Abstract
Regulators of G protein signalling (RGS) are a family of proteins classically known to accelerate the intrinsic GTPase activity of G proteins, which results in accelerated inactivation of heterotrimeric G proteins and inhibition of G protein coupled receptor signaling. RGS proteins play major roles in essential cellular processes, and dysregulation of RGS protein expression is implicated in multiple diseases, including cancer, cardiovascular and neurodegenerative diseases. The expression of RGS proteins is highly dynamic and is regulated by epigenetic, transcriptional and post-translational mechanisms. This review summarizes studies that report dysregulation of RGS protein expression in disease states, and presents examples of drugs that regulate RGS protein expression. Additionally, this review discusses, in detail, the transcriptional and post-transcriptional mechanisms regulating RGS protein expression, and further assesses the therapeutic potential of targeting these mechanisms. Understanding the molecular mechanisms controlling the expression of RGS proteins is essential for the development of therapeutics that indirectly modulate G protein signalling by regulating expression of RGS proteins.
Keywords: Regulator of G protein Signaling, RGS, epigenetics, transcription factors, post-translational modification, microRNA, proteasomal degradation, therapeutics
1. Introduction
Regulator of G protein signaling (RGS) proteins control signaling through heterotrimeric G proteins by accelerating the intrinsic GTPase activity of Gα subunits, typically resulting in an inhibition of downstream G protein signaling pathways [1, 2]. Due to the critical role of G protein signaling pathways in diverse cellular functions, it is unsurprising that RGS proteins are also essential in maintaining normal physiological processes and that dysregulation of RGS proteins is implicated in many pathologies. Like the more extensively studied G protein coupled receptors (GPCRs), which activate heterotrimeric G proteins, RGS proteins have emerged as attractive therapeutic targets [3]. However, RGS protein activity is typically regulated by control of expression, stability and localization rather than ligand binding, so RGS proteins are not as amenable to direct small molecule regulation as GPCRs. Therefore, to exploit RGS protein regulation of G protein pathways as a therapeutic target, a comprehensive understanding of the mechanisms that regulate the expression of RGS proteins is critical. In this review, we discuss the molecular mechanisms governing the expression and stability of RGS proteins, and evaluate the therapeutic potential of targeting these mechanisms.
2. RGS proteins in Pathophysiology
The established role of G proteins and GPCRs in the central nervous system, cardiovascular system, and in cancer biology naturally led to exploration of the physiologic role of RGS proteins in these systems. In this section, we will briefly discuss evidence demonstrating the roles and the regulation of RGS proteins in normal physiology and disease states in these systems. Also, it should be mentioned that in addition to their roles in the central nervous system, cancer and cardiovascular system, RGS proteins have important roles in multiple other systems, such as the immune system [4].
2.1 RGS proteins in the Central Nervous System
RGS proteins participate in multiple processes in the central nervous system, including synaptic plasticity [5], memory [6], and vision [7]. Therefore, predictably, dysregulation of RGS protein expression is evident and implicated in several CNS disorders [8, 9]. For example, RGS9 is a critical component of the phototransduction machinery of retinal neurons, and loss of RGS9 results in a visual disorder in which patients cannot adapt to changes in light [10, 11]; RGS7 and RGS9 critically regulate responses to dopamine [12] and opiate receptors [13] and are implicated in the development of tolerance and addiction [14]; and RGS14 has recently been identified as a critical control point for long term potentiation and memory [6]. In many cases, multiple RGS proteins contribute to the same CNS-related pathology, as is the case in Parkinson’s Disease (PD). RGS4 knockout animals display reduced motor symptoms in PD animal models [15], and inhibition of RGS4 improves symptoms of PD [16][17] suggesting that RGS4 contributes to the pathology of this disease. On the contrary, RGS2 [18], RGS6 [19], and RGS10 [20] protect dopaminergic neurons and delay Parkinson’s progression. These opposing roles in PD do not simply correlate with distinct G protein selectivity of GAP function or the activity of domains outside of the RGS domain, since RGS4 and RGS10 are both small Gi/o selective GAPs. This suggests that each RGS protein is finely tuned to a specific response, and subtle differences in regulation are critical in determining the physiologic role of RGS proteins. In addition to acting as classic GAPs, some RGS proteins modulate the pathogenesis of these diseases by GAP-independent mechanisms. For example, RGS2 protects neurons by directly binding and inhibiting LRRK2 in a mechanism that does not require RGS2 binding to G proteins [18]. This indicates that targeting GAP-independent functions of RGS proteins can be a beneficial approach in the treatment of some CNS diseases.
A common mechanism for regulation of G protein pathways is the modulation of RGS expression by upstream receptor agonists, and this regulation can result in feedback inhibition or feedforward activation. The expression of many RGS encoding genes and protein levels is highly sensitive to CNS-targeted drugs, with distinct mechanisms and time courses. For example, examination of human tissues revealed that RGS4 protein level is increased in the prefrontal cortex of long term opiate abusers with no change in short term users, while RGS10 protein level is decreased in short term opioid abuse but shows no change in long term users [21]. The increase in RGS4 expression is recapitulated in a rat model following chronic exposure to morphine [21]. Both RGS4 and RGS10 proteins have been shown to modulate μOR signaling, suggesting that μOR-induced regulation of RGS protein levels may mediate, at least partially, some tolerance to opioid agonists. Similarly, complex regulation of RGS expression occurs in psychosis and anti-psychotic treatment. RGS4 transcript levels are decreased in the prefrontal cortex of individuals with schizophrenia [22], and RGS4 immunoreactivity is higher in subjects treated anti-psychotics [23]. The antipsychotic drug olanzapine, which primarily targets 5-HT2A serotonin receptors, has been shown to increase RGS7 protein levels, and this effect is mediated by a Jak/Stat dependent pathway [24]. Therefore, changes in RGS protein expression levels are associated with both the pathology and therapeutic responses in several CNS diseases.
2.2 RGS Proteins in Cancer
In the past two decades, the role of GPCRs and heterotrimeric G proteins in cancer initiation and progression has been established [25], which has led to great interest in the regulatory role of RGS proteins in cancers [26]. Studies have provided an abundance of evidence implicating RGS proteins in multiple cancers, where they may either promote or inhibit cancer progression, depending on the type of cancer and RGS protein involved. For example, RGS2 [27], RGS4 [28], RGS6 [29], and RGS16 [30] suppress various aspects of breast cancer progression, whereas RGS20 promotes breast cancer carcinogenesis [31]. Even the same RGS protein can have opposing effects on cancers derived from different tissue. RGS17 is associated with inhibited cell growth and improved responses to chemotherapeutic drugs in ovarian cancer cells [32–34], while RGS17 has been shown to promote lung and prostate cancer growth [35]. Given the diversity of effects on different cancer types, it is unsurprising that not all RGS proteins mediate their effects through a simple G protein GAP activity. For example, while RGS4 actions in breast cancer are mediated by classic GAP activity, RGS6 and RGS16 inhibit breast cancer via GAP-independent mechanisms [36]. The fact that RGS proteins employ different mechanisms has therapeutic implications. For example, targeting the RGS-G protein interaction would selectively inhibit the GAP-dependent effects of RGS4 in breast cancer cells, while strategies targeting expression would impact both GAP-dependent and -independent functions of RGS4, RGS6 and RGS16.
Aberrant expression of RGS transcripts and proteins is also commonly observed in cancers. In breast cancer cells, RGS2, RGS4, and RGS6–which suppress growth–are down-regulated compared to normal cells [27–29], while RGS20–which promotes growth–is up-regulated in cancer [31]. Thus, in both cases, the changes in RGS expression may contribute to progression of disease. This is also observed in prostate cancer cells, where RGS2–which suppresses prostate cancer cell growth–is reduced [33], while expression of RGS17–which promotes prostate cancer cells growth–is elevated [37, 38]. Finally, RGS protein expression is also modulated by chemotherapeutic drugs [32, 39, 40], suggesting that RGS regulation of cancer cell growth continues to be modified through disease progression and therapy. Together, these observations demonstrate that RGS proteins are important regulators of cancer cell growth and survival, and dysregulation of RGS protein expression in cancer cells can modify disease progression.
2.3 RGS Proteins in Cardiovascular disease
Both GPCRs and G proteins are essential mediators of critical cardiovascular functions, and GPCRs are primary targets for many cardiovascular drugs [41]. RGS proteins also regulate multiple essential cardiac processes, and abnormal changes in their expression often results in cardiovascular system dysfunctions. For instance, loss of RGS2 amplifies angiotensin II (AngII) type 1 (AT1) receptor signaling, which leads to hypertension [42], and cardiac remodeling is regulated by RGS2 [43] and RGS14 [44]. RGS proteins also play important functions in heart failure and drug-induced cardiac injury, among other conditions (reviewed [45, 46]).
Changes in RGS expression levels have been reported in cardiovascular disease, suggesting that abnormal expression of RGS proteins may contribute to pathogenesis. In particular, it appears that RGS proteins and GPCRs participate in bi-directional regulatory mechanisms, in which RGS proteins regulate GPCR activity and GPCR activation in turn alters the expression of RGS proteins. For example, the AT1 receptor regulates the expression of RGS2 [47], RGS10 [48] and RGS14 [44], which regulate AT1 receptor-induced effects. Similarly, the β1 and β2 adrenoceptor agonist isoproterenol induces RGS5 expression [49], and RGS2 and RGS16 expression is regulated by lysophospholipid Sphingosine 1-phosphate (S1P) receptor activation in vascular smooth muscle cells [50].
Based on these diverse studies that have defined the role of RGS proteins in various pathophysiologies and the dynamic regulation of RGS expression during disease progression and treatment, several common observations can be made. First, it is evident that RGS proteins are critically important regulators of physiology and disease in these systems. Second, expression of RGS proteins is often dysregulated in disease states. Third, several drugs used in treatment of these diseases also alter the expression of RGS genes or protein levels. These observations suggest that changes in RGS expression may contribute directly to disease initiation, disease progression, treatment efficacy, tolerance, and unwanted side effects. Therefore, approaches targeted to manipulate the expression of RGS proteins can potentially be utilized for treating diseases in different systems, and also enhance the effectiveness or lower the toxicity of a number of drugs. To this end, understanding the molecular mechanisms that regulate the expression of RGS proteins will lay the groundwork for future development of effective and safe RGS-targeted therapies.
