Abstract
The aim of this study was to examine the presence of genes responsible for resistance to quaternary ammonium compounds (QACs) and the association of qac genes with class 1 integrons in Proteus mirabilis isolated from cooked meat products. A total of 52 P. mirabilis isolates (29.2%) were detected from 178 samples, and their minimum inhibitory concentrations (MICs) of benzalkonium chloride (BC) ranged from 4 to >32 μg/mL. The isolates with BC MICs of 24 μg/mL were observed most frequently. PCR assays indicated that mdfA, ydgE/ydgF, qacE, qacEΔ1, emrE, sugE(c), and sugE(p) were commonly present (32.7%–100%) in these isolates, but qacH was less prevalent (3.8%). Five groups of resistance gene cassettes were identified in 10 intI1-positive isolates. An unusual gene cassette array dfrA32-ereA-aadA2 was found in one foodborne isolate of P. mirabilis. Two isolates harbored qacH- and sul3- associated non-classic integrons: aadA2-cmlA1-aadA1-qacH-IS440-sul3 and a new arrangement dfrA32-ereA1-aadA2-cmlA1-aadA1-qacH-IS440-sul3, which is first reported in P. mirabilis. Non-classic class 1 integrons were located on conjugative plasmids of 100 kb in two tested isolates. Our data showed that the QAC resistance genes were commonly present among P. mirabilis isolates from cooked meats and qacH was associated with non-classic class 1 integrons. The creation of transconjugants demonstrated that qacH-associated non-classic class 1 integrons were located on conjugative plasmids and therefore could facilitate the co-dissemination of disinfectant and antimicrobial resistance genes among bacteria, an increasing area of concern.
Keywords: Proteus mirabilis, qac genes, integron, benzalkonium chloride, resistance
Introduction
Proteus mirabilis, widely distributed in the natural environment, is a member of Enterobacteriaceae family. As an opportunistic pathogen, P. mirabilis can cause urinary tract and wound infections. In addition, it can also contaminate meat (Kim et al., 2005; Wong et al., 2013), vegetables (Uzeh et al., 2009), and seafood (González-Rodríguez et al., 2002), and has been associated with food poisoning (Wang et al., 2010). Previous studies focused on the antimicrobial resistance and the distribution of resistance genes among foodborne P. mirabilis (Kim et al., 2005; Wong et al., 2013). Unlike other foodborne bacteria (Zou et al., 2014; Zhang et al., 2016), little data have been reported on the disinfectant resistance and molecular mechanisms underlying resistance in foodborne P. mirabilis. Disinfectants have the ability to co-select for antimicrobial resistance when these traits are genetically linked (Chapman, 2003). Moreover, much evidence has shown that disinfectant and antimicrobial resistance genes can be co-transferred between bacteria via horizontal gene transfer (Zhao et al., 2009; Call et al., 2010; Sáenz et al., 2010; Partridge et al., 2012), which poses a risk to public health.
Benzalkonium chloride (BC), an important representative of quaternary ammonium compounds (QACs), is used extensively as a disinfectant in the food processing environment to prevent the growth of microorganisms and to ensure the microbiological safety of food products. Its widespread use, however, may impose a selective pressure for resistant strains of bacteria (Cruz and Fletcher, 2012). In the past decades, resistance to BC (resistant breakpoints for BC were >30 μg/mL in Sidhu et al. (2002) and ≥20 μg/mL in Meier et al. (2017)) has been reported in many bacterial isolates from food and food processing plants (Sidhu et al., 2002; Meier et al., 2017).
Efflux pumps, QacE, QacEΔ1, QacF, QacG, QacH/I, and SugE(p), which contribute for BC resistance, have been identified in Gram-negative bacteria (Zou et al., 2014). They are members of the small multidrug resistance (SMR) family and are generally located on mobile genetic elements, such as integrons and plasmids (Zou et al., 2014). qac are closely associated with class 1 integrons. qacE is located in the 3′-conserved segment (CS) of class 1 integrons and qacEΔ1, a deletion derivative of qacE, confers increased resistance to BC (Kazama et al., 1998). qacF shows a high degree of similarity (67.8% identity) with qacE. Class 1 integrons carrying qacG have been found in Gram-negative bacteria (Chu et al., 2001; Partridge et al., 2012). qacH, which was first identified in Staphylococcus (Heir et al., 1998), has been observed frequently in Gram-negative bacteria (Hegstad et al., 2010) and it confers a broader resistance phenotype compared with qacG (Heir et al., 1998). qacH is usually recognized as an important component of class 1 integron that lacks the normal 3′-CS region (i.e., a “non-classic class 1 integron”), which has been detected in many species of Enterobacteriaceae (Antunes et al., 2007; Chang et al., 2009, 2011; Sáenz et al., 2010; Farkas et al., 2016). Several studies found qacH gene and β-lactamase genes (blaIMP-15, blaGES-1, blaGES-5, and blaOXA-2) linked to class 1 integrons from Pseudomonas aeruginosa clinical isolates (Garza-Ramos et al., 2008, 2010; Viedma et al., 2009). qacH-aadB has been reported from environmental bacteria, including Paracoccus versutus, Brevundimonas diminuta, Brachymonas denitrificans, Stenotrophomonas acidaminiphila, and Psychrobacter spp. (Li et al., 2009) and qacH-aadA8 from Vibrio cholerae (Ceccarelli et al., 2006). Additionally, qacH located on a novel transposon Tn6188 was found in Listeria monocytogenes (Müller et al., 2013). In several studies, qacH in Enterobacteriaceae was renamed as qacI to distinguish it from qacH from Staphylococcus (Naas et al., 2001; Curiao et al., 2011). For this paper, we still use the gene name “qacH”. qacH exhibits 91.6% similarity with the sequence of qacF. Finally, sugE(p) is frequently present on multidrug resistance plasmids, that have been reported in Escherichia coli and Salmonella (Zhao et al., 2009; Call et al., 2010).
Five chromosome-encoded efflux pump genes (sugE(c), emrE, mdfA, and ydgE/ydgF) have been reported to confer resistance to BC (Bay and Turner, 2009; Zou et al., 2014). In addition to mdfA encoding a multidrug resistance efflux pump belonging to the major facilitator superfamily (MFS), the remaining genes encode the SMR family efflux pumps.
The aims of this study were to assess the BC resistance and investigate the presence of disinfectant resistance genes and the association of qac genes with class 1 integrons among the P. mirabilis isolates from cooked meat products in China.
