Abstract
Background
MicroRNA (miRNA) expression is correlated with tumor histology, differentiation, invasiveness and treatment outcome. We aimed to identify miRNAs whose differential expression might enable early diagnosis of lung adenocarcinoma in patients presenting with ground-glass nodules (GGNs).
Methods
To identify potential miRNAs of interest, we analyzed the miRNA expression profile of tumor and adjacent non-para-tumor tissue in three participants by next-generation sequencing (NGS). We then assessed the expression levels of the miRNAs of interest in 73 lung adenocarcinomas presenting with GGNs with matched adjacent non-tumor tissue by quantitative real-time polymerase chain reaction (qRT-PCR). We also detected the miRNA panel in 66 lung benign diseases and 66 lung adenocarcinomas presenting with GGN lesion tissues by qRT-PCR. Target genes of our selected miRNA panel were predicted using Miranda with default parameters.
Results
Twenty-three miRNAs showed differential expression between tumor and adjacent non-tumor tissue by NGS. Five miRNAs exhibited higher expression in tumor tissue compared to adjacent non-tumor tissue (P<0.05); 18 miRNAs demonstrated lower expression in tumor tissue versus adjacent non-tumor tissue (P<0.05). When qRT-PCR was performed for the 23 miRNAs identified by NGS in the pilot stage, seven were found to have statistically significant expression in tumor versus adjacent non-tumor tissue (P<0.05). The sensitivity and specificity of seven-miRNA panel were 86.4% and 60.6%, respectively.
Conclusion
The predicted targets of our miRNAs of interest are frequently associated with cancer signaling pathways. We developed a miRNA panel that could potentially predict the presence of lung adenocarcinoma in patients presenting with GGNs.
Keywords: microRNA, miRNA, ground-glass nodules, GGNs, next-generation sequencing, NGS, lung adenocarcinoma, early diagnosis
Introduction
Lung cancer is one of the leading causes of cancer deaths worldwide. Adenocarcinoma represents over 40% of diagnosed lung cancer cases.1,2 At least in part due to the lack of an effective method for early diagnosis, lung adenocarcinoma is usually locally advanced or metastatic when discovered, and the prognosis is poor (5-year survival rate of 15%).2 Therefore, an effective, dependable, non-invasive tool for the early diagnosis of lung cancer would go very far to improve the prognosis and reduce the mortality of patients.
On computed tomography (CT) scan images, pulmonary ground-glass nodules (GGNs) appear as hazy lesions overlying bronchial structures or pulmonary vessels.3 The presence of GGNs, which are being detected with increasing frequency, is strongly suggestive of lung cancer, particularly lung adenocarcinoma. However, GGNs can also represent an entirely benign process.4 The clinical management of pulmonary GGNs remains controversial and complex. There is a crucial need for better biomarkers to identify which patients presenting with GGNs are at high risk for harboring lung adenocarcinoma and therefore require aggressive and speedy intervention to increase the likelihood of successful treatment.
MicroRNAs (miRNAs) are a category of small non-coding RNAs between 18 and 24 nucleotides in length5 that play a role in various pathologic processes including oncogenesis and tumor metastasis. Abnormal miRNA expression and function have been found in multiple human cancers. MiRNAs have the potential both as biomarkers and new drug targets for cancer patients.6–8 For example, differential expression of the precursors of miRNA-155, mature miRNA-15 and miRNA-16 was detected in malignant lymphomas.7 Furthermore, Yanaihara et al10 found 43 aberrantly expressed miRNAs in a comparison of 104 paired primary lung cancer and non-tumor tissues. In the lung cancer samples, expression levels of 28 miRNAs were lower and 15 miRNAs were higher, demonstrating that the comprehensive analysis of miRNA expression can identify miRNAs associated with cancer.
Next-generation sequencing (NGS) is an effective tool for miRNA analysis that quantifies miRNA levels by detecting the exact nucleotide sequence. MiRNA sequencing data for lung cancer has been reported in several studies.6–9 However, this is the first study to report the use of NGS to systematically characterize miRNAs associated with the presence of adenocarcinoma in pulmonary GGNs, which could facilitate earlier diagnosis and more successful treatment of patients with lung cancer.
