Abstract
Sublancin is a 37-amino acid antimicrobial peptide belonging to the glycocin family of natural products. It contains two helices that are held together by two disulfide bonds, as well as an unusual S-glucosidic linkage to a Cys in a loop connecting the helices. We report the reconstitution of the biosynthetic pathway to this natural product in Escherichia coli. This technology enabled the evaluation of the structure-activity relationships of the solvent-exposed residues in the helices. The biosynthetic machinery proved tolerant of changes in both helices, and the bioactivity studies of the resulting mutants show that two residues in helix B are important for bioactivity, Asn31 and Arg33.
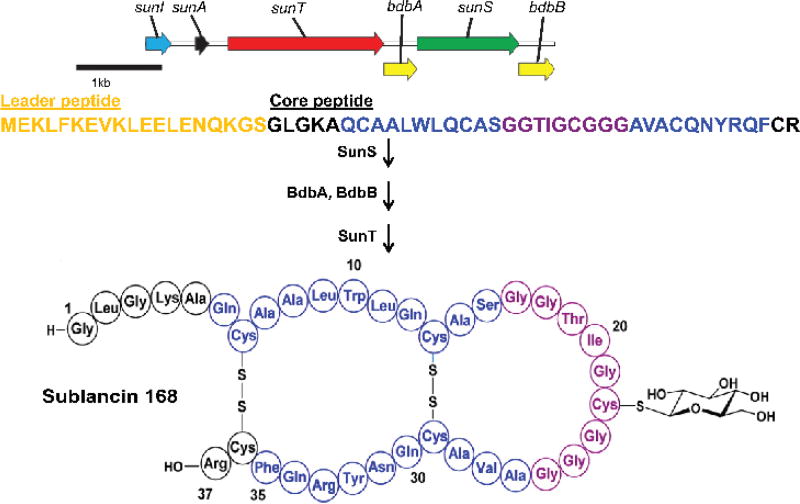
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a fast expanding class of natural products that display a wide range of interesting biological activities.1, 2 Glycocins (glycopeptide bacteriocins) are a family of RiPPs that are characterized by one or two sugars installed on serine, threonine or cysteine residues, and two nested disulfide bonds that serve to stabilize a hairpin structure characterized by a helix-loop-helix.3 The S-linked glycocin sublancin is produced by the Gram-positive soil bacterium Bacillus subtilis 168. Its mechanism of antimicrobial activity is currently not known and only very limited structure-activity (SAR) studies have been reported. Sublancin is encoded on the SPβ prophage as a pre-peptide by the sunA gene.4 It is produced biosynthetically from a gene cluster made up of five additional genes (Figure 1). Two thiol-disulfide oxidoreductases, encoded by bdbA and bdbB, are responsible for generating the two disulfide bonds of sublancin, which connect four of the five cysteine residues in the mature peptide.5 The fifth cysteine residue, Cys22, undergoes glucosylation of the sulfur atom by the glycosyltransferase encoded by the sunS gene.6 sunT is responsible for the export of modified SunA and the proteolytic removal of a leader sequence,6 and an immunity protein is encoded by the gene sunI.7
Figure 1.
Biosynthesis of sublancin in B. subtilis 168. SunS must precede disulfide bond formation,6 but the order of oxidative folding and leader peptide removal is not known. Leader peptide is depicted in gold. Residues that form the helices are colored blue while the residues that form the loop region are depicted in purple.13
Sublancin has antimicrobial activity against several Gram-positive bacteria including methicillin resistant Staphylococcus aureus (MRSA).8, 9 The mechanism by which sublancin kills sensitive strains is still a matter of investigation. Based on previous studies the mscL gene,10 the alternative sigma factor σW,11 and the phosphoenolpyruvate:sugar phosphotransferase system (PTS)12 play a major role in sensitivity to sublancin. The NMR structure of the peptide has been elucidated (Figure 2),13 and in this study we used this structural information to investigate the importance of the solvent exposed residues in the helices of sublancin for antimicrobial activity. In order to do this, we first established a system that allows sublancin and analogs to be produced in Escherichia coli.
