Abstract
Genome sequencing and the application of omic techniques are driving many important advances in the field of microbial natural products research. Despite these gains, there remain aspects of the natural product discovery pipeline where our knowledge remains poor. These include the extent to which biosynthetic gene clusters are transcriptionally active in native microbes, the temporal dynamics of transcription, translation, and natural product assembly, as well as the relationships between small molecule production and detection. Here we touch on a number of these concepts in the context of continuing efforts to unlock the natural product potential revealed in genome sequence data and discuss nomenclatural issues that warrant consideration as the field moves forward.
Introduction
Natural products (NPs) and their derivatives comprise about half of the clinically approved medicines including many of our most important antibiotics[1–3]. Despite the considerable gains made from studying this resource, industry’s investment in NP research waned in recent decades. This paradigm shift can be attributed to a number of issues including the continued rediscovery of known compounds and the alternative use of combinatorial libraries as a platform for discovery[4]. Enthusiasm for NPs research was renewed however following the revelation that only a small fraction of the biosynthetic potential observed in microbial genome sequences could be linked to the small molecules whose biosynthesis they encode[5]. This observation, coupled with the development of new omic-based technologies, has re-invigorated the field and spawned the concept of genome mining as an alternative method for NP discovery[6].
Natural product genome mining takes a bioinformatics first approach to small molecule discovery. It provides opportunities to specifically target biosynthetic gene clusters (BGCs) that can be predicted to encode interesting new compounds or new derivatives within structural classes of interest. Genome mining has benefited from a better understanding of the biosynthetic logic of natural products[7,8] and enhanced computational approaches that automate the process by which BGCs are detected and characterized[9,10]. These advances led to the development of the widely used web tool antiSMASH[5], while others such as PRISM (Prediction Informatics for Secondary Metabolome)[11] and IMG/ABC[12] represent more recent additions. Access to these tools has brought NP analyses into the mainstream, making it possible for anyone with a genome sequence to make an informed bioinformatic assessment of specialized metabolism in their organism of choice. Other recent tools such as plantiSMASH[13] and fungiSMASH[14] improve the analysis of BGCs in plants and fungi, respectively. The MIBiG repository of experimentally characterized BGCs (http://mibig.secondarymetabolites.org/index.html) represents another important milestone and provides a straightforward platform to perform comparative BGC analysis and metabolite prediction.
While there remains considerable excitement about the discovery opportunities afforded by genome mining, our ability to identify BGCs from sequence data continues to far exceed our ability to identify the NPs they encode. This bottleneck results from the fact that only certain types of BGCs, such as NRPSs and modular, type I PKSs[15], are generally amenable to bioinformatic linkage with their products. Many others require considerably more investment, usually in the form of a genetic knock-out or heterologous expression. Both of these experimental approaches have unique challenges that vary with the organism being studied. Nonetheless, progress has been made in the automation of heterologous expression systems[16] and in retro-biosynthetic approaches to link orphan BGCs to known compounds[17] thus suggesting that this bottleneck may ultimately be overcome through advances in synthetic and computational biology. Meanwhile, the relatively few BGCs that have been experimentally characterized (1,393 BGCs in MIBiG as of May 2017) relative to the considerably larger number of microbial NPs discovered over the last 50-plus years (52,395 microbial NPs[18]) raises the generally understated possibility that much of the apparent genetic potential observed in bacterial genome sequences has already been realized.
The many innovative approaches that have been applied to NP genome mining[9,19,20], including the activation of silent BGCs[21–24], have been extensively reviewed in recent years. Here we highlight a number of gaps that remain in our basic understanding of the relationships between BGCs and the NPs whose biosynthesis they encode. We also discuss a number of nomenclatural issues relevant to genome mining, which at present lacks a unified lexicon.
The lexicon of genome mining
The large number of BGCs observed in genome sequence data has generated considerable interest in developing new techniques to find their products. This has also led to the use of a variety of terms to describe those BGCs that cannot readily be linked to their products. Over the last decade, these unassigned BGCs have been called “silent”, “cryptic”, or “orphan”, all of which have been applied with the same intended meaning. While the term orphan was proposed 10 years ago by Harold Gross[25], the term cryptic has also come into common usage[24]. An NCBI PubMed search of the terms orphan and cryptic biosynthetic gene clusters suggests that the former is only slightly favored. Given the appropriate context, (i.e., a BGC) the implications of orphan or cryptic seem readily apparent. While there is clear precedent for the use of both terms to describe BGCs that have yet to be linked to the small molecules they encode, we have followed the original suggestion by Gross for the use of orphan[25]. Complicating the matter is the alternative use of the term orphan to describe compounds whose cognate BGC has not been identified[26]. This later usage appears fitting given the more colloquial interpretation of orphan as a lack of parents or affiliation. Thus, an orphan compound can be considered one that lacks affiliation with its parent BGC (Figure 1).
