Abstract
Male infertility is a major and growing problem and, in most cases, the specific root cause is unknown. Here we show that the transcription factor SOX30 plays a critical role in mouse spermatogenesis. Sox30-null mice are healthy and females are fertile, but males are sterile. In the absence of Sox30 meiosis initiates normally in both sexes but, in males, germ cell development arrests during the post-meiotic round spermatid period. In the mutant testis, acrosome and axoneme development are aberrant, multinucleated germ cells (symplasts) form and round spermatids unable to process beyond step 3 of spermiogenesis. No elongated spermatids nor spermatozoa are produced. Thus, Sox30 represents a rare example of a gene for which loss of function results in a complete arrest of spermatogenesis at the onset of spermiogenesis. Our results suggest that SOX30 mutations may underlie some instances of unexplained non-obstructive azoospermia in humans.
Introduction
Spermatogenesis, the process whereby diploid spermatogonia differentiate into mature haploid spermatozoa, is highly complex and involves many precisely coordinated steps. In the post-natal phase spermatogonia undergo several rounds of mitotic division to produce primary spermatocytes which, in turn, duplicate their DNA content then undergo two rounds of nuclear division (meiosis I and meiosis II) resulting in formation of haploid round spermatids. During the post-meiotic developmental phase, known as spermiogenesis, round spermatids undergo a complicated restructuring program that includes compaction of DNA, ejection of cytoplasm and formation of the acrosome and flagellum1,2. Finally, spermatids are released from the epithelium via a process known as spermiation. Several thousand genes are expressed during spermatogenesis, although the function of the majority is unknown3–5.
In the mouse, spermiogenesis is divided into four phases characterized by the presence of round spermatids, elongating spermatids, condensing spermatids and condensed spermatids. In the elongating and condensing spermatids there is dramatic remodelling of chromatin structure that involves the sequential replacement of histones with transition nuclear proteins (TNPs) and eventually by protamines (PRMs), both of which are germ cell specific chromatin proteins. Transcription is most active in late spermatocytes and round spermatids6; this is the time point when Crem (cAMP-responsive element modulator), which encodes a key transcriptional activator CREMτ (CREM-tau), is expressed at highest levels7,8. Subsequently, transcription ceases during the spermatid elongation phase because PRMs compact the DNA into an inactive state.
Members of the SOX (S ry-related High Motility Group (HMG) box) gene family encode transcription factors that are highly conserved and are important for a range of developmental processes, including sex determination, neuronal development and regulation of stem cell pluripotency9. Within the SOX family SOX30 is the sole representative of SOX group H and is a relatively divergent member, with an HMG domain only 46% identical to the SOX-HMG box consensus sequence10,11. Apart from the HMG domain, SOX30 has no significant homology to any known protein functional domains. Perhaps due to its singular classification and lack of other known domains, Sox30 has remained under-characterised to date. In mice, it has been reported that the expression of Sox30 is specific to adult male germ cells and that the timing of expression correlates to spermatocytes undergoing meiosis11. A recent analysis of publicly-available RNA and protein profiling and interaction network data predicted a role for Sox30 in spermatogenesis in both humans and mouse12.
Here we describe the expression pattern of Sox30 in mouse fetal ovary and pubertal testis demonstrating that, in both sexes, Sox30 expression begins in germ cells shortly after expression of the pre-meiotic marker, Stra8. This expression pattern suggested that SOX30 might be essential for successful meiosis in both sexes but, counter to this, we observe no meiotic abnormality in either sex in a Sox30-null mouse model. Instead, our results indicate that SOX30 is a key transcription factor necessary for normal progression of spermatogenesis at the post-meiotic spermiogenesis phase. The clear-cut infertility phenotype we reveal, including a complete lack of elongated spermatids, demonstrates that this protein is essential for male fertility in mice.
Results
Expression of Sox30 follows that of Stra8 in both fetal ovary and postnatal testis
Previously we conducted an Affymetrix microarray analysis using fetal gonadal tissue of both sexes and at several time points13 and found that the Sox30 transcript was more highly expressed in the developing ovary than in developing testis (data not shown). At 13.5 dpc, we found by qRT-PCR that Sox30 was expressed in the fetal gonads, with significantly higher levels in ovaries than testes (Fig. 1a). There was very little expression in the other fetal organs tested, but high Sox30 expression was observed in the adult testis, as noted previously11. We purified fetal germ cells from the Oct4ΔPE:eGFP mouse line14, using FACS, and found that the expression of Sox30 in both the fetal ovaries and testis was exclusive to germ cells (Fig. 1b).
Figure 1.
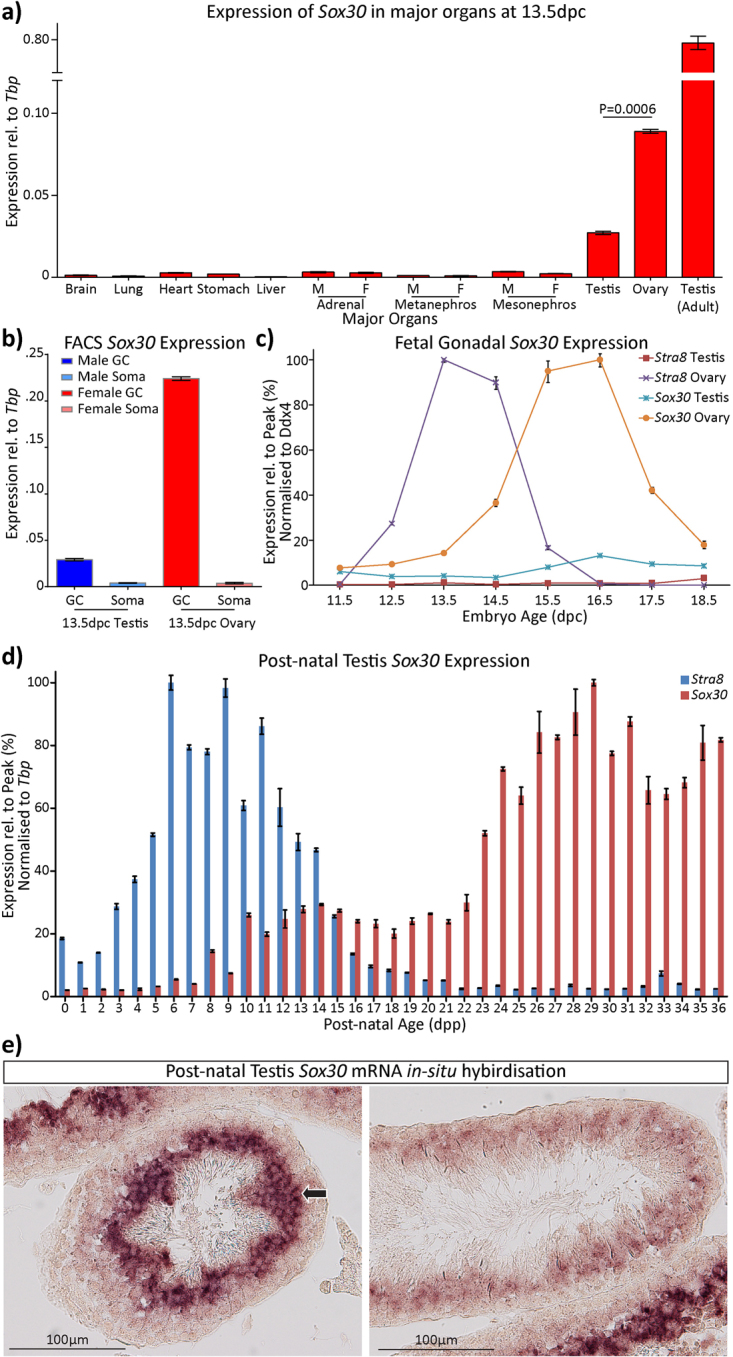
Expression of Sox30 is specific to the germ cells of the gonads at 13.5 dpc and follows the expression of Stra8 in both fetal ovaries and postnatal testis. (a) qRT-PCR was performed on various tissues of 13.5 dpc embryos, and adult testis, to assess Sox30 expression. Robust Sox30 expression in embryonic tissue is observed only in the gonads, with significantly higher levels detected in the fetal ovary than the fetal testis. Sox30 expression is ~8 fold higher in the adult testis than in the fetal ovary (pools of 4~8 individuals per column; two-tailed unpaired t-test). (b) qRT-PCR analysis of flow-sorted (FACS) populations of fetal gonadal tissues reveals that Sox30 transcript is enriched in the germ cell compartment in both the fetal ovary and testis at 13.5 dpc (cells isolated from pools of 4 to 8 gonad pairs). (c) qRT-PCR analysis in fetal gonads (from 11.5 to 18.5 dpc) demonstrates that the expression of Sox30 in the fetal ovary increases after the expression of Stra8 (pools of 4 to 8 gonad pairs). (d) qRT-PCR analysis demonstrates that expression of Sox30 follows expression of Stra8 in the postnatal testis (each time point represents one individual sample). All error bars represent S.E.M. of technical replicates. Ddx4 is expressed specifically in germ cells and is used as a normalizer for germ cell-specific genes when whole fetal gonadal tissue is analysed. (e) We verified by mRNA in situ hybridisation the previously described germ cell specific expression of Sox30 in the post-natal testis11. Transcripts for Sox30 were detected beginning at mid-pachytema (right panel), peaking in round and elongating spermatids (left panel, arrow). Elongated spermatids have diminished expression and no signal was detected in sperm just prior to spermiation.
