Abstract
In addition to its conventional role for cell proliferation and survival, the Raf/MEK/Extracellular signal-regulated kinase (ERK) pathway can also induce growth arrest and death responses, if aberrantly activated. Here, we determined a molecular basis of ERK1/2 signaling that underlies these growth inhibitory physiological outputs. We found that overexpression of ERK1 or ERK2 switches ΔRaf-1:ER-induced growth arrest responses to caspase-dependent apoptotic death responses in different cell types. These death responses, however, were reverted to growth arrest responses upon titration of cellular phospho-ERK1/2 levels by the MEK1/2 inhibitor AZD6244. These data suggest that a cellular threshold for active ERK1/2 levels exists and affects the cell fate between death and growth arrest. We also found that death-mediating ability of ERK2 is abolished by the catalytic site-disabling Lys52Arg replacement or significantly attenuated by the F-site recruitment site-disabling Tyr261Asn replacement, although unaffected by the mutations that disable the common docking groove or the dimerization interface. Therefore, ERK1/2 mediates death signaling dependently of kinase activity and specific physical interactions. Intriguingly, Tyr261Asn-replaced ERK2 could still mediate growth arrest signaling, further contrasting the molecular basis of ERK1/2-mediated growth arrest and death signaling. These data reveal a mechanism underlying the role of ERK1/2 as a focal point of Raf/MEK/ERK-mediated growth arrest and death signaling.
Keywords: Raf, MEK1/2, ERK1/2, cell death, growth arrest
1. Introduction
The Raf/MEK/Extracellular signal-regulated kinase (ERK) pathway is a highly specific three-layered kinase cascade that consists of the Ser/Thr kinase Raf (A-Raf, B-Raf, and C-Raf-1), the highly homologous dual-specificity kinases, MEK1 and MEK2 (collectively referred to as MEK1/2), and the ubiquitously expressed serine/threonine kinase ERK1 and its homologue ERK2 (collectively referred to as ERK1/2) [1, 2]. Upon activation, Raf phosphorylates MEK1/2, which in turn activate ERK1/2 by sequentially phosphorylating Tyr and Thr in their activation loop. ERK1/2 then activate/inactivate a variety of targets, including transcription factors, other kinases, phosphatases, cytoskeletal proteins, scaffolds, receptors and signaling components.
The Raf/MEK/ERK pathway has pivotal roles in cell cycle progression and cell survival, and its deregulation often causes tumorigenesis [3–5]. However, paradoxically, sustained activation of this pathway can also induce cell cycle arrest or death in various cell types (reviewed in [6–8]). We have also demonstrated growth inhibitory Raf/MEK/ERK signaling in different cell types and investigated the underlying molecular mechanisms, particularly at the level of MEK/ERK [9–17]. The growth inhibitory effects of Raf/MEK/ERK have been suggested to be biologically significant and, most notably, are appreciated as an innate tumor suppressive mechanism triggered upon aberrant pathway activation [18]. As such, better understanding of the molecular mechanisms underlying growth inhibitory Raf/MEK/ERK signaling is important.
As a focal point of the Raf/MEK/ERK pathway, ERK1 and ERK2 have the central roles in determining the physiological outputs of pathway signaling. The specificity of ERK1/2 signaling is mainly determined by the magnitude and duration of their catalytic activity, spatio-temporal regulation, as well as monomeric vs. dimeric status [1, 19]. These processes usually require ERK physical interactions with different effectors, regulators, and isoforms via specific domains and motifs [1, 19]. Although ERK1 and ERK2 are supposed to have largely overlapping biological functions [20, 21], recent evidences suggest that these enzymes also have unique functions [22, 23]. Moreover, although kinase activity of ERK1/2 is central for their canonical effects, recent studies including ours suggest that ERK1/2 also have non-kinase effects [11, 24–28]. These characteristics should be carefully considered when determining the mechanisms of ERK1/2 signaling.
In this study, we asked whether Raf/MEK/ERK-induced growth arrest and death signals can bifurcate at ERK1/2 levels and, if so, what characteristics of ERK1/2 are important for the bifurcation. We demonstrate that overexpression of ERK1 or ERK2 can switch Raf-induced growth arrest responses to apoptotic death responses in different cell lines. We then determine whether these effects require a high magnitude catalytic activity, specific physical interactions, and dimerization using a series of ERK2 mutants. Our study provides evidence that a cellular threshold for active ERK1/2 levels exists and affects the cell fate between growth arrest and death when the Raf/MEK/ERK pathway is aberrantly activated.
2. Materials and methods
2.1. Cell culture and reagents
LNCaP-ΔRaf-1:ER, HEK293-ΔRaf-1:ER, and U251-ΔRaf-1:ER were generated by stably transducing LNCaP, HEK293 and U251 with the lentiviral pHAGE vector containing ΔRaf-1:ER and selecting for puromycin resistance, as previously described [11]. ΔRaf-1:ER was activated with 1 µM 4-hydroxytamoxifen (Sigma, St. Louis, MO) as previously described [29]. LNCaP-ΔRaf-1:ER cells were maintained in phenol red-deficient RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 U of penicillin and 100 µg of streptomycin per ml. HEK293-ΔRaf-1:ER, and U251-ΔRaf-1:ER cells were maintained in minimal essential medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. The ERK1/2 stable knockdown cell line, LNCaP-shERK1/2, was generated by lentiviral pLL3.7-shRNA systems targeting ERK1 (nucleotides 801–819) and ERK2 (nucleotides 851–869), and was previously described [11]. AZD6244 was obtained from Selleck Chemicals (Houston, TX).
2.2. Cell survival/death and cell cycle analyses
The colorimetric 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT, Sigma) assay was performed as previously described [11]. Briefly, cells in 96 well-plates were incubated with 100 µl of MTT (0.5 mg/ml) in phenol red-free medium for 2 hours at 37°C, switched into 200 µl of dimethyl-sulfoxide, and shaken for 5 min at room temperature before measuring absorbance at 540 nm. Cell death was measured by counting trypan blue-stained cells using the hemocytometer. Annexin V/propidium iodide (Invitrogen) was stained, as previously described [30].
The cell-cycle profile was analyzed as previously described [11]. Briefly, cells were washed with ice-cold 0.2% bovine serum albumin in phosphate-buffered saline, and resus-pended in 250 mM sucrose/40 mM citrate buffer (pH 7.6) containing 0.5% dimethylsulfoxide. Nuclei were prepared, stained with propidium iodide, and analyzed using the Guava EasyCyte flowcytometry system (MilliporeSigma, Billerica, MA, USA) with a gate that selects single nuclei within a normal size range. The cell-cycle parameters from 5,000 gated nuclei were determined, and subsequent analysis was performed using FCS EXPRESS software (De Novo Software, Los Angeles, CA, USA).
2.3. Viral infection
The lentiviral expression vector pHAGE was used as previously described [31, 32]. Briefly, for viral production, pHAGE was co-transfected with packaging vectors into 293T cells and the resulting supernatant was collected after 48 hours. Viral titers were determined by infecting the recipient cell lines with serially diluted viral supernatants mixed with polybrene (Sigma) at 8 µg/ml and scoring cells expressing GFP at 48 hour post-infection. Cells were infected overnight and were switched into fresh culture media.