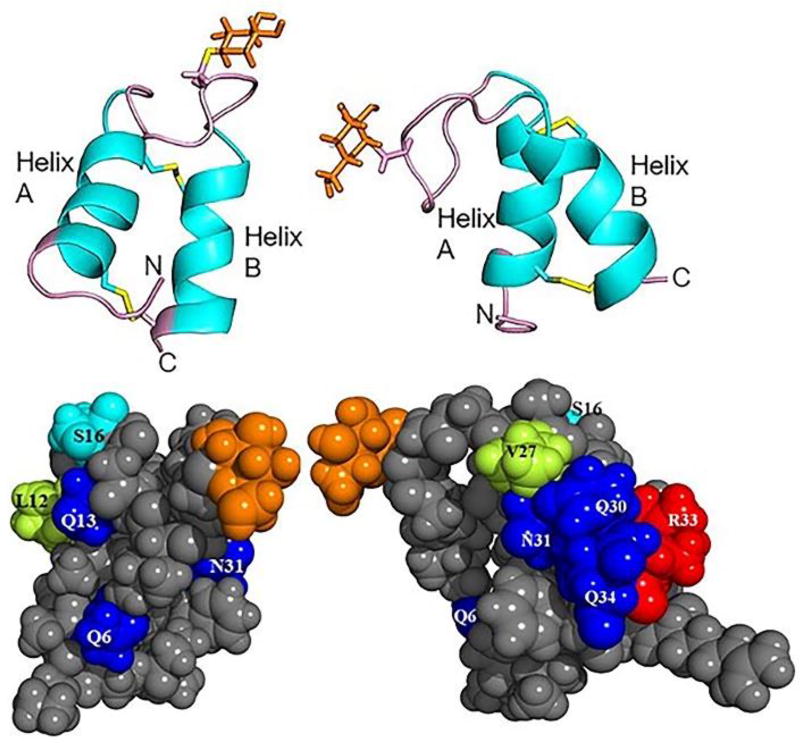
Figure 2.
(top) Ribbon diagram (PDB:2MIJ) of two views of sublancin with the loop, N- and C-termini colored in pink and the glucose in orange.13 (bottom) Surface diagram depicting the helices of sublancin. Solvent exposed non-alanine residues in the helices are color coded. Gln and Asn are in blue, Leu and Val are in lime, Ser is in cyan, Arg is in red, and all other residues are in gray. Glucose is depicted in orange. Left panels show view of helix A, right panels show view of helix B.
At present all analogs of sublancin have been generated in vitro by enzymatic modification of the precursor peptide or via chemical synthesis.6, 14, 15 Although these strategies work, they are time-consuming and do not result in readily renewable materials. Based on previous success with producing other classes of RiPPs in Escherichia coli, we first constructed a pRSFDuet-1 vector encoding His6-SunA-Xa (Xa refers to the factor Xa protease cleavage site, which has been engineered in the leader peptide of sublancin by replacing the last four amino acids with IEGR, the recognition sequence for factor Xa).6 His6-SunA-Xa was encoded in multiple cloning site I (MCSI) and the glycosyltransferase SunS in MCSII. E. coli BL21 (DE3) cells were transformed with the plasmid, and protein expression was induced by isopropyl β-D-1-thiogalacto-pyranoside (IPTG). After purification by immobilized metal affinity chromatography (IMAC), successful formation of glycosylated SunA was confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Figure 3a–b). Since we previously demonstrated that glycosylated peptide can be oxidatively folded in vitro followed by leader peptide removal using Factor Xa, a system for preparing sublancin analogs was in hand.