The increased abundance of Sox30 mRNA in the fetal ovary coincided with the peak expression of pre-meiotic marker Stra8. Sox30 expression was highest at 15.5 to 16.5 dpc, just after Stra8 is downregulated (Fig. 1c). Levels of Sox30 expression remained low during fetal testicular development. In the postnatal testis Sox30 mRNA became more abundant at around 8 dpp, which is coincident with the appearance of preleptotene spermatocytes, and continued to increase, peaking at approximately 29 dpp (Fig. 1d). In the early postnatal testis, Sox30 mRNA was also first detected during the peak expression of Stra8, but highest Sox30 mRNA abundance was observed after Stra8 expression diminished. The fact that Sox30 expression closely follows expression of the pre-meiotic marker Stra8 15 in both the fetal ovaries and postnatal testis led us to hypothesise that Sox30 plays a role during germ cell meiosis in both sexes.
We attempted to visualize SOX30 protein in fetal ovaries and postnatal testis tissues but we were unsuccessful using commercially-available antibodies (See Supplementary Figure 1). In lieu of a suitable antibody, in-situ hybridisation for Sox30 mRNA was used to confirm the germ cell specific expression of Sox30 in the post-natal testis and the pattern of its expression. Sox30 mRNA was first detected during mid-pachynema before being dramatically up-regulated in round spermatids and elongating spermatids. Expression of Sox30 mRNA was subsequently decreased in elongated spermatids and was not detected in sperm just prior to spermiation (Fig. 1e).
Sox30 deletion results in male infertility
We produced Sox30-heterozygous null mice using ES cells imported from KOMP (see Materials and Methods) and bred them to homozygosity. No gross phenotypic or behavioural abnormalities were observed. At 8 weeks of age, three male and two female homozygous Sox30-null mice were mated with wildtype C57BL/6 partners for test breeding. All males exhibited normal mating behaviour, and copulatory plugs were observed. However, over three months, none of the Sox30-null studs impregnated their partners. The females were replaced once during this period and the females removed from breeding were monitored for at least another 19 days for potential pregnancies. In contrast, Sox30-null females were normally fertile, disproving our initial hypothesis that SOX30 plays a critical role during meiosis in both sexes.
Absence of Sox30 does not impact on oocyte development during fetal life
Although Sox30 female mice were fertile, we found the transient expression of Sox30 shortly after Stra8 expression in fetal ovarian germ cells striking (Fig. 1c). We noted that SOX30 is also expressed in a female-specific manner during human fetal gonad development16. Therefore, we conducted further studies to determine if a subtle defect in meiotic progress in ovarian fetal germ cells could be identified. We checked for abnormalities in expression of meiosis-related genes in the Sox30-null fetal ovary. By qRT-PCR we saw lower than normal expression of Stra8 in Sox30-null ovaries at 15.5 dpc but expression of later meiotic markers Spo11, Sycp3, Dmc1 and Rec8 remained relatively unaffected or was even slightly enhanced (Supplementary Figure 2a). Histological analysis by immunofluorescence for meiotic marker SYCP3 at 16.5 dpc and H&E staining did not highlight any significant abnormalities in Sox30-null fetal ovaries (Supplementary Figure 2b,c). After considering these results, and the fertility of Sox30-null female mice, we concluded that SOX30 is not required for female fertility.
Sox30 deletion led to spermatogenic arrest at round spermatids
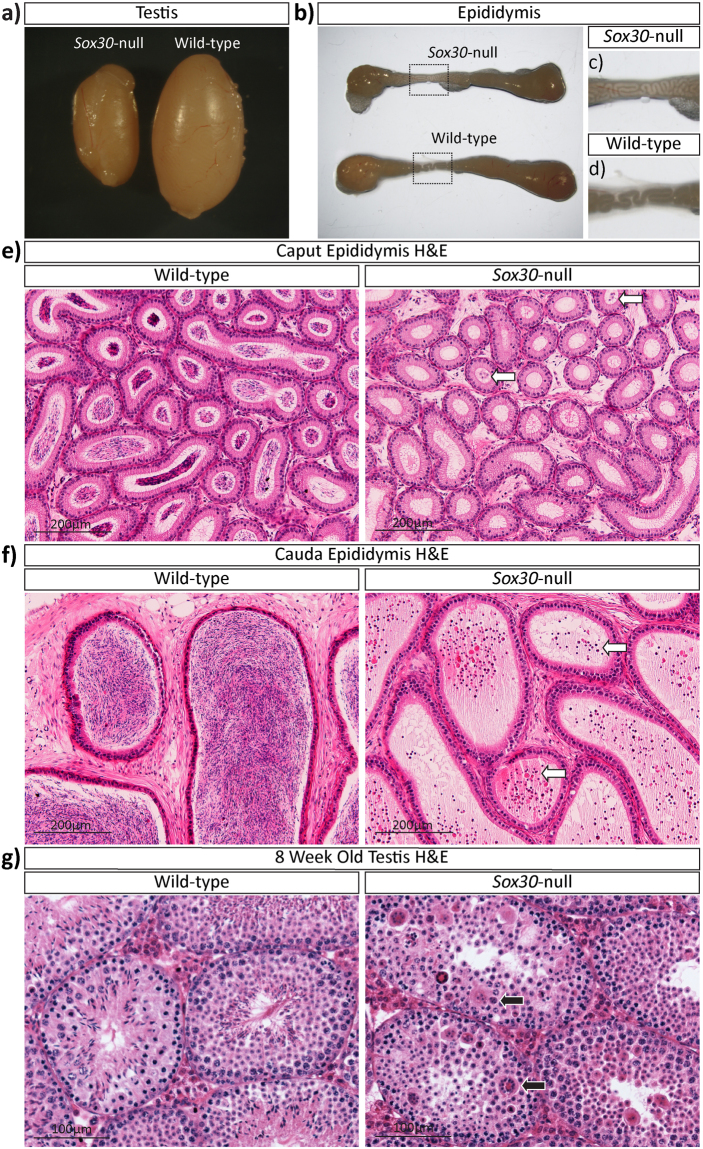
Following test breeding, Sox30-null studs were sacrificed and their testes collected for analysis. All three Sox30-null males had visibly smaller testes (P-value = 0.004) with an average length (5.921 mm, SD +/− 0.64, n = 3) approximately 72.4% of that of their wildtype (8.178 mm, SD +/− 0.13, n = 3) or heterozygous littermates (Fig. 2a). Sox30-null epididymal tubules appeared devoid of sperm when viewed using a light microscopy (Fig. 2b–d). Some abnormal germ cells (white arrows), but no sperm, were found in both the caput and cauda epididymides of Sox30-null males revealing that in the absence of SOX30 a significant number of haploid germ cells are precociously released from the seminiferous epithelium (Fig. 2e,f).