3. Mechanisms regulating RGS levels
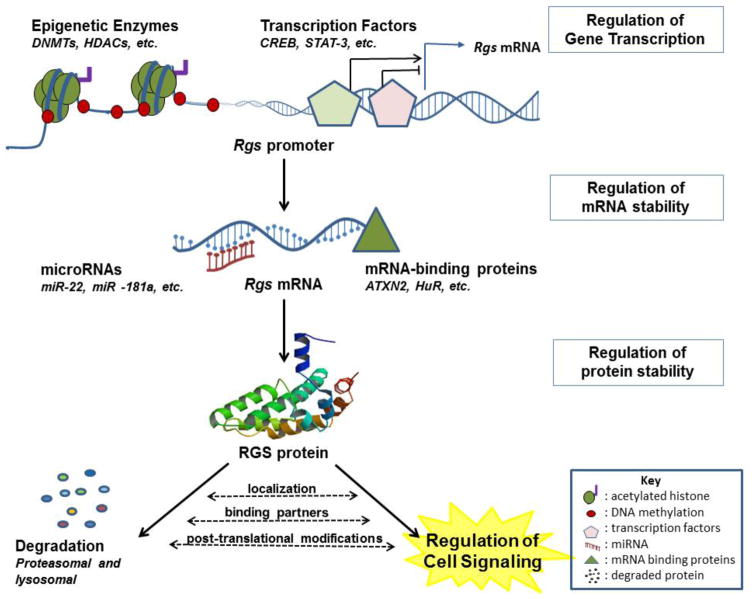
RGS proteins are primarily regulated by mechanisms that control local concentration of the protein at the site of signaling, either by regulating subcellular localization, protein stability, transcriptional regulation or epigenetic regulation (Figure 1). These combined mechanisms allow acute and chronic regulation of RGS levels in response to multiple signals. They also provide multiple potential points of intervention.
Figure 1. RGS gene expression and protein stability are regulated by multiple mechanisms.
Multiple regulatory mechanisms participate in determining the level of RGS proteins. Epigenetic modifications, mainly histone deacetylation and DNA methylation that are mediated by histone deacetylases (HDACs) or DNA methyltransferases (DNMTs), tighten the chromatin structure at RGS genes promoters, thereby obstructing the access of transcription factors and other proteins that are essential for transcription initiation, ultimately resulting in suppression of RGS gene expression. Multiple transcription factors directly bind RGS genes promoters to activate or repress transcription, adding another layer of regulation to the expression of RGS genes. In addition to transcription regulation, RGS mRNAs are targeted by both microRNAs and mRNA-binding proteins to either degrade or stabilize the respective mRNA, which critically determines the final levels of translated RGS proteins. Finally, active RGS proteins participate in various G protein dependant and independent signalling pathways in different cellular compartments. The activity and stability of RGS proteins is influenced by post-translational modifications such as phosphorylation, association with specific binding partners, and cellular localization of RGS proteins. Many RGS proteins undergo proteasomal degradation while some are degraded via lysosomal degradation. Regardless, this degradation is a critical step of regulation that ultimately governs the level of active cellular RGS proteins at a given time.
3.1 Epigenetic Regulation of RGS expression
Epigenetic modifications regulate gene expression by altering the structure of chromatin, which consists of histone proteins tightly wrapped by DNA [51]. Both DNA and histones can be epigenetically modified to influence the accessibility of transcription factors by loosening or tightening the chromatin complex, to ultimately activate or repress gene expression [51]. DNA (de)methylation and histone (de)acetylation are classic examples of epigenetic modifications. DNA methylation at cytosine is mediated by DNA methyltransferase enzymes (DNMTs) and results in gene repression [52]. Histones can be acetylated by Histone acetyltransferase (HATs) to activate gene expression, or deacetylated by histone deacetylases (HDACs) to repress gene expression [53]. DNA methylation and histone deacetylation are critical epigenetic mechanisms that regulate RGS gene expression in cancer, central nervous system, and cardiovascular systems (Figure 1).
Several studies provide evidence of epigenetic-mediated regulation of RGS genes in cancer. An early example of epigenetic regulation of RGS expression was a report of an increase in the methylation of the RGS16 promoter in breast cancer tumors, which correlated with a reduction in RGS16 expression [54]. Similarly, epigenetic regulation of RGS2 expression by DNA methylation has been reported in prostate cancer, where the suppression of RGS2 in prostate tumors was accompanied by an increase in the methylation of its promoter [38]. Inhibition of DNA methylation restored the expression of RGS2 in prostate cancer, providing additional evidence that expression of RGS2 is reversibly regulated by methylation of the promoter (Figure 2) [38]. Similarly, enhancement of methylation at the RGS2 promoter by the multifunctional protein Ubiquitin-like with PHD and ring-finger domain 1 (UHRF1) suppressed RGS2 expression and induced progression of bladder cancer [55]. In ovarian cancer, there is an increase in the methylation of RGS10 promoters in chemoresistant ovarian cancer cells, compared to their chemosensitive counterparts [32]. This hypermethylation, which was mediated by DNMT enzymes, correlated with suppressed expression of RGS10 in chemoresistant cells [32, 56]. However, in ovarian cancer cells, DNA methylation is not the only epigenetic mechanisms that have been shown to be involved in regulating RGS10 expression. Histone deacetylation, induced by HDAC enzymes, also mediates RGS10 suppression in chemoresistant cells [57]. Collectively, these studies demonstrate that epigenetic mechanisms contribute to the regulation of RGS genes in different cancers.
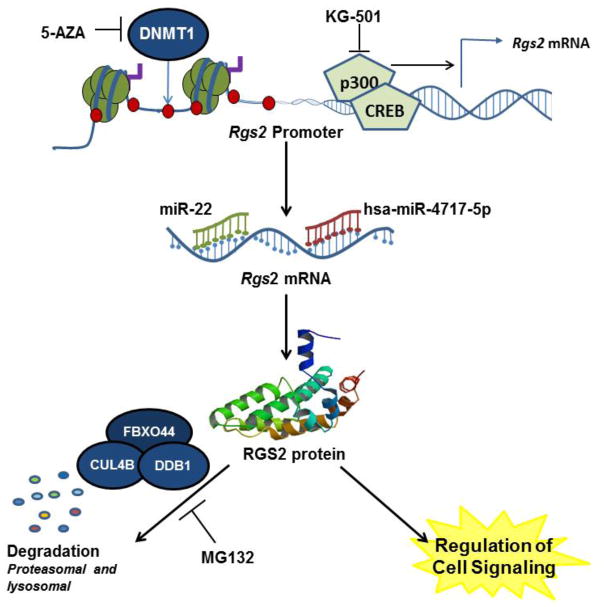
Figure 2. RGS2 protein levels are determined by multiple regulatory mechanisms, several of which can be manipulated by small molecules.
RGS2 transcription is suppressed by the DNA methyltransferase 1 (DNMT1) enzyme and activated by the transcription factor CREB. Accordingly, the DNMT1 inhibitor 5-AZA enhances RGS2 transcription, whereas inhibiting CREB-mediated transcription using the small molecule KG-501 results in suppressed transcription. In addition to transcriptional regulation, RGS2 is also regulated at the mRNA levels, by miR-22 and has-miR-4717-5p, and at the protein level, by proteasome-mediated degradation with the assistance of other proteins such as FOXO44, CUL4B, and DDB1. Because many of these regulatory mechanisms are usually not selective and can influence other proteins, manipulating RGS2 expression by combining drugs that targets multiple mechanisms of regulation is potentially more advantageous.
Several studies demonstrate that RGS genes can also be regulated by epigenetic mechanisms in the central nervous system. DNA methylation influences RGS expression in human neural progenitors [58]. During neuronal progenitor cell differentiation, low levels of DNMT enzymes correspond with increased expression of RGS proteins, which suggests that DNA methylation plays a role in regulating RGS protein expression in these cells [58]. Indeed, inhibition of DNMT enhanced the expression of many RGS proteins in human neural progenitors, including RGS2 and RGS10 [58]. In microglia, the resident macrophages of the central nervous system, inflammatory stimulators induce suppression of RGS10, a protein that has been shown to play a major anti-inflammatory function in these cells [20]. Activation of Toll-like receptors 4 (TLR4) stimulated association of HDAC enzymes with RGS10 promoters, which resulted in deacetylation of RGS10 promoters [59]. The suppression of RGS10 by this HDAC-induced mechanism was blocked by the HDAC inhibitor TSA [59]. TSA is also shown to induce anti-inflammatory effects in microglia [60]; therefore, it will be of interest to test whether the anti-inflammatory effect of TSA is due to its ability to restore RGS10 expression in microglia. DNMT inhibition had no effect on TLR4-induced suppression of RGS10 in microglia, suggesting that DNA methylation does not play a major role in regulating RGS10 expression in microglia [59]. The fact that DNA methylation suppressed RGS10 expression in ovarian cancer cells and neuronal progenitors but not in microglia suggests cell-type specific effects of epigenetic mechanisms in regulating RGS gene expression. Also, since HDAC enzymes are strongly implicated in mediated silencing of RGS10 during inflammation, it will be interesting to test whether HAT enzymes, which acetylate histones, would counteract HDAC and block suppression of RGS10.
Similar to the mechanisms described above, DNA methylation has also been implicated in regulating RGS5 expression in carotid arteries by enhanced methylation on CpG dinucleotides at its promoter [61], suggesting that epigenetic mechanisms contribute to the regulation of RGS genes in the cardiovascular system as well. Collectively, these studies provide evidence that RGS genes can be regulated in cancer, the central nervous system and the cardiovascular system by epigenetic mechanisms (Figure 1).
3.2 Transcription Factors regulating RGS expression
Detailed studies of RGS gene promoters have revealed binding sites for several transcription factors, indicating that multiple transcription factors regulate RGS transcription directly (Table 1). Isolation and characterization of the mouse RGS2 promoter revealed a highly conserved cAMP response element (CRE) site [62]. Mutation of the CRE site suppressed activation of the RGS2 promoter, suggesting that the CRE site is functionally required for efficient RGS2 transcription [62]. CRE sites were also found at the promoters of RGS4 [63] and RGS5 [64], indicating that this site is commonly participating in the transcription of multiple RGS genes. Interestingly, although CRE sites are present at the promoters of different RGS genes, they affect RGS transcription differently depending on the transcription factor occupying the CRE site. Binding of the transcription factor CRE-binding protein (CREB) to the CRE site in the RGS2 promoter activates RGS2 gene transcription (Figure 2), while binding of the related factor CRE-modulator (CREM) at the same site supresses RGS5 transcription, which suggests that CREB and CREM may compete and counteract each other at the CRE sites of RGS gene promoters to dynamically regulate expression.
