Abstract
Background
The microtubule-associated protein tau accumulates into toxic aggregates in multiple neurodegenerative diseases. We found previously that loss of D2-family dopamine receptors ameliorated tauopathy in multiple models including a Caenorhabditis elegans model of tauopathy.
Methods
To better understand how loss of D2-family dopamine receptors can ameliorate tau toxicity, we screened a collection of C. elegans mutations in dopamine-related genes (N=45) for changes in tau transgene-induced behavioral defects. These included many genes responsible for dopamine synthesis, metabolism, and signaling downstream of the D2 receptors.
Results
We identified one dopamine synthesis gene, DOPA decarboxylase (DDC), as a suppressor of tau toxicity in tau transgenic worms. Loss of the C. elegans DOPA decarboxylase gene, bas-1, ameliorated the behavioral deficits of tau transgenic worms, reduced phosphorylated and detergent-insoluble tau accumulation, and reduced tau-mediated neuron loss. Loss of function in other genes in the dopamine and serotonin synthesis pathways did not alter tau-induced toxicity; however, their function is required for the suppression of tau toxicity by bas-1. Additional loss of D2-family dopamine receptors did not synergize with bas-1 suppression of tauopathy phenotypes.
Conclusions
Loss of the DDC bas-1 reduced tau-induced toxicity in a C. elegans model of tauopathy, while loss of no other dopamine or serotonin synthesis genes tested had this effect. Because loss of activity upstream of DDC could reduce suppression of tau by DDC, this suggests the possibility that loss of DDC suppresses tau via the combined accumulation of dopamine precursor L-DOPA and serotonin precursor 5-hydroxytryptophan.
Keywords: Neurodegeneration, tau, dopamine, serotonin, aromatic amino acid decarboxylase, DOPA decarboxylase
Introduction
Tauopathies are neurodegenerative diseases characterized by the toxic accumulation of abnormal conformers of the microtubule-associated protein tau (1). Alzheimer’s disease and frontotemporal lobar degeneration (FTLD) are the most commonly occurring tauopathies (2). Various clinical strategies are being pursued to reduce tau toxicity, including reducing tau phosphorylation, tau cleavage, tau fibrillization, and tau expression levels (2). However, many candidate therapies have issues of non-specificity and safety. There are currently no treatments that halt or reverse the accumulation of pathological tau in human patients (2).
We previously identified D2-like receptor antagonist drugs ameliorating tauopathy phenotypes using a whole animal Caenorhabditis elegans model of tau toxicity (3). Pan-neuronal expression of human tau in C. elegans causes neurotoxicity, including a significant accumulation of tau aggregates, behavioral dysfunction, progressive neuron loss, and shortened lifespan (4). We demonstrated that loss of dop-2 and dop-3, the two D2-like dopamine receptors in C. elegans, significantly ameliorated tau-induced toxicity in tau transgenic C. elegans (3). Multiple studies have shown that overexpression of tau alters dopamine signaling by causing loss of dopaminergic neurons (5–6). In addition, activation of D1 dopamine receptors has been shown to increase tau phosphorylation (7). One study addressed how blocking D2-family dopamine receptors may suppress tauopathy using the D2 antagonist haloperidol (8); however, this drug is not specific to dopamine receptors (9). Therefore to better understand how loss of D2-like dopamine receptors suppresses tauopathy phenotypes, we performed a genetic screen of dopamine-related genes in tau transgenic C. elegans. We identified bas-1, the C. elegans homolog of DOPA decarboxylase (DDC), as a suppressor of tau-induced toxicity.
Materials and Methods
C. elegans strains and transgenics
C. elegans strains used are listed in Table S1. All strains were maintained at 20°C on standard NGM plates containing OP50 Escherichia coli as previously described (10). Worms were grown on NGM plates containing 5x more peptone prior to collection for protein studies. The Pbas-1::bas-1::GFP transgenic plasmid was a gift from Dr. Shi-Qing Cai. Pbas-1::bas-1::GFP and Pelt-2::mCherry were injected into N2 worms at 100 and 20 ng/μL, and chromosomally integrated using ~3500R of gamma rays from a Cesium source.
Behavioral analysis
Worms were synchronized by timed egg lays and grown at 20°C for 4 days (approximately day 1 of adulthood for tau transgenic C. elegans). Swimming behavior was quantified as described previously (3) with a few modifications. A single worm was transferred to a shallow well in a Teflon-coated glass slide filled with M9 buffer. After allowing the worm to adjust to liquid for 10 seconds, thrashes (body bends) were counted for 1 minute. A thrash was considered a large movement of the head or tail of the worm that causes a significant displacement of the middle third. For initial behavior screening (see Table 1), at least 15 animals were assayed per group. For all other comparisons, 100 animals were assayed per group.
Table 1.
Genes that modified behavior deficits in tau transgenic C. elegans
| Gene [Human Homolog] | Allele (mutation) | Motor Function Mutation alone |
Motor Function Tau Background |
|---|---|---|---|
| bas-1 [DDC] | ad446 (deletion) tm351 (deletion) |
105% (p=0.11) 98% (p = 0.61) |
254% (p = 9.1E-21) 202% (p = 7.6E-11) |
| egl-8 [PLCB1] | n488 (deletion) sa47 (Q85Stop) |
42% (p = 1.5E-10) 67% (p = 6.2E-16) |
32% (p = 3.2E-5) 51% (p = 3.0E-5) |
| egl-10 [RGS] | n692 (W418Stop) | 9% (p = 5.9E-10) | 8% (p = 1.5E-4) |
| grk-2 [GRK2] | gk268 (deletion) | 30% (p = 1.4E-51) | 50% (p = 5.6E-5) |
| unc-43 [CAMK2D] | n1186 (Q67Stop) n498x (E108K) |
29% (p = 6.0E-10) 5% (p = 1.2E-6) |
23% (p = 5.0E-10) 28% (p = 6.5E-4) |
| tax-6 [PPP3CC] | p675 (D259N) | 34% (p = 2.5E-4) | 38% (p = 4.7E-3) |
gain of function allele
Motor function was assessed via liquid thrashing assays. C. elegans carrying the mutation alone were compared to N2 (wild-type) C. elegans, while C. elegans strains carrying the mutation and the tau transgene were compared to tau transgenic C. elegans. At least 15 animals were assessed per strain and Student’s t-tests were used to determine significance.
Lifespan analysis
Lifespan assays were performed as described previously (11) with a few modifications. 100–150 L4 stage worms per strain were transferred to 35 mm NGM plates (25 worms/plate) seeded with 75 μL of 10X OP50 Escherichia coli and 0.05 mg/mL fluorodeoxyuridine (FUdR). Worms were maintained at 25°C. Dead worms were counted every day. A worm was considered dead when it did not respond to repeated poking by a platinum wire. Worms that crawled off the plate or died from unusual causes were censored from analysis.