Figure 1. The lexicon of omics-based natural product discovery.
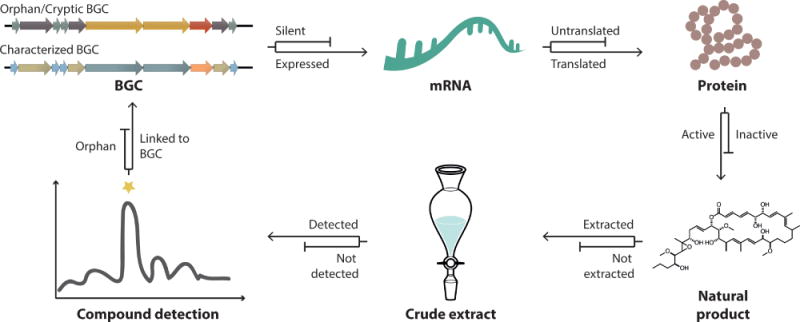
Biosynthetic gene clusters (BGCs) can be orphan or cryptic, i.e., they have not been linked to the small molecules they encode. BGC characterization can be accomplished based on bioinformatic prediction or experimental validation of the cognate products. BGCs may be transcribed into mRNA (expressed) at levels sufficient to yield detectable compound. If transcription falls below these levels, the BGC is considered to be silent. Assuming the mRNA is translated into functional protein, NP assembly will depend on the availability of precursors and co-factors required for protein activity. Once they are produced, many NPs are likely missed because they are either not extracted or not detected with the methods employed. The vast majority of microbial NPs that have been discovered over the last 50-plus years have yet to be linked to their respective BGC and thus the molecules remain orphan.
As mentioned, “silent” has also been commonly used to describe orphan BGCs. However, the term silent also carries the connotation of transcriptional inactivity. Surprisingly, there’s very little data to support the frequently cited suggestion that orphan BGCs are transcriptionally silent. Yet transcriptional silencing is only one of many possible reasons why so many BGC remain orphan. Alternative explanations can be found at the levels of translation, functional protein assembly, and small molecule extraction and detection (Figure 1). Furthermore, the products of many orphan BGCs are readily detectable yet there is simply no practical method to link them to the appropriate gene cluster. This failure to connect the two data points leaves both compound and BGC orphaned. BGCs have also been termed silent when their encoded NPs are known but not detected in the native organism. In this case, the implied silence could be due to a failure at any step in the gene to small molecule pipeline. Thus, as with the term orphan, the use of silent is most meaningful when appropriately qualified. E.g., it is clear that a transcriptionally silent BGC is one that is expressed below the levels where the cognate small molecules can be detected. It is much less clear how to interpret a BGC labelled as silent in the absence of transcriptome, proteome, or metabolome data. Here we reserve the term silent to describe BGCs for which there is evidence of ineffectual transcriptional activity.
Transcriptomics
To what extent are orphan BGCs expressed? Surprisingly, this remains largely unknown despite extensive efforts being devoted to their activation[24,27]. Global expression analyses have rarely been applied to distinguish between silent and expressed BGCs in a given organism. In cases where this has been addressed, methods such as PCR probing of cDNA have been used[28]. Microarrays have also been used to assess BGC expression. In the case of S. coelicolor, this approach revealed that eight of 22 BGCs were expressed at higher levels following the transition from exponential to stationary phase[29,30]. Microarrays also revealed that 12 of 18 BGCs were expressed in wild-type Myxococcus xanthus, thus suggesting that many orphan BGCs may not be transcriptionally silent[31]. More recently, next-generation sequencing was employed in the global transcriptome analysis of S. coelicolor[32]. This study provided important new insight into the relationships between transcription and translation, something that remains poorly understood in the context of NP biosynthesis. Despite these advances, it can be argued that we still have a generally poor understanding of how many BGCs are transcriptionally silent under any given set of cultivation conditions.