Figure 2.
Adult Sox30-null testes are abnormally small and spermatids and spermatozoa are not produced. (a) Testes are smaller in Sox30-null males with (b,c,d) empty epididymal tubules compared to wildtype littermates (n = 3). (e,f) Sperm are found in the caput and cauda epididymides of wildtype males, but are absent from Sox30-null epididymides (white arrows, H&E staining). (g) 8-week-old testes show an absence of elongating spermatids and the presence of multinucleated giant cells (black arrows) in Sox30-null testis.
Sectioning and histological analysis of Sox30-null testes at 8 weeks of age revealed round spermatid arrest: no cells progress to elongating spermatids and later forms (Fig. 2g). Multinucleated giant cells (symplasts)17 were consistently present in testis cords (Fig. 2g, black arrows) and these abnormal cells were first detected at 21 dpp, an age characterized by completion of meiosis and formation of round spermatids (Fig. 3a, white arrows). The abnormal cells stain positive with periodic acid-Schiffs reagent (PAS) suggesting that proacrosomal granules have begun to form and, therefore, that they arise from early round spermatids. Such multinucleated cells, also noted in a number of other spermatogenesis mutants18,19, are believed to represent abnormal syncytia and form when intercellular bridges are not maintained and so widen, open and then collapse resulting in spherical structures made up of fused spermatids20,21. In single round spermatids of the Sox30-null testis, PAS staining revealed that pro-acrosomal granules form and coalesce to form a single large granule, as is normal, but that the granule never flattens to cap the nuclear surface (Fig. 3a, insets). Hence, in the absence of SOX30 round spermatids are unable to progress beyond step 3 of spermiogenesis22.
Figure 3.
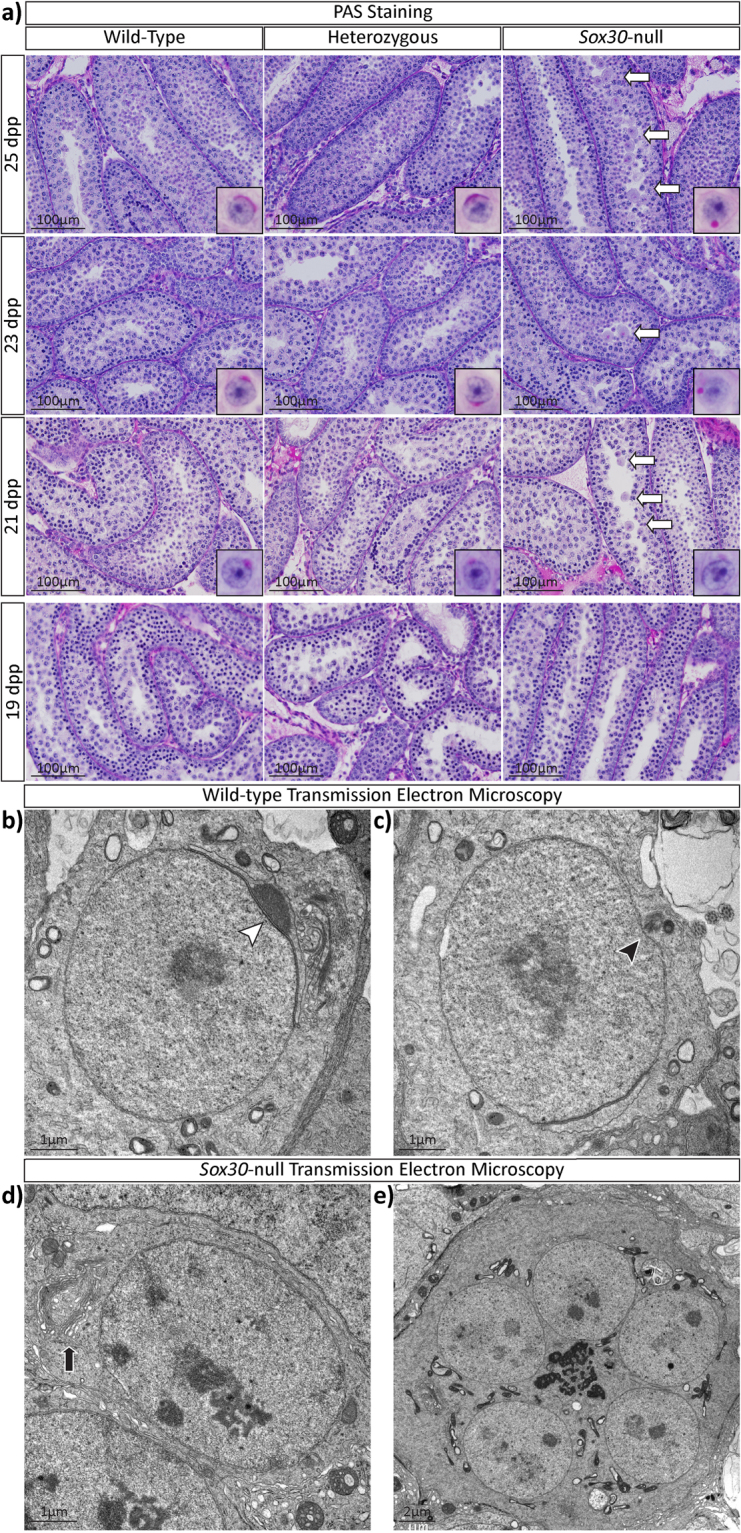
Sox30-null males are infertile with spermatogenesis blocked at the round spermatid stage. (a) PAS staining of 19, 21, 23 and 25 dpp testes reveals that abnormal germ cells (white arrows) are first seen in 21 dpp Sox30-null testes, a timpoint characterized by flattening of the pro-acrosome; most advanced appearance of round spermatids is shown in insert for each genotype and age. At 19 dpp no distinct granules are observed in any of the three genotypes. TEM examination of wildtype samples showing two ultrastructural features characteristic of step 3–4 spermatids: (b) acrosome capping of the nucleus (white arrowhead) and (c) docking of basal body to the nuclear membrane (black arrowhead). (d) These two features were not observed in Sox30-null round spermatids, nor were proacrosomal granules observed at the center of Golgi bodies (black arrow). (e) Nuclei within symplasts do not display any evidence of acrosome formation or tail development.