Materials and Methods
P. mirabilis Isolates
Between January and September 2015, 178 samples (250 g) of cooked meat products, including roasted meats (n = 67) and sauced meats (n = 111), were purchased from supermarkets and cooked meat shops in Xinxiang, a city of Henan. Six local supermarkets and ten cooked meat shops were randomly selected and each site was visited once. Thirteen samples were collected from each supermarket and ten samples from each cooked meat shops. Samples were transported to the laboratory in an icebox and were processed immediately for bacterial isolation. According to the previous study, the traditional Salmonella isolation protocol was modified to isolate P. mirabilis (Wong et al., 2013). Briefly, a rinse was performed by adding 25 g of sample to 225 mL of buffer peptone water (BPW; Huankai Ltd., Guangzhou, Guangdong, China) in sterile lateral filter bags with thorough mixing by using a homogenizer (BagMixer lab blender 400; Interscience, Saint-Nom-La-Breteche, France). These samples were then incubated at 37°C for 18 h. Pre-enriched sample (1 mL) was inoculated into 10 mL of tetrathionate broth base (TTB; Huankai) and incubated at 42°C for 24 h. A loop of inoculum was streaked onto xylose lysine deoxycholate agar (XLD; Huankai) and incubated for 24 h at 37°C. Three to four Salmonella-like colonies (pink with or without black center on XLD; and yellow with or without black center colonies on XLD were also considered as suspected colonies) were picked and re-streaked on nutrient agar (NA). The isolates with swarming phenotype were identified by using the API 20E bacterial identification system (BioMerieux, Marcy l’Etoile, France). All isolates designated as P. mirabilis were additionally analyzed by PCR-based 16S rDNA sequencing using a pair of universal primers 27F/1492R (Table 1). Only one isolate from each sample was selected for further characterization.
Table 1.
Primers used in this study.
| Target gene(s) or region | Primer | Sequence (5′–3′) | Annealing temperature (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| 16S rDNA | 27F | AGAGTTTGATCCTGGCTCAG | 55 | 1466 | Moreno et al., 2002 |
| 1492R | GGTTACCTTGTTACGACTT | ||||
| qacE | qacE-F | AGCCCCATACCTACAAAG | 55 | 194 | Gillings, 2010 |
| qacE-R | AGCTTGCCCCTTCCGC | ||||
| qacEΔ1 | qacEΔ1-F | AAGTAATCGCAACATCCG | 49 | 140 | Jiang et al., 2017 |
| qacEΔ1-R | ATAAGCAACACCGACAGG | ||||
| qacF | qacF-F | TTCCTTCCGTTGTAGTTGT | 52 | 228 | Jiang et al., 2017 |
| qacF-R | CCTTGGATAGCAGGTTTAG | ||||
| qacG | qacG-F | GTCGCTGACACTCAAATCG | 52 | 133 | Jiang et al., 2017 |
| qacG-R | GACACCAACAAATCCCCAC | ||||
| qacH | qacH-F | TTTGGTGAGGTCGTCGCA | 54 | 162 | Jiang et al., 2017 |
| qacH-R | GCCAGCCCAAACAGCATA | ||||
| sugE(p) | sugE(p)-F | CAATCGCCCAGACAACTT | 51 | 103 | Jiang et al., 2017 |
| sugE(p)-R | GCAAACGCTTCTTTCACC | ||||
| sugE(c) | sugE(c)-F | CTGCTGGAAGTGGTATGGG | 55 | 226 | Zou et al., 2014 |
| sugE(c)-R | GCATCGGGTTAGCGGACT | ||||
| emrE | emrE-F | CCTGTTATGGGCGGTAGAC | 54 | 310 | Jiang et al., 2017 |
| emrE-R | TTCGTGCTCACCTTTCCTT | ||||
| mdfA | mdfA-F | GTCAGGCGTTACTTTTCC | 54 | 596 | Jiang et al., 2017 |
| mdfA-R | GTCACGACCGAGTTCTTT | ||||
| ydgE | ydgE-F | GGCAATCGTGCTGGAAAT | 54 | 184 | Jiang et al., 2017 |
| ydgE-R | GGCGGCAATACCAAACCC | ||||
| ydgF | ydgF-F | ATTACCTTGTTTAGCGTTTT | 49 | 139 | Jiang et al., 2017 |
| ydgF-R | GGTTCACCTCCAGTTCAG | ||||
| intI1 | intI1-F | ACGAGCGCAAGGTTTCGGT | 50 | 565 | Li et al., 2006 |
| intI1-R | GAAAGGTCTGGTCATACATG | ||||
| qacEΔ1-sul1 region | qac-F | TAGCGAGGGCTTTACTAAGC | 55 | 632 | Jiang et al., 2017 |
| sul1-R | CAGTCCGCCTCAGCAATATC | ||||
| variable region | InF | GGCATACAAGCAGCAAGC | 52 | Sandvang et al., 1998 | |
| InB | AAGCAGACTTGACCTGAT | ||||
| aadA2 | aadA2-F | CATCCCGTGGCGTTATCC | 56 | 370 | Jiang et al., 2017 |
| aadA2-R | CTGGGCAGGTAGGCGTTT | ||||
| cmlA1 | cmlA1-F | CGCCACGGTGTTGTTGTTAT | 57 | 694 | Jiang et al., 2017 |
| cmlA1-R | TTGCCTGCCCATCATTAGTC | ||||
| aadA1 | aadA1-F | CGTAAGGCTTGATGAAACA | 52 | 141 | Jiang et al., 2017 |
| aadA1-R | GGATAACGCCACGGAATG | ||||
| IS440 | IS440-F | TATTCCGATTGAACACCTT | 53 | 299 | Jiang et al., 2017 |
| IS440-R | TATTGGCGTTGATTACAGC | ||||
| sul3 | sul3-F | CGAGATTTCACATCGGTTCC | 50 | 208 | Jiang et al., 2017 |
| sul3-R | TTGCTGCTTTAGTTGAGGCT |
Determination of Minimum Inhibitory Concentrations (MICs) for BC
The MICs of BC for P. mirabilis were determined using the agar dilution method recommended by the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute, 2012). BC was tested in concentration range of 4–32 μg/mL. P. mirabilis suspensions were adjusted to a turbidity equivalent to that of a 0.5 McFarland standard with sterilized saline solution (0.9%) and delivered to the Mueller–Hinton (MH; Huankai) agar containing different concentrations of BC (Aladdin Biochemical Technology Co., Ltd., Shanghai, China). The plates were incubated at 37°C for 24 h. The lowest concentration of BC that prevented growth was considered as the MIC. Each of the tests was done in triplicate. In cases in which not all three replicates had the same results, the MICs were determined once more. Escherichia coli ATCC 10536 (a gift from Lijun Zhou, Navy General Hospital, Beijing, China) was used as a quality control strain (the MIC of this strain for BC was 16 μg/mL).