2.4. Recombinant lentiviral constructs and RNA interference
Generation of pHAGE-puro-Raf:ER, pHAGE-GFP-ERK2wt, pHAGE-GFP-ERK2-K52R was previously described [11]. pHAGE-GFP-ERK1 was constructed by ligating N-terminal HA-tagged human ERK1 into the NotI/BamHI sites of the lentiviral pHAGE vector. pHAGE-GFP-ERK2-HE-L4A and pHAGE-GFP-ERK2-Y261N were generated by switching SacII/XbaI fragment of rat wild type ERK2 in pHAGE with the corresponding fragments from pCMV-Myc-ERK2-HE-L4A [33] and pCMV-FLAG-ERK2-Y261N [34], respectively. pHAGE-GFP-ERK2-D316/319A was generated by mutagenesis of pHAGE-GFP-ERK2wt using the Quickchange site-directed mutagenesis kit (Agilent Tech, La Jolla, CA) and the primers TGGAGCAGTATTATGCCCCAAGTGCTGAGCCCATTGCTG and CAGCAATGGGCTCAGCACTTGGGGCATAATACTGCTCCA.
2.5. Immunoblot analysis
Cells harvested at various times were lysed in 62.5 mM Tris (pH 6.8)-2% SDS mixed with the protease inhibitor cocktail (Sigma) that contains 4-(2-aminoethyl) benzenesulfonyl fluoride, pepstatin A, E-64, bestatin, leupeptin, and aprotinin, and briefly sonicated before determining the protein concentration using the BCA reagent (Pierce, Rockford, IL). 50 µg of protein was resolved by SDS-PAGE, transferred to a polyvinylidene difluoride membrane filter (Bio-Rad, Hercules, CA), and stained with Fast Green reagent (Thermo Fisher Scientific, Waltham, MA). Membrane filters were then blocked in 0.1 M Tris (pH 7.5)-0.9% NaCl-0.05% Tween 20 with 5% nonfat dry milk, and incubated with appropriate antibodies. Antibodies were diluted as follows: ERK1/2, 1:2,500; phospho-ERK1/2 (Thr202/Tyr204 for ERK1 and Thr183/Tyr185 for ERK2), 1:2,500; phospho-p90RSK (Thr359/Ser363), 1:2,500; phospho-ELK1 (Ser383), 1:2,000; phospho-Rb (Ser780), 1:2,500; glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 1:5,000; poly (ADP-ribose) polymerase (PARP), 1:2,000; cleaved lamin A, 1:2,000; caspase-9, 1:2,000; cleaved caspase-3 (Asp175), 1:2,000 (Cell Signaling Technology, Danvers, MA); E2F1, 1:1,000; c-MYC, 1:1,000 (Thermo Fisher Scientific); ERK2 (C-14), 1:2,000; p21CIP1, 1:2,500 (Santa Cruz Biotech, Santa Cruz, CA); p16INK4A, 1:2,500; Rb, 1:1,000; caspase-8, 1:2,000 (BD Bioscience, San Jose, CA); cyclin D1, 1:2,000; β-actin, 1:5,000 (Sigma); phospho-caspase-9, 1:2,000 (Assay Biotech, Sunnyvale, CA). The Supersignal West Pico and Femto chemiluminescence kits (Pierce) were used for visualization of the signal. For densitometry, immunoblots were scanned and analyzed using LabWorks™ (UVP BioImaging Systems, Upland, CA).
2.6. Statistical analysis
Unless otherwise specified, two-tailed unpaired student’s t-test was used to assess the statistical significance of two data sets. P values of < 0.05 were considered statistically significant.
3. Results
3.1. Overexpression of ERK1/2 switches growth arrest responses to cell death responses in ΔRaf-1:ER-activated cells
To determine cellular responses to different magnitudes of ERK1/2 activity, we examined the effects of ERK1 or ERK2 overexpression in different human tumor or immortalized normal cell lines that stably express the tamoxifen-inducible ΔRaf-1:ER (i.e., LNCaP-ΔRaf-1:ER, HEK293-ΔRaf-1:ER, and U251-ΔRaf-1:ER). ΔRaf-1:ER is the CR3 catalytic domain of Raf-1 fused to the hormone binding domain of the estrogen receptor [29]. Its Raf kinase activity can thus be precisely regulated by the estrogen analogue, 4-hydroxytamoxifen (4-HT). Indeed, we previously demonstrated that 4-HT induces stoichiometric activation of MEK/ERK in ΔRaf-1:ER-expressing cells and maximal ERK1/2 phosphorylation can be maintained for 24 – 48 hours when 4-HT is used in 100 – 1000 nM range (Fig. 2 of Ref [35]).
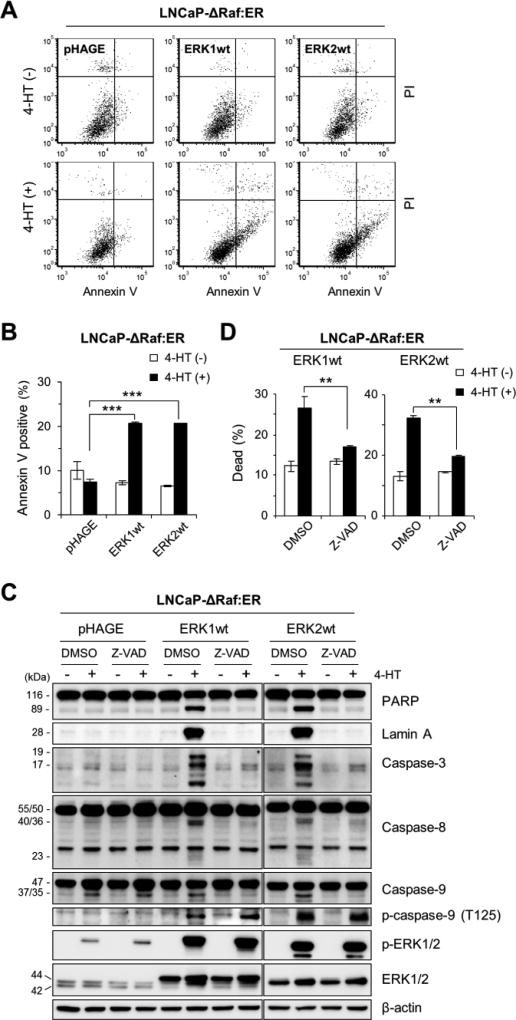
Fig 2. Overexpression of ERK1 or ERK2 induces caspase-dependent apoptotic cell death in ΔRaf-1:ER-activated cells.
(A and B) LNCaP-ΔRaf-1:ER cells, infected with the lentiviral pHAGE-ERK1, pHAGE-ERK2, or pHAGE, were treated with 1 µM 4-HT for 24 hours prior to annexin V/propidium iodide (PI) staining. The graphs (B) indicate annexin V-positive cell populations. (C and D) LNCaP-ΔRaf-1:ER cells, infected with the lentiviral pHAGE-ERK1, pHAGE-ERK2, or pHAGE, were treated with 1 µM 4-HT for 1 day in the presence of the pan-caspase inhibitor, Z-VAD(OMe)-FMK (20 µM). Cells were examined for expression of the indicated proteins by Western blot analysis (C) and viability by trypan blue exclusion analysis (D). Caspase-3 is cleaved into 17 and 19 kDa peptides. Caspase-8 (50/55 kDa doublets) is cleaved into 40/36 kDa doublets and 23 kDa peptides. Caspase-9 (47 kDa) is cleaved into 37/35 kDa peptides. β-actin is the control for equal protein loading. Data (mean ± SEM) are from a representative experiment performed in triplicate. **P < 0.01; ***P < 0.001.