The above conclusions were strengthened by an analysis using transmission electron microscopy (TEM). In wildtype testes, we were readily able to observe both the docking of the basal body and the attachment of the acrosomal vesical to the nucleus, characteristic of spermatids that have progressed beyond step 3 (Fig. 3b,c). However, these features were completely absent in both the round spermatids of normal appearance and multinucleated symplasts from Sox30-null samples (Fig. 3d,e and Supplementary Figure 3, 4). We also noted that the Golgi bodies in Sox30-null spermatids were notably smaller that those in wildtype spermatids and do not appear to be producing many proacrosomal granules (Fig. 3d, black arrow and Supplementary Figure 4). As indicated in Fig. 3e (and Supplementary Figure 4) the symplasts clearly contain nuclei from multiple spermatids within a single plasma membrane. The presence of greater than four spermatid nuclei in a single plasma membrane suggests that the origin of the symplasts was not due to the failure of cytokinesis in meiosis I and meiosis II alone; wherein you would expect a maximum of 4 nuclei per plasma membrane. Rather, it suggests that symplast formation results from the coalescence of round spermatids derived from multiple spermatocytes. It is of note that, unlike somatic cells, in germ cells from spermatogonia onwards cytokinesis is incomplete and sister germ cells retain cytoplasmic bridges which are thought to be important for intracellular communication23. Collectively, these data show that in the absence of SOX30, spermatogenesis is unable to proceed beyond step 3 of spermiogenesis.
Sox30 acts cell-autonomously in spermatogenesis
Although Sox30 mRNA appears to be specific to germ cells of the fetal (Fig. 1b) and adult testis11,12, it is theoretically possible that deletion of Sox30 in somatic cells, in our model, influences germ cell development. To formally exclude this possibility, we generated a Sox30 conditional mouse line (Sox30 flox/flox) by crossing our Sox30-null line with a FLPe recombinase-expressing line24,25. Subsequently, we used the Vasa-Cre transgenic line26 to specifically delete Sox30 in germ cells. Female Sox30 flox/flox mice were mated with Sox30 flox/+;Vasa-Cre studs to produce Sox30 Δ/Δ, Sox30 flox/flox, Sox30 +/Δ and Sox30 flox/+ pups. As expected, germ line specific ablation of Sox30 in Sox30 Δ/Δ pups produced the same testicular phenotype as observed in Sox30-null mice (Supplementary Figure 5), confirming that SOX30 acts cell autonomously in testicular germ cells.
Some key markers of spermiogenesis are lost in the absence of Sox30
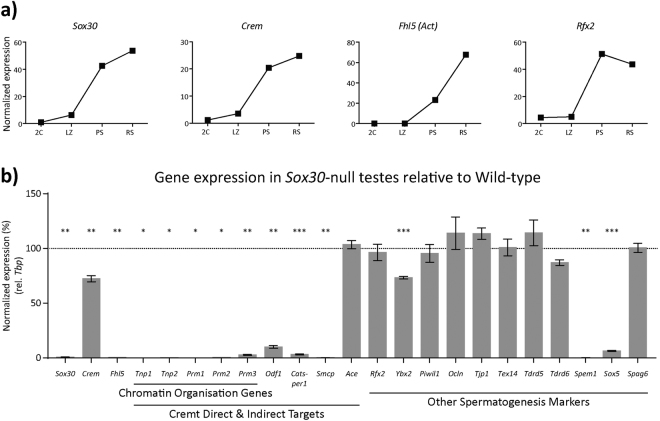
Using recently published RNA-Seq data27, we compared the Sox30 mRNA transcript profile, during the first wave of spermatogenesis, with profiles of known key spermatogenic marker genes (Fig. 4a). This analysis revealed that Sox30 mRNA was detected in a pattern virtually indistinguishable from that of Crem. CREMτ (Crem-tau) isoform mRNA starts to accumulate during pachynema, is the only isoform of Crem expressed in the male germline at that time, and encodes a master regulator of spermiogenesis8,28. The Crem-null phenotype is similar to that of Sox30 in that mice are healthy, females are fertile and males are sterile with spermatogenesis blocked at the post-meiotic round spermatid period. Further, as is the case for the Sox30-null, multinucleated cells (symplasts) are observed in the Crem-null testis cords. We also considered, in particular, the mRNA expression profiles of genes encoding other known transcriptional regulators. Fhl5 and Rfx2 mRNAs, encoding FHL5 (four and a half LIM domains protein 5, also known as ACT), a coactivator of CREM τ29, and RFX2, which like CREMτ is considered a master regulator of spermiogenesis19 were detected slightly later than Sox30 and Crem (Fig. 4a). mRNA detection profiles for other key genes that underlie various features of spermiogenic progression (also extracted from published RNA-seq data)27 are shown (Supplementary Figure 6).
Figure 4.
Expression of key markers of spermatogenesis in wildtype and Sox30-null testes. (a) Dynamic expression patterns of gene Sox30, Crem, Fhl5 and Rfx2, which encode transcriptional activators, in purified cell populations of the mouse testis: 2 C = heterogeneous cell population containing spermatogonia and somatic cells; LZ = leptotene and zygotene spermatocytes; PS = pachytene spermatocytes; RS = round spermatids. Data was obtained by RNAseq and is publically available27. (b) qRT-PCR analysis of expression of selected spermatogenesis genes in Sox30-null testes collected at 25 dpp, shown relative to expression in wildtype testis. Expression is normalized to TBP (n = 3, two-tailed unpaired t-test; error bars represent +/− S.E.M.). *p < 0.05; **p < 0.01, ***p < 0.001.
Based on these mRNA expression profiles, and information currently available about gene expression during spermiogenesis, we selected a number of marker genes and examined their expression in Sox30-null testes. This analysis involved qRT-PCR of wildtype and Sox30 null testicular tissue collected at 25 dpp. This time point was chosen so as to avoid gross differences in the cell composition of tissues between genotypes: the elongation phase does not begin until step 8 and elongating spermatids are not observed in significant numbers until 28 dpp2,30. Extensive differences were observed in expression of spermatogenesis-related genes in the absence of SOX30 when examined at 25 dpp (Fig. 4b). Although Crem mRNA abundance was diminished by about 25% when analysed at 25 dpp, it was unchanged at 19 and 23 dpp (Supplementary Figure 7a,b) suggesting that Crem is not a direct or indirect target of SOX30. In contrast, Fhl5 mRNA was not detected in Sox30-null testes at 25 dpp (Fig. 4b). This suggests that SOX30 is required for Fhl5 mRNA expression and that any CREMτ activity that is dependent on FHL5 co-activation will be lost in the absence of Sox30.
We next analysed expression of genes considered to be direct, or indirect, CREMτ targets. Although deletion of Fhl5 has no effect on expression of CREMτ targets Tnp1 (transition protein 1), Prm1 and −2 (protamine 1 and −2)31,32, expression of these genes, and related genes Tnp2 and Prm3, were essentially lost in the Sox30-null testis (Fig. 4b). Expression of additional CREMτ target genes Odf1 33, Catsper1 34 and Smcp 35 was lost in the Sox30-null testis but expression of direct CREMτ target Ace 36 was not affected (Fig. 4b). These results indicate that Fhl5 is not the only important target of SOX30 and that SOX30 and CREMτ may share some, but not all, transcriptional targets.