More commonly, expression analyses have been used in association with searching for the products of BGCs following either the genetic manipulation of the wild-type strain or the introduction of the BGC into a heterologous host. Recent efforts using the former technique include micro-array based comparative transcription profiling, which led to the discovery of a series of polyketides encoded by a BGC that was silent in the wild-type strain but expressed in a ΔbldM mutant[33]. Expression analyses have also been used to demonstrate that silent BGCs can be activated in a heterologous host following genetic reconstruction[34]. While synthetic biology holds great promise for NP discovery[35], it’s not always clear if an orphan BGC was transcriptionally silent in the native organism prior to genetic manipulation. This is unfortunate as evidence of compound production in the wild-type helps establish the relationships between orphan and transcriptionally silent BGCs and can be used to guide future BGC activation efforts.
While transcriptome analyses can provide important distinctions between silent and expressed BGCs, the levels of gene expression that correlate with NP detection also remain largely unknown. These levels will undoubtedly vary with each BGC depending upon numerous factors including the structure of the NP and the detection methods employed. Furthermore, translation efficiencies can vary and thus expression alone is unlikely to be an adequate predictor of protein or NP production. For example, a recent study in S. coelicolor revealed that the transcription of several BGCs increased throughout growth yet their translation efficiency decreased at certain times[32]. It is also interesting to consider how the abundance of various tRNAs can affect translation efficiencies[36] and how a mutation in bldA, which is predicted to encode a misfolded Leu-tRNA molecule, can have a profound effect on BGC expression[37].
Proteomics
As with transcription, we know little about genome level BGC translation. Early reports describe the detection of proteins associated with six orphan BGCs in M. xanthus[38]. More recently, the Kelleher group has pioneered the application of proteomics in NP discovery by capitalizing on the properties and size of non-ribosomal peptide synthetases and polyketide synthases (often >200 kDa) and their unique marker ions derived from phosphopantetheinyl cofactors[39,40]. The PrISM (Proteomic Investigation of Secondary Metabolism) platform (not to be confused with the more recent PRISM genome mining tool[11]) was developed to exploit this concept and represents a ‘protein-first’ strategy that is complementary to bioassay-guided and genomic approaches[41]. The more recent “Genome-Enabled” PrISM (GE-PrISM) incorporates draft genome sequences in the analysis and expands on the types of biosynthetic proteins that can be detected in a process termed “proteogenomics”[42]. Despite the successful applications of proteome-based genome mining, DNA-based approaches remain more widely accessible and employed.
Translation also requires another level of scrutiny as some key biosynthetic enzymes require post-translational modification for activity. PKSs and NRPSs are prime examples, in that they are modified by phosphopantetheinyl transferases (PPTases), which post-translationally install a 4′-phosphopantetheine arm on the respective acyl or peptidyl carrier proteins[43]. These prosthetic groups are derived from coenzyme A and are required to convert inactive apo-synthases to active holo-synthases. This concept was recently exploited by overexpressing PPTases in actinomycetes, which led to an increased production of metabolites in 70% of the strains studied[44]. To the best of our knowledge, this is the first example in which the regulation of protein activity was used to elicit NP production.
Metabolomics
Improved analytical techniques have made it possible to more effectively assess the complex NP mixtures present in most crude extracts. While NMR has been used for metabolomic analyses, mass spectrometry remains the instrument of choice[45]. Driving these advances are approaches such as Global Natural Product Social Molecular Networking (GNPS), which allows for the visualization of complex metabolomic datasets based on MS/MS fragmentation patterns[46]. Other new tools such as DEREPLICATOR allow users to rapidly identify novel peptidic NPs in their datasets[47]. While metabolomic analyses have historically been performed independently of any knowledge of the associated BGCs, it has become increasingly possible to integrate these datasets to generate bioinformatic links between NPs and their associated BGCs. Comparing BGC distributions with metabolomic profiles has been termed “pattern-based genome mining” and used to compare large numbers of closely related bacteria[48,49]. Similar approaches were used to identify the rimosamide, detoxin, and tambromycin NPs and their associated BGCs[26,50], with the later approach termed metabologenomics. Genomics and metabolomics have also been coupled to discover new NPs from cyanobacteria[51]. Other metabolomic approaches include peptidogenomics[52] and glycogenomics[53], where MS/MS fragmentation patterns are used to establish bioinformatic links between NPs and their associated BGCs based on adenylation domain amino acid specificity or glycosyltransferase substrate specificity, respectively. These informal links can be valuable in terms of prioritizing NPs and BGCs for study, yet don’t eliminate the bottleneck that remains with experimental validation.