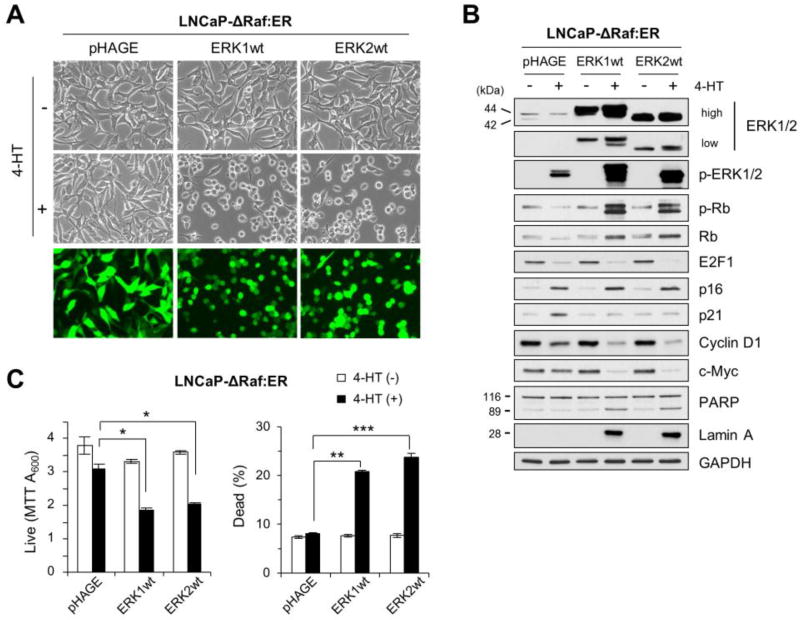
Upon transduction at higher than 95% efficiency, as determined by visualizing GFP (Fig. 1A), the pHAGE lentivirus expressing ERK1 and ERK2 (pHAGE-ERK1 and pHAGE-ERK2 hereafter) substantially increased corresponding ERK protein levels in LNCaP-ΔRaf-1:ER cells, as determined by Western blotting (Fig. 1B). This overexpression however did not increase the Western blot signal specific to phosphorylation of ERK activation loop (i.e., Thr202/Tyr204 for ERK1 and Thr183/Tyr185 for ERK2), indicating that these cells maintain very low basal signals for ERK1/2 activation (Fig. 1B). ERK1/2 overexpression also did not affect cell proliferation and survival (Fig. 1C). Therefore, if not activated, overexpressed ERK1/2 proteins do not induce any significant cellular effects.
Fig 1. Overexpression of ERK1 or ERK2 switches ΔRaf-1:ER-induced growth arrest responses to death responses.
LNCaP-ΔRaf-1:ER cells, infected with the lentiviral pHAGE-ERK1, pHAGE-ERK2, or the control pHAGE, were treated with 1 µM 4-hydroxytamoxifen (4-HT) for 24 hours. Cells were examined for morphological changes (A), expression of the indicated proteins by Western blot analysis (B), and viability by MTT assay and trypan blue exclusion analysis (C). Similar infection efficiency was verified by GFP expression (bottom panels in A). p-ERK1/2 indicates ERK1/2 phosphorylated at Thr202/Tyr204 (ERK1) and Thr183/Tyr185 (ERK2). p-Rb indicates Rb phosphorylated at Ser780. GAPDH is the control for equal protein loading. Data (means ± SE) are from a representative experiment performed in triplicate. *, P < 0.05; ***, P < 0.001.
We previously demonstrated that sustained activation of the Raf/MEK/ERK pathway can induce growth arrest by altering expression of several key regulators of cell proliferation and survival, e.g., cyclin-dependent kinase inhibitors p16INK4A and p21CIP1, the S-phase transcription factor E2F1 and its regulator Rb, cyclin D1, and the pleiotropic transcription factor c-MYC [11, 13, 14]. Consistent with our previous observation, upon 24 hour treatment of 1 µM 4-HT, the control pHAGE-infected LNCaP-ΔRaf-1:ER cells exhibited robust induction of p16INK4A and p21CIP1 as well as downregulation of E2F1, Rb, cyclin D1, and c-MYC (Fig. 1B), although c-MYC was upregulated while cyclin D1 was not downregulated until 8 hour post 4-HT treatment (Supplemental Fig. S1). However, in stark contrast, ERK1- or ERK2-overexpressing LNCaP-ΔRaf-1:ER cells exhibited remarkably different responses upon ΔRaf-1:ER activation. First, these cells underwent dramatic morphological changes (Fig. 1A). Second, as opposed to the control, p21CIP1 induction did not occur in these cells although ERK1/2 phosphorylation was substantially increased (Fig. 1B). Third, Rb phosphorylation was highly increased and the rates of cyclin D1 and c-MYC downregulation were accelerated, although the rates of p16INK4A induction and E2F1 downregulation were similar as those in the control (Fig. 1B). Fourth, these cells exhibited markedly increased the cleavage of PARP and lamin A (Fig. 1B), which are markers of caspase-dependent apoptotic cell death [36]. Indeed, MTT and trypan blue exclusion assays revealed that these cells were undergoing death robustly (Fig. 1C). These data demonstrate that increased ERK1/2 levels can switch cellular responses to ΔRaf-1:ER activation from growth arrest to death, wherein some downstream targets are sensitive to increased ERK1/2 activity while others are not.
3.2. Overexpression of ERK1/2 triggers apoptotic cell death in ΔRaf-1:ER-activated cells
We next determined the nature of ERK1/2-mediated cell death by conducting Annexin V/propidium iodide staining and by profiling different caspases. Consistent with the PARP and lamin A cleavages in Fig. 1B, the Annexin V/propidium iodide staining indicated that ERK1- or ERK2-overexpression markedly increased apoptotic populations in ΔRaf-1:ER-activated LNCaP cells (Fig. 2A and 2B). ERK1/2 overexpression also increased the cleavage of caspase-3 and caspase-8, but not caspase-9, in these cells (Fig. 2C). We found that the broad spectrum caspase inhibitor Z-VAD(OMe)-FMK inhibited cleavages of these caspases, PARP, and lamin A (Fig. 2C) while significantly rescuing cells from death (Fig. 2D). Importantly, Z-VAD(OMe)-FMK mediated these effects without affecting ERK1/2 protein levels or phosphorylation (Fig. 2C), which supports the specificity of its effects. Of note, ERK1/2 overexpression highly increased caspase-9 phosphorylation at Thr125, an ERK1/2-regulated inhibitory phosphorylation [37], in ΔRaf-1:ER-activated cells (Fig. 2C). This phosphorylation was not affected by Z-VAD(OMe)-FMK, suggesting that it is not a feedback response to cell death (Fig. 2C). These data suggest that ERK1 and ERK2 can trigger robust apoptotic death responses by activating caspase-8 and caspase-3, while they can also produce a parallel inhibitory signal for caspase-9.