Table 1.
Transcription factors shown to regulate RGS gene transcription
| Transcription Factor | RGS gene Regulated | Cells | Type of Regulation | References |
|---|---|---|---|---|
| CREB | RGS2 RGS4 |
vascular smooth muscle cells Cortical neurons |
Activation Activation |
[62] [136] |
| AP-1 | RGS4 | Colonic muscle cells | Repression | [66] |
| NF-κB | RGS4 RGS16 |
colonic muscle cells B lymphocytes |
Activation Activation |
[68] [72], [71] |
| STAT3 | RGS2 RGS7 |
cardiac myocytes A1A1v cortical cells |
Activation Activation |
[70] [24] |
| P53 | RGS16 RGS13 |
human EB1 colon cancer cells B cells, mast cells |
Activation Repression |
[73] [67] |
| CREM | RGS5 | vascular smooth muscle cells | Repression | [64] |
| GATA-6 | RGS4 | Colonic muscle cells | Activation | [69] |
| Phox2b | RGS4 | cranial motor and sensory neurons | Activation | [157] |
| YY1 | RGS16 | neonatal rat cardiac myocytes | Repression | [158] |
| Bcl6 | RGS4 | neuron-like PC6 cells | Repression | [63] |
| C/EBPβ | RGS4 | neuron-like PC6 cells | Activation | [63] |
| NF-YA | RGS4 | neuron-like PC6 cells | Activation | [63] |
In addition to the CRE site, promoters of RGS genes also harbour NF-κB [65], AP-1 [66] and P-53 binding sites [67], and these sites regulate transcription of RGS genes (Table 1). These sites can be present at the promoter of the same RGS gene allowing for several transcription factors to co-regulate the transcription of one RGS gene. For example, multiple transcription factors influence IL1β-induced upregulation of RGS4; NF-κB [68] and GATA-6 [69] activate RGS4 transcription following IL1β treatment, whereas AP-1 suppresses RGS4 transcription in colonic muscle cells [66]. In addition to RGS4, other examples of RGS genes that are regulated by different transcription factors include RGS2, which is activated by both CREB [62] and STAT3 transcription factors [70], and RGS16, which is regulated by both NF-κB [71, 72] and P-53 [73]. The participation of multiple transcription factors to regulate the same RGS gene allows for more complex regulation of expression in response to different physiologic signals, and provides multiple options in targeting these mechanisms to control the expression of RGS proteins. As the same RGS can be regulated by different transcription factors, the same transcription factor can also regulate different RGS genes. For example, NF-κB activates the transcription of both RGS16 [71] and RGS4 [68] while STAT3 enhances the transcription of RGS2 [70] and RGS7 [24].
Strikingly, the transcription factor P-53 increases the transcription of RGS16 in human EB1 colon cancer cells [73] but acts as suppressor of RGS13 transcription in mast cells and B lymphocytes [67], suggesting that the same transcription factor can regulate RGS gene transcription differently depending on the RGS gene being regulated and the type of cells involved. Transcriptional regulators typically function in multi-protein complexes to cooperatively regulate gene expression. Given that the expression of each component of these regulatory complexes may vary in different tissues and cell types, it is likely that there is significant variation in the effect of a specific transcription factor on RGS expression in different tissues. Further, many transcription factors regulate RGS gene transcription downstream of very specific and defined signalling pathways that tend to be active in a specific type of tissue. For example, the previous studies show that NF-κB, AP-1, and GATA-6 transcription factors acts on RGS4 transcription downstream of IL1β-induced signaling in colonic muscle cells, showing that this pathway in these cells is specifically regulated by multiple transcription factors. Such cell-type and pathway-specificity requires extensive investigation and characterization to fully exploit regulation of RGS expression, but also provides opportunities for more selective therapeutic approaches to regulate RGS gene expression with strategic pathway-specific approaches.
Evidence also suggests bi-directional regulation between RGS proteins and transcription factors, as several RGS proteins directly interact with transcription factors and regulate their function. For example, RGS2 directly binds STAT3 and inhibits STAT3-mediated transcription [74]. Similarly, RGS13 inhibits CREB-mediated transcription by translocating to the nucleus and forming a complex with CREB and CBP/P300 [75]. Since both STAT3 and CREB directly regulate RGS gene transcription, it is possible that some RGS proteins participate in auto-regulatory transcriptional feedback mechanisms.
3.3 Regulation of RGS mRNA stability by microRNA and RNA binding proteins
MicroRNAs are a family of short endogenous non-coding RNA sequences between 18–23 nucleotides in length that play an important role in posttranscriptional regulation of gene expression by targeting the 3′ untranslated regions of mRNAs, resulting in degradation of mRNAs and subsequent silencing of the encoded proteins [76]. Due to their crucial functions in many cellular processes, aberrant levels and/or mutations of microRNAs are implicated in various diseases [76].
As the evidence of microRNA involvement in carcinogenesis accumulates, numerous studies have identified the target genes of these microRNAs. Among the target genes, several RGS transcripts have been identified. For example, RGS16 has been shown to be targeted by miR-181a in chondrosarcoma, a cancer of bones and joints [77]. Genetically modifying miR-181a expression correlated with RGS16 mRNA expression in chondrosarcoma cell lines [77]. Moreover, RGS16 overexpression blocked miR-181a-induced production of VEGF and MMP1, suggesting that miR-181a ability to promote angiogenesis and metastasis is mediated, at least partially, by silencing RGS16 [77]. Activation of the G protein coupled receptor CXCR4 promotes production of VEGF, and it was previously shown that RGS16 regulates CXCR4 activity [78, 79]. Therefore, a possible mechanism of miR-181a-induced production of VEGF is via RGS16 silencing and subsequent enhancement of CXCR4 signaling. The participation of CXCR4 signaling in microRNA-induced RGS16 regulation is also reported in apoptotic bodies, where miR-126 suppressed RGS16 expression and amplified CXCR4 activity [80]. In addition to RGS16, miR-181a also targets RGS4 and induces its suppression during osteoplastic differentiation [81]. Another example of miRNA-induced regulation of RGS proteins is the Hsa-miR-182-induced suppression of RGS17 in lung cancer cells [82]. RGS17 overexpression blocked Hsa-miR-182-induced inhibition of lung cancer cell proliferation, suggesting that Hsa-miR-182 anti-proliferative actions are mediated through silencing of RGS17 [82]. MiRNA profiling revealed that RGS17 is also a target of miR-363 [83]. Taken together, these studies provide evidence that RGS proteins can be regulated by miRNA in cancer (Figure 1), and that RGS silencing contributes to miRNAs actions. Whether the result of miRNA-induced silencing of RGS proteins is to promote or inhibit cancer progression depends on the cell type and the target RGS protein. Future studies that aim to understand the molecular mechanisms and specificity of miRNAs in cancer will potentially identify additional miRNA-regulated RGS proteins.
In addition to cancer, studies have also investigated whether RGS proteins are regulated by miRNAs in the central nervous system. In in vitro models of Huntington’s disease (HD), miR-22 induces RGS2 silencing and results in a neuroprotective effect [84]. Interestingly, miR-22 [85] and hsa-miR-4717-5p [86] target RGS2 (Figure 2) and are associated with panic and anxiety related disorders. This suggests that RGS regulation by microRNA not only occurs in the central nervous system, but also plays a role in the etiology of CNS-related diseases. Attempts to therapeutically target microRNA-induced RGS protein regulation in the CNS should be preceded by comprehensive studies to evaluate the overall effects of this regulation in different brain regions that might result in unwanted CNS-related side effects.
Following transcription, mRNA stability is also controlled by specific RNA-binding proteins [87]. For example, Ataxin-2 (ATXN2) binds and regulates steady-state levels of RGS8 mRNA [88]. Furthermore, RGS4 mRNA is stabilized by binding to human antigen R (HuR), which is required for IL1β-induced upregulation of RGS4 in colonic smooth muscle cells [89]. IL1β also increases transcription of RGS4 via NF-κB, indicating that the same signal may employ multiple mechanisms to regulate the same RGS protein [68]. In addition to HuR, RGS4 mRNA is also regulated by the splicing factor transformer-2β (Tra2β), which possibly mediates morphine-induced up-regulation of RGS4 in the brain [90], and by the RNA-binding protein staufen2 (Stau2) in neurons [91]. Taken together, these data demonstrate that RGS4 mRNA is a common target of RNA-binding proteins, and that mRNA stability of RGS proteins can be affected by both miRNAs and RNA-binding proteins (Figure 1).
To date, there are considerably fewer studies reporting regulation of RGS mRNA stability by miRNA or RNA binding proteins compared to regulation by other mechanisms such as protein degradation. However, due to growing evidence for key roles of RGS proteins, miRNAs, and RNA binding proteins, identifying additional mRNA-targeted mechanisms to control RGS expression in both cancer and the central nervous system is expected. Future studies should also be expanded to the cardiovascular system, where both RGS proteins and miRNAs play many crucial roles [46, 92], to determine the mechanisms by which many important cardiovascular RGS proteins are regulated, and to determine whether some miRNA effects in the cardiovascular systems are mediated by targeting RGS proteins.
3.4 Protein Stability
Degradation of proteins is an essential mechanism employed by cells to control the levels of stable and functional proteins. This degradation commonly occurs via either lysosomal proteolysis or the ubiquitin-proteasome pathway [93, 94]. Lysosomes engulf proteins and utilize digestive enzymes to induce proteolysis [94]. The other pathway for protein degradation is the ubiquitin-proteasome pathway, where the target protein is polyubiquitinated [93]. The polyubiquitinated proteins are recognized by the proteasome complex, which subsequently binds and eventually degrades the target protein [93]. This process requires more energy compared to lysosomal degradation and is mediated by multiple enzymes, including ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3)[95].