Considerable advances have been made in the activation of silent BGCs[22,24,54,55] using techniques such as co-cultivation[56] or the addition of small molecule inducers[57,58]. While these methods have proven useful, it’s also possible that many BGCs are active yet their small molecule products are simply missed due to the extraction methods or analytical techniques employed (Figure 1). For example, recent metabolomics analyses have shown that extraction solvents had a major impact on the metabolites detected[59,60]. As we learn more about the relationships between orphan BGCs and their products, it is likely that much of the apparent biosynthetic potential observed in microbial genomes will ultimately be linked to compounds that are in fact produced but simply missed or ignored because they do not possess the properties that allow them to be detected or make them attractive targets for discovery.
Conclusions and Future perspectives
The extant number of NPs that await discovery is unknown and will undoubtedly remain so. However, a large reservoir of unexplored chemical space can be predicted[18], thus supporting the concept that NP research will continue to yield important new discoveries. One clear concept to emerge from the post-genomic era is that a majority of BGCs detected in genome sequences are orphan, i.e., they have yet to be linked to the NPs whose biosynthesis they encode. We also know that the number of microbial NPs discovered to date far exceeds the number of BGCs that have been experimentally characterized. Thus, the jury remains out on how many orphan BGCs will yield structurally unique NPs vs. known compounds or new derivatives thereof. This knowledge gap will be filled as the bottleneck in BGC characterization is resolved and remains fundamentally important for future NP drug discovery research. A seldom discussed unknown is the rate at which Nature creates new chemical diversity. While this rate may not meet our demanding pipelines for new drugs, a major goal of synthetic biology is to speed up these processes through genetic engineering. While there have been successes in this regard[61], we undoubtedly still have a great deal to learn from the microbes themselves, which are the true masters of NP biosynthesis.
A surprising unknown is the distinction between transcriptionally silent and expressed BGCs in wild-type strains. Without global transcriptome data, it is difficult to distinguish between silent BGCs and those that are expressed yet fail to be connected to their small molecule products. Thus, it’s possible that some discovery efforts may be better focused at the post-transcriptional level. Additional unknowns relate to the temporal dynamics of NP biosynthesis. For example, it is unclear what level of gene expression correlates with protein or NP detection. Also the time frame between gene expression and NP assembly is poorly understood, as are the half-lives of active biosynthetic enzymes. Furthermore, expression levels may not mirror protein assembly due to differences in translational efficiencies or post-translational regulation.
Despite these significant unknowns, there are numerous examples where orphan BGCs have yielded new compounds[34,40,62–64], thus supporting the concept that genome mining and associated omic approaches hold considerable promise for NP discovery. The continued development and application of these technologies will help fill the knowledge gaps discussed above and allow for the better exploitation of NP resources. As this field moves forward, having a unified lexicon to describe BGCs and the NPs whose biosynthesis they encode provides the best mechanism to effectively communicate among those working in this field and with the scientific community at large.
Highlights.
A lexicon for biosynthetic gene clusters and the small molecules they encode
Omic approaches advance natural product discovery
Knowledge gaps in the discovery process
The temporal dynamics of natural product biosynthesis
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM085770. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li JW-H, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–5. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 2.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 3.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 4.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, et al. antiSMASH 3.0–a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zerikly M, Challis GL. Strategies for the discovery of new natural products by genome mining. Chembiochem. 2009;10:625–33. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
- 7.Walsh CT. Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat Prod Rep. 2016;33:127–135. doi: 10.1039/c5np00035a. [DOI] [PubMed] [Google Scholar]