Neurodegeneration assays
tgT337;unc-47::GFP and tgT337;unc-47::GFP;bas-1(ad446) worms were synchronized by timed egg lays. Worms were mounted on 2% agarose pads containing 0.1% sodium azide as a paralytic at 4 or 7 days of age. The number of GABAergic neurons in the ventral nerve cord were counted with a DeltaVision microscope (Applied Precision) using 60X magnification. Twenty animals were counted per strain per time point.
Immunoblotting
Protein samples were diluted with 5x sample buffer (0.046 M Tris, 0.005 M EDTA, 0.2 M dithiothreitol, 50% sucrose, 5% sodium dodecyl sulfate, 0.05% bromophenol blue), boiled for 5 min., and centrifuged at 13,000 × g for 5 min. prior to being loaded onto 4–15% pre-cast Criterion sodium dodecyl sulfate polyacrylamide gel electrophoresis gradient gels and transferred to PVDF membranes as recommended by the manufacturer (Bio-Rad). 5–10 μL of diluted sample was loaded for each analysis. Primary antibodies used were rabbit monoclonal anti-tau antibody (Rockland) at 1:5000, mouse anti-phospho-tau antibody CP13 (Peter Davies) at 1:1000, mouse anti-phospho-tau antibody PHF-1 (Peter Davies) at 1:1000, rabbit anti-phospho-tau antibody pS422 (Abcam) at 1:500, goat anti-GFP antibody (Rockland) at 1:5000, and mouse anti-tubulin antibody E7 (Developmental Studies Hybridoma Bank) at 1:5000. Secondary antibodies used were anti-rabbit HRP (Jackson Immuno Research), anti-mouse HRP (Jackson Immuno Research), and anti-goat HRP (Rockland), all at 1:5000. ECL substrate (Bio-Rad) was added to the membrane and chemiluminescence signals were detected with ChemiDoc-It Imager (UVP) and measured with UVP Software.
Tau protein extraction
Staged young adult tau transgenic C. elegans were grown from eggs at 20°C for 3 days on 5XPEP plates, washed off plates in M9 buffer, and collected by centrifugation. Worms were snap frozen in liquid nitrogen and stored at −70°C. Tau fractions were obtained as described previously (4). 2 μL of RAB Hi-Salt buffer (0.1 M MES, 1 mM EGTA, 0.5 mM MgSO4, 0.75 M NaCl, 0.02 M NaF, pH 7.0) containing PMSF and protease inhibitors was added per mg of worm pellet and homogenized by sonication. A portion of the sample was saved for immunoblotting while the rest was centrifuged at 40,000 × g for 40 min. The supernatant was the RAB/soluble fraction. The pellet was extracted by adding 1 μL of RIPA buffer (50 mM Tris, 150 mM NaCl, 1% Nonidet P-40, 5 mM EDTA, 0.5% deoxycholate, 0.1% SDS, pH 8.0) containing PMSF and protease inhibitors per mg of original worm pellet weight and centrifuged at 40,000 × g for 20 min. The supernatant was the RIPA/detergent soluble fraction. The pellet was extracted by adding 1 μL 70% formic acid (FA) per mg of original worm pellet weight and centrifuged at 13,000 × g for 15 min. The supernatant was the FA/detergent insoluble fraction.
Catecholamine measurements
Staged young adult C. elegans were grown from eggs at 20°C for 3 days on 5XPEP plates, washed off plates in M9 buffer, and collected by centrifugation. ~150 μL of packed worms were snap frozen in liquid nitrogen and stored at −70°C. Pellets were extracted with 0.1M perchloric acid. 50 μl was used for Pierce BCA protein determination. The remainder was centrifuged and the supernatant frozen at −70°C until assayed. The acid extract was put through an alumina extraction procedure before injection onto HPLC. Catecholamine levels were measured using electrochemical detection by a Dionex Choulochem III HPLC system. The data was analyzed using the Chromeleon 7 software package (Dionex). The catecholamine levels were normalized to the total protein.
Results
Identification of dopamine-related genes involved in tauopathy phenotypes
Previous work in our lab demonstrated that the combined loss of dop-2 and dop-3, D2-like dopamine receptors, suppresses tauopathy phenotypes in our C. elegans tauopathy model (3). D2-like dopamine receptors are G-protein coupled receptors that activate Gi/o-mediated signaling pathways and also act presynaptically to regulate dopamine release (12). Therefore, we crossed tau transgenic C. elegans with strains containing loss of function alleles in dopamine-related genes including genes involved in dopamine synthesis, metabolism, and downstream signaling (Table S1). We assayed the resulting strains for behavioral deficits using liquid thrashing assays, which measure C. elegans responses to immersion in liquid (Table S2). A few genes significantly modulated the swimming response of tau transgenic worms (Table 1). Most of these genes enhanced the tau-induced behavioral deficit. However, these enhancers of tau toxicity also caused significant impairment of locomotion in wildtype animals that do not carry the tau transgene, demonstrating that they influence behavior independent of tau.
Loss of DOPA decarboxylase suppresses tau-induced behavioral dysfunction
bas-1, the C. elegans gene for DOPA decarboxylase (DDC), was the only suppressor of tau related phenotypes identified in our screen. We tested two independent deletion alleles of bas-1, ad446 and tm351, which both rescued the behavioral dysfunction of tau transgenic C. elegans without altering behavior in wild-type C. elegans (Figure 1A–B; Table 1; Figure S1). The allele ad446 contains a deletion that removes almost the entire coding sequence of bas-1, including the catalytic site encoded by exon 4, while the allele tm351 deletes only exon 2. Both alleles caused increases in L-DOPA levels and decreases in dopamine levels in C. elegans, which suggests they are both functionally null (Table S3). The allele ad446 additionally deletes basl-1, a gene predicted to encode a nonfunctional paralog of DDC (13). We assayed the effect of deleting basl-1 alone by examining basl-1(ok703) and found it did not alter the behavior of tau transgenic C. elegans (Table S2).
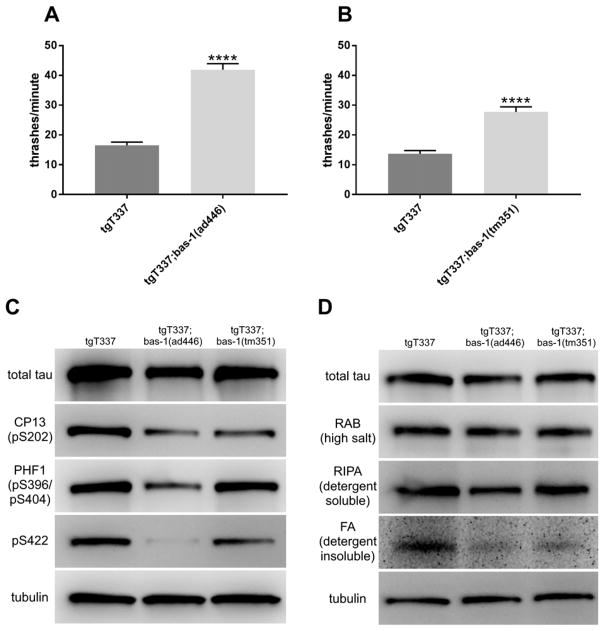
Figure 1.