Detection of QAC Resistance Genes
All isolates were screened by PCR for the presence of qac genes, including qacE, qacEΔ1, qacF, qacG, qacH, sugE(p), sugE(c), emrE, mdfA, and ydgE/ydgF (Table 1). Colonies were transferred to an Eppendorf tube filled with water and boiled to prepared DNA template (Wang et al., 2008). The PCR mixture consisted of 2.5 μL of boiled lysate DNA, 0.6 μM (each) primer, 200 μM deoxynucleoside triphosphate (Takara Bio Inc., Otsu, Shiga, Japan), 1× PCR buffer (Takara), 0.5 U Taq DNA polymerase (Takara) in a total volume of 25 μL. The PCR conditions were as follows: initial denaturation at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing at different temperatures (between 49 and 55°C depending on the primer set) for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 7 min. The purified PCR products were sequenced and DNA sequence data were analyzed using the BLAST program1.
Characterization of Class 1 Integrons
All isolates were screened for intI1 (Table 1). Because the occurrence of sulfonamide resistance gene (sul1) and quaternary ammonium compounds resistance gene (qacEΔ1) is often associated with classic class 1 integrons, presence of qacEΔ1-sul1 region in all intI1-positive isolates was tested by using the primers qac-F and sul1-R (Table 1). The presence of gene cassettes in variable regions were characterized by PCR using specific primers (Table 1). Primers InF and InB were used for classic class 1 gene cassettes amplifications; and primers InF and aadA2-R were used for non-classic class 1 integron amplifications. To determine the genetic structure of non-classic class 1 integrons, a wide variety primers were designed. PCR “primer-walking” strategy was used to amplify overlapped individual fragments using Takara LA Taq DNA polymerase to get the complete arrangement. All the obtained amplicons were sequenced on both strands.
Genetic Locations of qacH-Associated Class 1 Integrons
Plasmid DNA was isolated from two qacH-positive isolates using TIANpure Mini Plasmid Kit (TIANGEN Biotech Co., Ltd., Beijing, China). To determine the number and size of plasmids, genomic DNA from qacH-positive isolates was performed by S1 nuclease (Takara) digestion prior to PFGE. S1-PFGE fragments were transferred onto membranes (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) by Southern blot and hybridized with specific probes of intI1 and qacH genes. The probes were made with the DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche Applied Science, Mannheim, Germany).
Conjugation Experiments
The transfer of plasmids carrying qacH-associated class 1 integrons was studied by performing conjugation experiments as described previously (Koo and Woo, 2011). E. coli J53Azr (a sodium azide resistant strain) was used as the recipient and isolates containing non-classic class 1 integron severed as donors. Briefly, donor and recipient cells were mixed with each other at 1:10 ratio and incubated at 37°C overnight. Transconjugants were selected on trypticase soy agar (TSA; Huankai) plates containing sodium azide (150 μg/mL; Sinopharm Chemical Reagent Co., Ltd., Shanghai, China), streptomycin (50 μg/mL; Sigma–Aldrich, St. Louis, MO, United States) and chloramphenicol (16 μg/mL; Sigma–Aldrich). PCR was used to confirm that the transconjugants carried the same resistance genes as their donors.
Statistical Analysis
The statistical package SPSS 15.0 (SPSS Inc., Chicago, IL, United States) was used, and the two-tailed paired Student’s t-test was applied to determine the significance of differences. A P-value < 0.05 was considered statistically significant for comparisons.
Nucleotide Sequence Accession Numbers
The nucleotide sequences of the qacH-carrying integrons have been submitted to GenBank under accession numbers KY662007 for aadA2-cmlA1-aadA1-qacH-IS440-sul3 and KY426918 for dfrA32-ereA1-aadA2-cmlA1-aadA1-qacH-IS440-sul3.
Results
Isolation of P. mirabilis
Colony characteristics of P. mirabilis on XLD agar were similar to those of Salmonella. In this study, Salmonella-like colonies were found in 88 samples (Supplementary Table S1). Among these samples, fifty-six samples were positive for isolates with swarming phenotype (Supplementary Table S1). These suspected P. mirabilis isolates were identified by the biochemical tests and PCR-based 16S rDNA sequencing. Fifty-two samples were positive for P. mirabilis, five samples were positive for P. vulgaris, and one sample was positive for Salmonella (Supplementary Table S1).
Susceptibility of P. mirabilis Isolates to BC
A total of 52 (29.2%) P. mirabilis isolates were recovered from 178 cooked meat samples (Supplementary Table S1). For these isolates, the MICs of BC ranged from 4 to >32 μg/mL (Figure 1 and Supplementary Table S2). The isolates with BC MICs of 24 μg/mL (n = 17, accounting for 32.7% of all isolates) were observed most frequently, followed by the isolates with MICs of 32 μg/mL (n = 11, 21.1%).
FIGURE 1.
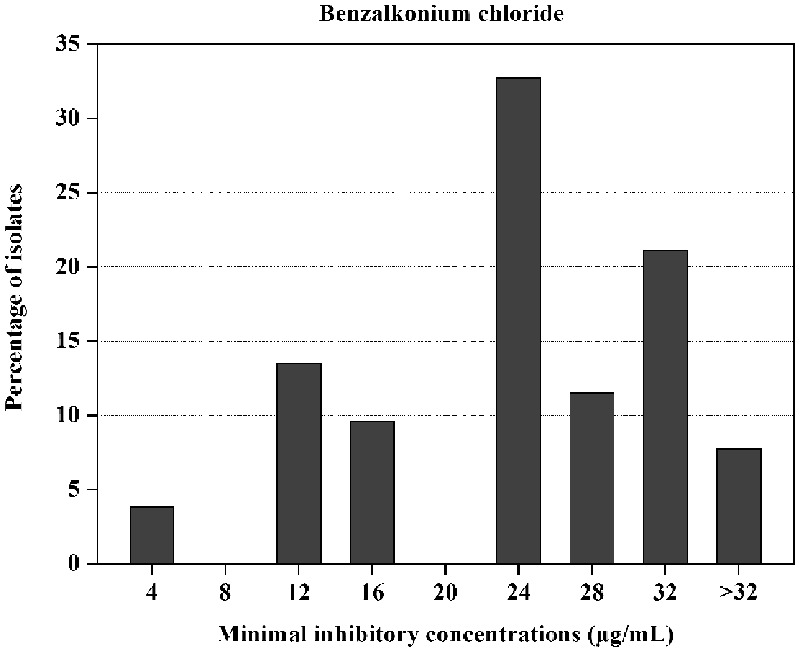
Distribution of the MICs of benzalkonium chloride for 52 Proteus mirabilis isolates.