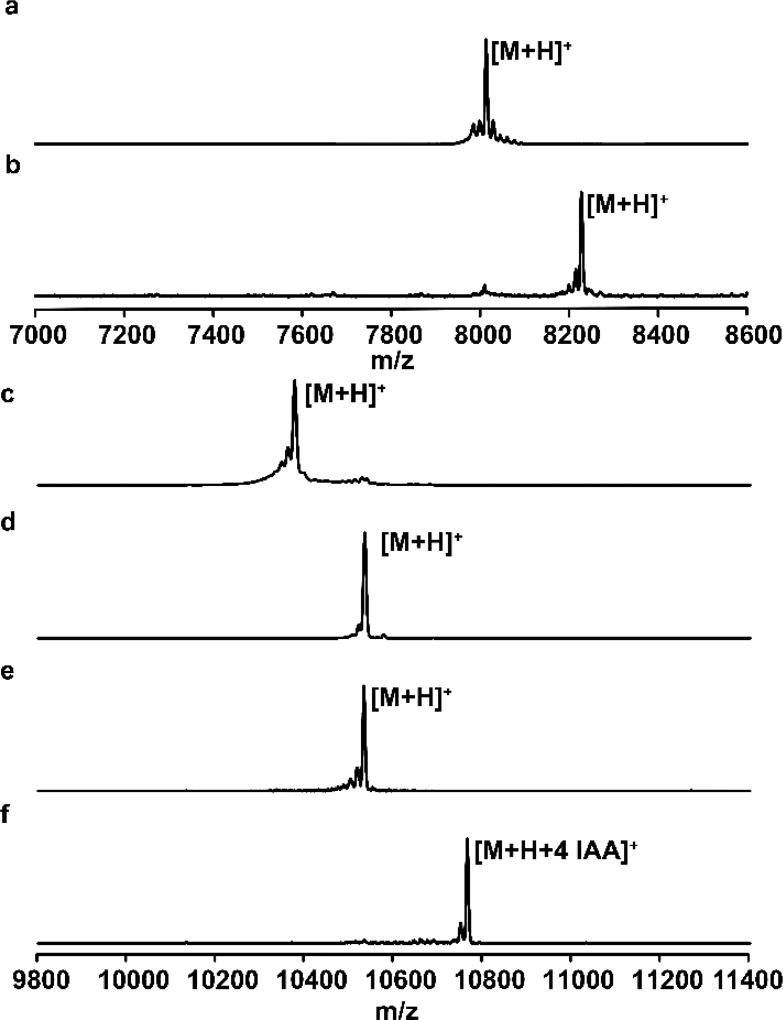
Figure 3.
Representative MALDI-TOF mass spectra of different variants of SunA produced in E. coli. (a) SunA-Xa-WT; (b) Glycosylated SunA-Xa-WT; (c) Unmodified His6-HalA2LPSunACP (d) Glycosylated His6-HalA2LPSunACP; (e) Iodoacetamide (IAA) treated glycosylated His6-HalA2LPSunACP. The lack of alkylation indicates the absence of a free Cys, and hence confirms oxidative folding of the Cys residues. (f) Sample from panel (e) treated with tris-(carboxyethyl) phosphine (TCEP) and IAA. TCEP reduces the cysteines involved in disulfide formation allowing for alkylation by IAA. For observed and expected masses, see Table S2.
Having established that we could successfully glycosylate SunA in E. coli, an alanine scanning mutagenesis approach was chosen to assess which of the residues might be important for bioactivity. We focused on residues with surface exposed side chains (Figure 2) as these might make interactions that are important for activity. Thus, four of the residues in helix A (Gln6, Leu12, Gln13 and Ser16) and five of the residues in helix B (Val27, Gln30, Asn31, Arg33 and Gln34) were chosen for mutagenesis to Ala (Figure S1). Analyses with the software PSIPRED16 indicated that the alanine mutants would not lead to disruption of helicity.
The nine SunA mutants were generated using site-directed mutagenesis and the mutant peptides were co-expressed in E. coli with SunS. For a representative mutant, SunA-Xa-R33A, the peptide was isolated using IMAC followed by MALDI-TOF MS analysis indicating that the sublancin mutant expressed in E. coli was modified with a hexose moiety (Figure S2). Unfortunately, the yields of glycosylated products were <0.01 mg/L for all mutants, which was insufficient for SAR studies. We considered two changes to the strategy. First, we reasoned that the poor yields might be related to degradation of the linear glycosylated products. Oxidatively folded sublancin is very stable4 and hence we decided to attempt oxidative folding in vivo. We chose E. coli SHuffle T7 Express cells, which constitutively express the disulfide bond isomerase DsbC aiding oxidative folding.17 All co-expressions in SHuffle were initially performed with SunS encoded in pETDuet-1 and His6-SunA-Xa in pRSFDuet-1, thus increasing the substrate to enzyme ratio. MALDI-TOF MS analysis of the peptides produced confirmed that they were glycosylated (Figure S3). Yields of peptides after purification IMAC and HPLC were about 100 µg/L of folded sublancin with the leader peptide still attached.