Many studies have focused on RGS4 as a target for proteasomal degradation and the mechanisms have been well defined. RGS4 is targeted by the N-end rule pathway, a pathway that tags proteins for degradation based on the presence of certain residues at their N-termini [96]. Inhibitors of this pathway prevent degradation and ubiquitination of RGS4 in the reticulocyte lysate system [96]. In addition, the proteasome inhibitor MG132 blocked degradation and enhanced the levels of polyubiquitinated RGS4, suggesting that RGS4 is subject to ubiquitination and proteasome degradation in accordance to the N-end rule pathway [96]. Studies also revealed that the arginylation of the cysteine 2 residue (Cys2) at the N-terminus of RGS4 is the trigger for N-end rule pathway activation and subsequent degradation [96]. To determine whether this pathway targets RGS4 in intact mammalian cells, a subsequent study tested this mechanism in embryos and embryonic fibroblasts (EF) isolated from wild type or ATE1−/− animals [97]. ATE1 encodes Arg-transferase, the enzyme that mediates arginylation resulting in RGS4 proteasome degradation [97]. Proteasome inhibition or ATE1 depletion significantly increased levels of RGS4 in EF, indicating that arginylation and proteasome degradation regulates RGS4 levels in this cell model as well [97]. Additionally, knockout of ubiquitin ligases UBR1 and UBR2, which recognize and bind N-terminal Arginine, stabilized RGS4 expression, suggesting that UBR1 and/or UBR2 mediates ATE1-triggered degradation of RGS4 [97].
Nitric Oxide also contributes to RGS4 degradation by aiding in oxidizing the N-terminal cysteine residue required for arginylation [98]. RGS4 proteasomal degradation and the crucial role of Cys2 in destabilizing RGS4 were also confirmed in HEK293 cells, in which using the proteasome inhibitor MG132 or replacing Cys2 with serine resulted in the stabilization of RGS4 [99]. Interestingly, palmitoylation of Cys2 by acyltransferases protected RGS4 against proteasomal degradation, establishing this residue as an important determinant of RGS4 fate [100]. An alternative pathway that exclusively targets cytoplasmic RGS4 for degradation and is mediated by the ubiquitin-like modifier activating enzyme UBA6 has also been described [101]. In addition to RGS4, other RGS proteins are targeted by the N-end rule pathway, including RGS2, RGS5, and RGS16 [97–99].
In addition to N-terminal arginylation, the N-end rule pathway can also be initiated via N-terminal acetylation [102]. In contrast to the RGS4 mechanism, RGS2 proteasomal degradation was found to be stimulated by N-terminal acetylation [103]. Furthermore, unlike RGS4, RGS2 proteasomal degradation does not require ATE-1 but instead depends on a complex of proteins consisting of cullin 4B (CUL4B), F-box 44 (FBXO44) and DNA damage binding protein 1 (DDB1) (Figure 2) [104]. Therefore, proteasomal degradation of RGS proteins can be mediated by different pathways, which provides opportunities to selectively target the degradation of RGS proteins.
RGS protein degradation and stability is regulated by multiple post-translational modifications and protein-protein interactions. Members of the R7 family of RGS proteins, including RGS9 and RGS7, uniquely depend on the binding partner Gβ5 for stabilization [105]. Additionally, RGS9 Anchor Protein (R9AP) is another binding partner that is essential for RGS9-1-Gβ5 complex stability and membrane association in the retina [106]. Similarly, R7 family binding protein (R7BP) binds and stabilizes RGS9-2 in striatal neurons (as well as other R7 family members) [107]. The association between R7BP and RGS9-2 prevents the binding of RGS9-2 and Hsc70 (Heat shock cognate protein 70), a protein that mediates RGS9-2 degradation [108]. Interestingly, cysteine protease inhibitors that block lysosomal degradation prevented RGS9-2 degradation, whereas proteasome inhibitors had no effect on RGS9-2 proteolysis, indicating that RGS9-2, unlike many other RGS proteins, is mainly regulated by lysosomal rather than proteasomal degradation [109]. Collectively, these findings indicate that association with different binding partners is a common method by which the stability of RGS7 family members is controlled.
Phosphorylation is a common post-translational modification that affects the activity, localization, and stability of many proteins, including RGS proteins. In fact, some RGS proteins are phosphorylated at multiple sites, leading to specific effects on the activity and stability of RGS proteins. For example, RGS16 is constitutively phosphorylated at serine 194 and is dynamically phosphorylated at serine 53 site by α2A-adrenoceptor activation. Both phosphorylation events result in the suppression of RGS16 GAP activity. On the other hand, phosphorylation at RGS16 Tyr 168 enhances both GAP activity [110] and stability [111], indicating that RGS16 function and levels can be tightly regulated by phosphorylation at multiple sites. Moreover, the same kinase can induce multiple regulatory effects on RGS proteins. For example, protein-kinase A (PKA) was shown to trigger nuclear localization of RGS13, which facilitates the inhibitory effect of RGS13 on CREB-induced transcription in the nucleus [75]. Additionally, PKA-induced phosphorylation at Thr residue 41 inhibits the proteasome degradation of RGS13, which indicates that PKA influences both the nuclear function as well as the stability of RGS13 [112]. Interestingly, RGS10 nuclear localization was also shown to be enhanced by PKA activation, indicating that PKA may affect other RGS proteins in a similar manner [113]. Finally, the cGMP-dependent protein kinase cGKIα regulates the stability, GAP activity, and localization of RGS2 [114]. In addition to phosphorylation, RGS proteins have also been shown to undergo other post-translational modifications such as palmitoylation, but the effects on activity and stability are not fully defined [115].
In summary, protein degradation is a critical regulatory mechanism utilized by cells to maintain physiological levels of RGS proteins (Figure 1), and degradation-mediated changes in RGS protein levels are implicated in the pathogenesis of several diseases. Further, the previous studies reveal that degradation of RGS proteins is the final result of a complex processes orchestrated by a network of enzymes, binding partners, and post-translational modifications offering multiple prospects for effective and selective therapeutic approaches to control RGS proteins levels.
4. Therapeutic Potential of targeting RGS expression
4.1 Why RGS proteins?
GPCRs are the most common molecular targets of clinically approved drugs, and these receptors can be modulated with extracellular agonists, antagonists, and biased agonists to carefully control downstream G protein pathways [116]. Given the enormous clinical success and versatility of therapeutics targeting GPCRs, it is reasonable to ask what benefits can be obtained by targeting RGS proteins that cannot be obtained by targeting GPCRs. A few reasons can be proposed based on the mechanistic differences between GPCRs and RGS proteins in regulating G protein signaling. First, it should be noted that the success of targeting GPCRs is not without limitations. Activation of GPCRs can trigger, in addition to G protein mediated pathways, other pathways mediated by β-arrestins that can induce unintended adverse effects [117]. Manipulating RGS protein activity or expression could be used as a strategy to differentially regulate G protein mediated versus G protein independent pathways downstream of GPCRs. For example, morphine-induced activation of G proteins downstream of opioid receptors produces analgesic effects, but morphine also activates the β-arrestin pathway causing desensitization and tolerance [118, 119]. RGS proteins selectively target G protein signaling downstream of opioid receptors, but not β-arrestin mediated signaling [120]. Therefore, combining modest receptor agonist treatment with inhibition of RGS proteins would amplify G protein mediated analgesic effects with no added effect on the β-arrestin-mediated responses, which would theoretically allow for lower doses of morphine without compromising effectiveness. In contrast, in other systems, β-arrestin mediated signaling is actually desired, whereas G proteins mediate undesirable effects. For example, the AT1 receptor improves cardiac function via β-arrestin-mediated signalling [121], but also causes hypertension through G protein activation [122]. Loss of RGS2 causes an enhancement of AT1 receptor-induced G protein activation, resulting in hypertension [42]. Therefore, in theory, the combination of AT1 receptor agonists and induction of RGS2 expression should improve cardiac function while suppressing hypertension.
Second, many RGS proteins display highly tissue-specific expression patterns, such as RGS9-1 in the retina [10]. This suggests that RGS modifying approaches could be coupled with sub-threshold doses of drugs that target more widely expressed GPCRs to achieve greater regional specificity of action and, again, reduce the required dose of the GPCR targeted drug and associated side effects. Finally, it is now well established that many RGS proteins are more than just GTPase-activating proteins (GAPs), which means that targeting RGS proteins can affect pathways that go beyond G protein signaling, providing yet another advantage over targeting GPCRs alone [123]. These advantages have fuelled efforts to develop and identify small molecule inhibitors of RGS proteins [3, 124]. Aside from directly targeting the activity of RGS proteins, an alternative approach is to target the expression and stability of RGS proteins, which would regulate the GAP-dependent and GAP-independent activities of RGS proteins. Unfortunately, the mechanisms that regulate RGS expression and post-translational stability are generally non-specific, so an ongoing challenge is to establish a therapeutic approach with sufficient specificity. Regardless of these challenges, multiple therapeutics targeting epigenetic, transcriptional, and post-translational regulation have entered the drug discovery pipeline, some with surprising success.
4.2 Targeting Epigenetic mechanisms
Due to the role of epigenetic-mediated mechanisms in disease progression, efforts are underway in several disease models to therapeutically target epigenetic enzymes to restore the expression of dysregulated genes [125]. In cancer, DNA methyl transferase enzymes (DNMTs) mediate silencing of RGS2 and RGS10 in prostate cancer and ovarian cancer, respectively. Restoring the expression of RGS2 or RGS10 can be beneficial, as the suppression of RGS2 is implicated in the progression of prostate cancer [38], and RGS10 silencing contributes to the development of chemoresistance in ovarian cancer cells [32]. Inhibition of DNMT by 5-azacytidine successfully restored the expression of RGS2 in prostate cancer and 5-azacytidine-induced inhibition of prostate cancer growth was mediated partially be restoring the expression of RGS2 [38]. Likewise, both 5-azacytidine and the HDAC inhibitor TSA restored the expression of RGS10 in chemoresistant ovarian cancer cells [57]. These studies suggest that targeting epigenetic processes to restore the expression of RGS proteins may be a future therapeutic approach to slow or stop cancer progression.