- 8.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chemie - Int Ed. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 9.Ziemert N, Alanjary M, Weber T. The evolution of genome mining in microbes - a review. Nat Prod Rep. 2016;33:988–1005. doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]
- 10••.Medema MH, Fischbach MA. Computational approaches to natural product discovery. Nat Chem Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. This perspective paper presents the computational strategies available to capitalize on genomic information and predict the products encoded therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, Webster ALH, Wyatt MA, Magarvey NA. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM) Nucleic Acids Res. 2015;43:9645–9662. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wasmuth EV, Lima CD. IMG-ABC: new features for bacterial secondary metabolism analysis and targeted biosynthetic gene cluster discovery in thousands of microbial genomes [Internet] Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kautsar SA, Suarez Duran HG, Blin K, Osbourn A, Medema MH. plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters [Internet] Nucleic Acids Res. 2017;320:543–547. doi: 10.1093/nar/gkx305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de los Santos ELC, Kim HU, Nave M, et al. antiSMASH 4.0–improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 16.Luo Y, Enghiad B, Zhao H. New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat Prod Rep. 2016;33:174–182. doi: 10.1039/c5np00085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dejong CA, Chen GM, Li H, Johnston CW, Edwards MR, Rees PN, Skinnider MA, Webster ALH, Magarvey NA. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat Chem Biol. 2016;12:1007–1014. doi: 10.1038/nchembio.2188. [DOI] [PubMed] [Google Scholar]
- 18.Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG. Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1614680114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler MS, Blaskovich MA, Owen JG, Cooper MA. Old dogs and new tricks in antimicrobial discovery. Curr Opin Microbiol. 2016;33:25–34. doi: 10.1016/j.mib.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Weber T. In silico tools for the analysis of antibiotic biosynthetic pathways. Int J Med Microbiol. 2014;304:230–5. doi: 10.1016/j.ijmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Scherlach K, Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms [Internet] Org Biomol Chem. 2009;7:1753. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 22•.Zarins-Tutt JS, Barberi TT, Gao H, Mearns-Spragg A, Zhang L, Newman DJ, Goss RJM. Prospecting for new bacterial metabolites: a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat Prod Rep. 2016;33:54–72. doi: 10.1039/c5np00111k. This review provides an overview of the main approaches available to unlock the biosynthetic potential of bacteria. It covers bioinformatics (genome mining), the alteration of culturing conditions (eg. co-culturing, chemical elicitors), genetic engineering, and heterologous expression. [DOI] [PubMed] [Google Scholar]
- 23.Yuan H, Zhao W, Zhong Y, Wang J, Qin Z, Ding X, Zhao GP. Two genes, rif15 and rif16, of the rifamycin biosynthetic gene cluster in Amycolatopsis mediterranei likely encode a transketolase and a P450 monooxygenase, respectively, both essential for the conversion of rifamycin SV into B. Acta Biochim Biophys Sin (Shanghai) 2011;43:948–956. doi: 10.1093/abbs/gmr091. [DOI] [PubMed] [Google Scholar]
- 24.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 25.Gross H. Strategies to unravel the function of orphan biosynthesis pathways: Recent examples and future prospects. Appl Microbiol Biotechnol. 2007;75:267–277. doi: 10.1007/s00253-007-0900-5. [DOI] [PubMed] [Google Scholar]
- 26.McClure RA, Goering AW, Ju K-S. Elucidating the Rimosamide-Detoxin Natural Product Families and Their Biosynthesis Using Metabolite/Gene Cluster Correlations. ACS Chem Biol. 2016 doi: 10.1021/acschembio.6b00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdelmohsen UR, Grkovic T, Balasubramanian S, Kamel MS, Quinn RJ, Hentschel U. Elicitation of secondary metabolism in actinomycetes. Biotechnol Adv. 2015;33:798–811. doi: 10.1016/j.biotechadv.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Qu X, Lei C, Liu W. Transcriptome mining of active biosynthetic pathways and their associated products in Streptomyces flaveolus. Angew Chemie - Int Ed. 2011;50:9651–9654. doi: 10.1002/anie.201103085. [DOI] [PubMed] [Google Scholar]