In this study, we investigated aberrations of miRNAs in resected GGNs to design a miRNA panel that predicts lung adenocarcinoma in patients presenting with GGNs.
Materials and methods
Patients and clinical specimens
Patients enrolled in the study were diagnosed with lung adenocarcinoma and had one or more intrapulmonary pure or partially solid GGNs. In total, 76 pairs of tumor samples from adenocarcinomas located in GGNs and adjacent non-tumor samples were surgically resected from the enrolled patients at Shanghai Pulmonary Hospital between May 2013 and June 2015. A total of 66 lung benign diseases and 66 lung adenocarcinomas presenting with GGNs lesion tissues were surgically resected from the enrolled patients at Shanghai Pulmonary Hospital between September 2014 and June 2015. Immediately following resection, fresh tissue samples were flash frozen with dry ice and then stored at −80°C. All clinical investigations were performed in accordance with the tenets of the Declaration of Helsinki. All participants provided written informed consent to participate prior to inclusion in the study. The protocol was approved by the Shanghai Pulmonary Hospital, Tongji University (issue number 13-786, ethical number 13-768). All participants were competent to provide their consent.
Next-generation sequencing
MiRNA expression in both tumor and adjacent non-tumor tissue was evaluated by NGS in three patients to identify whether it is higher or lower in tumor. Extraction of total RNA was performed by TRIzol solution (Thermo Fisher Scientific), and the RNA quality was measured by Agilent BioAnalyzer2100. To achieve optimal tissue miRNA profiles, we carried out high-throughput NGS. The adaptor sequence was cut by the primer. After the quality inspection and the length screening, the basic sequencing fragments were selected. Subsequently, miRNAs were reverse transcribed and sequenced using a single-ended 50 bp sequencing model on the platform.
To identify miRNAs differentially expressed between lung cancer tissues and normal controls, we applied transcripts per million (TPM) to normalize the expression of miRNA in two groups. Then we calculated fold change (FC) and P-value via double sample t-test, and corrected P-value into false discovery rate (FDR). FDR ≤0.05 and |log2FC| ≥2 were set as the cutoffs to screen out differentially expressed miRNAs.
Validation of the NGS by qRT-PCR
The subset of miRNAs found to be differentially expressed in tumor versus non-tumor analysis by NGS was assayed by quantitative real-time polymerase chain reaction (qRT-PCR) in 73 lung adenocarcinoma patients who presented with GGNs. Total RNA in tumor and adjacent non-tumor tissue was extracted with an RNA Extraction Kit (SLNco, Cinoasia, Shanghai, People’s Republic of China), and cDNA was synthesized by Prime Script RT reagent Kit (TaKaRa Biotechnology, Kusatsu, Japan). Primers for the U6 gene and 23 miRNAs were obtained from Cinoasia (miR-Quant), validated with PRIMER 5.0 (ABI, Foster City, CA, USA) and produced by Generay (Shanghai, People’s Republic of China).
The cDNA was reverse transcribed using the ReverTra Ace® qRT-PCR RT Kit (FSQ-101; Toyobo, Osaka, Japan) according to the manufacturer’s protocol (incubation for 15 minutes at 37°C and then 5 seconds at 85°C). qRT-PCR was performed on a Real time Thermo Cycler (FTC3000; Funglyn, Ontario, Canada) with SYBR Green Real-time qRT-PCR Master Mix (QPK-201; Toyobo) as follows: 5 minutes at 95°C, followed by 45 cycles of 15 seconds at 95°C and 1 minute at 60°C. The specificity of qRT-PCR product was assessed by melting-curve analysis. Relative Ct values were normalized using the U6 Ct value. Data were analyzed with the 2−ΔΔCt formula, where ΔCt = (CTmiRNA − CTU6). Each reaction was performed in triplicates.