3.3. Titration of the level of active ERK2 by MEK1/2 inhibition reverts ΔRaf-1:ER-induced cell death to growth arrest responses in ERK2-overexpressing cells
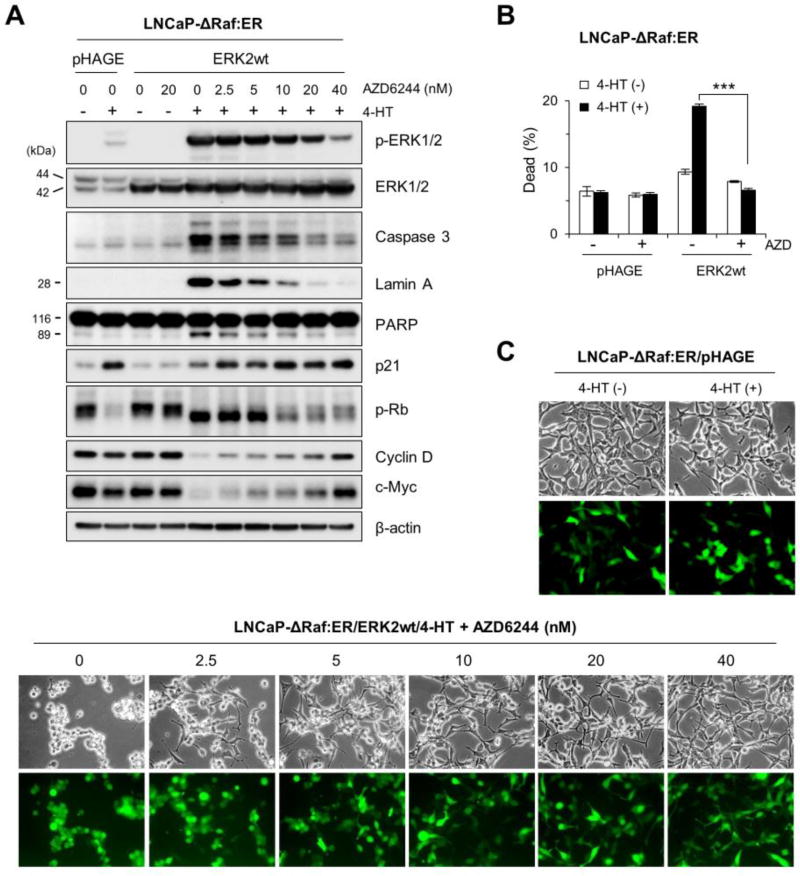
Our data above indicated that there may be a cellular threshold for ERK1/2 levels that affects cellular decision to undergo growth arrest or death in the face of aberrant Raf/MEK/ERK activation. To elaborate this possibility, we titrated the levels of active ERK1/2 in ERK2-overexpressing LNCaP-ΔRaf-1:ER cells using different doses of AZD6244, a highly selective MEK1/2 inhibitor [38], and monitored subsequent effects on the surrogate markers of growth arrest and cell death. AZD6244 gradually decreased exogenous phospho-ERK2 levels in a dose-dependent manner without affecting total ERK2 levels. At 40 nM, AZD6244 decreased exogenous phospho-ERK2 level quite close to endogenous phospho-ERK1/2 levels (Fig. 3A). Remarkably in these cells, cleavages of caspase 3, lamin A, and PARP were gradually decreased in proportion to decreasing phospho-ERK2 levels and were no longer detected when cells were treated with 40 nM AZD6244 (Fig. 3A). Indeed, 40 nM AZD6244 completely blocked cell death, as determined by trypan blue exclusion analysis (Fig. 3B). Along with these effects, Rb hyper-phosphorylation, suppressed p21CIP1 induction, augmented cyclin D1 and c-MYC downregulation, and morphology changes were also abrogated by AZD6244 in a dose-dependent manner (Fig. 3A and 3C). Therefore, titration of phospho-ERK2 levels reverted cell death responses to growth arrest responses in these cells. These data demonstrate that a threshold for active ERK1/2 levels exists in cells and determines the cell fate between growth arrest and death.
Fig 3. AZD6244 reverts ERK2 overexpression-mediated cell death responses to growth arrest responses in ΔRaf-1:ER-activated cells.
LNCaP-ΔRaf-1:ER cells, infected with the lentiviral pHAGE-ERK2 or pHAGE, were treated with 1 µM 4-HT for 24 hours in the presence of the MEK1/2 inhibitor AZD6244. Cells were examined for expression of the indicated proteins by Western blot analysis (A), viability by trypan blue exclusion analysis (B), and morphological changes (C). Similar infection efficiency was verified by GFP expression (bottom panels in C). Caspase-3 is cleaved into 17 and 19 kDa peptides. β-actin is the control for equal protein loading. Data (means ± SE) are from a representative experiment performed in triplicate. ***, P < 0.001.
3.4. Kinase activity is necessary for ERK2 to mediate ΔRaf-1:ER-induced cell death
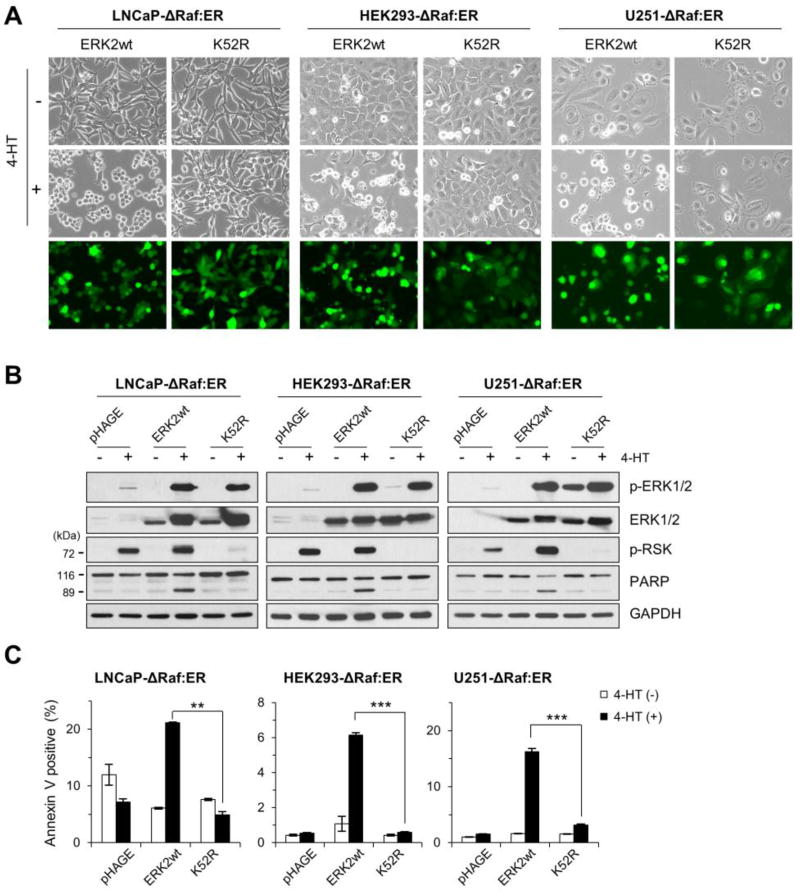
We next determined whether kinase activity is necessary for ERK1/2 to mediate cell death by examining the effects of Lys52Arg (K52R) replacement in the active site of ERK2 in LNCaP-ΔRaf-1:ER, HEK293-ΔRaf-1:ER, and U251-ΔRaf-1:ER cells. The ERK2-K52R mutant is kinase-deficient due to the disabled active site but has an intact activation loop [39]. Therefore, it can be used to increase the levels of ERK2 in active conformation without increasing ERK catalytic activity in cells.