Previous in vitro studies have indicated that the leader peptide of sublancin is not essential for its recognition by the modifying enzyme SunS.6 Therefore, to further improve the yields, we replaced the leader of sublancin with the leader peptide of haloduracin β, a lanthipeptide that has been produced at higher levels in E. coli.18 We designed a chimeric gene encoding a hexa-histidine tag, the leader peptide (LP) of the haloduracin β precursor HalA2 followed by a spacer sequence (NDVNPE) cleavable with endoproteinase GluC or LicP,19 and then the core peptide (CP) of sublancin. This chimeric peptide indeed expressed much better from pRSFDuet-1 than the SunA-Xa peptide, but when it was co-expressed with SunS encoded on pETDuet-1, much of the product was not glycosylated. This indicated that oxidative folding outcompeted glycosylation, since previous studies have shown that SunS cannot glycosylate the oxidatively folded peptide.6 Therefore, the gene encoding the chimeric substrate peptide was inserted into MCSI of pRSFDuet-1 with sunS inserted into MCSII, with the aim of expressing more SunS from the high copy number pRSF-Duet-1 plasmid. Co-expression of His6-HalA2LPSunACP with SunS in E. coli Shuffle indeed led to much improved yields of the completely glycosylated oxidatively folded peptide (Figure 3c–f). After removal of the leader peptide by endoproteinase GluC (which worked more efficiently than LicP with the folded peptide) and HPLC purification, the yields of sublancin and its mutants were sufficient for SAR studies (e.g. 2 mg/L for the wild type).
Because in vitro the glycosyltransferase SunS displays high substrate tolerance for nucleotide sugars other than UDP-glucose,6 it was essential to establish the identity of the hexose moiety incorporated on sublancin produced in E. coli. The isolated core peptide was subjected to acid-catalyzed hydrolysis to release the sugar, which was then subjected to chemical derivatization of the hydroxyl groups to the corresponding trimethylsilyl ethers.6 Gas chromatography-mass spectrometry (GC-MS) analysis was then performed. Comparison to a series of authentic standards as well as native sublancin established the identity of the sugar modification on sublancin isolated from E. coli as glucose (Figure S4). Iodoacetamide treatment under native and reducing conditions and chymotrypsin digests and tandem MS analysis established that wild type sublancin and its mutants had the correct topology of the disulfides and installation of the glucose on Cys22 (Figure S5). To the best of our knowledge, this is the first report of production of a glycocin in E. coli.
With the expression system in hand, we successfully isolated all the desired mutant peptides (Figure S6). All mutants were correctly glycosylated and oxidatively folded (Figure S7), indicating the tolerance of the biosynthetic system. We next evaluated the bioactivity of sublancin and the mutant peptides using agar diffusion assays (Figure 4), and determined the minimum inhibitory concentration (MIC) in liquid culture of each peptide against a sensitive Bacillus subtilis Δspβ strain (Figure S8). Most mutations did not significantly affect the bioactivity of sublancin with the exception of two residues near its C-terminus. Mutation of either Asn31 or Arg33 in helix B to Ala led to a large decrease in bioactivity, suggesting that these two residues might have an important role in interacting with the target or delivery to the target. Chymotrypsin digests of these two peptides and MS analysis of the fragments indicated that the mutant peptides had the same native disulfide topology as wild type sublancin (Figure S9). We also made the R33K mutant to evaluate the importance of positive charge. This variant displayed activity similar to that of native sublancin. A sequence alignment with the core peptides of different putative glycocins that display similar disulfide connectivity as sublancin demonstrates that residues at equivalent positions in these peptides bear a positive charge in nearly all other glycocins (Figure S10).3
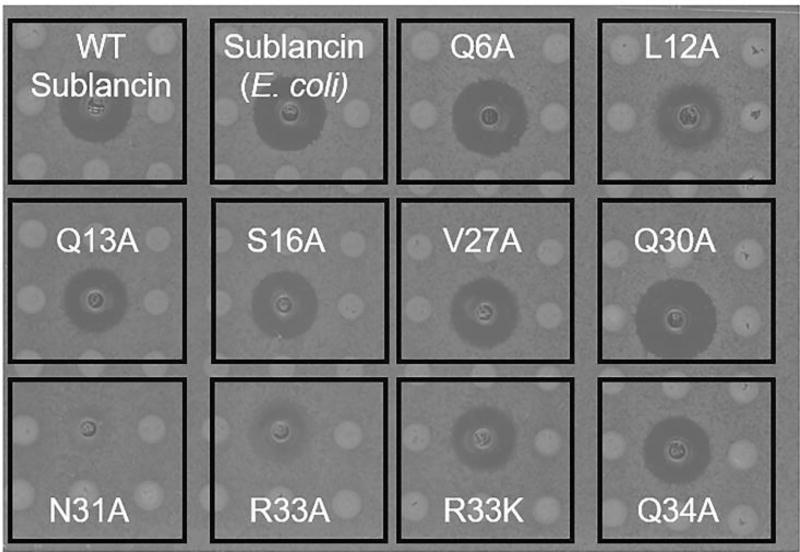
Figure 4.