Presence of QAC Resistance Genes
The presence of QAC resistance genes among P. mirabilis isolates are presented in Supplementary Table S2. The mdfA gene was the most widespread QAC resistance gene, being found in 100% of isolates, followed by ydgE/ydgF (90.4%, n = 47), qacE (53.8%, n = 28), qacEΔ1 (53.8%, n = 28), emrE (44.2%, n = 23), sugE(c) (40.4%, n = 21), sugE(p) (32.7%, n = 17), and qacH (3.8%, n = 2). The qacF and qacG genes were not detected in any of the isolates. The top two QAC resistance genotypes were qacE-qacEΔ1-mdfA-ydgE/ydgF (26.9%) and mdfA-ydgE/ydgF (13.5%) (Table 2).
Table 2.
Different resistance gene combinations in Proteus mirabilis isolates.
| Gene combination | Number of isolates with MIC (μg/mL) as follows |
Total | ||||||
|---|---|---|---|---|---|---|---|---|
| 4 | 12 | 16 | 24 | 28 | 32 | >32 | ||
| qacE-qacEΔ1-mdfA-ydgE/ydgF | 4 | 1 | 9 | 14 | ||||
| mdfA-ydgE/ydgF | 2 | 3 | 2 | 7 | ||||
| sugE(p)-qacE-qacEΔ1-emrE-mdfA-ydgE/ydgF | 1 | 1 | 4 | 6 | ||||
| sugE(c)-sugE(p)-qacE-qacEΔ1-emrE-mdfA-ydgE/ydgF | 2 | 2 | 4 | |||||
| sugE(c)-qacE-qacEΔ1-emrE-mdfA-ydgE/ydgF | 3 | 1 | 4 | |||||
| emrE-mdfA-ydgE/ydgF | 2 | 1 | 1 | 4 | ||||
| sugE(c)-sugE(p)-mdfA | 1 | 3 | 4 | |||||
| sugE(c)-emrE-mdfA-ydgE/ydgF | 2 | 1 | 3 | |||||
| sugE(c)-sugE(p)-emrE-mdfA-ydgE/ydgF | 2 | 2 | ||||||
| sugE(c)-mdfA-ydgE/ydgF | 1 | 1 | 2 | |||||
| sugE(c)-qacH-mdfA-ydgE/ydgF | 1 | 1 | ||||||
| sugE(c)-sugE(p)-qacH-mdfA | 1 | 1 | ||||||
Our results showed that qacE always occurred simultaneously with qacEΔ1, but the MICs of BC were not significantly different (P > 0.05) between qacE-qacEΔ1-positive and -negative isolates. The presence of sugE(p) was significantly associated with the higher MICs of BC (P < 0.05). Among the 17 sugE(p)-positive isolates, 82.4% (n = 14) had the MICs of ≥32 μg/mL. High MICs of BC (>32 μg/mL) were also observed in the two isolates that carried qacH.
Genetic Structure of Class 1 Integrons
The 52 P. mirabilis isolates were subjected to the PCR screening for the expected integrase gene, and the 565-bp corresponding amplicon was detected in 10 isolates, consistent with the presence of the class 1 integrase gene. Five groups of resistance gene cassettes, named as type I-V, were identified in these isolates (Figure 2), including: dfrA17-aadA5, dfrA5, dfrA1-orfC, dfrA32-ereA1-aadA2, and aadA2 (Table 3).
FIGURE 2.
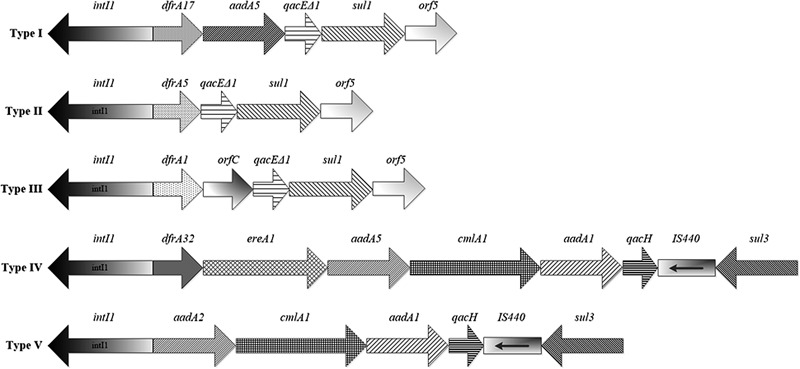
Schematic representation of the variable regions of class 1 integrons identified in P. mirabilis isolates. Integron type with different cassette arrays (Type I to V) are arranged as identified in Table 3.
Table 3.
Characteristics of isolates carrying integrons.
| Isolate | Class 1 integrons |
||||
|---|---|---|---|---|---|
| Size (bp) | Gene cassette | Typea | qacEΔ1-sul1 | Classic or non-classicb | |
| PM1 | 1664 | dfrA17-aadA5 | I | + | Classic |
| PM4 | 2900 | dfrA32-ereA-aadA2 | IV | - | Non-classic |
| PM7 | 721 | dfrA5 | II | + | Classic |
| PM9 | 1664 | dfrA17-aadA5 | I | + | Classic |
| PM13 | 1664 | dfrA17-aadA5 | I | + | Classic |
| PM19 | 792 | aadA2 | V | - | Non-classic |
| PM21 | 1163 | dfrA1-orfC | III | + | Classic |
| PM24 | 721 | dfrA5 | II | + | Classic |
| PM35 | 1664 | dfrA17-aadA5 | I | + | Classic |
| PM39 | 1664 | dfrA17-aadA5 | I | + | Classic |
aType I-V represented five different integron structures in our study (Figure 2).
bIn this study, class 1 integrons with qacEΔ1 and sul1 genes in their 3′-CS were named as classic class 1 integrons. Accordingly class 1 integrons without the normal 3′-CS were named as non-classic class 1 integrons.