When transduced by the lentivirus expressing wild type ERK2 and treated with 4-HT, HEK293-ΔRaf-1:ER and U251-ΔRaf-1:ER cells exhibited substantial morphological changes (Fig. 4A), PARP cleavage (Fig. 4B), and apoptotic population (Fig. 4C), which are consistent with the aforementioned effects in LNCaP-ΔRaf-1:ER cells. When transduced by lentiviral ERK2-K52R at a similar efficiency (Fig. 4A), all three cell lines expressed similar levels of exogenous ERK2 proteins (Fig. 4B). ERK2-K52R by itself did not induce any notable effects in these cells. However, upon ΔRaf-1:ER activation, ERK2-K52R was highly phosphorylated and acted as a competitively inhibitor of endogenous ERK1/2, as determined by ribosomal S6 kinase (p90RSK) phosphorylation (Fig. 4B). p90RSK phosphorylation is a bona fide readout of in vivo ERK1/2 kinase activity [40]. Moreover, as opposed to wild type ERK2, ERK2-K52R did not induce any morphological changes (Fig. 4A), PARP cleavage (Fig. 4B), or apoptosis (Fig. 4C) in ΔRaf-1:ER-activated cells. These data indicate that kinase activity is necessary for ERK1/2 to mediate death signaling.
Fig 4. Kinase-deficient ERK2 cannot mediate cell death signaling.
LNCaP-ΔRaf-1:ER, HEK293-ΔRaf-1:ER, and U251-ΔRaf-1:ER cells, infected with the lentiviral pHAGE-ERK2, pHAGE-ERK2-K52R, or pHAGE, were treated with 1 µM 4-HT for 24 hours. Cells were then examined for morphological changes (A), expression of the indicated proteins by Western blot analysis (B), and apoptosis by annexin V/propidium iodide staining (C). Similar infection efficiency was verified by GFP expression (bottom panels in A). p-RSK indicates p90RSK phosphorylated at Thr359/Ser363. GAPDH is the control for equal protein loading. Data (means ± SE) are from a representative experiment performed in triplicate. **, P < 0.01; ***, P < 0.001.
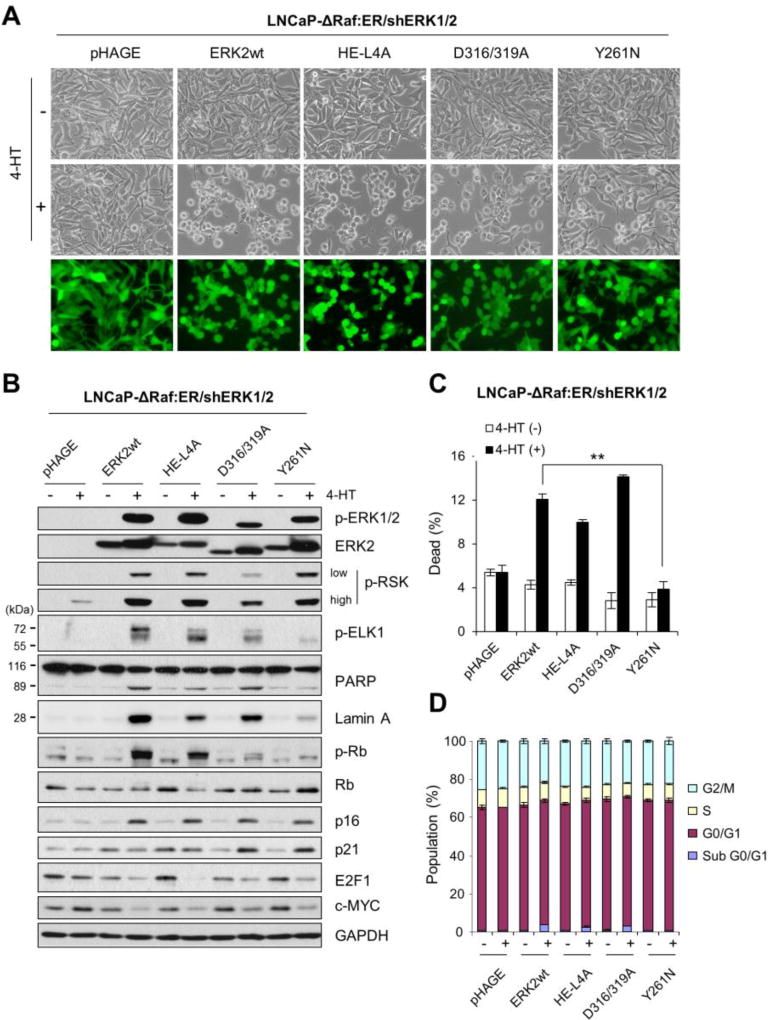
3.5. Defective F-site recruitment site attenuates the ability of ERK2 to mediate cell death signaling
The specificity of ERK signaling is mainly determined via its different domains and motifs [41]. Among these, the common docking (CD) groove and the F-site recruitment site (FRS) are the two major docking sites on ERK, which enable ERK to specifically interact with the proteins containing “D (docking) site” and “F-site (also called Docking site for ERK, FXF [DEF])”, respectively [42]. The specificity of ERK signaling is also determined by its monomeric vs. dimeric status [43]. To further understand the mechanism by which ERK1/2 mediates cell death, we examined CD groove-defective ERK2-D316/319A, FRS-defective ERK2-Y261N, and dimerization-defective ERK2-H/E-L4A (H176E combination with L333, 336, 341, 344A) mutants [33, 34, 43, 44] in LNCaP-ΔRaf-1:ER cells in which endogenous ERK1/2 expression was stably attenuated by RNA interference. ERK1/2 knockdown abrogates ΔRaf-1:ER-induced growth arrest in these cells [11], thereby reducing the potential interference of endogenous ERK1/2 in this reconstitution experiment (knockdown efficiency shown in Supplemental Fig. S2).
When transduced at a similar efficiency by lentivirus expressing these ERK2 mutants (Fig. 5A), LNCaP-ΔRaf-1:ER/shERK1/2 cells expressed similar levels of the exogenous proteins, which were substantially phosphorylated upon 24 hour 4-HT treatment (Fig. 5B). Consistent with the requirement of the CD groove for ERK1/2 interaction with p90RSK [44], ERK2-D316/319A did not induce p90RSK phosphorylation as effectively as other ERK2 constructs (Fig. 5B). Likewise, consistent with the requirement of FRS for ERK1/2 interaction with ELK1 [42], ERK2-Y261N did not induce ELK1 phosphorylation as effectively as other ERK2 constructs (Fig. 5B). ELK1, a member of the ternary complex transcription factor subfamily, is activated via ERK1/2-mediated Ser383 phosphorylation [45].
Fig 5. FRS-disabled ERK2 cannot mediate cell death signaling.
LNCaP-ΔRaf-1:ER/shERK1/2 cells, infected with the lentiviral pHAGE-ERK2, pHAGE-ERK2-HE/L4A, pHAGE-ERK2-D316/319A, pHAGE-ERK2-Y261N, or pHAGE, were treated with 1 µM 4-HT for 24 hours. Cells were examined for morphological changes (A), expression of the indicated proteins by Western blot analysis (B), viability by trypan blue exclusion analysis (C), and cell cycle analysis (D). Similar infection efficiency was verified by GFP expression (bottom panels in A). p-ELK1 indicates ELK1 phosphorylated at Ser383. GAPDH is the control for equal protein loading. Data (means ± SE) are from a representative experiment performed in triplicate. **, P < 0.01.