Agar diffusion bioactivity assays of sublancin and its mutants against B. subtilis Δspβ. An aliquot of 10 µL of 20 µM of each peptide was spotted in each well. Q30A might be slightly more active than wild type in this assay, but this is not seen in liquid medium (Figure S8).
Using the developed robust production system in E. coli, we decided to revisit the importance of the sugar moiety for the bioactivity of sublancin. Previous investigations suggested that the sugar on sublancin might not be necessary for its bioactivity.20 This previous study was performed with non-glycosylated sublancin analogs obtained in vitro, which resulted in small quantities because of low yields in the oxidative folding step. We first produced the oxidatively folded C22S mutant that in this work could be obtained in much higher quantities such that it could be HPLC-purified. Chymotrypsin digest and MALDI-TOF MS analysis established that the oxidatively folded mutant peptide had the same disulfide topology as native sublancin (Figure S11). Agar diffusion growth inhibition assays showed that the C22S mutant of sublancin was inactive (Figure S12). This result was in contrast to our previously reported observation, and hence we investigated this result in more detail. Perhaps the best test of the importance of the sugar would be to produce the aglycone without any mutations. Previous attempts in vitro did not succeed,20 but expression of HalA2LPSunACP in the SHuffle strain and subsequent removal of the leader peptide produced the aglycone. Chymotrypsin digests of the expressed peptide showed fragments corresponding to correctly folded non-glycosylated sublancin (Figure S13). Evaluation of its bioactivity showed the absence of any zone of growth inhibition (Figure S14), consistent with the data obtained with C22S in this work. We then investigated the different components present in our previous in vitro oxidative folding conditions that could have led to the previous conclusion that the C22S mutant was active. We suspect that EDTA present in the in vitro oxidative folding conditions was likely responsible for the observed bioactivity (Figure S12). Collectively, our results indicate that sublancin is ‘glycoactive’.3
In summary, we have developed a general method for the heterologous expression of glycocins in E. coli that reliably installs all the post-translational modifications. A single in vitro step of proteolytic removal of the leader peptide gives the natural product. This method provided a platform for carrying out structure-activity relationship studies of the amino acid residues in sublancin to complement the previous evaluation of the importance of the identity of the sugar.6 These SAR studies demonstrate that two residues near the C-terminus of sublancin are important for bioactivity. One of these, Arg33, is at a position where positive charge appears important based on sequence conservation. Bioinformatic analysis has revealed putative glycocin gene clusters in a number of other organisms,3 and the current approach enables the future characterization of glycocins from bacteria that cannot be easily cultured under laboratory conditions or that do not produce the glycocin in culture.
METHODS
LC-ESI-Q/TOF MS and MSMS analyses
Data acquisition and analysis were performed as described previously, with minor modification:6 samples were injected on a Phenomenex C4 column (5 µm, 100 Å, 50 mm × 2.0 mm) and the gradient was run for 10 min.