Finally, we examined expression of several additional marker genes of spermatogenesis. Rfx2, which encodes a recently discovered transcription factor now considered a master transcriptional regulator during spermiogenesis19, was expressed normally in the Sox30-null testis while Ybx2 (also known as Msy2)37, which marks specific transcripts for cytoplasmic storage, was only slightly downregulated. Similarly, loss of Sox30 had no effect on expression of Piwil1 (also known as Miwi)38 and no changes in expression were observed for Ocln and Tjp1, involved in construction and maintenance of the blood-testis barrier39,40 and Tex14, critical for the formation of intercellular bridges41. Tdrd5 and Tdrd6, encoding Tudor domain-containing proteins involved in retrotransposon silencing and chromatoid body assembly during spermiogenesis4,42,43, were also expressed normally. Expression of Spem1, encoding a protein important for the removal of cytoplasm during spermiogenesis44, was lost in the Sox30-null testis. We examined expression of Sox5, another member of the SOX family of transcription factors that is relatively closely related to Sox30 9. The short 2 kb transcript of Sox5 is detected in the post-meiotic germ cells of the adult testis as well as in the human brain45,46 and it has been suggested that this isoform is involved in ciliogenesis47. Sox5 expression was virtually lost in the absence of SOX30, a result in keeping with our expectations based on their relative temporal expression profiles (Supplementary Figure 6). Surprisingly, Spag6 which encodes a protein of the central axoneme of the sperm flagellum and is believed to be a direct target of SOX547, was expressed normally. Overall these findings demonstrate that SOX30 is responsible, directly or indirectly, for expression of some known key participants in spermiogenesis. The full repertoire of SOX30-dependent genes, and the overlap of this set with genes downstream of CREMτ, remains to be determined.
Discussion
Our findings demonstrate that the SOX family member SOX30 has an indispensable role in spermatogenesis, in mice. Homozygous deletion of Sox30 results in male sterility with spermatogenesis completely blocked in early round spermatids. Staining of testis sections with PAS and further investigation with TEM showed that the round spermatids fail to process beyond step 3 of spermiogenesis, a transition characterised by the attachment and flattening of the pro-acrosomal granule around the nucleus and the initiation of spermatid tail growth and attachment22. In addition to the spermatogenic block, a large number of multinucleated giant cells, also known as symplasts, were present: these are likely to be of early round spermatid origin as the cells contain multiple scattered granules of various sizes that stained positively for PAS.
Despite our initial hypothesis, that SOX30 is involved in meiosis, we did not find any evidence that this is so, in either sex. In the fetal ovary, Sox30 expression is initiated very shortly after Stra8 yet, in the Sox30-null model, we found no evidence that meiosis is perturbed. In the male, meiosis appears to proceed unimpeded as spermatocytes are able to develop into post-meiotic early round spermatids. It is not until later, at the round spermatid phase, that abnormalities are first observed. The production of Sox30 mRNA transcripts well before the protein is required is in agreement with observations for spermatogenesis-associated genes in general: it was recently concluded that genes initiating at pachynema tend to function in post-meiotic rather than meiotic processes27. This is in line with evidence that many mRNAs are produced, but then subjected to translational delays, until the relevant protein is required later in spermiogenesis48,49.
The exact role that SOX30 plays in spermiogenesis is not yet clear. What we know thus far is that, in the absence of SOX30, Crem mRNA is still expressed relatively normally. The Cremτ isoform encodes a master transcriptional regulator that induces transcription of numerous post-meiotic genes in round spermatids50,51 and we reveal here that its temporal expression profile during spermatogenesis is extremely similar to that of Sox30. Homozygous Crem mutants have a phenotype strikingly similar to that of Sox30 null mutants, with a complete spermiogenic block at round spermatids and the formation of symplasts18,52. The expression of several genes considered direct or indirect targets of CREMτ is essentially lost in the Sox30-null testis (Tnp1 and −2, Prm1, −2 and −3, Odf1, Catsper1) whilst at least one other is expressed normally (Ace). Thus, it seems likely that SOX30 works either cooperatively or in parallel with CREMτ to drive transcription that is necessary for round spermatid maturation. Because these changes are observed at 25 dpp, before elongating spermatids normally emerge, we can conclude that the transcriptional changes observed reflect the loss of direct or indirect targets of SOX30 rather than absence of a particular cell type. Similar trends were also observed at 23 dpp but with lower robustness due to the normally low expression of these genes at this time point (Supplementary Figure 7b)
Publicly available data suggest that SOX30 is unlikely to be a direct or indirect target of CREMτ: no change in Sox30 expression was detected in the Crem-null testis32,53. Although a half Cre site was identified 87 bp upstream of the Sox30 transcription start site in ChIP-seq studies, this was one of more than 6700 genomic loci found to be bound by CREMτ in mouse male haploid germ cells54. It is thought that round spermatids have a particularly accessible chromatin environment which possibly explains the large number of CREMτ-bound loci, many of which correspond to genes that are never expressed during spermiogenesis54. Given that CREMτ clearly occupies many more promoters that it actually regulates it seems plausible that co-regulation by SOX30, which is expressed in a highly cell type-restricted manner11,12 this study, lends specificity to transactivation instigated by CREMτ. The details of any physical or functional interactions between CREMτ and SOX30 remain to be determined.
Expression of Flh5, which encodes a coactivator for CREMτ, is lost in the absence of Sox30. Despite strong evidence for the coactivator function of FLH529 the spermatogenic phenotype of the Fhl5-null (fewer mature germ cells and abnormal tail and head morphology) is considerably less severe than that of the Crem-null (complete block in spermiogenesis)55. Presumably this means that FLH5 functions as coactivator for only a subset of CREMτ post-meiotic target genes31,55. Given the Fhl5 null phenotype, loss of Fhl5 expression in the Sox30-null testis is not sufficient, by itself, to account for the severe spermatogenic block we observe.
RFX2 is considered a second master transcriptional regulator of spermiogenic gene expression19. Rfx2-null mice have a phenotype resembling that of the Sox30-null, including defective spreading/adhesion of the acrosomal cap and the formation of giant multinucleated cells. RFX2 seems to be particularly important for the transcription of genes involved in cilia assembly and function and the set of RFX2 target genes is distinct from those regulated by CREMτ19. Although the onset of Rfx2 expression looks to be slightly later than that for Sox30 (Fig. 4a), Rfx2 expression is not affected by Sox30 deletion and, reciprocally, our examination of publicly-available data shows that the expression of Sox30 is unaffected in the Rfx2-null19. Hence it seems that Rfx2 and Sox30 are independently regulated.
Other than Crem-null and Rfx2-null models, described above, mouse mutants showing complete blockage of spermatogenesis at the round spermatid phase are rare56. One additional genetic mutant that arrests in early spermiogenesis is the homozygous deletion of Piwil1 (piwi-like RNA-mediated gene silencing 1, also known as Miwi)38. In the Piwil1-null round spermatids are blocked at a similar developmental period to those in the Sox30-null testis, with only a small number of spermatids proceeding to step 4 of spermiogenesis. Spermatids with pyknotic and fragmented nuclei were observed in Piwil1 mutant gonads, however, the presence of multinucleated cells was not mentioned. As is the case for the Sox30 mutant, Piwil1-null females are fertile. The cytoplasmic RNA binding protein PIWIL1 binds to Fhl5 mRNA as well as to mRNAs of target genes of CREMτ although, in Piwil1-null mice, expression of Crem itself is not affected. We found that Piwil1 expression was normal in the Sox30-null testis.