Validation of miRNA panel by qRT-PCR
We chose miRNA with significant difference and the same trend for further detection. We also detected the miRNA panel in 66 lung benign diseases and 66 lung adenocarcinomas presenting with GGN lesion tissue by qRT-PCR. Total RNA was extracted with an RNA Extraction Kit (SLNco, Cinoasia), and cDNA was synthesized by Prime Script RT reagent Kit (TaKaRa Biotechnology). Primers for the U6 gene and seven miRNAs were obtained from Cinoasia (miR-Quant), validated with PRIMER 5.0 (ABI) and produced by Generay.
The cDNA was reverse transcribed using the ReverTra Ace® qRT-PCR RT Kit (FSQ-101; Toyobo) kit according to the manufacturer’s protocol (incubation for 15 minutes at 37°C and then 5 seconds at 85°C). qRT-PCR was performed on a Real time Thermo Cycler (FTC3000; Funglyn) with SYBR Green Real-time qRT-PCR Master Mix (QPK-201; Toyobo) as follows: 5 minutes at 95°C, followed by 45 cycles of 15 seconds at 95°C and 1 minute at 60°C. The specificity of qRT-PCR product was assessed by melting-curve analysis. Relative Ct values were normalized using the U6 Ct value. Data were analyzed with the 2−ΔΔCt formula, where ΔCt = (CTmiRNA − CTU6). Each reaction was performed in triplicate.
In silico analysis
The sequencing adapters were clipped from the 3′ end of reads, and short reads of less than 18 bp were filtered. Low quality reads were filtered if the average base quality score was less than 20. The clean reads were aligned to the hg19 reference human genome using Bowtie2, which does not allow any mismatches in the 15 bp seed regions and discards reads mapped to 10 or more genomic regions. Mapped reads were annotated using BED Tools with miRBase database V21. The read counts were transformed to tags per million miRNA alignments (TPM) for normalization, and then the TPM values were log transformed.
Analysis of the differential expression of miRNAs detected by NGS was performed by calculating the FCs of miRNA expression and t-test P-value between three tumor and three normal samples. We defined significantly different expression levels for a miRNA as a FC ≥1.5 and a P-value <0.05. We used TarBase v7.0 to identify target genes of miRNAs with significantly different tumor expression. The TarBase database collects published experimentally validated miRNA/gene interactions. Pathway analysis was performed with the genes that we found to be targeted by differentially expressed miRNAs.
Statistical analysis
Results were presented as mean ± standard error of the mean. Graph Pad Prism 5.0 (GraphPad Software, San Diego, CA, USA) was used for statistical analysis. All P-values were two sided, and P-values <0.05 were considered significant.
Results
Patient characteristics
Three patients analyzed by NGS were all 55 years old, females and non-smokers. Participants’ characteristics, which were analyzed by qRT-PCR, are presented in Table 1. The median age was 55 years (range: 24–82 years). The characteristics of the 73 patients were as follows: 54 were female, 55 were never smokers, 78.1% had mixed GGN (median GGN size on CT was 12 mm [range, 4–30 mm]), 54 exhibited one lesion, 17 had two GGNs, and the remaining two patients had three GGNs. Participants’ characteristics with 66 lung benign diseases and 66 lung adenocarcinomas presenting with GGNs are presented in Table 2.
Table 1.
Characteristics of 73 patients
| Characteristics | n (%) |
|---|---|
| Sex, n (%) | |
| Male | 19 (26.0) |
| Female | 54 (74.0) |
| Age (years), mean | 55 |
| Smoker, n (%) | 18 (24.7) |
| Performance status (ECOG), n (%) | |
| 0 | 41 (56.2) |
| 1 | 31 (42.5) |
| 2 | 1 (1.4) |
| Solid component, n (%) | |
| Pure GGN | 16 (21.9) |
| Mixed GGN | 57 (78.1) |
| Lesion diameter, mm (%) | |
| ≤15 | 53 (72.6) |
| 15 | 20 (27.4) |
| Number of GGNs, n (%) | |
| 1 | 54 (74.0) |
| 2 | 17 (23.3) |
| 3 | 2 (2.7) |
Abbreviations: ECOG, Eastern Cooperative Oncology Group; GGNs, ground-glass nodules.