Analysis of Plasmids Carrying qacH-Associated Class 1 Integrons
According to the bands obtained by S1-PFGE of qacH-positive isolates, the number and size of their plasmids were determined (Figure 3). PM4 contained a plasmid of 100 kb and PM19 contained two large plasmids of 100 and 150 kb. Both specific probes for intI1 and qacH genes hybridized with the plasmids of 100 kb in the two isolates (Figure 3). Two transconjugants named PM4T and PM19T were obtained after conjugation experiments. Both of them exhibited resistance to streptomycin and chloramphenicol. However, increased MICs of BC were not observed in transconjugants (data not shown). PCR experiments confirmed that the transconjugants harbored the same gene structure of integrons as their donors (PM4T with dfrA32-ereA1-aadA2-cmlA1-aadA1-qacH-IS440-sul3 and PM19T with aadA2-cmlA1-aadA1-qacH-IS440-sul3). All the above indicated results confirmed that qacH-associated class 1 integrons in the two studied isolates were located on conjugative plasmids of 100 kb.
FIGURE 3.
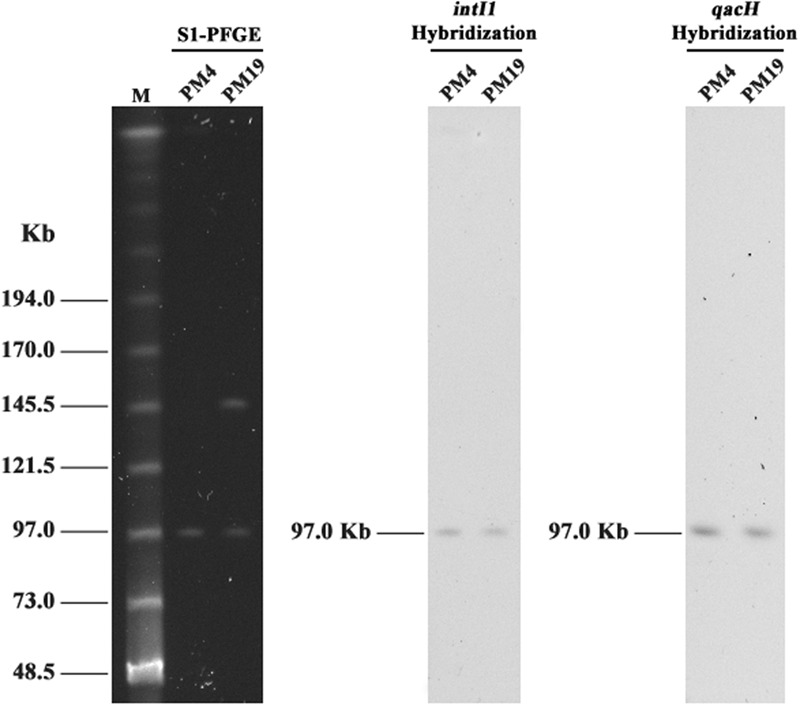
S1-PFGE and southern hybridization of intI1 and qacH on two qacH-positive isolates. M, MidRange PFG Marker; PM4 and PM19 are qacH-positive P. mirabilis isolates.
Discussion
In the present study, fifty-two isolates of P. mirabilis were isolated from cooked meat samples. According to Wong et al. (2013), the rate of P. mirabilis in raw chicken carcass samples in Hong Kong was 86.2% (50/58), which was much higher than that (29.2%) in our study. Cooking procedures kill most microorganisms that colonize raw meats, however, survival of microorganisms due to improper processing or cross-contamination of food after cooking may occur. This could be an explanation for the lower incidence of P. mirabilis in cooked meat products compared to raw meat samples.
Proteus mirabilis isolates in our study showed the MICs of BC ranging from 4 to >32 μg/mL, with MICs of 24 μg/mL most frequently. As there is no standard resistant breakpoint of BC for P. mirabilis, it is difficult to classify the isolates as BC susceptible, intermediate or resistance in this study. Although data on the BC susceptibility in P. mirabilis were scarce, several studies have reported the MICs of BC in Enterobacteriaceae isolates from different sources (Sidhu et al., 2002; Aarestrup and Hasman, 2004; Zhang et al., 2016). In the study of Zhang et al. (2016), E. coli isolated from retail meat showed the MICs of BC in the range of 16–64 μg/mL. Aarestrup and Hasman (2004) have shown that the MICs of BC for Salmonella (n = 156) and E. coli (n = 202) from food animals ranged from 64 to 256 μg/mL and from 16 to 128 μg/mL, respectively. Compared with the studies mentioned above, P. mirabilis isolates in our study showed relatively lower MICs to BC. In another survey of Enterobacteriaceae from food, almost half of the isolates exhibited the MICs of BC with <10 μg/mL (Sidhu et al., 2002). Actually, the user concentrations of BC in food industry are usually 200–1000 μg/mL (Møretrø et al., 2017), which are much higher than the MICs of P. mirabilis in this study. In practical application, BC is commonly rinsed from surfaces (equipment, machines, floor etc.) with water after disinfection. This rinsing step, however, is not sufficient to remove all BC residues from surfaces and consequently, bacteria are likely to be exposed to diluted, sub-lethal BC concentrations (Buffet-Bataillon et al., 2012). Repeated exposure to sub-lethal BC concentrations may facilitate the development of resistance (Buffet-Bataillon et al., 2012). Therefore, it was not surprising that P. mirabilis isolates in the present study exhibited low-level of BC MICs.
In this study, the presence of QAC resistance genes was investigated. Our results showed that mdfA and ydgE/ydgF were the most prevalent among P. mirabilis, which was in agreement with the similar studies of E. coli (Zou et al., 2014; Zhang et al., 2016). Among the isolates tested in our study, qacE always occurred simultaneously with qacEΔ1. It was noted that the presence of sugE(p) was significantly associated with the higher MICs of BC (P < 0.05). Two qacH-positive isolates also exhibited relatively high MICs of BC (>32 μg/mL). In previous research, the higher MICs of BC were associated with plasmid-encoded genes (Zou et al., 2014). Because each isolate harbored more than one QAC resistance gene, it is difficult to assess what level of BC resistance was contributed by each QAC resistance gene.