Loss of bas-1 ameliorates behavioral deficits and reduces phosphorylated and insoluble tau in tau transgenic C. elegans. [A] Effect of bas-1(ad446) on liquid thrashing rate of tau transgenic (tgT337) worms. [B] Effect of bas-1(tm351) on liquid thrashing rate of tgT337 worms. 4-day-old C. elegans were placed in M9 buffer and thrashes were counted for 1 minute. Experiments were performed 5 times for a total of 100 worms per group. Data is displayed as mean ± standard error (S.E.M.). Student’s t-test was used to compare tgT337 to tgT337;bas-1 worms. **** p = 9.1E-21 for ad446 and 7.6E-11 for tm351. [C] Effects of bas-1 alleles on phosphorylation of tau in tau transgenic C. elegans. Representative immunoblots for tau phosphorylation at different sites. CP13 detects phosphorylation at Ser202. PHF-1 detects phosphorylation at Ser396/Ser404. pS422 detects phosphorylation at Ser422. [D] Effects of bas-1 alleles on tau solubility in tau transgenic C. elegans. Representative immunoblots for tau after sequential extraction of tau transgenic worms (tgT337) and bas-1 mutants (tgT337;bas-1(ad446) and tgT337;bas-1(tm351)). Total tau is tau in lysates prior to sequential extraction. The RAB fraction is soluble tau, the RIPA fraction is detergent soluble tau, and the formic acid (FA) fraction is detergent insoluble tau. Tubulin was used as a load control. Chemiluminscence signals were quantified for 4–6 replicates by Kruskal-Wallis with post-hoc Dunn’s multiple comparisons to determine significance. See Table 2 for values and statistics.
Loss of DOPA decarboxylase decreases tau phosphorylation
Phosphorylation of tau is a normal mechanism for regulation of tau activity, but hyperphosphorylation of tau, commonly seen in tauopathies, is thought to be toxic and precede the formation of insoluble aggregates (14). We examined whether loss of bas-1 modulated total tau levels or tau phosphorylation in tau transgenic C. elegans. We assessed tau phosphorylation with three different antibodies against phosphorylated tau: CP13 (pSer202), PHF-1 (pSer396/pSer404), and pSer422 (15). Phosphorylation of tau at these three sites is elevated in Alzheimer’s disease patient brains (16). Loss of bas-1 did not alter total tau levels significantly but generally reduced the phosphorylation at all three sites probed (Figure 1C; Table 2).
Table 2.
Effect of bas-1 alleles on tau levels.
| Comparison to tgT337 levels | tgT337;bas-1(ad446) | tgT337;bas-1(tm351) |
|---|---|---|
| total tau/tubulin (n = 6) | 88% (S.E.M. 9%; p = 0.88) | 125% (S.E.M. 23%; p = 0.88) |
| RAB (soluble) tau/total tau (n = 5) | 99% (S.E.M. 14%; p = 1) | 107% (S.E.M. 29%; p = 1) |
| RIPA (detergent soluble) tau/total tau (n = 5) | 43% (S.E.M. 10%; p = 0.01) | 76% (S.E.M. 16%; p = 0.26) |
| FA (detergent insoluble) tau/total tau (n = 5) | 44% (S.E.M. 13%; p = 0.07) | 39% (S.E.M. 20%; p = 0.05) |
| CP13 tau/total tau (n = 5) | 52% (S.E.M. 8% p = 0.02) | 55% (S.E.M. 7% p = 0.01) |
| PHF-1 tau/total tau (n = 5) | 71% (S.E.M. 8% p = 0.04) | 76% (S.E.M. 14% p = 0.10) |
| pS422/total tau (n = 4) | 31% (S.E.M. 8% p = 0.04) | 53% (S.E.M. 15% p = 0.39) |
For each replicate, the measured chemiluminescence signals were normalized to that for tau transgenic alone (tgT337). 4–6 replicates were used for each analysis. Data was analyzed by Kruskal-Wallis with post-hoc Dunn’s multiple comparisons used to determine significance.
Loss of DOPA decarboxylase decreases tau aggregation
The deposition of detergent insoluble aggregates of tau protein is a hallmark of human tauopathies (2). Detergent insoluble tau increases in tau transgenic C. elegans with age and with disease-causing mutations in tau (4). To determine whether loss of bas-1 affected the pathological aggregation of tau, we subjected tau transgenic C. elegans with or without bas-1 null alleles to extraction with buffers of increasing solubilizing strength. We found that loss of bas-1 did not change tau levels in soluble (RAB) fractions but reduced tau in detergent-soluble (RIPA) and detergent-insoluble (FA) fractions (Figure 1D; Table 2).
Loss of DOPA decarboxylase rescues neurodegeneration, but not longevity of tau transgenic C. elegans
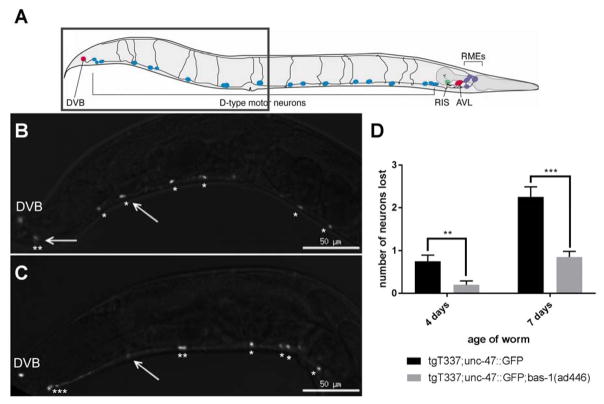
Neuronal loss occurs during the brain organ failure that causes premature death in tauopathy patients. Progressive neuronal loss and shortened lifespan are phenotypes seen in this tau transgenic C. elegans model (4). To determine whether loss of bas-1 affected the neurodegenerative phenotype seen in C. elegans, we crossed tau transgenic C. elegans carrying the bas-1 allele ad446 with the transgenic strain EG1285. EG1285 carries a GFP reporter transgene (Punc-47::GFP) which marks the cell bodies and processes of motor neurons within the ventral nerve cord of C. elegans (17). The resulting strain carries the bas-1 null allele, the tau transgene, and the GFP reporter. We counted the number of GABAergic motor neurons and observed a significant rescue of degenerating GABAergic neurons in bas-1 mutants at both 4 and 7 days of age (Figure 2).
Figure 2.