Table 2.
Participants’ characteristics with 66 lung benign diseases and 66 lung adenocarcinomas presenting with GGNs
| Participants’ characteristics | Benign pulmonary diseases | Pulmonary malignant |
|---|---|---|
| Sex, n (%) | ||
| Male | 34 (51.5) | 27 (40.9) |
| Female | 32 (48.5) | 39 (59.1) |
| Age (years), median | 50 | 56 |
| Smoker, n (%) | 20 (30.3) | 27 (40.9) |
| Lung GGNs/nodule size, mean | ||
| ≤8 mm, n (%) | 37 (56.1) | 30 (45.5) |
| 8 mm, n (%) | 29 (43.9) | 36 (754.5) |
| Number of lung GGNs, n (%) | ||
| ≤3 | 58 (87.9) | 60 (90.9) |
| 3 | 8 (12.1) | 6 (9.1) |
Abbreviation: GGNs, ground-glass nodules.
Next-generation sequencing
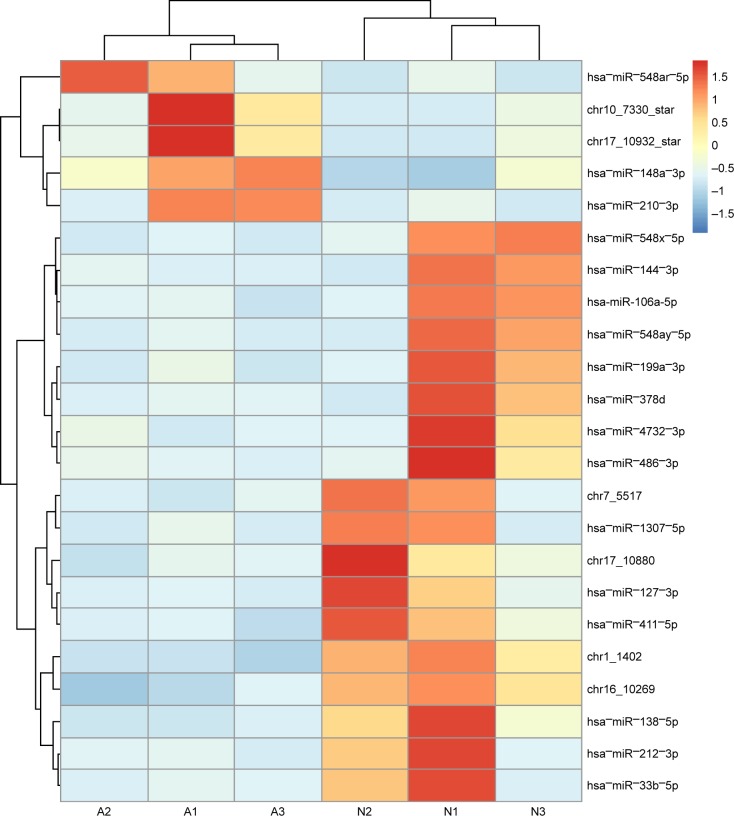
We analyzed expression of 2,231 miRNAs in tumor and paired control tissues in three patients, as shown in Figure 1. Of these, five miRNAs (hsa−miR−548ar−5p, chr10_7330_star, chr17_10932_star, hsa−miR−148a−3p and hsa−miR−210−3p) exhibited higher expression in the adenocarcinoma samples than that in the paired control non-tumor tissues (P<0.05), and 18 miRNAs (hsa−miR−548x−5p, hsa−miR−144−3p, hsa-miR-106-a-5p, hsa−miR−548ay−5p, hsa−miR−199a−3p, hsa−miR−378d, hsa−miR−4732−3p, hsa−miR−486−3p, chr7_5517, hsa−miR−1307−5p, chr17_10880, hsa−miR−127−3p, hsa−miR−411−5p, chr1_1402, chr16_10269, hsa−miR−138−5p, hsa−miR−212−3p and hsa−miR−33b−5p) demonstrated lower expression in adenocarcinoma samples than that in the paired control non-tumor tissues.