A previous study reported that aadA1 gene cassette was observed most commonly among the P. mirabilis isolates from retail meat products (Kim et al., 2005). In contrast, our results showed that dfrA17-aadA5 was the most common cassette array, which is similar to other reports of foodborne P. mirabilis isolates in China (Shen et al., 2011). Interestingly, an uncommon integron gene cassette array dfrA32-ereA1-aadA2 was found in one P. mirabilis isolate in this study. To the best of our knowledge, this is the first report of dfrA32-ereA1-aadA2 in foodborne P. mirabilis. The integron gene cassette array dfrA32-ereA1-aadA2 in our study showed 99.6% identity to that of Salmonella enterica (Krauland et al., 2010; GenBank accession number GU067642), 99.7% identity to that of Laribacter hongkongensis (Feng et al., 2011; GenBank accession number GU726907) and 99.7% identity to that of Aeromonas hydrophila (unpublished; GenBank accession number KJ543558). Recently, this cassette array was detected in clinical P. mirabilis isolates in Zhejiang Province of China (Wei et al., 2014), which had 99.6% identity to our sequence. The high similarity of dfrA32-ereA1-aadA2 indicated that the class 1 integrons carrying this cassette array may be located on conjugative plasmids.
Classic class 1 integrons are composed of qacEΔ1 and sul1 genes in their 3′-CS. Recently, class 1 integrons without the typical 3′-CS have been found in some species of Enterobacteriaceae, including Escherichia coli, Salmonella, Shigella sonnei and Klebsiella pneumoniae (Antunes et al., 2007; Chang et al., 2009, 2011; Sáenz et al., 2010). In these previous studies, the 3′-CS of integrons was associated with qacH and sul3. Xu et al. (2009) reported that two clinical isolates of Pseudomonas aeruginosa carried a class 1 integron lacking 3′-CS, however, the genetic structure of these non-classic integrons was unclear. In our study, non-classic class 1 integrons lacking qacEΔ1 and sul1 genes were found in 2 of the 10 intI1-positive isolates (PM4 with dfrA32-ereA1-aadA2 and PM19 with aadA2). PCR amplifications revealed that the gene cassettes in the two isolates were followed by an unusual 3′-CS linked to qacH and sul3. The gene cassette organization dfrA32-ereA1-aadA2-cmlA1-aadA1-qacH-IS440-sul3 and aadA2-cmlA1-aadA1-qacH-IS440-sul3 were identified in PM4 and PM19, respectively. Both of them contained cmlA1 coding for chloramphenicol resistance, aadA1 coding for streptomycin resistance, qacH coding for QAC resistance, insertion element IS440, and sul3 coding for sulfonamide resistance. It seems that sul3 had replaced sul1 in addition to the replacement of qacEΔ1 by qacH in the 3′-CS region. Although the gene structure cmlA1-aadA1-qacH-IS440-sul3 was commonly found in non-classic integrons (Antunes et al., 2007; Sáenz et al., 2010), the combination of dfrA32-ereA1-aadA2-cmlA1-aadA1-qacH-IS440-sul3 was reported for the first time. This novel arrangement detected in P. mirabilis isolate from food has been included in GenBank with the accession number KY426918. A similar gene cassette array of qacH-dfrA32-ereA1-aadA2-cmlA1-aadA1 was detected in Salmonella enterica serovar Stanley isolates from Taiwan (Krauland et al., 2010). Both of the two arrays carried the same gene cassettes, but the position of qacH was different. PM4 and PM19 had similar qacH-IS440-sul3-integron platforms with different gene cassette arrays, suggesting evolution of the genetic background by different recombinatorial events.
Our results demonstrated that non-classic class 1 integrons were located on conjugative plasmids of 100 kb in two tested isolates. Previous studies showed that the class 1 integrons lacking the normal 3′-CS and associated with qacH and sul3 genes were usually located on large plasmids of different size (between 70 and 240 kb) and the conjugative plasmids of 100 kb were the most disseminated among the different isolates (Antunes et al., 2007; Sáenz et al., 2010). Pal et al. (2015) also found that plasmids with co-selection potential for resistance to disinfectants and antimicrobials tended to be large and conjugative. Conjugation experiments confirmed that the disinfectant and antimicrobial resistance genes of non-classic class 1 integrons in PM4 and PM19 were co-existed on the same conjugative plasmids and could be co-transferred to E. coli. When theses isolates are exposed to QACs, selection pressure from disinfectants could increase risks for the spread of QAC and antimicrobial resistance genes among the bacteria. Notably, two transconjugants containing qacH showed the same MICs of BC as the recipient in this study. Similar observations that the presence of QAC resistance genes didn’t increase the MICs of BC in transconjugants has also been reported in a previous study (Zhang et al., 2016). There was the possibility that the agar dilution method used for susceptibility testing was not sensitive enough to detect the differences of BC MICs between the recipient and transconjugants.
Conclusion
Quaternary ammonium compounds resistance genes, including mdfA, ydgE/ydgF, qacE, qacEΔ1, emrE, sugE(c), and sugE(p) were found in foodborne P. mirabilis isolates in this study. Our data demonstrated the presence of non-classic class 1 integrons with the gene structure qacH-IS440-sul3 among the isolates. Moreover, qacH-associated non-classic class 1 integrons were located on conjugative plasmids and therefore could constitute an effective way for co-dissemination of antimicrobial and disinfectant resistance genes.
Author Contributions
TY, HW, and LS designed and supervised the study. XJ, LL, YL, and KZ performed the experiments. XJ analyzed data. XJ and TY drafted the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Minggui Wang, Huashan Hospital, Fudan University, for kindly providing the E. coli J53Azr.