Loss of bas-1 function ameliorates tau-induced neuron loss. Tau transgenic (tgT337) and bas-1 mutant (tgT337;bas-1(ad446)) C. elegans were crossed with EG1285, a reporter transgenic strain expressing GFP in GABAergic neurons (unc-47::GFP). [A] Schematic showing the 19 D-type motor neurons, adapted from Schuske et al. (36). The gray box indicates the approximate region of the worm imaged in B and C [B] Representative image of a tgT337;unc-47::GFP worm (tail end) at 7 days. The stars indicate the neurons that were counted. The arrows indicate missing neurons. [C] Representative image of tgT337;unc-47::GFP;bas-1(ad446) at 7 days. [D] Quantification of GABAergic neuron loss in tgT337;unc-47::GFP and tgT337;unc-47::GFP;bas-1(ad446) worms. The D-type GABAergic motor neurons were counted in 4-day-old and 7-day-old worms. A total of 20 worms were analyzed for each group at each time point. Data is displayed as mean ± standard error (S.E.M.). Student’s t-test was used to compare tgT337;unc-47::GFP to tgT337;unc-47::GFP;bas-1(ad446) at each time point. ** p = 0.003 *** p = 1.8E-5
Tau transgenic C. elegans have shortened lifespans when compared to WT worms (4). However, we previously found that azaperone, a chemical suppressor of tau toxicity in tau transgenic C. elegans, was unable to extend lifespan (3). Similarly, loss of bas-1 did not extend lifespan significantly in tau transgenic C. elegans (Figure S2; Table S4).
Overexpression of DOPA decarboxylase worsens tau-induced phenotypes
Since loss of DDC gene bas-1 suppresses tau toxicity in our tau transgenic C. elegans, we investigated whether overexpression of BAS-1 protein would modulate tau toxicity. We crossed tau transgenic C. elegans with two different strains overexpressing BAS-1 fused to GFP under the bas-1 promoter (Pbas-1::bas-1::gfp). Overexpression of BAS-1::GFP fusion protein has been shown to rescue age-dependent decreases in serotonin and dopamine levels in C. elegans, suggesting that the GFP fusion does not interfere with BAS-1 protein activity (18). We found significant enhancement of the tau-induced behavioral dysfunction with overexpression of BAS-1 (Figure 3A–C). However, overexpressing BAS-1 impaired the behavior of wild-type C. elegans to a similar extent. This suggests that overexpression of BAS-1 may induce behavioral dysfunction independently of tau.
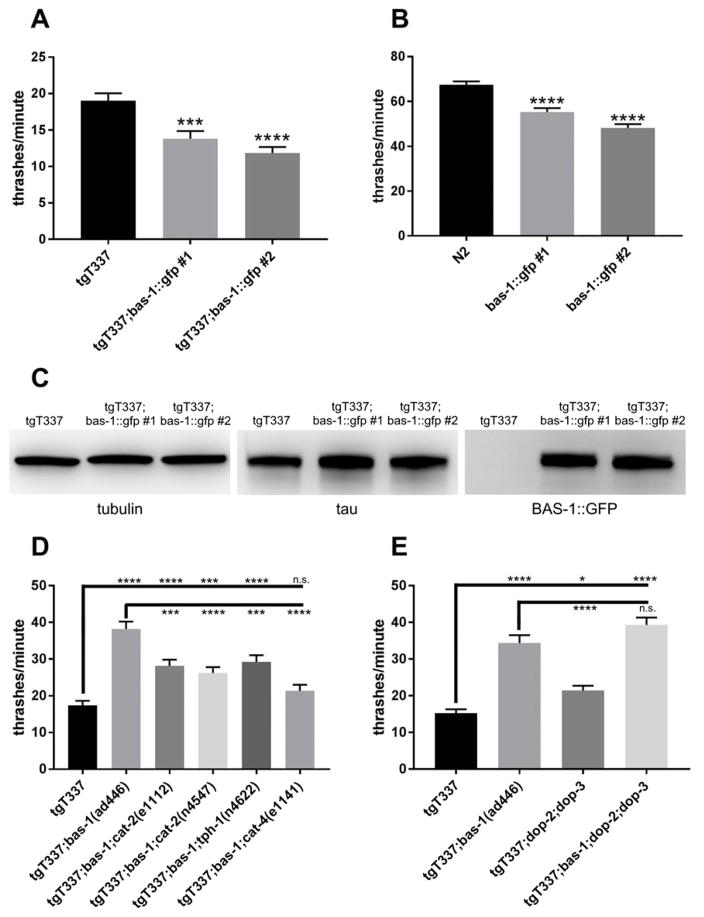
Figure 3.
Overexpression of BAS-1::GFP increases behavioral dysfunction in tau transgenic C. elegans. [A] Effect of BAS-1::GFP overexpression on liquid thrashing rate of tau transgenic (tgT337) worms. [B] Effect of BAS-1::GFP overexpression on liquid thrashing rate of wild-type (N2) worms. For A and B, 4-day-old C. elegans were placed in M9 buffer and thrashes were counted for 1 minute. The experiment was performed 5 times for a total of 100 worms per group. Data is displayed as mean ± standard deviation. One-way ANOVA and multiple pairwise comparisons (Dunnett’s) were used to compare tgT337;bas-1::GFP worms to tgT337 and bas-1::GFP worms to N2. *** p = 0.0004 **** p = 0.0001 [C] Western blot for total tau and bas-1::GFP for tau transgenic and double transgenic worms. The rabbit monoclonal antibody (Rockland) was used to detect total tau protein. GFP antibody was used to detect BAS-1::GFP (~80 kDa). Tubulin was used as a load control. [D] Loss of function in cat-2 or tph-1 partially blocks bas-1 suppression of tau-induced phenotypes while loss of function in cat-4 completely blocks suppression, as measured by liquid thrashing assay. [E] Loss of function in dop-2 and dop-3 does not alter bas-1 suppression of tau-induced phenotypes. Mutant worms had alleles dop-2(vs105), dop-3(vs106), and/or bas-1(ad446). For D and E, 4-day-old C. elegans were placed in M9 buffer and thrashes were counted for 1 minute. Experiments were performed 7 times (for D) or 5 times (for E) for a total of 100 worms per group. Data is displayed as mean ± standard error (S.E.M.). Data was analyzed by two-way ANOVA. Multiple pairwise comparisons (Tukey’s) were performed between tau transgenic C. elegans (tgT337) or tgT337;bas-1(ad446) and the other strains. Average, S.E.M., and p values are listed in Table S5 (for D) and Table S6 (for E). * p < 0.05 *** p < 0.005 **** p < 0.0001.