Intriguingly, ERK2-Y261N and ERK2-D316/319A induced differential effects on the surrogate makers of growth arrest and cell death upon ΔRaf-1:ER activation. For example, ERK2-Y261N did not significantly induce morphological changes (Fig. 5A), cleavages of PARP and lamin A (Fig. 5B), trypan blue staining (Fig. 5C), and sub-G0/G1 population (Fig. 5D) whereas ERK2-D316/319A induced all these effects. Nevertheless, both ERK mutants induced p16INK4A and p21CIP1, and downregulated E2F1 at similar levels (Fig. 5B). At this 24 h time-point, we did not however observe any significant differences in G0/G1, S, or G2/M phases among these ERK constructs (Fig. 5D). This is because it takes at least more than two days of ΔRaf-1:ER activation before LNCaP cells exhibit clear profiles of G0/G1 cell cycle arrest, as we previously demonstrated [11, 13–15]. 2 day of ΔRaf-1:ER activation killed almost all cells expressing ERK2-D316/219A mutants, hindering a side-by-side comparison with ERK2-Y261N. These data suggest that FRS, but not the CD groove, is required for ERK1/2 to mediate death signaling although both domains may not be necessary for ERK1/2 to mediate growth arrest signaling. Meanwhile, ERK2-H/E-L4A and wild type ERK2 induced almost identical effects (Fig. 5B to 5D), suggesting that dimerization is not required for ERK1/2 to mediate growth inhibitory signaling.
4. Discussion
Although it has long been noted that ERK1/2 can mediate growth arrest and death in different cell types, the underlying mechanisms of ERK1/2 signaling require better understanding. While cell-specific availability of an ERK1/2 effector would account for different physiological outputs of ERK1/2 activation, our data in this report suggest that cellular levels of activated ERK1/2 are also a determinant of cell fate between growth arrest and death. Of note, we previously demonstrated that overexpression of active MEK1/2 or B/C-Raf induced only growth arrest, but not death, in the same cell lines used in this study [11, 14]. Therefore, ERK1/2 are unique among the molecular switches in the Raf/MEK/ERK pathway in the context of our cellular death responses.
ERK1 and ERK2 are highly homologous and have overlapping functions. Nevertheless, studies in mice have shown distinct effects of ERK1 and ERK2 ablation at different stages of development, including stem cell lineage commitment [46, 47], T cell development [48], thymocyte maturation [49], and trophoblast development [50]. The significance of ERK2 over ERK1 for cell proliferation and survival has also been demonstrated in NIH3T3 cells using RNA interference [22]. However, studies also suggest that functional redundancy of ERK1 and ERK2 is evolutionarily conserved [20], and that differentially regulated expression of ERK1 and ERK2 mainly drive their biological differences [21]. It is therefore important to determine ERK1 and ERK2 functions not only by gene depletion approaches but also by ectopic gene expression approaches. Indeed, in a recent study [23], ectopically expressed ERK1 and ERK2 exhibited distinct effects on epithelial-mesenchymal transition. Our previous [11] and current studies using both approaches strongly suggest that ERK1 and ERK2 have overlapping functions in mediating the growth inhibitory signaling of Raf/MEK/ERK.
In many studies, mainly in the context of cell survival and proliferation, kinase activity has been characterized as the key biochemical property that accounts for ERK effects. However, recent studies including ours suggest that ERK1/2 also have kinase-independent effects [11, 24–28]. Of note, we previously demonstrated that overexpression of active site-disabled, but activation loop-intact, ERK2 mutants (i.e., ERK2-K52R and ERK2-D147A) can restore Raf-induced growth arrest responses in ERK1/2-knocked down cells [11], suggesting that a high magnitude kinase activity is not necessary for ERK1/2 to mediate growth arrest signaling. In contrast, our current data suggest that a high magnitude kinase activity is necessary for ERK1/2 to mediate death signaling. This conclusion is also supported by our recent demonstration that overexpression of ERK2-L73P/S151D, an auto-phosphorylation mutant with intrinsically low kinase activity, induced only growth arrest but not cell death [15]. Taken all, a key mechanistic distinction in Raf/MEK/ERK-mediated growth arrest vs. death signaling is determined at ERK1/2 levels.
By what mechanism ERK1/2 mediate death signaling? The CD groove and FRS are two major domains for physical interactions of ERK1/2. These domains are almost independent from each other with respect to ERK catalysis, and occupancy of one domain does not affect the ERK physical interaction via the other domain [51]. Our data suggest that ERK1/2 require the FRS/F-site interaction to mediate death signaling. The F-site signature “Phe-Xaa-Phe-Pro” are relatively less frequent than the D-site and are found in certain ERK1/2 substrates, including the cell-proliferative transcription factors ELK1, c-Fos, Fra1, and c-Myc, as well as the anti-apoptotic BH3-only protein BimEL [52–54] Indeed, many F-site containing proteins promote cell proliferation and survival but, intriguingly, our data suggest that ERK1/2 might also interact with their death effectors via FRS. Given our observation that the molarity and activity threshold of ERK1/2 determines death signaling, it is conceivable that death-specific ERK1/2 effectors might have relatively low affinity to ERK1/2 and, thus, limited access to the kinase under normal condition. However, when the levels of active ERK1/2 increase beyond a threshold, these effectors would gain access to ERK1/2 for activation.
Of note, although our data suggest that the CD groove or the dimerization interface is not necessary for ERK1/2 to mediate death signaling, it should be noted that PEA-15, an adaptor for ERK1/2-mediated senescence [55], interacts with the CD groove of ERK1/2 [56] and that senescence responses are sometimes associated with death in certain cell types [8]. If PEA-15 has ability to directly promote the senescence-associated death, it would suggest an involvement of the CD groove in ERK1/2-mediated death signaling. Therefore, although our data suggest the significance of the FRS/F-site interaction for ERK1/2-mediated death signaling, we cannot exclude the possibility that this mechanism is specific to a subset of certain cell types rather than being universal.
Based upon our observation, it is conceivable that the stoichiometric balance between ERK1/2 and their death effectors should be important for cells to avoid unwanted death in the face of aberrant Raf/MEK/ERK activation. However, ΔRaf-1:ER induces a much greater level of ERK1/2 activation than many biological stimuli, which cells would never achieve in a normal physiological setting. One may thus question the physiological relevance of our findings. Of note, MAPK1/3 (encoding ERK1/2) amplification is not detected in BRAF- or NRAS-mutated melanomas although MAPK3 amplification is detected in BRAF/NRAS wild type melanomas [57], while ectopic ERK1/2 expression induces robust cell death in a subset of human B-RafV600E melanoma lines [58]. Given these, it may be conceivable that the combination of ERK1/2 overexpression with a strong upstream signal is disadvantageous to tumorigenesis. Our report supports this possibility by demonstrating the toxicity produced upon combining ERK1/2 overexpression with ΔRaf-1:ER and by uncovering an ERK motif involved in this death signaling. We speculate that RAS/RAF-mutated tumor cells may need a mechanism to avoid too high basal ERK1/2 levels even if they depend upon ERK1/2 for survival and proliferation and that an optimal ERK1/2 level in tumor cells is determined partly based upon the magnitude of an upstream signal. Would it then be possible to design a strategy to trigger cell death in RAS/RAF-mutated tumor cells by increasing ERK1/2 activity above the death threshold? Intriguingly, the idea to manipulate the intensity and duration of EKR1/2 activity using the inhibitors targeting ERK1/2-specific phosphatases has been proposed recently [59]. Our findings are consistent with this emerging context while adding additional layer of complexity to the regulation of ERK1/2-mediated growth inhibitory signaling.