- 29.Karoonuthaisiri N, Weaver D, Huang J, Cohen SN, Kao CM. Regional organization of gene expression in Streptomyces coelicolor. Gene. 2005;353:53–66. doi: 10.1016/j.gene.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 30••.Huang J, Lih C-J, Pan K-H, Cohen SN. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 2001;15:3183–3192. doi: 10.1101/gad.943401. This study describes a global analysis of the transcriptome and translatome of the model antibiotic-producer Streptomyces coelicolor. It applies differential RNA-sequencing and ribosome profiling to evaluate transcription and translation over different growth phases. Results indicate there is a decoupling between transcription and translation efficiencies for biosynthetic gene clusters in this organism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bode HB, Ring MW, Schwär G, Altmeyer MO, Kegler C, Jose IR, Singer M, Müller R. Identification of additional players in the alternative biosynthesis pathway to Isovaleryl-CoA in the myxobacterium Myxococcus xanthus. ChemBioChem. 2009;10:128–140. doi: 10.1002/cbic.200800219. [DOI] [PubMed] [Google Scholar]
- 32.Jeong Y, Kim J-N, Kim MW, Bucca G, Cho S, Yoon YJ, Kim B-G, Roe J-H, Kim SC, Smith CP, et al. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2) Nat Commun. 2016;7:11605. doi: 10.1038/ncomms11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thanapipatsiri A, Gomez-Escribano JP, Song L, Bibb MJ, Al-Bassam M, Chandra G, Thamchaipenet A, Challis GL, Bibb MJ. Discovery of unusual biaryl polyketides by activation of a silent Streptomyces venezuelae biosynthetic gene cluster. ChemBioChem. 2016;17:2189–2198. doi: 10.1002/cbic.201600396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, Huang H, Liang J, Wang M, Lu L, Shao Z, Cobb RE, Zhao H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4:1–8. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim E, Moore BS, Yoon YJ. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat Chem Biol. 2015;11:649–659. doi: 10.1038/nchembio.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gingold H, Pilpel Y. Determinants of translation efficiency and accuracy. Mol Syst Biol. 2011;7 doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Kalan L, Gessner A, Thaker MN, Waglechner N, Zhu X, Szawiola A, Bechthold A, Wright GD, Zechel DL. A cryptic polyene biosynthetic gene cluster in Streptomyces calvus is expressed upon complementation with a functional bldA gene. Chem Biol. 2013;20:1214–1224. doi: 10.1016/j.chembiol.2013.09.006. The authors present evidence that complementation with a functional Leu-tRNA led to the expression of a cryptic biosynthetic gene cluster in Streptomyces calvus. This finding brings awareness to post-transcriptional events in the expression of biosynthetic gene clusters and the biosynthesis of natural products. [DOI] [PubMed] [Google Scholar]
- 38.Schley C, Altmeyer MO, Swart R, Müller R, Huber CG. Proteome analysis of Myxococcus xanthus by off-line two-dimensional chromatographic separation using monolithic poly-(styrene-divinylbenzene) columns combined with ion-trap tandem mass spectrometry. J Proteome Res. 2006;5:2760–2768. doi: 10.1021/pr0602489. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Unger M, Ntai I, McClure RA, Albright JC, Thomson RJ, Kelleher NL. Gobichelin A and B: mixed-ligand siderophores discovered using proteomics. Medchemcomm. 2013;4:233–238. doi: 10.1039/C2MD20232H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, McClure RA, Zheng Y, Thomson RJ, Kelleher NL. Proteomics guided discovery of flavopeptins: Anti-proliferative aldehydes synthesized by a reductase domain-containing non-ribosomal peptide synthetase. J Am Chem Soc. 2013;135:10449–10456. doi: 10.1021/ja4031193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bumpus SB, Evans BS, Thomas PM, Ntai I, Kelleher NL. A proteomics approach to discovering natural products and their biosynthetic pathways. Nat Biotechnol. 2009;27:951–6. doi: 10.1038/nbt.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albright JC, Goering AW, Doroghazi JR, Metcalf WW, Kelleher NL. Strain-specific proteogenomics accelerates the discovery of natural products via their biosynthetic pathways. J Ind Microbiol Biotechnol. 2014;41:451–459. doi: 10.1007/s10295-013-1373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beld J, Sonnenschein EC, Vickery CR, Noel JP, Burkart MD. The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat Prod Rep. 2014;31:61–108. doi: 10.1039/c3np70054b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Zhang B, Tian W, Wang S, Yan X, Jia X, Pierens GK, Chen W, Ma H, Deng Z, Qu X. Activation of natural products biosynthetic pathways via a protein modification level regulation. ACS Chem Biol. 2017 doi: 10.1021/acschembio.7b00225. This study shows how protein activity can be used to enhance the biosynthesis of new metabolites. Increasing protein modification by overexpression of Pptase genes led to the increased production of metabolites in 70% of the strains used. [DOI] [PubMed] [Google Scholar]