Enzymes upstream of DOPA decarboxylase are required for suppression of tau-induced phenotypes
DDC catalyzes the second step in the dopamine and serotonin synthesis pathways. Loss of DDC gene bas-1 significantly reduces dopamine and serotonin levels in C. elegans (18–20). Loss of function in the genes encoding tyrosine hydroxylase (cat-2) or tryptophan hydroxylase (tph-1) causes significant reduction in dopamine or serotonin levels respectively (20–21), but neither had an effect on tau-induced behavior dysfunction (Table S2). In addition, loss of cat-4 or cat-1, two other genes that regulate dopamine and serotonin levels in C. elegans (22), did not suppress tau-induced behavior dysfunction (Table S2). One way loss of bas-1 could suppress tau-induced toxicity is via accumulation of dopamine and serotonin synthesis intermediates L-DOPA and 5-hydroxytryptophan (5-HTP). Accumulation of these intermediates requires the activity of upstream enzymes tyrosine hydroxylase and tryptophan hydroxylase. Thus, to test whether suppression of tau by loss of DDC is modulated by upstream enzymatic activity, we generated tau transgenic C. elegans carrying the bas-1 null allele ad446 and a loss of function allele in cat-2, tph-1, or cat-4. We compared the behavior of these strains to that of tau transgenic C. elegans (tgT337) and to tau transgenic C. elegans carrying the bas-1 null allele (tgT337;bas-1(ad446)) (Figure 3D; Table S5).
Loss of cat-2 should decrease L-DOPA synthesis but not affect 5-HTP synthesis. We found that tau transgenic C. elegans carrying null alleles in both bas-1 and cat-2 still had significantly improved behavior compared to tau transgenic C. elegans but were significantly worse than tau transgenic C. elegans only carrying a null allele in bas-1, suggesting loss of cat-2 partially blocked suppression of tau by loss of bas-1.
Similarly, loss of tph-1 should decrease 5-HTP synthesis but not affect L-DOPA synthesis. We found that tau transgenic C. elegans carrying null alleles in both bas-1 and tph-1 had significantly improved behavior compared to tau transgenic C. elegans but were significantly worse than tau transgenic C. elegans only carrying a null allele in bas-1, suggesting that loss of tph-1 also partially blocks tau suppression by loss of bas-1.
Loss of cat-4 should decrease both L-DOPA and 5-HTP synthesis. Tetrahydrobiopterin is a cofactor for aromatic amino acid hydroxylases such as tyrosine hydroxylase and tryptophan hydroxylase and cat-4 encodes the C. elegans gene for GTP cyclohydrolase, the first enzyme in tetrahydrobiopterin synthesis (23). We found that tau transgenic C. elegans carrying loss of function alleles in both bas-1 and cat-4 had similar behavior dysfunction as tau transgenic C. elegans. This suggests that loss of cat-4 completely blocks the suppression of tau toxicity by loss of bas-1. Because loss of cat-4 reduced the L-DOPA accumulation in bas-1 null C. elegans to the same extent as loss of cat-2 (Table S3), loss of cat-4 does not block suppression of tau toxicity by bas-1 solely through lowering L-DOPA but must involve other molecular mechanisms.
Loss of DOPA decarboxylase and loss of D2-like dopamine receptors suppress tau-induced behavior via a shared mechanism
We initially tested bas-1 and other dopamine-related genes due to the fact that loss of D2-like dopamine receptor genes suppressed tau-induced toxicity in tau transgenic C. elegans (3). To see if loss of DDC and loss of D2-like dopamine receptors suppress tau via a shared mechanism, we created tau transgenic C. elegans with loss of function alleles in bas-1, dop-2 and dop-3. We compared the behavior of tau transgenic C. elegans with loss of bas-1 only, tau transgenic C. elegans with loss of dop-2 and dop-3, and tau transgenic C. elegans with loss of bas-1, dop-2, and dop-3 (Figure 3E; Table S6). We found that all three strains exhibited significantly suppressed tau-induced toxicity, but there was no significant additive suppression of tauopathy in animals carrying all three dopamine pathway mutations. This indicates that bas-1 and dop-2/dop-3 suppression of tauopathy share a common signaling mechanism.
Discussion
We have used our transgenic C. elegans model of tau toxicity to evaluate dopamine-related genes in the genesis of tauopathy phenotypes. In this screen, we found several candidate enhancers of tau mediated behavioral phenotypes, but none appeared to specifically modify tauopathy. We identified bas-1, the C. elegans homolog of DOPA decarboxylase (DDC), as a suppressor of tau toxicity in C. elegans. We found that loss of bas-1 ameliorated the behavioral deficits caused by tau, reduced the phosphorylation level of tau, reduced the accumulation of insoluble tau, and reduced neurodegeneration. Interestingly, no suppressors were identified among the screened genes that encode proteins downstream of the dopamine receptors in signaling pathways. Whether this is due to the essential nature of downstream genes or their redundant functions in signaling remains unclear. Alternatively, there may be signaling pathways downstream of dopamine receptors that mediate suppression which were not screened, such as the pathway from dopamine receptors to AMP-activated protein kinase (AMPK) inactivation (8).
Regardless, the enhancers we identified included genes whose human homologs have been reported to modulate tau including tax-6 (human homolog PPP3CC) and unc-43 (human homolog CAMK2D) (24–25). In addition, all of the enhancer genes caused profound behavioral deficits in C. elegans even in the absence of tau transgenic expression. The enhancement in behavioral dysfunction observed when these genes are lost may be due to an independent, additive effect on behavior. For instance, both loss and gain of function alleles of unc-43 caused significant behavioral dysfunction with or without tau transgenic expression. Therefore, it is difficult to interpret the effect of identified enhancers on tau. However, tau suppression by bas-1 is interpretable because loss of bas-1 on its own does not overtly impact motor function. Interestingly, overexpression of BAS-1 affected behavior in both WT and tau transgenic C. elegans to a similar extent. This suggests the possibility that any behavioral enhancement seen with BAS-1 overexpression in tau transgenic C. elegans is due to two independent mechanisms. Perhaps we did not observe a synergistic effect between BAS-1 and tau overexpression on behavior because the effect of BAS-1 activity on motor phenotypes is already maximal.
DDC catalyzes the second step in the two-step synthesis process for both dopamine and serotonin. Loss of DDC gene bas-1 causes a reduction in dopamine and serotonin as well as the accumulation of precursors L-DOPA and 5-HTP (18–20; see Table S3). Our results indicate that loss of dopamine and serotonin cannot be the mechanism of suppression because loss of function mutations in other genes required for their biosynthesis did not suppress tau-induced toxicity.