Selectivity remains a major concern, as an epigenetic drug can potentially regulate multiple unintended genes that may produce side effects [125]. Despite these concerns, a number of epigenetic drugs have been approved or reached clinical trials. The DNMT inhibitor azacytidine for Myelodysplastic syndrome (MDS) [126] and additional drugs targeting DNA methylation have been developed and entered clinical trials to treat cancer [127, 128]. HDAC inhibitors have also been intensely studied as a potential cancer therapy, and the HDAC inhibitor suberanilohydroxamic acid (SAHA) was FDA approved for cutaneous T-cell lymphoma [129]. Combination therapy of DNMT and HDAC inhibitors is also being studied with encouraging results [130]. Even with this success in developing effective epigenetic drugs, the side effects that they produce due to the dysregulation of unintended genes remain a disadvantage that necessitates better approaches [131]. One approach is to carefully delineate which specific genes mediate the epigenetic drug’s effects, and combining other non-epigenetic drugs that target these genes, which allows reducing the dose of the epigenetic drug without compromising the expression of the intended genes. This concept can be applied to RGS proteins as well, and future studies that gauge the extent of RGS protein contribution to the overall effects of epigenetic drugs should aid in developing more selective and safer epigenetic therapies.
4.3 Targeting Transcription Factors
Attempts to control the activity of transcription factors has included small molecules targeted against upstream proteins such as kinases to indirectly influence the activity of transcription factors [132], as well as directly preventing DNA binding of transcription factors to target transcription factors therapeutically [133]. However, lack of specificity is a major barrier, given that one transcription factor can influence multiple target genes. An alternative approach is to target specific protein-protein interactions in transcriptional regulatory complexes [134]. Targeting a transcriptional regulator’s ability to bind to a specific complex rather than inhibiting a transcription factor activity altogether will potentially enhance specificity and minimize side effects. Employing small molecules to target protein-protein interactions in transcriptional complexes has shown promise in influencing RGS gene expression. For example, preventing the interaction between CREB and the co-factor P300 using the small molecule KG-501 suppressed CREB-induced RGS2 gene expression (Figure 2) [135]. Additionally, inhibiting CREB and the co-factor CBP interaction resulted in a reduction in RGS4 expression in cortical neurons [136]. These approaches provide proof of concept for targeting protein-protein interaction of transcription factors to control RGS protein gene expression. In addition to CREB, protein-protein interactions of other transcription factors such as NF-κB, STAT3, and P53 have been targeted successfully by small molecules (reviewed in [137]); it remains to be determined, however, whether the expression of RGS proteins regulated by STAT3 or P53 is affected by these small molecules. Although promising, targeting protein-protein interactions is an extremely complex approach and much work remains to validate targets and establish this as a viable therapeutic strategy. Thus, more detailed structural studies and an overall better understanding of transcription factor interactions at RGS promoters are required for the development of effective and safe therapeutics to control the transcription of RGS genes.
4.4 Targeting mRNA stability
The development of microRNA-based therapy is hampered by a plethora of specificity concerns as well as difficulties in ensuring the delivery of stable microRNAs to target tissues and cells [138]. Yet, quite a few miRNA-targeted therapies are showing promise and some have reached clinical trials [139]. This success continuously ignites the drive to develop strategies to overcome challenging technical difficulties. The effectiveness of microRNA-based therapies, compared to traditionally targeting an individual protein, is attributed to the broad range of effects a single microRNA can have on several targets involved in the same disease. This is clearly demonstrated in different cancers as the expression of microRNAs influence many genes involved in regulating apoptosis and cell cycle, which effectively induces cell death [140, 141]. Interestingly, microRNA-induced regulation of RGS proteins is often accompanied by regulating other relevant genes commonly implicated in the same disease. For example, in addition to RGS2, miR-22 down-regulates HDAC4 and Rcor1, and all three genes are implicated in Huntington’s disease [84, 142–144]. Similarly, miR-181a targets TGFBI (Tgf-beta induced), TβR-I/Alk5 (TGF-β type I receptor) and Gata6 alongside regulating RGS4, which together promote osteoblastic differentiation [81]. This suggests that utilizing microRNAs to target RGS proteins for treatment dictates a more in-depth investigation of both the global effects of microRNAs as well as the role that the RGS protein plays within the network of targets implicated in the disease.
Recently, an interesting study tested the effect of an antisense oligonucleotide (ASO7) targeted against the RNA binding protein ataxin 2 (ATXN2) in mouse models of spinocerebellar ataxia type 2 (SCA2) [145]. ASO7 suppressed ATXN2 mRNA and increased RGS8 mRNA and other genes implicated in SCA2, which significantly delayed the development of SCA2 [145]. This study provides additional evidence that RGS8 mRNA is targeted by the RNA-binding protein ATXN2, and further suggests that targeting this mechanism can be therapeutically beneficial. Overall, targeting stability of RGS mRNAs is indeed promising, but more studies to identify additional miRNAs and RNA-binding proteins that regulate RGS mRNAs are needed to seriously advance mRNA-targeted therapeutic approaches.
4.5 Targeting Protein Stability
The N-end rule protein degradation pathway has been established as a key regulatory process of RGS4 expression, follow up studies have aimed to determine the effect of inhibiting RGS4 proteasomal degradation on cellular processes and/or disease progression. RGS4 suppresses breast cancer cell migration and invasion in-vitro, as well as tumor invasiveness in vivo [28]. More importantly, RGS4 protein levels are suppressed in breast cancer cells and tissues compared to normal cells [28]. Interestingly, the proteasome inhibitor MG132 inhibited breast cancer cell invasion and migration, but this inhibition was blocked by RGS4 knockdown, indicating that MG132 inhibition of breast cancer cell migration and invasion is due to its ability to block RGS4 degradation [28]. A similar study also tested the effect of pristimerin, a natural compound that has been reported to inhibit proteasome function, on breast cancer cell migration and invasion [146]. Pristimerin inhibited breast cancer migration and invasion, and this inhibition was also lost by RGS4 knockdown [146]. Similarly, blocking RGS4 proteasomal degradation also affects progression of renal dysfunction. RGS4 −/− animals display impaired renal functions and are more prone to renal injuries [147]. Treating the animals experiencing renal injury with the proteasomal inhibitor MG132 significantly improved renal functions in WT, and not in RGS4 KO animals, again suggesting that inhibiting proteasomal degradation of RGS4 can influence disease pathogenesis [147].
These studies provide encouraging results indicating that manipulating protein stability to restore dysregulated expression of RGS proteins can be considered as a viable therapeutic approach. However, proteasomal degradation is a common regulatory mechanism for numerous proteins and it was unsurprising that the first FDA approved proteasome inhibitor caused dose-limiting side effects [148]. Proteasomal degradation of proteins is the final result of multiple events mediated by different proteins, each of which can be targeted for more selective approaches [149]. In the case of RGS4, for example, instead of inhibiting global proteasome degradation to restore RGS4 expression, an alternative approach is to inhibit the N-end rule pathway that directly triggers the degradation of RGS4. Indeed, the N-end rule pathway inhibitor RF-C11 significantly stabilized RGS4 in mammalian cells with no significant cytotoxicity [150]. The substituted amphetamine compound para-chloroamphetamine has been shown to cross the blood brain barrier, where it inhibits arginylation and the N-end rule pathway, thus stabilizing RGS4 expression in the central nervous system [151]. Added selectivity can be achieved by identifying specific proteins that mediate degradation of only a specific subset of proteins that undergo N-end rule mediated degradation. An example is arginyltransferase (ATE-1), which mediates the degradation of RGS4 [97] but not RGS2 [104]. Tannic acid and merbromin, small molecule inhibitors of ATE-1, stabilized RGS4 expression in cells, suggesting that selective inhibition of RGS protein proteasome degradation is possible [152]. Taken together, the previous studies suggest that stabilizing RGS protein expression in vitro and in vivo by targeting the N-end rule pathway is not only achievable, but also moderately safer and more selective compared to inhibiting global proteasome degradation. Future studies aiming to identify additional proteins that differentially regulate the RGS protein degradation, such as ATE-1, will potentially provide more targets for selective drugs.
Several drugs regulate RGS protein expression by influencing proteasome degradation, which can possibly mediate the drugs’ intended or unintended effects. For example, opioid receptor agonists and subsequent Gαo/i activation down-regulate RGS4 [153] and RGS20 [154] by activating the proteasome degradation process. Additionally, proteasome degradation of RGS proteins can be affected by several other stimulants, including inflammatory molecules [155] and cardiotonic steroids [156]. As discussed earlier, phosphorylation by some kinases, such as Src [111] and PKA [112], also regulate proteasome degradation of RGS proteins. Therefore, inhibitors of these kinases can potentially be utilized to influence proteasome degradation of RGS proteins. Identifying drugs that control RGS protein expression by influencing their proteasome degradation will possibly provide more therapeutic options in conditions where controlling RGS protein expression is needed. Alternatively, recognizing that certain drugs influence RGS protein degradation might explain some of the drugs’ side effects and aid in developing approaches to mitigate these unwanted effects.
5. Summary and Conclusions
Targeting RGS proteins shows promise in vitro and in vivo, and RGS-targeted approaches can be used in combination with GPCR-targeted drugs or other drugs to minimize side effects or enhance effectiveness. However, the development of agents targeting RGS protein activity is understandably slow due to the difficulty of targeting RGS interactions with G proteins. Because the expression of RGS proteins is often dysregulated in diseases, an alternative approach is to manipulate the expression of RGS proteins, which allows for tuning their expression to the desired levels. Several epigenetic, transcriptional, and post-translational mechanisms control the ultimate level of cellular RGS proteins, offering multiple opportunities for targeting (Figure 1). Indeed, many studies that tested the effect of targeting these mechanisms on RGS levels and disease progression reported promising results on both activity and safety. However, each of these mechanisms is inherently non-selective, and targeting any single regulatory mechanism is unlikely to provide sufficient selectivity to be therapeutically viable. Rather, strategies to regulate RGS expression and protein levels may be most effective when combined with complementary receptor-targeted approaches, or when multiple mechanisms targeting expression of a single RGS are simultaneously targeted for synergistic regulation. For example, the ultimate level of RGS2 protein in a cell reflects regulation by epigenetic enzymes, transcription factors, miRNAs, and proteasomal degradation (Figure 2). This suggests that targeting a combination of these mechanisms would enhance the effectiveness of regulating RGS2 levels, and simultaneously allow for lower doses, which will potentially reduce toxicity. Thus, it is essential to understand the network of regulatory mechanisms that ultimately control the expression of RGS proteins in disease states in order to design the appropriate interventions.