HalA2LP-SunACP and SunS Co-expression
A synthetic gene encoding HalA2leader-NDVNPE was inserted in pET15b (see Supporting Information, SI). The gene was sub-cloned using primers HalA2LP_F and HalA2LP_R (Table S1) with Phusion polymerase and purified using a 2% agarose gel. An oligonucleotide fragment encoding SunACP and the vector backbone was amplified by PCR with primers SunACP_F and SunACP_R (Table S1) using SunA-Xa_pRSFDuet-1 (see SI) as the template and Q5 as the DNA polymerase, followed by purification on a 1.0% agarose gel. The two fragments were used in a Gibson assembly using a vector:insert ratio of 1:10. The resulting plasmid (HalA2LP-SunACP_pRSFDuet-1) was used to transform E. coli (DH10B) cells. Plasmids were isolated and sequenced as described previously.6
HalA2LP-SunACP_pRSFDuet-1 was digested with XhoI and EcoRV. sunS was then amplified using PCR with SunS_pETDuet-1 (see SI) as the template and the primers pRSFSunS_F and pRSFSunS_R (Table S1). Then a Gibson assembly was performed with a vector:insert ratio of 1:5. Transformation of E. coli (DH10B) cells and isolation of plasmids was performed as described previously.6 The sequence of the resulting plasmid (HalA2LP-SunACP_SunS_pRSFDuet-1) was confirmed by DNA sequencing.
Mutagenesis
Site-directed mutagenesis of HalA2LP-SunACP_SunS_pRSFDuet-1 was performed by a standard QuikChange protocol using either Phusion or Q5 as the DNA polymerase. Methylated non-mutated DNA (i.e. the template plasmid) was digested with DpnI for 2–3 h. Transformation of E. coli (DH10B) cells and isolation and sequencing of the resultant plasmids was carried out as described previously.6
Expression and purification of His6HalA2LP-SunACP co-expressed with SunS
E. coli SHuffle T7 Express cells were transformed via electroporation with HalA2LP-SunACP_SunS_pRSFDuet-1. Peptide purification was performed as described previously, with a few modifications:6 Terrific Broth (TB) was used as the medium. Following isolation of the peptide by IMAC, purification was performed by solid phase extraction using a VydacC4 Column.
Leader peptide removal
Wild type sublancin or mutants were obtained using endoproteinase GluC. His6HalA2CP-SunACP obtained after solid phase extraction was dissolved in water, and incubated in 10 mM Tris buffer (pH 8.0), followed by the addition of GluC at a concentration of 1:100 (w/w) with respect to the peptide and incubation overnight at 37 °C. Extent of cleavage was monitored using MALDI-TOF MS. The peptides were purified using a C5-Phenomenex column (5 µm, 100 Å, 250 mm × 10 mm) with a Shimadzu Prep-HPLC Instrument and eluted at 19–21 min with a gradient of 2% solvent B to 100% solvent A (solvent A = 0.1% TFA in water, solvent B = 0.0866% TFA in 80% ACN/20% water) over 39.0 min at a flow rate of 6.0 mL /min. UV detection identified fractions likely containing peptide which was confirmed by MALDI-TOF MS. The fractions containing the desired peptides were lyophilized and then redissolved in water for bioactivity assays and subsequent experiments.
Antimicrobial activity assays of sublancin and its mutants
Ninety-six well agar plates were prepared as described previously.6 Peptides (10 µL of 20 µM stock) were added to each well. Plates were evaluated for antibacterial activity as described previously.6
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by the Howard Hughes Medical Institute (to W.A.V.). C.G.D.G was supported by a NIGMS-NIH Chemistry-Biology Interface Training Grant (5T32-GM070421). L.M.R. was supported by a grant from the National Institutes of Health (F32 GM108275). A Bruker UltrafleXtreme MALDI TOF/TOF mass spectrometer was purchased in part with a grant from the National Institutes of Health (S10 RR027109 A).
The authors thank A. Ulanov in the Carver Biotechnology Center at UIUC for assistance with the GC-MS analysis.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Description of all molecular biology procedures, protein purifications, and supporting figures. This information is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
W.A.V., S.B., and C.G.D.G. designed the study, S.B. and C.G.D.G. produced sublancin in E. coli, L.M.R. introduced the haloduracin leader peptide, and S.B. carried out all mutagenesis and bioactivity studies.