Using GeneChip RNA profiling data and mass spectrometry, others have shown human SOX30 expression is highly specific to the adult testis12. Expression of SOX30 was not detected above baseline levels in any adult tissue except the testis whilst SOX30 protein was found at high levels only in adult testis in data obtained by mass spectrometry. Those authors hypothesized that SOX30 is important in spermatogenesis and that it might act as a potential hub protein as it is predicted to have at least 13 interacting proteins. Given this apparent specificity of expression, and our findings in mice - that deletion of Sox30 results in healthy individuals with fertile females and sterile males - we speculate that mutations in SOX30 could easily be retained in the population and, therefore, underlie some cases of male infertility. It is interesting to note that despite the apparent restriction of SOX30 expression to the germ cell lineage it has been suggested that SOX30 is ectopically expressed in lung cancer and acts as a tumour suppressor, by virtue of its ability to directly activate transcription of Trp53, a major cell-cycle regulator57.
Approximately 10–15% of couples are infertile and a male factor is involved in almost half of these cases. Male infertility encompasses a wide variety of disorders but the most severe presentation is nonobstructive azoospermia (NOA), a condition where spermatogenic failure has occurred and spermatozoa are completely lacking58. NOA is found in about 5 to 10% of male infertility patients and cannot be overcome by use of approaches such as in vitro fertilization and intracytoplasmic sperm injection. Thus, there is a compelling need to gain a better understanding of the molecular mechanisms that underlie normal mammalian sperm production: such knowledge will inform the development of new diagnostics, targeted therapies and the production of gametes in vitro as well as efforts to develop male-based nonhormonal contraceptive methods for humans and animals. Given our findings, that we speculate that SOX30 mutations may underlie some of the cases of idiopathic round spermatid arrest in humans.
Methods
Animal Ethics
All procedures involving animals and their care were carried out in accordance with institutional, state and national guidelines. This study was approved by the University of Queensland and Monash University Animal Ethics Committees.
Mice
For analysis of fetal and postnatal expression of Sox30, gonadal tissue samples were obtained from an X-linked-eGFP mouse line59 maintained on a Swiss Quackenbush background. An Oct4ΔPE:eGFP mouse line (OG2)14 on a C56BL/6 J background was used to segregate germ and somatic cells via fluorescence activated cell sorting (FACS) as previously described60. Sox30tm1a(KOMP)Wtsi targeted embryonic stem (ES) cells were obtained from Knockout Mouse Project (KOMP) 25repository (UC Davis) and Sox30-null mice were produced, using these cells, by the Australian Phenomics Network (APN, Monash node)61. The modified Sox30 allele carried by the Sox30 knockout mice consists of a gene trap cassette inserted into the first intron of Sox30. This cassette includes a mouse En2 splice acceptor followed by lacZ reporter transgene with a SV40 polyadenylation sequence: this creates a null allele. This initial null allele also contains FRT sites which are positioned so as to allow excise of the gene trap cassette upon exposure to FLP recombinase, leaving behind loxP sites flanking the second and third exon of Sox30 (thus generating a Sox30 conditionally null allele). Thus, we generated a conditional knockout (Sox30 flox/flox) mouse line by crossing Sox30-null mice with FLPeR mice24. For experimental analysis Sox30 flox/flox mice were bred with VASA-Cre transgenic mice, which express Cre recombinase in germ cells from approximately 15.5 dpc onwards26. Genotyping of mice was performed by PCR using the primers indicated in Supplementary Table S1 online with the exception of the X-linked-eGFP mice which were genotyped by virtue of their green fluorescence under UV light.
Timed matings and tissue collection
X-linked-eGFP and homozygous Oct4ΔPE:eGFP studs were housed with wildtype females (Swiss Quackenbush and C57BL/6 J respectively) for timed matings with noon of the day when the copulatory plug was observed designated as 0.5 dpc. To investigate the Sox30-null phenotype, males and females heterozygous for the Sox30-null allele were mated. For germ cell-specific ablation of Sox30, Sox30 flox/flox females were mated with Sox30 flox/+;VASA-Cre+ males. The sex of the embryos was identified by visual inspection of dissected gonads or, in the case of X-linked-eGFP, by presence or absence of fluorescence of the whole embryo. Ube1 genotyping using tail tissue62 was retrospectively preformed to reconfirm the sex of all collected samples. For tissues to be analysed by qRT-PCR, gonad pairs were dissected free of the adjacent mesonephric tissue and the gonads were placed immediately in RNAlater (QIAGEN, #76106) for storage. When collected for qRT-PCR postnatal testes were first decapsulated before storage in RNAlater. For embryonic tissues to be analysed by histology, whole embryos (minus tail used for genotyping) were collected, fixed in 4% paraformaldehyde in phosphate buffered saline (PFA/PBS), dehydrated and embedded in paraffin for sectioning. When collected for histological analysis the capsule of postnatal testes was pierced in multiple locations with a 30 G needle and the samples were fixed with Bouin’s solution (Sigma, #HT10132) or 4% PFA/PBS before being processed and embedded in paraffin for sectioning.
Quantitative reverse transcriptase PCR (qRT-PCR)
Total RNA was extracted from individual gonads or gonad pairs using RNeasy Micro Kit (Qiagen, #74004) or RNeasy Mini Kit (Qiagen, #74106) including on-column DNase treatment. Total RNA-containing eluate was immediately used for cDNA synthesis by reverse transcription using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, #4368813). For fetal gonadal samples and the time course of Sox30 and Stra8 expression in the postnatal testis qRT-PCR was performed on a Viia7TM Real-Time PCR System (Applied Biosystems). Expression levels of Dmc1 (Mm00494485_m1), Rec8 (Mm00490939_m1), Sox30 (Mm00557681_m1), Stra8 (Mm00486473_m1) and Sycp3 (Mm00488519_m1) relative to Ddx4 (Mm00802445t_m1) or Tbp (Mm01277045_m1) were quantified using Taqman Gene Expression Assays and Universal Taqman Master Mix (Applied Biosystems, #4318157). To investigate the effects of Sox30 ablation in 19 dpp and 25 dpp testis, qRT-PCR was performed on a Quantstudio6 and Quantstudio7 (Applied Biosystems) respectively using primers listed in Supplementary Table S2 online and SYBR Green PCR Master Mix (Applied Biosystems, #4309155). Relative cDNA levels were determined by the 2−ΔCT method. All kits and assays were performed according to instructions supplied by the manufacturer.
Histological sectioning and staining
Whole embryos and postnatal testes embedded in paraffin were sectioned at 7 µm and dewaxed by immersion into xylene twice for 10 minutes and then rehydrated through an ethanol series ranging from 100% to 35% ethanol (v/v) in water. Histological staining of Bouin’s fixed samples was performed with either hematoxylin and eosin (H&E, Sigma, #HHS32 and #HT110332, respectively) or periodic acid-Schiff reagents (PAS, Sigma, #395B-1KT). All staining procedures were carried out as per manufacturers’ instructions. In our attempts to visualize SOX30 protein by immunofluorescence we used SOX30 antibodies ab26024 and ab71033 (Abcam), sc-20104 and sc-390333 (Santa Cruz Biotechnology) and 13017-AP (Proteintech) without success.