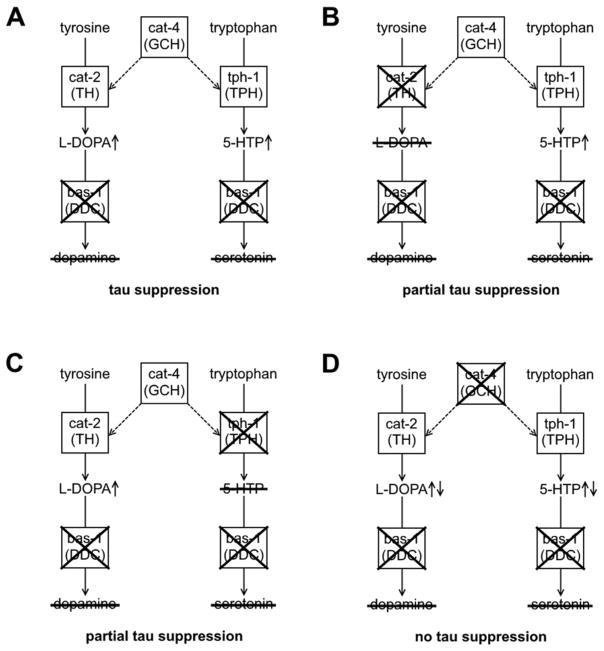
Our results support a possible mechanism for suppression of tau toxicity by the combined accumulation of L-DOPA and 5-HTP (Figure 4). Loss of cat-2, the C. elegans gene for tyrosine hydroxylase, only reduces levels of L-DOPA and not 5-HTP. Conversely, loss of tph-1, the C. elegans gene for tryptophan hydroxylase, only reduces the levels of 5-HTP and not L-DOPA. Loss of either cat-2 or tph-1 partially blocked the suppression of tau toxicity by loss of bas-1. Loss of cat-4, the C. elegans gene for GTP cyclohydrolase, reduces the synthesis of tetrahydrobiopterin (BH4), an essential cofactor for both tyrosine hydroxylase and tryptophan hydroxylase. Loss of cat-4 function, which reduces both L-DOPA and 5-HTP levels, completely blocked the suppression of tau toxicity by bas-1 loss of function. Because loss of cat-2 and loss of cat-4 caused similar reductions in L-DOPA levels in bas-1 null C. elegans, accumulation of L-DOPA cannot solely explain the mechanism of suppression. Similarly, since loss of tph-1 only partially blocked suppression of tau toxicity by bas-1, accumulation of 5-HTP also cannot solely explain the mechanism of suppression. The combined loss of tyrosine hydroxylase and tryptophan hydroxylase activity and subsequent reduction in both L-DOPA and 5-HTP levels may explain how loss of cat-4 blocks suppression of tau toxicity by loss of bas-1. BH4 is also a cofactor for phenylalanine hydroxylase and alkylglycerol monooxygenase (23). Phenylalanine hydroxylase activity is necessary for melanin synthesis in C. elegans, while alkylglycerol monooxygenase activity is necessary for cuticle integrity. Neither of these enzymes is expressed in neurons and loss of these enzymes does not affect serotonin or dopamine levels. It seems unlikely that loss of cat-4 would block suppression of tau toxicity by bas-1 via enzymes that are not expressed in the same cells or modulate the same pathways.
Figure 4.
A possible model for suppression of tau by loss of bas-1. [A] Loss of bas-1 (DOPA decarboxylase) leads to the accumulation of L-DOPA and 5-hydroxytryptophan (5-HTP). L-DOPA is synthesized from tyrosine by cat-2 (tyrosine hydroxylase) and 5-HTP is synthesized from tryptophan by tph-1 (tryptophan hydroxylase). cat-4 (GTP cyclohydrolase) synthesizes the cofactor needed for cat-2 and tph-1 activity. [B] Additional loss of cat-2 reduces L-DOPA levels without affecting 5-HTP and partially blocks suppression of tau by loss of bas-1. [C] Additional loss of tph-1 reduces 5-HTP levels without affecting L-DOPA and partially blocks suppression of tau by loss of bas-1. [D] Additional loss of cat-4 reduces both L-DOPA and 5-HTP levels and completely blocks suppression of tau by loss of bas-1. Altogether this suggests that the accumulation of both L-DOPA and 5-HTP are necessary to see the full effect of loss of bas-1 on tau toxicity.
The mechanism of suppression of tauopathy by bas-1 loss of function must be cell non-autonomous because GABAergic neurons are protected despite the fact they do not normally express bas-1. At the molecular level, L-DOPA and 5-HTP could mediate tauopathy suppression by directly activating receptors (26), or derivate compounds such as tetrahydroisoquinolines could mediate neurotoxicity (27). Alternatively, loss of bas-1 could suppress tauopathy by reducing consumption of its coenzyme, pyridoxal 5′-phosphate (PLP), a limiting cofactor for many PLP-dependent enzymes (28). This hypothetical mechanism explains why overexpression of BAS-1 exacerbates tauopathy phenotypes, but does not reconcile how loss of tyrosine hydroxylase or tryptophan hydroxylase activity could reduce tau suppression by loss of bas-1.
Interestingly, exogenous L-DOPA administration increased phosphorylation of tau in mice (29–31). This is the opposite of what we observed in tau transgenic C. elegans lacking the DOPA decarboxylase gene bas-1. Multiple mechanisms could explain this discrepancy. First, exogenous L-DOPA can be converted to dopamine in any cell with DDC expression, including serotonergic neurons (26). The metabolism of L-DOPA may also differ when DDC activity is lost compared to administration of exogenous L-DOPA. Second, the manner in which L-DOPA was administered affected tau phosphorylation; pulsatile administration but not continuous infusion was found to increase tau phosphorylation (29–30). Third, exogenous L-DOPA administration in mice increased plasma homocysteine and decreased S-adenosylmethionine, the methyl donor needed for methylation of the major tau phosphatase, PP2A (31). It is possible that this pathway is not conserved in C. elegans.
Another possible mechanism for suppression of tau toxicity by loss of DDC gene bas-1 could be modulation of expression levels of other related genes. For instance, loss of the dopamine transporter has been shown to change the expression of tyrosine hydroxylase (32). However, the expression levels of dopamine-related proteins, except for monoamine oxidase MAOA, were unchanged in mice with significantly reduced DDC expression (33). This suggests that a change in transcription of related genes is not likely to be the mechanism of bas-1 suppression.
Instead, our results support the possibility that loss of D2 family dopamine receptors suppresses tau by decreasing DDC activity. There is some evidence that D2-like dopamine receptors are expressed in serotonin neurons in C. elegans (34), which suggests that these receptors could modulate DDC in both dopamine and serotonin neurons. Suppression of tau-induced behavior by loss of DDC gene bas-1 was not altered by additional loss of D2-like dopamine receptor genes dop-2 and dop-3, indicating that DDC and D2 family dopamine receptors share a mechanism for suppression of tau. Interestingly, pharmacological agents that inhibit D2 receptors or antisense RNA against D2 receptors have been shown to increase DDC activity in rodents (35). This is the opposite of what we would expect based on our data. However, constitutive loss of D2 family dopamine receptors might have different effects on DDC activity in C. elegans compared to short-term or pharmacological loss of D2 family dopamine receptor activity in rodents.