- 45.Pérez-Victoria I, Martín J, Reyes F. Combined LC/UV/MS and NMR strategies for the dereplication of marine natural products. Planta Med. 2016;82:857–871. doi: 10.1055/s-0042-101763. [DOI] [PubMed] [Google Scholar]
- 46.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohimani H, Gurevich A, Mikheenko A, Garg N, Nothias L-F, Ninomiya A, Takada K, Dorrestein PC, Pevzner PA. Dereplication of peptidic natural products through database search of mass spectra. Nat Chem Biol. 2016;13:30–37. doi: 10.1038/nchembio.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duncan KR, Crüsemann M, Lechner A, Sarkar A, Li J, Ziemert N, Wang M, Bandeira N, Moore BS, Dorrestein PC, et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem Biol. 2015;22:460–471. doi: 10.1016/j.chembiol.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maansson M, Vynne NG, Klitgaard A, Nybo JL, Melchiorsen J, Nguyen DD, Sanchez LM, Ziemert N, Dorrestein PC, Andersen MR, et al. An integrated metabolomic and genomic mining workflow to uncover the biosynthetic potential of bacteria. mSystems. 2016;1:e00028–15. doi: 10.1128/mSystems.00028-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Goering AW. Metabologenomics: correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent Sci. 2016;2:99–108. doi: 10.1021/acscentsci.5b00331. This study reports the isolation and characterization of tambromycin, a new chlorinated nonribosomal peptide. It uses metabolomics and genomics to identify the biosynthetic gene cluster and as a guide for structure elucidation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleigrewe K, Almaliti J, Tian IY, Kinnel RB, Korobeynikov A, Monroe EA, Duggan BM, Di Marzo V, Sherman DH, Dorrestein PC, et al. Combining mass spectrometric metabolic profiling with genomic analysis: a powerful approach for discovering natural products from cyanobacteria. J Nat Prod. 2015;78:1671–1682. doi: 10.1021/acs.jnatprod.5b00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kersten RD, Yang Y-L, Xu Y, Cimermancic P, Nam S-J, Fenical W. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kersten RD, Ziemert N, Gonzalez DJ, Duggan BM, Nizet V, Dorrestein PC, Moore BS. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc Natl Acad Sci. 2013;110:E4407–E4416. doi: 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Netzker T, Fischer J, Weber J, Mattern DJ, König CC, Valiante V, Schroeckh V. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol. 2015;6:1–13. doi: 10.3389/fmicb.2015.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Traxler MF, Kolter R. Natural products in soil microbe interactions and evolution. Nat Prod Rep. 2015;32:956–70. doi: 10.1039/c5np00013k. [DOI] [PubMed] [Google Scholar]
- 56.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. 2013;4:1–12. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okada BK, Seyedsayamdost MR, Aedo S, Tomasz A, Ahmed S, Craney A, Pimentel-Elardo S, Amano S, Morota T, Kano Y, et al. Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol Rev. 2016;60:83–91. doi: 10.1093/femsre/fuw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoon V, Nodwell JR. Activating secondary metabolism with stress and chemicals. J Ind Microbiol Biotechnol. 2014;41:415–424. doi: 10.1007/s10295-013-1387-y. [DOI] [PubMed] [Google Scholar]
- 59.Floros DJ, Jensen PR, Dorrestein PC, Koyama N. A metabolomics guided exploration of marine natural product chemical space. Metabolomics. 2016;12:1–11. doi: 10.1007/s11306-016-1087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crüsemann M, O’Neill EC, Larson CB, Melnik AV, Floros DJ, da Silva RR, Jensen PR, Dorrestein PC, Moore BS. Prioritizing natural product diversity in a collection of 146 bacterial strains based on growth and extraction protocols. J Nat Prod. 2017;80:588–597. doi: 10.1021/acs.jnatprod.6b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Medema MH, Breitling R, Bovenberg R, Takano E. Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat Rev Microbiol. 2011;9:131–137. doi: 10.1038/nrmicro2478. [DOI] [PubMed] [Google Scholar]
- 62.Olano C, García I, González A, Rodriguez M, Rozas D, Rubio J, Sánchez-Hidalgo M, Braña AF, Méndez C, Salas JA. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol. 2014;7:242–256. doi: 10.1111/1751-7915.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo F, Xiang S, Li L, Wang B, Rajasärkkä J, Gröndahl-Yli-Hannuksela K, Ai G, Metsä-Ketelä M, Yang K. Targeted activation of silent natural product biosynthesis pathways by reporter-guided mutant selection. Metab Eng. 2015;28:134–142. doi: 10.1016/j.ymben.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 64.Sidda JD, Song L, Poon V, Al-Bassam M, Lazos O, Buttner MJ, Challis GL, Corre C. Discovery of a family of γ-aminobutyrate ureas via rational derepression of a silent bacterial gene cluster. Chem Sci. 2014;5:86–89. [Google Scholar]