In-situ hybridisation
The 3′ UTR of Sox30 was PCR amplified from postnatal testis cDNA using primers 5′-ccctttggctatggaaattttcc-3′ and 5′-caatgcataccaaatgggaaaga-3′ and cloned into pGEM-T-easy (Promega). Resulting clones were verified by Sanger sequencing and then used for DIG labelled RNA probe synthesis as previously described63. 4% PFA fixed postnatal testes embedded in paraffin were used for in-situ hybridisation using previously described protocol64 with adaptations to utilise and detect DIG-labelled probe. Briefly, DIG-labelled probes were hybridised at 1μg/ml and detected by blocking the slides with 10% heat-inactivated sheep serum (HISS) in NT buffer (50 mM Tris-HCl and 150 mM NaCl at pH 7.5) for at least 1 hour before incubating overnight with 1:2000 dilution of α-DIG-AP Fab fragments (Roche) in 1% HISS/NT at 4 °C. The Fab fragments were rinsed off with 3 washes using NT buffer and the slides were then placed in NTM buffer (100 mM Tris-HCl, 100 mM NaCl and 50 mM MgCl2 at pH 9.5) for 10 minutes to equilibrate. A staining solution consisting of 175μg/ml BCIP (Roche) and 350μg/ml NBT (Roche) in NTM was applied to the slides and colour development was check at regular intervals by placing the slides in NTM. Once the desired staining intensity has been reached the slides were fixed with 4% PFA and mounted with an aqueous mounting media.
Transmission electron microscopy
Freshly dissected testis were fixed in a solution of 100 mM Sodium Cacodylate buffer with 4% PFA, 5% glutaraldehyde and 0.02% picric acid for at least 3 hours then bisected for fixation overnight at 4 °C. After fixation the samples were washed three time with 100 mM Sodium Cacodylate buffer before preforming a secondary fixation with 2% Osmium tetroxide in 100 mM Cacodylate buffer for 2 hours. The samples were rinsed with 3 washes of MilliQ water before staining with 1% Uranyl Acetate for 90 minutes and then rinsed with another 3 washes of MilliQ water. Samples were dehydrated in an acetone series of 50%, 70%, 90% and three 100% acetone washes. Araldyte Epon resin was gradually introduced, first with a wash of 50:50 resin:acetone mix for 2 hours then a 70:30 resin:acetone mix overnight on rollers. The samples were soaked in three changes of 100% Araldyte Epon for 2 hours before being embedded and polymerisation at 60 °C for 48 hours. Samples were sectioned at 90 nm, mounted onto carbon coated copper grids and double stained with uranyl acetate and lead citrate.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
The Sox30tm1a(KOMP)Wtsi ESC used for this research project was generated by the trans-NIH Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository (www.komp.org). NIH grants to Velocigene at Regeneron Inc (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ESCs for 8500 genes in the KOMP Program and archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244). MKOB was supported by a fellowship from the National Health and Medical Research Council (NHMRC, APP1058356). PK was supported by a fellowship from the NHMRC (APP1059006). Work was supported by an Australian Research Council grant (JB; DP140104059) and an NHMRC grant (JB; APP1109502).
Author Contributions
C.-W.F., J.B. and P.K. conceived the project; C.-W.F., C.S. and D.J.M. performed experiments. All authors analysed the data. C.-W.F. and J.B. wrote the manuscript and all authors read and approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Josephine Bowles and Peter Koopman contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17854-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Oakberg EF. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat. 1956;99:391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- 2.O’Donnell L. Mechanisms of spermiogenesis and spermiation and how they are disturbed. Spermatogenesis. 2014;4:e979623. doi: 10.4161/21565562.2014.979623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nature medicine. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Massart A, Lissens W, Tournaye H, Stouffs K. Genetic causes of spermatogenic failure. Asian journal of andrology. 2012;14:40–48. doi: 10.1038/aja.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamsai D, O’Bryan MK. Mouse models in male fertility research. Asian journal of andrology. 2011;13:139–151. doi: 10.1038/aja.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sassone-Corsi P. Transcriptional checkpoints determining the fate of male germ cells. Cell. 1997;88:163–166. doi: 10.1016/S0092-8674(00)81834-6. [DOI] [PubMed] [Google Scholar]
- 7.Delmas V, van der Hoorn F, Mellstrom B, Jegou B, Sassone-Corsi P. Induction of CREM activator proteins in spermatids: down-stream targets and implications for haploid germ cell differentiation. Molecular endocrinology. 1993;7:1502–1514. doi: 10.1210/mend.7.11.8114765. [DOI] [PubMed] [Google Scholar]
- 8.Foulkes NS, Schlotter F, Pevet P, Sassone-Corsi P. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis. Nature. 1993;362:264–267. doi: 10.1038/362264a0. [DOI] [PubMed] [Google Scholar]
- 9.Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 10.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Developmental biology. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 11.Osaki E, et al. Identification of a novel Sry-related gene and its germ cell-specific expression. Nucleic acids research. 1999;27:2503–2510. doi: 10.1093/nar/27.12.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petit, F. G. et al. Combining RNA and Protein Profiling Data with Network Interactions Identifies Genes associated with Spermatogenesis in Mouse and Human. Biol Reprod, doi:10.1095/biolreprod.114.126250 (2015). [DOI] [PubMed]
- 13.Holt JE, et al. CXCR4/SDF1 interaction inhibits the primordial to primary follicle transition in the neonatal mouse ovary. Developmental biology. 2006;293:449–460. doi: 10.1016/j.ydbio.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Szabó PE, Hübner K, Schöler H, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mechanisms of development. 2002;115:157–160. doi: 10.1016/S0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- 15.Baltus AE, et al. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nature genetics. 2006;38:1430–1434. doi: 10.1038/ng1919. [DOI] [PubMed] [Google Scholar]
- 16.Houmard B, et al. Global gene expression in the human fetal testis and ovary. Biol Reprod. 2009;81:438–443. doi: 10.1095/biolreprod.108.075747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacGregor GR, et al. Symplastic spermatids (sys): a recessive insertional mutation in mice causing a defect in spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:5016–5020. doi: 10.1073/pnas.87.13.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nantel F, et al. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature. 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 19.Kistler WS, et al. RFX2 Is a Major Transcriptional Regulator of Spermiogenesis. PLoS genetics. 2015;11:e1005368. doi: 10.1371/journal.pgen.1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber JE, Russell LD. A study of intercellular bridges during spermatogenesis in the rat. Am J Anat. 1987;180:1–24. doi: 10.1002/aja.1001800102. [DOI] [PubMed] [Google Scholar]
- 21.Faridha A, Faisal K, Akbarsha MA. Aflatoxin treatment brings about generation of multinucleate giant spermatids (symplasts) through opening of cytoplasmic bridges: light and transmission electron microscopic study in Swiss mouse. Reprod Toxicol. 2007;24:403–408. doi: 10.1016/j.reprotox.2007.04.071. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Methods in molecular biology. 2009;558:263–277. doi: 10.1007/978-1-60761-103-5_16. [DOI] [PubMed] [Google Scholar]
