion tensor imaging: A study on neural fiber maps in human medulla oblongata. NeuroImage. 2014b;100:395–404. doi: 10.1016/j.neuroimage.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Benner T, Sorensen AG, Wedeen VJ. Proc Intl Soc Mag Reson Med. Berlin: 2007. Diffusion toolkit: a software package for diffusion imaging data processing and tractography. [Google Scholar]
- Wang RK, Elder JB. Propylene glycol as a contrasting agent for optical coherence tomography to image gastrointestinal tissues. Lasers Surg. Med. 2002;30:201–208. doi: 10.1002/lsm.10013. [DOI] [PubMed] [Google Scholar]
- Wang RK, Tuchin VV. Optical Tissue Clearing to Enhance Imaging Performance for OCT. In: Drexler W, Fujimoto JG, editors. Optical Coherence Tomography. Springer International Publishing; 2015. pp. 1455–1487. [Google Scholar]
- Wieser W, Biedermann BR, Klein T, Eigenwillig CM, Huber R. Multi-Megahertz OCT: High quality 3D imaging at 20 million A-scans and 45 GVoxels per second. Opt. Express. 2010;18:14685. doi: 10.1364/OE.18.014685. [DOI] [PubMed] [Google Scholar]
- Wieser W, Draxinger W, Klein T, Karpf S, Pfeiffer T, Huber R. High definition live 3D-OCT in vivo: design and evaluation of a 4D OCT engine with 1 GVoxel/s. Biomed. Opt. Express. 2014;5:2963. doi: 10.1364/BOE.5.002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang C, Xu J, Wong KKY, Tsia KK. Megahertz all-optical swept-source optical coherence tomography based on broadband amplified optical time-stretch. Opt. Lett. 2014;39:622. doi: 10.1364/OL.39.000622. [DOI] [PubMed] [Google Scholar]
- Yuan J, Gong H, Li A, Li X, Chen S, Zeng S, Luo Q. Visible rodent brain-wide networks at single-neuron resolution. Front. Neuroanat. 2015:70. doi: 10.3389/fnana.2015.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Avants BB, Ng L, Hawrylycz M, Burstein PD, Zhang H, Gee JC. 3D Mouse Brain Reconstruction from Histology Using a Coarse-to-Fine Approach. In: Pluim JPW, Likar B, Gerritsen FA, editors. Biomedical Image Registration, Lecture Notes in Computer Science. Springer; Berlin Heidelberg: 2006. pp. 230–237. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.