Funding. This work was supported by the National Natural Science Foundation of China (31601568), the Key Project of Natural Science of the Education Department of Henan Province, China (15A180006 and 16A180011), the Key Scientific and Technological Project of Xinxiang (ZG15007) and the Doctoral Scientific Research Foundation of Henan Normal University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02417/full#supplementary-material
References
- Aarestrup F. M., Hasman H. (2004). Susceptibility of different bacterial species isolated from food animals to copper sulphate, zinc chloride and antimicrobial substances used for disinfection. Vet. Microbiol. 100 83–89. 10.1016/j.vetmic.2004.01.013 [DOI] [PubMed] [Google Scholar]
- Antunes P., Machado J., Peixe L., Mora D. (2007). Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3′ conserved sequence region among Salmonella isolates. Antimicrob. Agents Chemother. 51 1545–1548. 10.1128/AAC.01275-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay D. C., Turner R. J. (2009). Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 9:140. 10.1186/1471-2148-9-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffet-Bataillon S., Tattevin P., Bonnaure-Mallet M., Jolivet-Gougeon A. (2012). Emergence of resistance to antibacterial agents: the role of quaternary ammonium compounds-a critical review. Int. J. Antimicrob. Agents 39 381–389. 10.1016/j.ijantimicag.2012.01.011 [DOI] [PubMed] [Google Scholar]
- Call D. R., Singer R. S., Meng D., Broschat S. L., Orfe L. H., Anderson J. M., et al. (2010). blaCMY -2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54 590–596. 10.1128/AAC.00055-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D., Salvia A. M., Sami J., Cappuccinelli P., Colombo M. M. (2006). New cluster of plasmid-located class 1 integrons in Vibrio cholerae O1 and a dfrA15 cassette-containing integron in Vibrio parahaemolyticus isolated in Angola. Antimicrob. Agents Chemother. 50 2493–2499. 10.1128/AAC.01310-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Fang Y., Tsai S., Chang L., Yu W. (2009). Characterization of class 1 integrons and gene cassettes in clinical isolates of Klebsiella pneumoniae from Taiwan. Diagn. Microbiol. Infect. Dis. 65 214–216. 10.1016/j.diagmicrobio.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Chang C., Lu P., Lin C., Lee T., Tsai M., Chang L. (2011). Integron types, gene cassettes, antimicrobial resistance genes and plasmids of Shigella sonnei isolates from outbreaks and sporadic cases in Taiwan. J. Med. Microbiol. 60 197–204. 10.1099/jmm.0.022517-0 [DOI] [PubMed] [Google Scholar]
- Chapman J. S. (2003). Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegrad. 51 271–276. 10.1016/S0964-8305(03)00044-1 [DOI] [Google Scholar]
- Chu Y. W., Afzal-Shah M., Houang E. T., Palepou M. I., Lyon D. J., Woodford N., et al. (2001). IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45 710–714. 10.1128/AAC.45.3.710-714.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2012). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement 2012 Vol. 32 Wayne, IL: CLSI [Google Scholar]
- Cruz C. D., Fletcher G. C. (2012). Assessing manufacturers’ recommended concentrations of commercial sanitizers on inactivation of Listeria monocytogenes. Food Control 26 194–199. 10.1016/j.foodcont.2012.01.041 [DOI] [Google Scholar]
- Curiao T., Cantón R., Garcillán-Barcia M. P., de la Cruz F., Baquero F., Coque T. M. (2011). Association of composite IS26-sul3 elements with highly transmissible IncI1 plasmids in extended-spectrum-β-lactamase-producing Escherichia coli clones from humans. Antimicrob. Agents Chemother. 55 2451–2457. 10.1128/AAC.01448-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas A., Crăciunaş C., Chiriac C., Szekeres E., Coman C., Butiuc-Keul A. (2016). Exploring the role of coliform bacteria in class 1 integron carriage and biofilm formation during drinking water treatment. Microb. Ecol. 72 773–782. 10.1007/s00248-016-0758-0 [DOI] [PubMed] [Google Scholar]
- Feng J. L., Yan H., Chowdhury N., Neogi S. B., Yamasaki S., Shi L., et al. (2011). Identification and characterization of integron-associated antibiotic resistant Laribacter hongkongensis isolated from aquatic products in China. Int. J. Food Microbiol. 144 337–341. 10.1016/j.ijfoodmicro.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Garza-Ramos J. U., Sanchez-Martinez G., Barajas J. M., Suarez S., Sanchez-Perez A., Rojas-Moreno T., et al. (2010). Variability of the blaIMP-15-containing integrons, highly related to In95, on an endemic clone of Pseudomonas aeruginosa in Mexico. Microb. Drug Resist. 16 191–195. 10.1089/mdr.2010.0017 [DOI] [PubMed] [Google Scholar]
- Garza-Ramos U., Morfin-Otero R., Sader H. S., Jones R. N., Hernández E., Rodriguez-Noriega E., et al. (2008). Metallo-β-lactamase gene blaIMP-15 in a class 1 integron, In95, from Pseudomonas aeruginosa clinical isolates from a hospital in Mexico. Antimicrob. Agents Chemother. 52 2943–2946. 10.1128/AAC.00679-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings M. R. (2010). Biocide use, integrons and novel genetic elements. Microbiol. Aust. 31 192–194. 10.1038/ismej.2008.98 [DOI] [PubMed] [Google Scholar]
- González-Rodríguez M., Sanz J., Santos J., Otero A., García-López M. (2002). Numbers and types of microorganisms in vacuum-packed cold-smoked freshwater fish at the retail level. Int. J. Food Microbiol. 77 161–168. 10.1016/S0168-1605(02)00048-X [DOI] [PubMed] [Google Scholar]
- Hegstad K., Langsrud S., Lunestad B. T., Scheie A. A., Sunde M., Yazdankhah S. P. (2010). Does the wide use of quaternary ammonium compounds enhance the selection and spread of antimicrobial resistance and thus threaten our health. Microb. Drug Resist. 16 91–104. 10.1089/mdr.2009.0120 [DOI] [PubMed] [Google Scholar]
- Heir E., Sundheim G., Holck A. L. (1998). The Staphylococcus qacH gene product: a new member of the SMR family encoding multidrug resistance. FEMS Microbiol. Lett. 163 49–56. 10.1111/j.1574-6968.1998.tb13025.x [DOI] [PubMed] [Google Scholar]
- Jiang X., Xu Y., Li Y., Zhang K., Liu L., Wang H., et al. (2017). Characterization and horizontal transfer of qacH-associated class 1 integrons in Escherichia coli isolated from retail meats. Int. J. Food Microbiol. 258 12–17. 10.1016/j.ijfoodmicro.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Kazama H., Hamashima H., Sasatsu M., Arai T. (1998). Distribution of the antiseptic-resistance gene qacE and qacEΔ1 in Gram-negative bacteria. FEMS Microbiol. Lett. 159 173–178. [DOI] [PubMed] [Google Scholar]