Highlights.
RGS proteins critically regulate cell physiology and pathophysiology.
RGS protein expression and stability are dysregulated in multiple diseases.
Regulation occurs by epigenetic, transcriptional, and post-translational mechanisms.
Modulation of RGS expression represents a novel therapeutic strategy.
Acknowledgments
Funding: This work was supported by funding from the National Institutes of Health [NS101161].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koelle MR. A new family of G-protein regulators - the RGS proteins. Curr Opin Cell Biol. 1997;9(2):143–7. doi: 10.1016/s0955-0674(97)80055-5. [DOI] [PubMed] [Google Scholar]
- 2.Watson N, et al. RGS family members: GTPase-activating proteins for heterotrimeric G-protein alpha-subunits. Nature. 1996;383(6596):172–5. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- 3.Kimple AJ, et al. Regulators of G-protein signaling and their Galpha substrates: promises and challenges in their use as drug discovery targets. Pharmacol Rev. 2011;63(3):728–49. doi: 10.1124/pr.110.003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie Z, Chan EC, Druey KM. R4 Regulator of G Protein Signaling (RGS) Proteins in Inflammation and Immunity. AAPS J. 2016;18(2):294–304. doi: 10.1208/s12248-015-9847-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerber KJ, Squires KE, Hepler JR. Roles for Regulator of G Protein Signaling Proteins in Synaptic Signaling and Plasticity. Mol Pharmacol. 2016;89(2):273–86. doi: 10.1124/mol.115.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans PR, Dudek SM, Hepler JR. Regulator of G Protein Signaling 14: A Molecular Brake on Synaptic Plasticity Linked to Learning and Memory. Prog Mol Biol Transl Sci. 2015;133:169–206. doi: 10.1016/bs.pmbts.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Chen CK. RGS Protein Regulation of Phototransduction. Prog Mol Biol Transl Sci. 2015;133:31–45. doi: 10.1016/bs.pmbts.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JK, Bou Dagher J. Regulator of G-protein Signaling (RGS)1 and RGS10 Proteins as Potential Drug Targets for Neuroinflammatory and Neurodegenerative Diseases. AAPS J. 2016;18(3):545–9. doi: 10.1208/s12248-016-9883-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neubig RR, Siderovski DP. Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discov. 2002;1(3):187–97. doi: 10.1038/nrd747. [DOI] [PubMed] [Google Scholar]
- 10.Chen CK, et al. Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9-1. Nature. 2000;403(6769):557–60. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- 11.Nishiguchi KM, et al. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature. 2004;427(6969):75–8. doi: 10.1038/nature02170. [DOI] [PubMed] [Google Scholar]
- 12.Rahman Z, et al. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38(6):941–52. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- 13.Zachariou V, et al. Essential role for RGS9 in opiate action. Proc Natl Acad Sci U S A. 2003;100(23):13656–61. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooks SB, Martemyanov K, Zachariou V. A role of RGS proteins in drug addiction. Biochem Pharmacol. 2008;75(1):76–84. doi: 10.1016/j.bcp.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 15.Lerner TN, Kreitzer AC. RGS4 is required for dopaminergic control of striatal LTD and susceptibility to parkinsonian motor deficits. Neuron. 2012;73(2):347–59. doi: 10.1016/j.neuron.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blazer LL, et al. Selectivity and anti-Parkinson’s potential of thiadiazolidinone RGS4 inhibitors. ACS Chem Neurosci. 2015;6(6):911–9. doi: 10.1021/acschemneuro.5b00063. [DOI] [PubMed] [Google Scholar]
- 17.Ko WK, et al. RGS4 is involved in the generation of abnormal involuntary movements in the unilateral 6-OHDA-lesioned rat model of Parkinson’s disease. Neurobiol Dis. 2014;70:138–48. doi: 10.1016/j.nbd.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Dusonchet J, et al. A Parkinson’s disease gene regulatory network identifies the signaling protein RGS2 as a modulator of LRRK2 activity and neuronal toxicity. Hum Mol Genet. 2014;23(18):4887–905. doi: 10.1093/hmg/ddu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bifsha P, et al. Rgs6 is required for adult maintenance of dopaminergic neurons in the ventral substantia nigra. PLoS Genet. 2014;10(12):e1004863. doi: 10.1371/journal.pgen.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JK, et al. Regulator of G-protein signaling 10 promotes dopaminergic neuron survival via regulation of the microglial inflammatory response. J Neurosci. 2008;28(34):8517–28. doi: 10.1523/JNEUROSCI.1806-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivero G, et al. Differential regulation of RGS proteins in the prefrontal cortex of short- and long-term human opiate abusers. Neuropharmacology. 2012;62(2):1044–51. doi: 10.1016/j.neuropharm.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Mirnics K, et al. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6(3):293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- 23.Rivero G, et al. Brain RGS4 and RGS10 protein expression in schizophrenia and depression. Effect of drug treatment. Psychopharmacology (Berl) 2013;226(1):177–88. doi: 10.1007/s00213-012-2888-5. [DOI] [PubMed] [Google Scholar]
- 24.Singh RK, et al. Olanzapine increases RGS7 protein expression via stimulation of the Janus tyrosine kinase-signal transducer and activator of transcription signaling cascade. J Pharmacol Exp Ther. 2007;322(1):133–40. doi: 10.1124/jpet.107.120386. [DOI] [PubMed] [Google Scholar]
- 25.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7(2):79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 26.Hurst JH, Hooks SB. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol. 2009 doi: 10.1016/j.bcp.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 27.Lyu JH, et al. RGS2 suppresses breast cancer cell growth via a MCPIP1-dependent pathway. J Cell Biochem. 2015;116(2):260–7. doi: 10.1002/jcb.24964. [DOI] [PubMed] [Google Scholar]
- 28.Xie Y, et al. Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res. 2009;69(14):5743–51. doi: 10.1158/0008-5472.CAN-08-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maity B, et al. Regulator of G protein signaling 6 is a novel suppressor of breast tumor initiation and progression. Carcinogenesis. 2013;34(8):1747–55. doi: 10.1093/carcin/bgt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang G, et al. RGS16 inhibits breast cancer cell growth by mitigating phosphatidylinositol 3-kinase signaling. J Biol Chem. 2009;284(32):21719–27. doi: 10.1074/jbc.M109.028407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, et al. Regulator of G protein signaling 20 enhances cancer cell aggregation, migration, invasion and adhesion. Cell Signal. 2016;28(11):1663–72. doi: 10.1016/j.cellsig.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 32.Hooks SB, et al. Regulators of G-Protein signaling RGS10 and RGS17 regulate chemoresistance in ovarian cancer cells. Mol Cancer. 2010;9:289. doi: 10.1186/1476-4598-9-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao X, et al. Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene. 2006;25(26):3719–34. doi: 10.1038/sj.onc.1209408. [DOI] [PubMed] [Google Scholar]
- 34.Yang SH, et al. Overexpression of regulator of G protein signaling 11 promotes cell migration and associates with advanced stages and aggressiveness of lung adenocarcinoma. Oncotarget. 2016;7(21):31122–36. doi: 10.18632/oncotarget.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodle CR, Mackie DI, Roman DL. RGS17: an emerging therapeutic target for lung and prostate cancers. Future Med Chem. 2013;5(9):995–1007. doi: 10.4155/fmc.13.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maity B, et al. Regulator of G protein signaling 6 (RGS6) induces apoptosis via a mitochondrial-dependent pathway not involving its GTPase-activating protein activity. J Biol Chem. 2011;286(2):1409–19. doi: 10.1074/jbc.M110.186700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James MA, et al. RGS17, an overexpressed gene in human lung and prostate cancer, induces tumor cell proliferation through the cyclic AMP-PKA-CREB pathway. Cancer Res. 2009;69(5):2108–16. doi: 10.1158/0008-5472.CAN-08-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolff DW, et al. Epigenetic repression of regulator of G-protein signaling 2 promotes androgen-independent prostate cancer cell growth. Int J Cancer. 2012;130(7):1521–31. doi: 10.1002/ijc.26138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J, et al. Regulator of G protein signaling 6 mediates doxorubicin-induced ATM and p53 activation by a reactive oxygen species-dependent mechanism. Cancer Res. 2011;71(20):6310–9. doi: 10.1158/0008-5472.CAN-10-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu P, et al. Combinational therapy of interferon-alpha and chemotherapy normalizes tumor vasculature by regulating pericytes including the novel marker RGS5 in melanoma. J Immunother. 2011;34(3):320–6. doi: 10.1097/CJI.0b013e318213cd12. [DOI] [PubMed] [Google Scholar]
- 41.Belmonte SL, Blaxall BC. Conducting the G-protein Coupled Receptor (GPCR) Signaling Symphony in Cardiovascular Diseases: New Therapeutic Approaches. Drug Discov Today Dis Models. 2012;9(3):e85–e90. doi: 10.1016/j.ddmod.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hercule HC, et al. Regulator of G protein signalling 2 ameliorates angiotensin II-induced hypertension in mice. Exp Physiol. 2007;92(6):1014–22. doi: 10.1113/expphysiol.2007.038240. [DOI] [PubMed] [Google Scholar]
- 43.Zhang W, et al. Selective loss of fine tuning of Gq/11 signaling by RGS2 protein exacerbates cardiomyocyte hypertrophy. J Biol Chem. 2006;281(9):5811–20. doi: 10.1074/jbc.M507871200. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, et al. Regulator of G protein signalling 14 attenuates cardiac remodelling through the MEK-ERK1/2 signalling pathway. Basic Res Cardiol. 2016;111(4):47. doi: 10.1007/s00395-016-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang P, Mende U. Regulators of G-protein signaling in the heart and their potential as therapeutic targets. Circ Res. 2011;109(3):320–33. doi: 10.1161/CIRCRESAHA.110.231423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart A, Huang J, Fisher RA. RGS Proteins in Heart: Brakes on the Vagus. Front Physiol. 2012;3:95. doi: 10.3389/fphys.2012.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grant SL, et al. Specific regulation of RGS2 messenger RNA by angiotensin II in cultured vascular smooth muscle cells. Mol Pharmacol. 2000;57(3):460–7. doi: 10.1124/mol.57.3.460. [DOI] [PubMed] [Google Scholar]
- 48.Miao R, et al. Regulator of G-Protein Signaling 10 Negatively Regulates Cardiac Remodeling by Blocking Mitogen-Activated Protein Kinase-Extracellular Signal-Regulated Protein Kinase 1/2 Signaling. Hypertension. 2016;67(1):86–98. doi: 10.1161/HYPERTENSIONAHA.115.05957. [DOI] [PubMed] [Google Scholar]