In conclusion, we have identified bas-1, the C. elegans homolog of DOPA decarboxylase (DDC), as a suppressor of tau toxicity in our transgenic C. elegans model of tauopathy. While we have determined that upstream enzymatic activity by tyrosine hydroxylase and tryptophan hydroxylase are necessary for observing the full suppression by DDC, the mechanism of tau suppression is still unclear. Since loss of bas-1 reduced phosphorylation of tau, there are likely kinases or phosphatases modulated by loss of bas-1 function; further study will be needed to identify them. In addition, further translational studies exploring whether loss of DDC suppresses tau toxicity in the mammalian brain will be required to understand whether DDC could serve as a point of intervention in authentic human tauopathy disorders.
Supplementary Material
Figure S1. Loss of bas-1 does not significantly alter swimming behavior in wild-type C. elegans. [A] Effect of bas-1(ad446) on liquid thrashing rate of wild-type (N2) worms. [B] Effect of bas-1(tm351) on liquid thrashing rate of N2 worms. 4-day-old C. elegans were placed in M9 buffer and thrashes were counted for 1 minute. Experiments were performed 5 times for a total of 100 worms per group. Data is displayed as mean ± standard error (S.E.M.). Student’s t-test was used to compare N2 to bas-1 worms. See Table 1 for exact p values.
Figure S2. Effects of loss of function bas-1 alleles on lifespan of tau transgenic C. elegans. Survival curves are shown for representative lifespan assay of tau transgenic (tgT337) and bas-1 mutant worms (tgT337;bas-1(ad446) and tgT337;bas-1(tm351)). Lifespan assays were performed at 25°C on 144–146 worms/strain.
Table S1. List of C. elegans strains and alleles used
Table S2. Dopamine-related genes examined for effects on behavior deficits in tau transgenic C. elegans
Table S3. L-DOPA and dopamine measurements in tau transgenic and dopamine synthesis pathway mutant C. elegans
Table S4. Effects of loss of function bas-1 alleles on lifespan of tau transgenic C. elegans.
Table S5. Comparison of swimming behavior of tgT337;bas-1(ad446);cat-2(e1112), tgT337;bas-1(ad446);cat-2(n4547), tgT337;bas-1(ad446);tph-1(n4622), and tgT337;bas-1(ad446);cat-4(e1141) worms to tau transgenic (tgT337) and tgT337;bas-1(ad446).
Table S6. Comparison of swimming behavior of tau transgenic worms (tgT337) vs. tgT337;bas-1(ad446), tgT337;dop-2(vs105);dop-3(vs106), and tgT337;bas-1(ad446);dop-2(vs105);dop-3(vs106).
Acknowledgments
We thank the reviewers for helpful comments and suggestions. We thank Elaine Loomis, Aleen Saxton, Kaili Chickering, and Susan Danner for outstanding technical assistance. We thank Peter Davies and Virginia Lee for tau antibodies and the Developmental Studies Hybridoma Bank (NICHD) for the β-tubulin antibody E7. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and the National Bioresource Project (Japan). This work was supported by grants from the Department of Veterans Affairs [Merit Review Grant #1147891 to B.K.] and Bright Focus Foundation [Grant #A2014438S]. RK was supported by the NIH NIGMS Medical Genetics Postdoctoral Training Program grant [T32-GM-007454].
Footnotes
Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang Y, Mandelkow E. Tau in physiology and pathology. Nat Neurosci. 2016;17:5–21. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 2.Khanna MR, Kovalevich J, Lee VM, Trojanowski JQ, Brunden KR. Therapeutic strategies for the treatment of tauopathies: Hopes and challenges. Alzheimers Dement. 2016;12:1051–1065. doi: 10.1016/j.jalz.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCormick AV, Wheeler JM, Guthrie CR, Liachko NF, Kraemer BC. Dopamine D2 Receptor antagonism suppresses tau aggregation and neurotoxicity. Biol Psychiatry. 2013;73:464–471. doi: 10.1016/j.biopsych.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraemer BC, Zhang B, Leverenz JB, Thomas JH, Trojanowski JQ, Schellenberg GD. Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy. PNAS. 2003;100:9980–9985. doi: 10.1073/pnas.1533448100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiba S, Takada E, Tadokoro M, Taniguchi T, Kadoyama K, Takenokuchi M, et al. Loss of dopaminoreceptive neuron causes L-dopa resistant parkinsonism in tauopathy. Neurobiol Aging. 2012;33:2491–2505. doi: 10.1016/j.neurobiolaging.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Wu TH, Lu YN, Chuang CL, Wu CL, Chiang AS, Krantz DE, et al. Loss of vesicular dopamine release precedes tauopathy in degenerative dopaminergic neurons in a Drosophila model expressing human tau. Acta Neuropathol. 2013;125:711–725. doi: 10.1007/s00401-013-1105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebel M, Patenaude C, Allyson J, Massicotte G, Cyr M. Dopamine D1 receptor activation induces tau phosphorylation via cdk5 and GSK3 signaling pathways. Neuropharmacology. 2009;57:392–402. doi: 10.1016/j.neuropharm.2009.06.041. [DOI] [PubMed] [Google Scholar]
- 8.Koppel J, Jimenez H, Adrien L, Greenwald BS, Marambaud P, Cinamon E, et al. Haloperidol inactivates AMPK and reduces tau phosphorylation in a tau mouse model of Alzheimer’s disease. TRCI. 2016;2:121–130. doi: 10.1016/j.trci.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyler MW, Zaldivar-Diez J, Haggarty SJ. Classics in Chemical Neuroscience: Haloperidol. ACS Chem Neurosci. 2017 doi: 10.1021/acschemneuro.7b00018. [DOI] [PubMed] [Google Scholar]
- 10.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liachko NF, Guthrie CR, Kramer BC. Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J Neurosci. 2010;30:16208–16219. doi: 10.1523/JNEUROSCI.2911-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaulieu JM, Espinoza S, Gainetdinov RR. Dopamine receptors – IUPHAR Review 13. Br J Pharmacol. 2015;172:1–23. doi: 10.1111/bph.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hare EE, Loer CM. Function and evolution of the serotonin-synthetic bas-1 gene and other aromatic amino acid decarboxylase genes in Caenorhabditis. BMC Evol Biol. 2004;4:24. doi: 10.1186/1471-2148-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider A, Biernat J, von Bergen M, Mandelkow E, Mandelkow EM. Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry. 1999;38:3548–3558. doi: 10.1021/bi981874p. [DOI] [PubMed] [Google Scholar]