Figure 1.
Heat map of expression data of the 23 miRNAs.
Notes: Heat map of expression data of the 23 miRNAs found to be significantly differentially expressed by next-generation sequencing. The three left columns represent the three adenocarcinoma tumor samples from patients 1, 2 and 3, respectively, and the three columns on the right represent the three adjacent normal (N) paired tissues from patients 1, 2 and 3, respectively. High expression levels are depicted by red and low by blue.
Abbreviation: miRNAs, microRNAs.
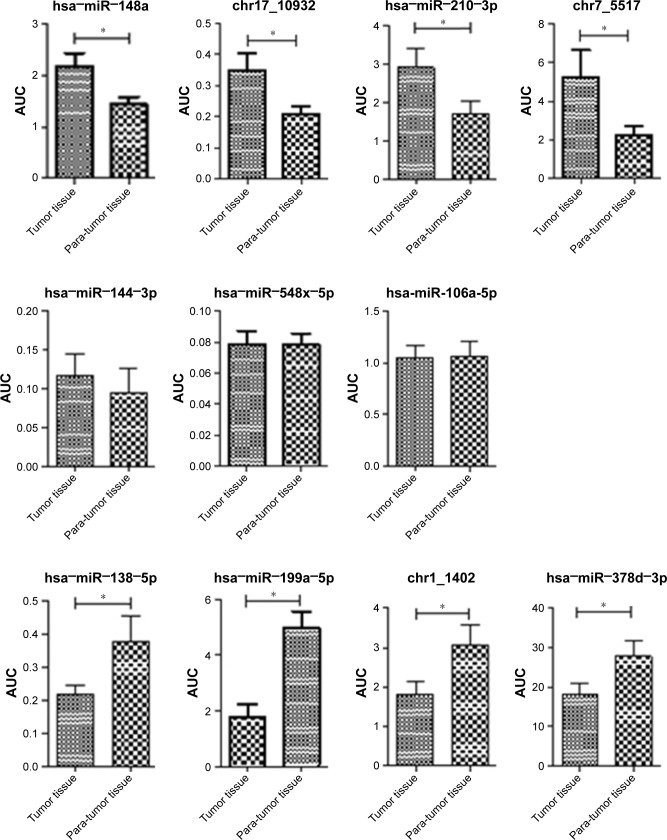
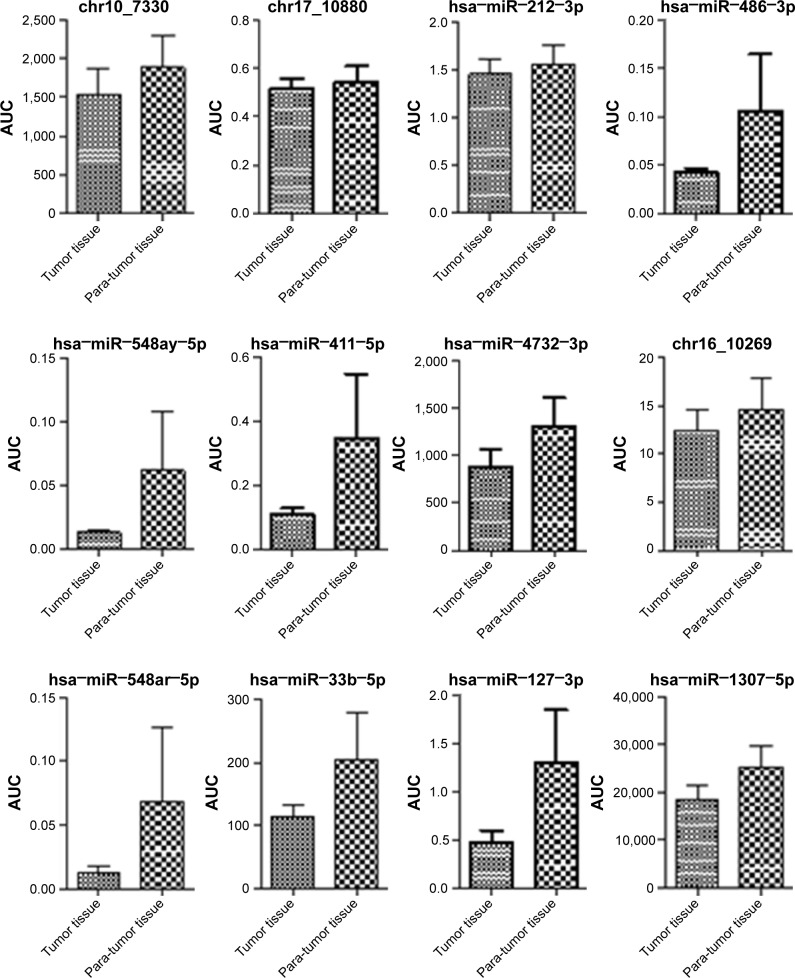
Validation of NGS results by qRT-PCR
In a validation study using qRT-PCR of 73 tumor and matched non-tumor samples, seven miRNAs (hsa−miR−199a−3p, chr17_10932, hsa−miR−148a−3p, hsa−miR−210−3p, chr1_1402, hsa−miR−378d and hsa−miR−138−5p) were confirmed to have significantly different expression (Figure 2). chr17_10932, hsa−miR−148a−3p and hsa−miR−210−3p exhibited higher expression in the tumor tissue than in adjacent non-tumor tissue (P<0.05). By contrast, hsa−miR−199a−3p, hsa−miR−378d, chr1_1402 and hsa−miR−138−5p demonstrated lower expression in tumor tissue than that in the adjacent non-tumor tissue (P<0.05; Figure 2).
Figure 2.
AUC of 23 different microRNAs in tumor and non-tumor tissue by qRT-PCR.
Note: *P<0.05.
Abbreviations: AUC, area under the curve; miRNAs, microRNAs; qRT-PCR, quantitative real-time polymerase chain reaction.
Validation of miRNA panel by qRT-PCR
We also detected the miRNA panel in 66 lung benign diseases and 66 lung adenocarcinomas presenting with GGN lesion tissue by qRT-PCR. We used more than 75% of the 2−ΔΔCt value in lung adenocarcinoma patients as the cutoff of chr17_10932, hsa−miR−148a−3p and hsa−miR−210−3p. We used less than 75% of the 2−ΔΔCt value in lung adenocarcinoma patients as the cutoff of hsa−miR−199a−3p, hsa−miR−378d, chr1_1402 and hsa−miR−138−5p. The sensitivity and specificity of miRNA panel were 86.4% and 60.6%, respectively.