- Kim S., Wei C., An H. (2005). Molecular characterization of multidrug-resistant Proteus mirabilis isolates from retail meat products. J. Food Prot. 68 1408–1413. 10.4315/0362-028X-68.7.1408 [DOI] [PubMed] [Google Scholar]
- Koo H. J., Woo G. J. (2011). Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int. J. Food Microbiol. 145 407–413. 10.1016/j.ijfoodmicro.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Krauland M., Harrison L., Paterson D., Marsh J. (2010). Novel integron gene cassette arrays identified in a global collection of multi-drug resistant non-typhoidal Salmonella enterica. Curr. Microbiol. 60 217–223. 10.1007/s00284-009-9527-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Yang M., Hu J., Zhang J., Liu R., Gu X., et al. (2009). Antibiotic-resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving river. Environ. Microbiol. 11 1506–1517. 10.1111/j.1462-2920.2009.01878.x [DOI] [PubMed] [Google Scholar]
- Li X., Shi L., Yang W., Li L., Yamasaki S. (2006). New array of aacA4-catB3-dfrA1 gene cassettes and a noncoding cassette from a class-1-integron-positive clinical strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50 2278–2279. 10.1128/AAC.01378-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A. B., Guldimann C., Markkula A., Pöntinen A., Korkeala H., Tasara T. (2017). Comparative phenotypic and genotypic analysis of Swiss and Finnish Listeria monocytogenes isolates with respect to benzalkonium chloride resistance. Front. Microbiol. 8:397. 10.3389/fmicb.2017.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno C., Romero J., Espejo R. T. (2002). Polymorphism in repeated 16S rRNA genes is a common property of type strains and environmental isolates of the genus Vibrio. Microbiology 148 1233–1239. 10.1099/00221287-148-4-1233 [DOI] [PubMed] [Google Scholar]
- Møretrø T., Schirmer B. C. T., Heir E., Fagerlund A., Hjemli P., Langsrud S. (2017). Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int. J. Food Microbiol. 241 215–224. 10.1016/j.ijfoodmicro.2016.10.025 [DOI] [PubMed] [Google Scholar]
- Müller A., Rychli K., Muhterem-Uyar M., Zaiser A., Stessl B., Guinane C. M., et al. (2013). Tn6188-a novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLOS ONE 8:e76835. 10.1371/journal.pone.0076835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Mikami Y., Imai T., Poirel L., Nordmann P. (2001). Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183 235–249. 10.1128/JB.183.1.235-249.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C., Bengtsson-Palme J., Kristiansson E., Joakim Larsson D. G. (2015). Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16:964. 10.1186/s12864-015-2153-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R., Ginn A. N., Paulsen I. T., Iredell J. R. (2012). pEl1573 carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob. Agents Chemother. 56 6029–6032. 10.1128/AAC.01189-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz Y., Vinué L., Ruiz E., Somalo S., Martínez S., Rojo-Bezares B. (2010). Class 1 integrons lacking qacEΔ1 and sul1 genes in Escherichia coli isolates of food, animal and human origins. Vet. Microbiol. 144 493–497. 10.1016/j.vetmic.2010.01.026 [DOI] [PubMed] [Google Scholar]
- Sandvang D., Aarestrup F. M., Jensen L. B. (1998). Characterization of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160 37–41. 10.1111/j.1574-6968.1998.tb12887.x [DOI] [PubMed] [Google Scholar]
- Shen J., Yang B., Xi M., Meng J. (2011). Class I integron and antibiotic resistance of Salmonella and Proteus mirabilis in retail meat from Shaanxi Province. Food Sci. 32 130–134. [Google Scholar]
- Sidhu M. S., Sørum H., Holck A. (2002). Resistance to quaternary ammonium compounds in food-related bacteria. Microb. Drug Resist. 8 393–399. 10.1089/10766290260469679 [DOI] [PubMed] [Google Scholar]
- Uzeh R. E., Alade F. A., Bankole M. (2009). The microbial quality of pre-packed mixed vegetable salad in some retail outlets in Lagos, Nigeria. Afr. J. Food Sci. 3 270–272. [Google Scholar]
- Viedma E., Juan C., Acosta J., Zamorano L., Otero J. R., Sanz F., et al. (2009). Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extend-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob. Agents Chemother. 53 4930–4933. 10.1128/AAC.00900-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Yang Y., Lu Q., Wang Y., Chen Y., Deng L., et al. (2008). Presence of qnr gene in Escherichia coli and Klebsiella pneumoniae resistant to ciprofloxacin isolated from pediatric patients in China. BMC Infect. Dis. 8:68. 10.1186/1471-2334-8-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang S., Yu J., Zhang H., Yuan Z., Sun Y., et al. (2010). An outbreak of Proteus mirabilis food poisoning associated with eating stewed pork balls in brown sauce, Beijing. Food Control 21 302–305. 10.1016/j.foodcont.2009.06.009 [DOI] [Google Scholar]
- Wei Q., Hu Q., Li S., Lu H., Chen G., Shen B., et al. (2014). A novel functional class 2 integron in clinical Proteus mirabilis isolates. J. Antimicrob. Chemother. 69 973–976. 10.1093/jac/dkt456 [DOI] [PubMed] [Google Scholar]
- Wong M. H. Y., Wan H. Y., Chen S. (2013). Characterization of multidrug-resistant Proteus mirabilis isolated from chicken carcasses. Foodborne Pathog. Dis. 10 177–181. 10.1089/fpd.2012.1303 [DOI] [PubMed] [Google Scholar]
- Xu Z., Li L., Shirtliff M. E., Alam M. J., Yamasaki S., Shi L. (2009). Occurrence and characteristics of class 1 and 2 integrons in Pseudomonas aeruginosa isolates from patients in southern China. J. Clin. Microbiol. 47 230–234. 10.1128/JCM.02027-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., He X., Meng Y., Guo L., Long M., Yu H., et al. (2016). Antibiotic and disinfectant resistance of Escherichia coli isolated from retail meats in Sichuan, China. Microb. Drug Resist. 22 80–87. 10.1089/mdr.2015.0061 [DOI] [PubMed] [Google Scholar]
- Zhao S., Blickenstaff K., Glenn A., Ayers S. L., Friedman S. L., Abbott J. W., et al. (2009). β-lactam resistance in Salmonella strains isolated from retail meats in the United States by the National antimicrobial resistance monitoring system between 2002 and 2006. Appl. Environ. Microbiol. 75 7624–7630. 10.1128/AEM.01158-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Meng J., McDermott P. F., Wang F., Yang Q., Cao G., et al. (2014). Presence of disinfectant resistance genes in Escherichia coli isolated from retail meats in the USA. J. Antimicrob. Chemother. 69 2644–2649. 10.1093/jac/dku197 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