- 49.Jean-Baptiste G, et al. Beta adrenergic receptor-mediated atrial specific up-regulation of RGS5. Life Sci. 2005;76(13):1533–45. doi: 10.1016/j.lfs.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Hendriks-Balk MC, et al. Sphingosine-1-phosphate regulates RGS2 and RGS16 mRNA expression in vascular smooth muscle cells. Eur J Pharmacol. 2009;606(1–3):25–31. doi: 10.1016/j.ejphar.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 51.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16(3):178–89. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 52.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204–20. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 53.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3(3):224–9. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiechec E, Overgaard J, Hansen LL. A fragile site within the HPC1 region at 1q25.3 affecting RGS16, RGSL1, and RGSL2 in human breast carcinomas. Genes Chromosomes Cancer. 2008;47(9):766–80. doi: 10.1002/gcc.20578. [DOI] [PubMed] [Google Scholar]
- 55.Ying L, et al. Epigenetic repression of regulator of G-protein signaling 2 by ubiquitin-like with PHD and ring-finger domain 1 promotes bladder cancer progression. FEBS J. 2015;282(1):174–82. doi: 10.1111/febs.13116. [DOI] [PubMed] [Google Scholar]
- 56.Ali MW, et al. Transcriptional suppression, DNA methylation, and histone deacetylation of the regulator of G-protein signaling 10 (RGS10) gene in ovarian cancer cells. PLoS One. 2013;8(3):e60185. doi: 10.1371/journal.pone.0060185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cacan E, et al. Inhibition of HDAC1 and DNMT1 modulate RGS10 expression and decrease ovarian cancer chemoresistance. PLoS One. 2014;9(1):e87455. doi: 10.1371/journal.pone.0087455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tuggle K, et al. Regulator of G protein signaling transcript expression in human neural progenitor differentiation: R7 subfamily regulation by DNA methylation. Neurosignals. 2014;22(1):43–51. doi: 10.1159/000362128. [DOI] [PubMed] [Google Scholar]
- 59.Alqinyah M, et al. Regulator of G Protein Signaling 10 (Rgs10) Expression Is Transcriptionally Silenced in Activated Microglia by Histone Deacetylase Activity. Mol Pharmacol. 2017;91(3):197–207. doi: 10.1124/mol.116.106963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsing CH, et al. Histone Deacetylase Inhibitor Trichostatin A Ameliorated Endotoxin-Induced Neuroinflammation and Cognitive Dysfunction. Mediators Inflamm. 2015;2015:163140. doi: 10.1155/2015/163140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, et al. Origin-specific epigenetic program correlates with vascular bed-specific differences in Rgs5 expression. FASEB J. 2012;26(1):181–91. doi: 10.1096/fj.11-185454. [DOI] [PubMed] [Google Scholar]
- 62.Xie Z, et al. Identification of a cAMP-response element in the regulator of G-protein signaling-2 (RGS2) promoter as a key cis-regulatory element for RGS2 transcriptional regulation by angiotensin II in cultured vascular smooth muscles. J Biol Chem. 2011;286(52):44646–58. doi: 10.1074/jbc.M111.265462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang J, et al. A novel mechanism involving coordinated regulation of nuclear levels and acetylation of NF-YA and Bcl6 activates RGS4 transcription. J Biol Chem. 2010;285(39):29760–9. doi: 10.1074/jbc.M110.121459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seidl MD, et al. Transcription factor cAMP response element modulator (Crem) restrains Pdgf-dependent proliferation of vascular smooth muscle cells in mice. Pflugers Arch. 2015;467(10):2165–77. doi: 10.1007/s00424-014-1652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li F, et al. Cloning and characterization of rabbit Rgs4 promoter in gut smooth muscle. Gene. 2010;451(1–2):45–53. doi: 10.1016/j.gene.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. MEKK1-MKK4-JNK-AP1 pathway negatively regulates Rgs4 expression in colonic smooth muscle cells. PLoS One. 2012;7(4):e35646. doi: 10.1371/journal.pone.0035646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iwaki S, et al. p53 negatively regulates RGS13 protein expression in immune cells. J Biol Chem. 2011;286(25):22219–26. doi: 10.1074/jbc.M111.228924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu W, et al. Interleukin-1beta up-regulates RGS4 through the canonical IKK2/IkappaBalpha/NF-kappaB pathway in rabbit colonic smooth muscle. Biochem J. 2008;412(1):35–43. doi: 10.1042/BJ20080042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, et al. Regulator of G protein signaling 4 is a novel target of GATA-6 transcription factor. Biochem Biophys Res Commun. 2017;483(3):923–929. doi: 10.1016/j.bbrc.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yue H, et al. Role of nuclear unphosphorylated STAT3 in angiotensin II type 1 receptor-induced cardiac hypertrophy. Cardiovasc Res. 2010;85(1):90–9. doi: 10.1093/cvr/cvp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie S, et al. IL-17 activates the canonical NF-kappaB signaling pathway in autoimmune B cells of BXD2 mice to upregulate the expression of regulators of G-protein signaling 16. J Immunol. 2010;184(5):2289–96. doi: 10.4049/jimmunol.0903133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J, et al. Novel NEMO/IkappaB kinase and NF-kappa B target genes at the pre-B to immature B cell transition. J Biol Chem. 2001;276(21):18579–90. doi: 10.1074/jbc.M100846200. [DOI] [PubMed] [Google Scholar]
- 73.Buckbinder L, et al. The p53 tumor suppressor targets a novel regulator of G protein signaling. Proc Natl Acad Sci U S A. 1997;94(15):7868–72. doi: 10.1073/pnas.94.15.7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee HK, et al. RGS2 is a negative regulator of STAT3-mediated Nox1 expression. Cell Signal. 2012;24(3):803–9. doi: 10.1016/j.cellsig.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 75.Xie Z, et al. RGS13 acts as a nuclear repressor of CREB. Mol Cell. 2008;31(5):660–70. doi: 10.1016/j.molcel.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nelson P, et al. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28(10):534–40. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 77.Sun X, et al. miR-181a Targets RGS16 to Promote Chondrosarcoma Growth, Angiogenesis, and Metastasis. Mol Cancer Res. 2015;13(9):1347–57. doi: 10.1158/1541-7786.MCR-14-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun X, et al. CXCR4-targeted therapy inhibits VEGF expression and chondrosarcoma angiogenesis and metastasis. Mol Cancer Ther. 2013;12(7):1163–70. doi: 10.1158/1535-7163.MCT-12-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berthebaud M, et al. RGS16 is a negative regulator of SDF-1-CXCR4 signaling in megakaryocytes. Blood. 2005;106(9):2962–8. doi: 10.1182/blood-2005-02-0526. [DOI] [PubMed] [Google Scholar]
- 80.Zernecke A, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2(100):ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 81.Bhushan R, et al. miR-181a promotes osteoblastic differentiation through repression of TGF-beta signaling molecules. Int J Biochem Cell Biol. 2013;45(3):696–705. doi: 10.1016/j.biocel.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 82.Sun Y, et al. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem Biophys Res Commun. 2010;396(2):501–7. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- 83.Mosakhani N, et al. MicroRNA profiling in chemoresistant and chemosensitive acute myeloid leukemia. Cytogenet Genome Res. 2013;141(4):272–6. doi: 10.1159/000351219. [DOI] [PubMed] [Google Scholar]
- 84.Jovicic A, et al. MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington’s disease-related mechanisms. PLoS One. 2013;8(1):e54222. doi: 10.1371/journal.pone.0054222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muinos-Gimeno M, et al. Human microRNAs miR-22, miR-138-2, miR-148a, and miR-488 are associated with panic disorder and regulate several anxiety candidate genes and related pathways. Biol Psychiatry. 2011;69(6):526–33. doi: 10.1016/j.biopsych.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 86.Hommers L, et al. MicroRNA hsa-miR-4717-5p regulates RGS2 and may be a risk factor for anxiety-related traits. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(4):296–306. doi: 10.1002/ajmg.b.32312. [DOI] [PubMed] [Google Scholar]
- 87.Glisovic T, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–86. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dansithong W, et al. Ataxin-2 regulates RGS8 translation in a new BAC-SCA2 transgenic mouse model. PLoS Genet. 2015;11(4):e1005182. doi: 10.1371/journal.pgen.1005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li F, et al. RNA-binding protein HuR regulates RGS4 mRNA stability in rabbit colonic smooth muscle cells. Am J Physiol Cell Physiol. 2010;299(6):C1418–29. doi: 10.1152/ajpcell.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li SJ, et al. Splicing factor transformer-2beta (Tra2beta) regulates the expression of regulator of G protein signaling 4 (RGS4) gene and is induced by morphine. PLoS One. 2013;8(8):e72220. doi: 10.1371/journal.pone.0072220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heraud-Farlow JE, et al. Staufen2 regulates neuronal target RNAs. Cell Rep. 2013;5(6):1511–8. doi: 10.1016/j.celrep.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 92.Papageorgiou N, et al. The role of microRNAs in cardiovascular disease. Curr Med Chem. 2012;19(16):2605–10. doi: 10.2174/092986712800493048. [DOI] [PubMed] [Google Scholar]
- 93.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17(7):1807–19. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 94.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6(1):79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 95.Myung J, Kim KB, Crews CM. The ubiquitin-proteasome pathway and proteasome inhibitors. Med Res Rev. 2001;21(4):245–73. doi: 10.1002/med.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Davydov IV, Varshavsky A. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J Biol Chem. 2000;275(30):22931–41. doi: 10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
- 97.Lee MJ, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci U S A. 2005;102(42):15030–5. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu RG, et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437(7061):981–6. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 99.Bodenstein J, Sunahara RK, Neubig RR. N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol Pharmacol. 2007;71(4):1040–50. doi: 10.1124/mol.106.029397. [DOI] [PubMed] [Google Scholar]