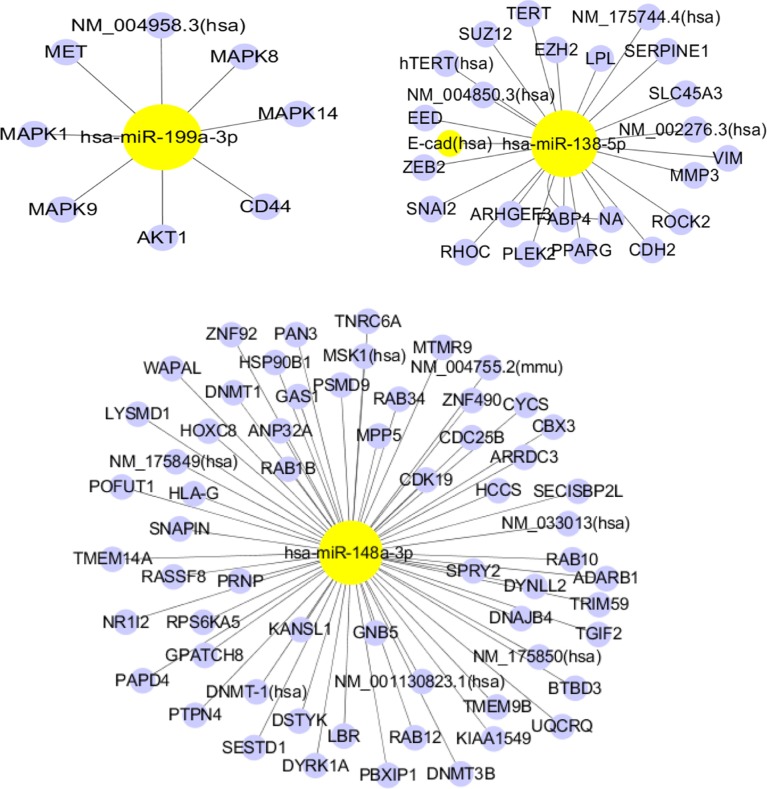
In silico analysis of potential targets of the seven validated differentially expressed miRNAs
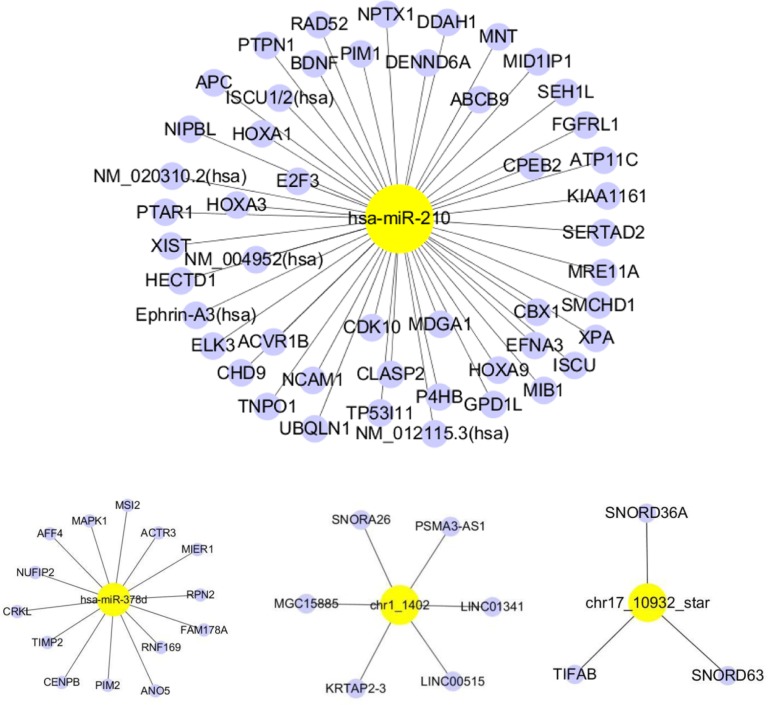
To consider the potential biological functions of the seven validated miRNAs, we used TarBase to predict the targeted mRNAs. We used the miRand algorithm to predict the targets of two novel miRNAs (chr1_1402 and chr17_10932). Figure 3 depicts the mRNAs predicted to be targeted by our miRNAs of interest. Has-miR-138-5p, has-miR-148a-3p and has-miR-210 were predicted to target the largest number of mRNAs. Pathway enrichment analysis was performed for each predicted target mRNA. The enriched pathways are listed in Figure 3, ordered according to the spell out FDR here adjusted fisher exact test P-value. Most of the enriched pathways were associated with cancer (pancreatic, colorectal, bladder and chronic myeloid leukemia) (Figure 3).
Figure 3.
miRNA target genes.
Note: Each network represents the interaction of a miRNA (yellow) and its target genes (blue).
Abbreviation: miRNA, microRNA.