Supplementary Material
Highlights.
ERK1/2 overexpression switches Raf-induced growth arrest to apoptotic death.
Titrating active ERK levels reverts the death responses to growth arrest responses.
Kinase activity is necessary for ERK1/2 to mediate death signaling.
Defective F-site recruitment site hinders ERK1/2-mediated death signaling.
Different ERK domains differentially regulate growth inhibitory signaling.
Acknowledgments
We thank Drs. Melanie Cobb and Natalie Ahn for ERK2 cDNAs, and Drs. Amy Hudson and Richard Mulligan for pHAGE. This work was supported by the National Cancer Institute (R01CA138441) to J.P.
Abbreviations
- 4-HT
4-hydroxytamoxifen
- CD
common docking
- ERK
extracellular signal-regulated kinase
- FRS
F-site recruitment site
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- MEK
mitogen-activated protein kinase kinase
- MTT
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide
- PARP
poly (ADP-ribose) polymerase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest for this article.
References
- 1.Roskoski R., Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Shaul YD, Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta. 2007;1773:1213–1226. doi: 10.1016/j.bbamcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Chambard JC, Lefloch R, Pouyssegur J, Lenormand P. ERK implication in cell cycle regulation. Biochim Biophys Acta. 2007;1773:1299–1310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Balmanno K, Cook SJ. Tumour cell survival signalling by the ERK1/2 pathway. Cell death and differentiation. 2009;16:368–377. doi: 10.1038/cdd.2008.148. [DOI] [PubMed] [Google Scholar]
- 5.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Montalto G, Cervello M, Nicoletti F, Fagone P, Malaponte G, Mazzarino MC, Candido S, Libra M, Basecke J, Mijatovic S, Maksimovic-Ivanic D, Milella M, Tafuri A, Cocco L, Evangelisti C, Chiarini F, Martelli AM. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget. 2012;3:954–987. doi: 10.18632/oncotarget.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park JI. Growth arrest signaling of the Raf/MEK/ERK pathway in cancer. Front Biol (Beijing) 2014;9:95–103. doi: 10.1007/s11515-014-1299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramaniam S, Unsicker K. ERK and cell death: ERK1/2 in neuronal death. FEBS J. 2010;277:22–29. doi: 10.1111/j.1742-4658.2009.07367.x. [DOI] [PubMed] [Google Scholar]
- 8.Cagnol S, Chambard JC. ERK and cell death: mechanisms of ERK-induced cell death--apoptosis, autophagy and senescence. FEBS J. 2010;277:2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 9.Park JI, Strock CJ, Ball DW, Nelkin BD. The Ras/Raf/MEK/extracellular signal-regulated kinase pathway induces autocrine-paracrine growth inhibition via the leukemia inhibitory factor/JAK/STAT pathway. Mol Cell Biol. 2003;23:543–554. doi: 10.1128/MCB.23.2.543-554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JI, Powers JF, Tischler AS, Strock CJ, Ball DW, Nelkin BD. GDNF-induced leukemia inhibitory factor can mediate differentiation via the MEK/ERK pathway in pheochromocytoma cells derived from nf1-heterozygous knockout mice. Exp Cell Res. 2005;303:79–88. doi: 10.1016/j.yexcr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 11.Hong SK, Yoon S, Moelling C, Arthan D, Park JI. Noncatalytic function of ERK1/2 can promote Raf/MEK/ERK-mediated growth arrest signaling. J Biol Chem. 2009;284:33006–33018. doi: 10.1074/jbc.M109.012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arthan D, Hong SK, Park JI. Leukemia inhibitory factor can mediate Ras/Raf/MEK/ERK-induced growth inhibitory signaling in medullary thyroid cancer cells. Cancer Lett. 2010;297:31–41. doi: 10.1016/j.canlet.2010.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Hong SK, Kim JH, Lin MF, Park JI. The Raf/MEK/extracellular signal-regulated kinase 1/2 pathway can mediate growth inhibitory and differentiation signaling via androgen receptor downregulation in prostate cancer cells. Exp Cell Res. 2011;317:2671–2682. doi: 10.1016/j.yexcr.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu PK, Hong SK, Veeranki S, Karkhanis M, Starenki D, Plaza JA, Park JI. A Mortalin/HSPA9-Mediated Switch in Tumor-Suppressive Signaling of Raf/MEK/Extracellular Signal-Regulated Kinase. Mol Cell Biol. 2013;33:4051–4067. doi: 10.1128/MCB.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu PK, Hong SK, Yoon SH, Park JI. Active ERK2 is sufficient to mediate growth arrest and differentiation signaling. FEBS J. 2015;282:1017–1030. doi: 10.1111/febs.13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong SK, Wu PK, Karkhanis M, Park JI. ERK1/2 can feedback-regulate cellular MEK1/2 levels. Cell Signal. 2015;27:1939–1948. doi: 10.1016/j.cellsig.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karkhanis M, Park JI. Sp1 regulates Raf/MEK/ERK-induced p21 transcription in TP53-mutated cancer cells. Cell Signal. 2015;27:479–486. doi: 10.1016/j.cellsig.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooi WJ, Peeper DS. Oncogene-induced cell senescence--halting on the road to cancer. N Engl J Med. 2006;355:1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 19.Wainstein E, Seger R. The dynamic subcellular localization of ERK: mechanisms of translocation and role in various organelles. Curr Opin Cell Biol. 2016;39:15–20. doi: 10.1016/j.ceb.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Busca R, Christen R, Lovern M, Clifford AM, Yue JX, Goss GG, Pouyssegur J, Lenormand P. ERK1 and ERK2 present functional redundancy in tetrapods despite higher evolution rate of ERK1. BMC Evol Biol. 2015;15:179. doi: 10.1186/s12862-015-0450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefloch R, Pouyssegur J, Lenormand P. Single and combined silencing of ERK1 and ERK2 reveals their positive contribution to growth signaling depending on their expression levels. Mol Cell Biol. 2008;28:511–527. doi: 10.1128/MCB.00800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vantaggiato C, Formentini I, Bondanza A, Bonini C, Naldini L, Brambilla R. ERK1 and ERK2 mitogen-activated protein kinases affect Ras-dependent cell signaling differentially. J Biol. 2006;5:14. doi: 10.1186/jbiol38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010;38:114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapiro PS, Whalen AM, Tolwinski NS, Wilsbacher J, Froelich-Ammon SJ, Garcia M, Osheroff N, Ahn NG. Extracellular signal-regulated kinase activates topoisomerase IIalpha through a mechanism independent of phosphorylation. Mol Cell Biol. 1999;19:3551–3560. doi: 10.1128/mcb.19.5.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, Long S, He X, Wade H, Blackshaw S, Qian J, Zhu H. Profiling the Human Protein-DNA Interactome Reveals ERK2 as a Transcriptional Repressor of Interferon Signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucharska A, Rushworth LK, Staples C, Morrice NA, Keyse SM. Regulation of the inducible nuclear dual-specificity phosphatase DUSP5 by ERK MAPK. Cell Signal. 2009;21:1794–1805. doi: 10.1016/j.cellsig.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez J, Calvo F, Gonzalez JM, Casar B, Andres V, Crespo P. ERK1/2 MAP kinases promote cell cycle entry by rapid, kinase-independent disruption of retinoblastoma-lamin A complexes. J Cell Biol. 2010;191:967–979. doi: 10.1083/jcb.201004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez J, Crespo P. Working without kinase activity: phosphotransfer-independent functions of extracellular signal-regulated kinases. Sci Signal. 2011;4:re3. doi: 10.1126/scisignal.2002324. [DOI] [PubMed] [Google Scholar]