- 100.Wang J, et al. DHHC protein-dependent palmitoylation protects regulator of G-protein signaling 4 from proteasome degradation. FEBS Lett. 2010;584(22):4570–4. doi: 10.1016/j.febslet.2010.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee PC, et al. Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol Cell. 2011;43(3):392–405. doi: 10.1016/j.molcel.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327(5968):973–7. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park SE, et al. Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science. 2015;347(6227):1249–52. doi: 10.1126/science.aaa3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sjogren B, Swaney S, Neubig RR. FBXO44-Mediated Degradation of RGS2 Protein Uniquely Depends on a Cullin 4B/DDB1 Complex. PLoS One. 2015;10(5):e0123581. doi: 10.1371/journal.pone.0123581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen CK, et al. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100(11):6604–9. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Keresztes G, et al. Absence of the RGS9.Gbeta5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse. J Biol Chem. 2004;279(3):1581–4. doi: 10.1074/jbc.C300456200. [DOI] [PubMed] [Google Scholar]
- 107.Anderson GR, et al. The membrane anchor R7BP controls the proteolytic stability of the striatal specific RGS protein, RGS9-2. J Biol Chem. 2007;282(7):4772–81. doi: 10.1074/jbc.M610518200. [DOI] [PubMed] [Google Scholar]
- 108.Posokhova E, Uversky V, Martemyanov KA. Proteomic identification of Hsc70 as a mediator of RGS9-2 degradation by in vivo interactome analysis. J Proteome Res. 2010;9(3):1510–21. doi: 10.1021/pr901022m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anderson GR, et al. Expression and localization of RGS9-2/G 5/R7BP complex in vivo is set by dynamic control of its constitutive degradation by cellular cysteine proteases. J Neurosci. 2007;27(51):14117–27. doi: 10.1523/JNEUROSCI.3884-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Derrien A, Druey KM. RGS16 function is regulated by epidermal growth factor receptor-mediated tyrosine phosphorylation. J Biol Chem. 2001;276(51):48532–8. doi: 10.1074/jbc.M108862200. [DOI] [PubMed] [Google Scholar]
- 111.Derrien A, et al. Src-mediated RGS16 tyrosine phosphorylation promotes RGS16 stability. J Biol Chem. 2003;278(18):16107–16. doi: 10.1074/jbc.M210371200. [DOI] [PubMed] [Google Scholar]
- 112.Xie Z, Yang Z, Druey KM. Phosphorylation of RGS13 by the cyclic AMP-dependent protein kinase inhibits RGS13 degradation. J Mol Cell Biol. 2010;2(6):357–65. doi: 10.1093/jmcb/mjq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Burgon PG, et al. Phosphorylation and nuclear translocation of a regulator of G protein signaling (RGS10) J Biol Chem. 2001;276(35):32828–34. doi: 10.1074/jbc.M100960200. [DOI] [PubMed] [Google Scholar]
- 114.Osei-Owusu P, et al. Regulation of RGS2 and second messenger signaling in vascular smooth muscle cells by cGMP-dependent protein kinase. J Biol Chem. 2007;282(43):31656–65. doi: 10.1074/jbc.M706360200. [DOI] [PubMed] [Google Scholar]
- 115.Hiol A, et al. Palmitoylation regulates regulators of G-protein signaling (RGS) 16 function. I. Mutation of amino-terminal cysteine residues on RGS16 prevents its targeting to lipid rafts and palmitoylation of an internal cysteine residue. J Biol Chem. 2003;278(21):19301–8. doi: 10.1074/jbc.M210123200. [DOI] [PubMed] [Google Scholar]
- 116.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5(12):993–6. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 117.Shenoy SK, Lefkowitz RJ. beta-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol Sci. 2011;32(9):521–33. doi: 10.1016/j.tips.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bohn LM, et al. Enhanced morphine analgesia in mice lacking beta-arrestin 2. Science. 1999;286(5449):2495–8. doi: 10.1126/science.286.5449.2495. [DOI] [PubMed] [Google Scholar]
- 119.Bohn LM, et al. Mu-opioid receptor desensitization by beta-arrestin-2 determines morphine tolerance but not dependence. Nature. 2000;408(6813):720–3. doi: 10.1038/35047086. [DOI] [PubMed] [Google Scholar]
- 120.Traynor J. mu-Opioid receptors and regulators of G protein signaling (RGS) proteins: from a symposium on new concepts in mu-opioid pharmacology. Drug Alcohol Depend. 2012;121(3):173–80. doi: 10.1016/j.drugalcdep.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ryba DM, et al. Long-Term Biased beta-Arrestin Signaling Improves Cardiac Structure and Function in Dilated Cardiomyopathy. Circulation. 2017;135(11):1056–1070. doi: 10.1161/CIRCULATIONAHA.116.024482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kawai T, et al. AT1 receptor signaling pathways in the cardiovascular system. Pharmacol Res. 2017 doi: 10.1016/j.phrs.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sethakorn N, Yau DM, Dulin NO. Non-canonical functions of RGS proteins. Cell Signal. 2010;22(9):1274–81. doi: 10.1016/j.cellsig.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Roman DL, Traynor JR. Regulators of G protein signaling (RGS) proteins as drug targets: modulating G-protein-coupled receptor (GPCR) signal transduction. J Med Chem. 2011;54(21):7433–40. doi: 10.1021/jm101572n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28(10):1069–78. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kaminskas E, et al. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10(3):176–82. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 127.Cheng JC, et al. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95(5):399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- 128.Brueckner B, et al. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65(14):6305–11. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 129.Duvic M, Vu J. Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs. 2007;16(7):1111–20. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- 130.Pathania R, et al. Combined Inhibition of DNMT and HDAC Blocks the Tumorigenicity of Cancer Stem-like Cells and Attenuates Mammary Tumor Growth. Cancer Res. 2016;76(11):3224–35. doi: 10.1158/0008-5472.CAN-15-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heerboth S, et al. Use of epigenetic drugs in disease: an overview. Genet Epigenet. 2014;6:9–19. doi: 10.4137/GEG.S12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kwiatkowski N, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511(7511):616–20. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Leung CH, et al. DNA-binding small molecules as inhibitors of transcription factors. Med Res Rev. 2013;33(4):823–46. doi: 10.1002/med.21266. [DOI] [PubMed] [Google Scholar]
- 134.Reiter F, Wienerroither S, Stark A. Combinatorial function of transcription factors and cofactors. Curr Opin Genet Dev. 2017;43:73–81. doi: 10.1016/j.gde.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 135.Best JL, et al. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proc Natl Acad Sci U S A. 2004;101(51):17622–7. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chekler EL, et al. Transcriptional Profiling of a Selective CREB Binding Protein Bromodomain Inhibitor Highlights Therapeutic Opportunities. Chem Biol. 2015;22(12):1588–96. doi: 10.1016/j.chembiol.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 137.Fontaine F, Overman J, Francois M. Pharmacological manipulation of transcription factor protein-protein interactions: opportunities and obstacles. Cell Regen (Lond) 2015;4(1):2. doi: 10.1186/s13619-015-0015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–38. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 139.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 140.Garzon R, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114(26):5331–41. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Seredenina T, Gokce O, Luthi-Carter R. Decreased striatal RGS2 expression is neuroprotective in Huntington’s disease (HD) and exemplifies a compensatory aspect of HD-induced gene regulation. PLoS One. 2011;6(7):e22231. doi: 10.1371/journal.pone.0022231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mielcarek M, et al. SAHA decreases HDAC 2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington’s disease. PLoS One. 2011;6(11):e27746. doi: 10.1371/journal.pone.0027746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zuccato C, et al. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J Neurosci. 2007;27(26):6972–83. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Scoles DR, et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature. 2017;544(7650):362–366. doi: 10.1038/nature22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mu XM, et al. Pristimerin inhibits breast cancer cell migration by up- regulating regulator of G protein signaling 4 expression. Asian Pac J Cancer Prev. 2012;13(4):1097–104. doi: 10.7314/apjcp.2012.13.4.1097. [DOI] [PubMed] [Google Scholar]
- 147.Siedlecki AM, et al. RGS4, a GTPase activator, improves renal function in ischemia-reperfusion injury. Kidney Int. 2011;80(3):263–71. doi: 10.1038/ki.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen D, et al. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11(3):239–53. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9(9):679–90. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jiang YXL, et al. Characterization of mammalian N-degrons and development of heterovalent inhibitors of the N-end rule pathway. Chemical Science. 2013;4(8):3339–3346. [Google Scholar]
- 151.Jiang Y, et al. A neurostimulant para-chloroamphetamine inhibits the arginylation branch of the N-end rule pathway. Sci Rep. 2014;4:6344. doi: 10.1038/srep06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Saha S, et al. Small molecule inhibitors of arginyltransferase regulate arginylation-dependent protein degradation, cell motility, and angiogenesis. Biochem Pharmacol. 2012;83(7):866–73. doi: 10.1016/j.bcp.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang Q, Traynor JR. Opioid-induced down-regulation of RGS4: role of ubiquitination and implications for receptor cross-talk. J Biol Chem. 2011;286(10):7854–64. doi: 10.1074/jbc.M110.160911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pagano M, et al. Galphao/i-stimulated proteosomal degradation of RGS20: a mechanism for temporal integration of Gs and Gi pathways. Cell Signal. 2008;20(6):1190–7. doi: 10.1016/j.cellsig.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Benzing T, et al. Upregulation of RGS7 may contribute to tumor necrosis factor-induced changes in central nervous function. Nat Med. 1999;5(8):913–8. doi: 10.1038/11354. [DOI] [PubMed] [Google Scholar]
- 156.Sjogren B, et al. Cardiotonic steroids stabilize regulator of G protein signaling 2 protein levels. Mol Pharmacol. 2012;82(3):500–9. doi: 10.1124/mol.112.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Grillet N, et al. Dynamic expression of RGS4 in the developing nervous system and regulation by the neural type-specific transcription factor Phox2b. J Neurosci. 2003;23(33):10613–21. doi: 10.1523/JNEUROSCI.23-33-10613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stuebe S, et al. Sphingosine-1-phosphate and endothelin-1 induce the expression of rgs16 protein in cardiac myocytes by transcriptional activation of the rgs16 gene. Naunyn Schmiedebergs Arch Pharmacol. 2008;376(5):363–73. doi: 10.1007/s00210-007-0214-2. [DOI] [PubMed] [Google Scholar]