- 15.Acker CM, Forest SK, Zinkowski R, Davies P, d’Abramo C. Sensitive quantitative assays for tau and phosphor-tau in transgenic mouse models. Neurobiol Aging. 2013;34:338–350. doi: 10.1016/j.neurobiolaging.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y, Run X, Liang Z, Li Y, Liu F, Liu Y, et al. Developmental regulation of tau phosphorylation, tau kinases, and tau phosphatases. J Neurochem. 2009;108:1480–1494. doi: 10.1111/j.1471-4159.2009.05882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eastman C, Horvitz HR, Jin Y. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J Neurosci. 1999;19:6224–6234. doi: 10.1523/JNEUROSCI.19-15-06225.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin JA, Liu XJ, Yuan J, Jiang J, Cai SQ. Longevity manipulations differentially affect serotonin/dopamine level and behavioral deterioration in aging Caenorhabditis elegans. J Neurosci. 2014;34:3947–3958. doi: 10.1523/JNEUROSCI.4013-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 20.Sze JW, Victor M, Loer C, Shi Y, Ruvkun G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- 21.Sulston J, Dew M, Brenner S. Dopaminergic neurons in the nematode Caenorhabditis elegans. J Comp Neurol. 1975;163:215–226. doi: 10.1002/cne.901630207. [DOI] [PubMed] [Google Scholar]
- 22.Loer CM, Kenyon CJ. Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans. J Neurosci. 1993;13:5407–5417. doi: 10.1523/JNEUROSCI.13-12-05407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loer CM, Calvo AC, Watschinger K, Werner-Felmayer G, O’Rourke D, Stroud D, et al. Cuticle integrity and biogenic amine synthesis in Caenorhabditis elegans require the cofactor tetrahydrobiopterin (BH4) Genetics. 2015;200:237–253. doi: 10.1534/genetics.114.174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reese LC, Taglialatela G. A role for calcineurin in Alzheimer’s disease. Current Neuropharmacology. 2011;9:685–692. doi: 10.2174/157015911798376316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sengupta A, Grundke-Iqbal I, Iqbal K. Regulation of phosphorylation of tau by protein kinases in rat brain. Neurochem Res. 2006;31:1473–1480. doi: 10.1007/s11064-006-9205-9. [DOI] [PubMed] [Google Scholar]
- 26.De Deurwaerdère P, Di Giovanni G, Millan MJ. Expanding the repertoire of L-DOPA’s actions: A comprehensive review of its functional neurochemistry. Prog Neurobiol. 2017;151:57–100. doi: 10.1016/j.pneurobio.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Larghi EL, Amongero M, Bracca ABJ, Kaufman TS. The intermolecular Pictet-Spengler condensation with chiral carbonyl derivatives in the stereoselective syntheses of optically-active isoquinoline and indole alkaloids. ARKIVOC. 2005;12:98–153. [Google Scholar]
- 28.Bertoldi M. Mammalian dopa decarboxylase: Structure, catalytic activity and inhibition. Arch Biochem Biophys. 2014;546:1–7. doi: 10.1016/j.abb.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 29.Lebel M, Chagniel L, Bureau G, Cyr M. Striatal inhibition of PKA prevents levodopa-induced behavioural and molecular changes in the hemiparkinsonian rat. Neurobiol Dis. 2010;38:59–67. doi: 10.1016/j.nbd.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 30.Xie CL, Wang WW, Zhang SF, Yuan ML, Che JY, Gan J, et al. Levodopa/bensarazide microsphere (LBM) prevents L-dopa induced dyskinesia by inactivation of the DR1/PKA/P-tau pathway in 6-OHDA-lesioned Parkinson’s rats. Sci Rep. 2014;4:7506. doi: 10.1038/srep07506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bottiglieri T, Arning E, Wasek B, Nunbhakdi-Craig V, Sontag JM, Sontag E. Acute administration of L-Dopa induces changes in methylation metabolites, reduced protein phosphatase 2A methylation, and hyperphosphorylation of Tau protein in mouse brain. J Neurosci. 2012;32:9173–9181. doi: 10.1523/JNEUROSCI.0125-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvatore MF, Calipari ES, Jones SR. Regulation of tyrosine hydroxylase expression and phosphorylation in dopamine transporter-deficient mice. ACS Chem Neurosci. 2016;7:941–951. doi: 10.1021/acschemneuro.6b00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee NC, Shieh YD, Chien YH, Tzen KY, Yu IS, Chen PW, et al. Regulation of the dopaminergic system in a murine model of aromatic l-amino acid decarboxylase deficiency. Neurobiol Dis. 2013;52:177–190. doi: 10.1016/j.nbd.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Zhang F, Bhattacharya A, Nelson JC, Abe N, Gordon P, Lloret-Fernandez C, et al. The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development. 2014;141:422–435. doi: 10.1242/dev.099721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadjiconstatinou M, Neff NH. Enhancing aromatic L-amino acid decarboxylase activity: implications for L-DOPA treatment in Parkinson’s Disease. CNS Neurosci Ther. 2008;14:340–351. doi: 10.1111/j.1755-5949.2008.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuske K, Beg AA, Jorgensen EM. The GABA nervous system in C. elegans. Trends Neurosci. 2004;27:407–414. doi: 10.1016/j.tins.2004.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Loss of bas-1 does not significantly alter swimming behavior in wild-type C. elegans. [A] Effect of bas-1(ad446) on liquid thrashing rate of wild-type (N2) worms. [B] Effect of bas-1(tm351) on liquid thrashing rate of N2 worms. 4-day-old C. elegans were placed in M9 buffer and thrashes were counted for 1 minute. Experiments were performed 5 times for a total of 100 worms per group. Data is displayed as mean ± standard error (S.E.M.). Student’s t-test was used to compare N2 to bas-1 worms. See Table 1 for exact p values.
Figure S2. Effects of loss of function bas-1 alleles on lifespan of tau transgenic C. elegans. Survival curves are shown for representative lifespan assay of tau transgenic (tgT337) and bas-1 mutant worms (tgT337;bas-1(ad446) and tgT337;bas-1(tm351)). Lifespan assays were performed at 25°C on 144–146 worms/strain.
Table S1. List of C. elegans strains and alleles used
Table S2. Dopamine-related genes examined for effects on behavior deficits in tau transgenic C. elegans
Table S3. L-DOPA and dopamine measurements in tau transgenic and dopamine synthesis pathway mutant C. elegans
Table S4. Effects of loss of function bas-1 alleles on lifespan of tau transgenic C. elegans.
Table S5. Comparison of swimming behavior of tgT337;bas-1(ad446);cat-2(e1112), tgT337;bas-1(ad446);cat-2(n4547), tgT337;bas-1(ad446);tph-1(n4622), and tgT337;bas-1(ad446);cat-4(e1141) worms to tau transgenic (tgT337) and tgT337;bas-1(ad446).
Table S6. Comparison of swimming behavior of tau transgenic worms (tgT337) vs. tgT337;bas-1(ad446), tgT337;dop-2(vs105);dop-3(vs106), and tgT337;bas-1(ad446);dop-2(vs105);dop-3(vs106).