- 23.Greenbaum MP, Iwamori T, Buchold GM, Matzuk MM. Germ cell intercellular bridges. Cold Spring Harb Perspect Biol. 2011;3:a005850. doi: 10.1101/cshperspect.a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (Flipper) mice. Genesis. 2000;28:106–110. doi: 10.1002/1526-968X(200011/12)28:3/4<106::AID-GENE30>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 25.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.da Cruz I, et al. Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC genomics. 2016;17:294. doi: 10.1186/s12864-016-2618-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foulkes NS, Mellstrom B, Benusiglio E, Sassone-Corsi P. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature. 1992;355:80–84. doi: 10.1038/355080a0. [DOI] [PubMed] [Google Scholar]
- 29.Fimia GM, De Cesare D, Sassone-Corsi P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature. 1999;398:165–169. doi: 10.1038/18237. [DOI] [PubMed] [Google Scholar]
- 30.Laiho A, Kotaja N, Gyenesei A, Sironen A. Transcriptome profiling of the murine testis during the first wave of spermatogenesis. Plos One. 2013;8:e61558. doi: 10.1371/journal.pone.0061558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotaja N, et al. Abnormal sperm in mice with targeted deletion of the act (activator of cAMP-responsive element modulator in testis) gene. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10620–10625. doi: 10.1073/pnas.0401947101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosir R, et al. Novel insights into the downstream pathways and targets controlled by transcription factors CREM in the testis. Plos One. 2012;7:e31798. doi: 10.1371/journal.pone.0031798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang K, et al. The small heat shock protein ODF1/HSPB10 is essential for tight linkage of sperm head to tail and male fertility in mice. Molecular and cellular biology. 2012;32:216–225. doi: 10.1128/MCB.06158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren D, et al. A sperm ion channel required for sperm motility and male fertility. Nature. 2001;413:603–609. doi: 10.1038/35098027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nayernia K, et al. Asthenozoospermia in mice with targeted deletion of the sperm mitochondrion-associated cysteine-rich protein (Smcp) gene. Molecular and cellular biology. 2002;22:3046–3052. doi: 10.1128/MCB.22.9.3046-3052.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard T, Balogh R, Overbeek P, Bernstein KE. Sperm-specific expression of angiotensin-converting enzyme (ACE) is mediated by a 91-base-pair promoter containing a CRE-like element. Molecular and cellular biology. 1993;13:18–27. doi: 10.1128/MCB.13.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, et al. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5755–5760. doi: 10.1073/pnas.0408718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Developmental cell. 2002;2:819–830. doi: 10.1016/S1534-5807(02)00165-X. [DOI] [PubMed] [Google Scholar]
- 39.Saitou M, et al. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. J Cell Biol. 1997;138:181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenbaum MP, Ma L, Matzuk MM. Conversion of midbodies into germ cell intercellular bridges. Developmental biology. 2007;305:389–396. doi: 10.1016/j.ydbio.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yabuta Y, et al. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol. 2011;192:781–795. doi: 10.1083/jcb.201009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Current biology: CB. 2009;19:630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng H, et al. Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6852–6857. doi: 10.1073/pnas.0701669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denny P, Swift S, Connor F, Ashworth A. An SRY-related gene expressed during spermatogenesis in the mouse encodes a sequence-specific DNA-binding protein. Embo J. 1992;11:3705–3712. doi: 10.1002/j.1460-2075.1992.tb05455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wunderle VM, Critcher R, Ashworth A, Goodfellow PN. Cloning and characterization of SOX5, a new member of the human SOX gene family. Genomics. 1996;36:354–358. doi: 10.1006/geno.1996.0474. [DOI] [PubMed] [Google Scholar]
- 47.Kiselak EA, et al. Transcriptional regulation of an axonemal central apparatus gene, sperm-associated antigen 6, by a SRY-related high mobility group transcription factor, S-SOX5. The Journal of biological chemistry. 2010;285:30496–30505. doi: 10.1074/jbc.M110.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–561. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 49.Braun RE. Post-transcriptional control of gene expression during spermatogenesis. Semin Cell Dev Biol. 1998;9:483–489. doi: 10.1006/scdb.1998.0226. [DOI] [PubMed] [Google Scholar]
- 50.De Cesare D, Fimia GM, Sassone-Corsi P. CREM, a master-switch of the transcriptional cascade in male germ cells. J Endocrinol Invest. 2000;23:592–596. doi: 10.1007/BF03343781. [DOI] [PubMed] [Google Scholar]
- 51.Krausz C, Sassone-Corsi P. Genetic control of spermiogenesis: insights from the CREM gene and implications for human infertility. Reprod Biomed Online. 2005;10:64–71. doi: 10.1016/S1472-6483(10)60805-X. [DOI] [PubMed] [Google Scholar]
- 52.Blendy JA, Kaestner KH, Weinbauer GF, Nieschlag E, Schutz G. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature. 1996;380:162–165. doi: 10.1038/380162a0. [DOI] [PubMed] [Google Scholar]
- 53.Beissbarth T, et al. Analysis of CREM-dependent gene expression during mouse spermatogenesis. Molecular and cellular endocrinology. 2003;212:29–39. doi: 10.1016/j.mce.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Martianov I, et al. Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC genomics. 2010;11:530. doi: 10.1186/1471-2164-11-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lardenois A, et al. Fhl5/Act, a CREM-binding transcriptional activator required for normal sperm maturation and morphology, is not essential for testicular gene expression. Reprod Biol Endocrinol. 2009;7:133. doi: 10.1186/1477-7827-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan W. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Molecular and cellular endocrinology. 2009;306:24–32. doi: 10.1016/j.mce.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han, F. et al. Epigenetic Regulation of Sox30 Is Associated with Testis Development in Mice. Plos One9, doi:ARTN e97203 DOI 10.1371/journal.pone.0097203 (2014). [DOI] [PMC free article] [PubMed]
- 58.Esteves SC. Novel concepts in male factor infertility: clinical and laboratory perspectives. J Assist Reprod Genet. 2016;33:1319–1335. doi: 10.1007/s10815-016-0763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Non-invasive sexing of preimplantation stage mammalian embryos. Nature genetics. 1998;19:220–222. doi: 10.1038/893. [DOI] [PubMed] [Google Scholar]
- 60.Spiller CM, et al. Endogenous Nodal signaling regulates germ cell potency during mammalian testis development. Development. 2012;139:4123–4132. doi: 10.1242/dev.083006. [DOI] [PubMed] [Google Scholar]
- 61.Cotton LM, et al. Utilising the resources of the International Knockout Mouse Consortium: the Australian experience. Mammalian genome: official journal of the International Mammalian Genome Society. 2015;26:142–153. doi: 10.1007/s00335-015-9555-1. [DOI] [PubMed] [Google Scholar]
- 62.Chuma S, Nakatsuji N. Autonomous transition into meiosis of mouse fetal germ cells in vitro and its inhibition by gp130-mediated signaling. Developmental biology. 2001;229:468–479. doi: 10.1006/dbio.2000.9989. [DOI] [PubMed] [Google Scholar]
- 63.Hargrave M, Bowles J, Koopman P. In situ hybridization of whole-mount embryos. Methods in molecular biology. 2006;326:103–113. doi: 10.1385/1-59745-007-3:103. [DOI] [PubMed] [Google Scholar]
- 64.Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods in enzymology. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-W. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.