- 29.Samuels ML, Weber MJ, Bishop JM, McMahon M. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human raf-1 protein kinase. Mol Cell Biol. 1993;13:6241–6252. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starenki D, Hong SK, Lloyd RV, Park JI. Mortalin (GRP75/HSPA9) upregulation promotes survival and proliferation of medullary thyroid carcinoma cells. Oncogene. 2015;34:4624–4634. doi: 10.1038/onc.2014.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc Natl Acad Sci U S A. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 33.Khokhlatchev AV, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb MH. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 34.Yazicioglu MN, Goad DL, Ranganathan A, Whitehurst AW, Goldsmith EJ, Cobb MH. Mutations in ERK2 binding sites affect nuclear entry. J Biol Chem. 2007;282:28759–28767. doi: 10.1074/jbc.M703460200. [DOI] [PubMed] [Google Scholar]
- 35.Kim JH, Hong SK, Wu PK, Richards AL, Jackson WT, Park JI. Raf/MEK/ERK can regulate cellular levels of LC3B and SQSTM1/p62 at expression levels. Exp Cell Res. 2014;327:340–352. doi: 10.1016/j.yexcr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosen A, Casciola-Rosen L. Macromolecular substrates for the ICE-like proteases during apoptosis. J Cell Biochem. 1997;64:50–54. doi: 10.1002/(sici)1097-4644(199701)64:1<50::aid-jcb8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 37.Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol. 2003;5:647–654. doi: 10.1038/ncb1005. [DOI] [PubMed] [Google Scholar]
- 38.Wu PK, Park JI. MEK1/2 Inhibitors: Molecular Activity and Resistance Mechanisms. Semin Oncol. 2015;42:849–862. doi: 10.1053/j.seminoncol.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robbins DJ, Zhen E, Owaki H, Vanderbilt CA, Ebert D, Geppert TD, Cobb MH. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J Biol Chem. 1993;268:5097–5106. [PubMed] [Google Scholar]
- 40.Lazar DF, Wiese RJ, Brady MJ, Mastick CC, Waters SB, Yamauchi K, Pessin JE, Cuatrecasas P, Saltiel AR. Mitogen-activated protein kinase kinase inhibition does not block the stimulation of glucose utilization by insulin. J Biol Chem. 1995;270:20801–20807. doi: 10.1074/jbc.270.35.20801. [DOI] [PubMed] [Google Scholar]
- 41.Chuderland D, Seger R. Protein-protein interactions in the regulation of the extracellular signal-regulated kinase. Mol Biotechnol. 2005;29:57–74. doi: 10.1385/MB:29:1:57. [DOI] [PubMed] [Google Scholar]
- 42.Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 43.Adachi M, Fukuda M, Nishida E. Two co-existing mechanisms for nuclear import of MAP kinase: passive diffusion of a monomer and active transport of a dimer. Embo J. 1999;18:5347–5358. doi: 10.1093/emboj/18.19.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burkhard KA, Chen F, Shapiro P. Quantitative Analysis of ERK2 Interactions with Substrate Proteins: ROLES FOR KINASE DOCKING DOMAINS AND ACTIVITY IN DETERMINING BINDING AFFINITY. J Biol Chem. 2011;286:2477–2485. doi: 10.1074/jbc.M110.177899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kortenjann M, Thomae O, Shaw PE. Inhibition of v-raf-dependent c-fos expression and transformation by a kinase-defective mutant of the mitogen-activated protein kinase Erk2. Mol Cell Biol. 1994;14:4815–4824. doi: 10.1128/mcb.14.7.4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamilton WB, Kaji K, Kunath T. ERK2 Suppresses Self-Renewal Capacity of Embryonic Stem Cells, but Is Not Required for Multi-Lineage Commitment. PLoS One. 2013;8:e60907. doi: 10.1371/journal.pone.0060907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saulnier N, Guihard S, Holy X, Decembre E, Jurdic P, Clay D, Feuillet V, Pages G, Pouyssegur J, Porteu F, Gaudry M. ERK1 Regulates the Hematopoietic Stem Cell Niches. PLoS One. 2012;7:e30788. doi: 10.1371/journal.pone.0030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer AM, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 2005;23:431–443. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 49.Pages G, Guerin S, Grall D, Bonino F, Smith A, Anjuere F, Auberger P, Pouyssegur J. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 1999;286:1374–1377. doi: 10.1126/science.286.5443.1374. [DOI] [PubMed] [Google Scholar]
- 50.Nadeau V, Guillemette S, Belanger LF, Jacob O, Roy S, Charron J. Map2k1 and Map2k2 genes contribute to the normal development of syncytiotrophoblasts during placentation. Development. 2009;136:1363–1374. doi: 10.1242/dev.031872. [DOI] [PubMed] [Google Scholar]
- 51.Lee S, Warthaka M, Yan C, Kaoud TS, Ren P, Dalby KN. Examining docking interactions on ERK2 with modular peptide substrates. Biochemistry. 2011;50:9500–9510. doi: 10.1021/bi201103b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy LO, MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol Cell Biol. 2004;24:144–153. doi: 10.1128/MCB.24.1.144-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 54.Ley R, Hadfield K, Howes E, Cook SJ. Identification of a DEF-type docking domain for extracellular signal-regulated kinases 1/2 that directs phosphorylation and turnover of the BH3-only protein BimEL. J Biol Chem. 2005;280:17657–17663. doi: 10.1074/jbc.M412342200. [DOI] [PubMed] [Google Scholar]
- 55.Callejas-Valera JL, Guinea-Viniegra J, Ramirez-Castillejo C, Recio JA, Galan-Moya E, Martinez N, Rojas JM, Ramon y Cajal S, Sanchez-Prieto R. E1a gene expression blocks the ERK1/2 signaling pathway by promoting nuclear localization and MKP up-regulation: implication in v-H-Ras-induced senescence. J Biol Chem. 2008;283:13450–13458. doi: 10.1074/jbc.M709230200. [DOI] [PubMed] [Google Scholar]
- 56.Callaway K, Abramczyk O, Martin L, Dalby KN. The anti-apoptotic protein PEA-15 is a tight binding inhibitor of ERK1 and ERK2, which blocks docking interactions at the D-recruitment site. Biochemistry. 2007;46:9187–9198. doi: 10.1021/bi700206u. [DOI] [PubMed] [Google Scholar]
- 57.Orouji E, Orouji A, Gaiser T, Larribere L, Gebhardt C, Utikal J. MAP kinase pathway gene copy alterations in NRAS/BRAF wild-type advanced melanoma. Int J Cancer. 2016;138:2257–2262. doi: 10.1002/ijc.29970. [DOI] [PubMed] [Google Scholar]
- 58.Goetz EM, Ghandi M, Treacy DJ, Wagle N, Garraway LA. ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors. Cancer Res. 2014;74:7079–7089. doi: 10.1158/0008-5472.CAN-14-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov. 2007;6:391–403. doi: 10.1038/nrd2289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.