References
- 1.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RE, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Ribosomally Synthesized and Post-translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Norris GE, Patchett ML. The glycocins: in a class of their own. Curr. Opin. Struct. Biol. 2016;40:112–119. doi: 10.1016/j.sbi.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Paik SH, Chakicherla A, Hansen JN. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 1998;273:23134–23142. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- 5.Dorenbos R, Stein T, Kabel J, Bruand C, Bolhuis A, Bron S, Quax WJ, Van Dijl JM. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J. Biol. Chem. 2002;277:16682–16688. doi: 10.1074/jbc.M201158200. [DOI] [PubMed] [Google Scholar]
- 6.Oman TJ, Boettcher JM, Wang H, Okalibe XN, van der Donk WA. Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat. Chem. Biol. 2011;7:78–80. doi: 10.1038/nchembio.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubois JY, Kouwen TR, Schurich AK, Reis CR, Ensing HT, Trip EN, Zweers JC, van Dijl JM. Immunity to the bacteriocin sublancin 168 Is determined by the SunI (YolF) protein of Bacillus subtilis. Antimicrob. Agents Chemother. 2009;53:651–661. doi: 10.1128/AAC.01189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Zeng XF, Wang QW, Zhu JL, Peng Q, Hou CL, Thacker P, Qiao SY. The antimicrobial peptide sublancin ameliorates necrotic enteritis induced by Clostridium perfringens in broilers. J. Anim. Sci. 2015;93:4750–4760. doi: 10.2527/jas.2015-9284. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Wang Q, Zeng X, Ye Q, Huang S, Yu H, Yang T, Qiao S. Use of the antimicrobial peptide sublancin with combined antibacterial and immunomodulatory activities to protect against methicillin-resistant Staphylococcus aureus infection in mice. J. Agric. Food Chem. 2017 doi: 10.1021/acs.jafc.7b02592. [DOI] [PubMed] [Google Scholar]
- 10.Kouwen TR, Trip EN, Denham EL, Sibbald MJ, Dubois JY, van Dijl JM. The large mechanosensitive channel MscL determines bacterial susceptibility to the bacteriocin sublancin 168. Antimicrob. Agents Chemother. 2009;53:4702–4711. doi: 10.1128/AAC.00439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butcher BG, Helmann JD. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol. Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- 12.Garcia De Gonzalo CV, Denham EL, Mars RA, Stulke J, van der Donk WA, van Dijl JM. The phosphoenolpyruvate:sugar phosphotransferase system is involved in sensitivity to the glucosylated bacteriocin sublancin. Antimicrob. Agents Chemother. 2015;59:6844–6854. doi: 10.1128/AAC.01519-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia De Gonzalo CV, Zhu L, Oman TJ, van der Donk WA. NMR structure of the S-linked glycopeptide sublancin 168. ACS Chem. Biol. 2014;9:796–801. doi: 10.1021/cb4008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katayama H, Asahina Y, Hojo H. Chemical synthesis of the S-linked glycopeptide, sublancin. J. Pept. Sci. 2011;17:818–821. doi: 10.1002/psc.1406. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh YS, Wilkinson BL, O'Connell MR, Mackay JP, Matthews JM, Payne RJ. Synthesis of the bacteriocin glycopeptide sublancin 168 and S-glycosylated variants. Org. Lett. 2012;14:1910–1913. doi: 10.1021/ol300557g. [DOI] [PubMed] [Google Scholar]
- 16.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 17.Lobstein J, Emrich CA, Jeans C, Faulkner M, Riggs P, Berkmen M. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 2012;11:56. doi: 10.1186/1475-2859-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, Yang X, Garg N, van der Donk WA. Production of lantipeptides in Escherichia coli. J. Am. Chem. Soc. 2011;133:2338–2341. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang W, Dong S-H, Repka LM, He C, Nair SK, van der Donk WA. Applications of the class II lanthipeptide protease LicP for sequence-specific, traceless peptide bond cleavage. Chem. Sci. 2015;6:6270–6279. doi: 10.1039/c5sc02329g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, van der Donk WA. Substrate selectivity of the sublancin S-glycosyltransferase. J. Am. Chem. Soc. 2011;133:16394–16397. doi: 10.1021/ja2075168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.