Discussion
Worldwide, lung cancer is the leading cause of cancer death. Lung cancer’s deadliness is due in large part to the fact that only 20% of lung cancer patients are diagnosed at an early stage.2 Effective biomarkers could permit earlier diagnosis, significantly improving the patient’s prognosis. MiRNAs are involved in a myriad of important biological processes including development, differentiation, apoptosis and proliferation.9 MiRNAs can act as tumor suppressors or oncogenes.9 Abnormal expression of miRNAs has been found to be an effective biomarker for both solid and hematopoietic tumors.6–8,10–12 For example, one previous study suggested that miR-125a-5p, miR-145 and miR-146a may be useful biomarkers for the clinical diagnosis of non-small-lung cancer (NSCLC). However, the data, especially regarding GGNs (a hazy opacity in the lung with the preservation of bronchial and vascular margins3), remain limited. GGNs can indicate both malignant and benign lesions, including lung adenocarcinoma, focal interstitial fibrosis, hemorrhage or infection.3 The difficulty associated with diagnosis involving GGN increases the need for and value of molecular markers to identify disease.
Several miRNA studies have been reported on various cancer types, including chronic lymphocytic leukemia,13 breast,14 glioblastoma,15 thyroid papillary carcinoma,16 hepatocellular carcinoma,17 lung10,18 and colon.19,20 The downregulation of miR-143 and miR-145 was reported in colorectal neoplasia.20 Garzon et al21 demonstrated that significant differences in miRNA levels can differentiate the stages of megakaryocytopoiesis. Volinia et al8 found distinct miRNA profiles in cancer cells compared with normal cells. In our study, a miRNA panel (hsa−miR−199a−3p, chr17_10932, hsa−miR−148a−3p, hsa−miR−210−3p, chr1_1402, hsa−miR−378d and hsa−miR−138−5p) shows potential as a tool for the early diagnosis for lung adenocarcinoma presenting with GGNs.
In a lung cancer study, Yanaihara et al10 found 43 miRNAs that were expressed at significantly different levels in lung cancer versus matched normal lung tissue. Also, serum concentration of miR-1254 and miR-574-5p could serve as an early diagnostic marker of lung cancer.22 Serum levels of miR-146b, miR-221, let-7a, miR-155, miR-17-5p, miR-27a and miR-106a were significantly decreased in NSCLC, while miR-29c levels were significantly increased.23 Shen et al24 were able to distinguish lung cancer patients from controls by detecting differences in miR-21, miR-126, miR-210 and miR-486-5p in plasma. It has also been demonstrated that the panel of miR-155, miR-197 and miR-182 in plasma can diagnose lung cancer patients.25 Another independent study found seven upregulated and eight downregulated miRNAs in lung cancer patients.26 A meta-analysis using the vote-counting strategy showed miR-210 to be expressed significantly differently in tumor samples in nine studies, and miR-21 to be differentially expressed in seven studies.27 A subsequent study of 250 smokers with CT-detected solitary pulmonary nodules was able to use plasma levels of miR-21, miR-210 and miR-486-5p to discriminate malignant from benign disease with 76% sensitivity and 85% specificity,28 indicating that miRNA biomarkers could potentially supplement and enhance the efficacy of CT screening. This collective body of evidence strongly suggests that miRNAs can be diagnostic and prognostic markers of lung cancer.22–29 However, we did not find data in the literature about miRNA expression profiles in lung adenocarcinoma presenting with GGNs, which are generally present in early-stage disease.
In order to address this lack of data, we employed NGS to identify all the miRNAs that were expressed in lung adenocarcinoma presenting with GGNs in three patients. Our deep sequencing analysis data identified 23 significantly differentially expressed miRNAs in adenocarcinoma tissue with GGNs compared with paired adjacent non-tumor tissue. Among the miRNAs of interest identified by NGS, expressions of hsa−miR−199a−3p, chr17_10932, hsa−miR−148a−3p, hsa−miR−210−3p, chr1_1402, hsa−miR−378d and hsa−miR−138−5p were validated to be statistically different by qRT-PCR in a larger independent group of 73 paired cancer/normal samples. We also detected the miRNA panel in 66 lung benign diseases and 66 lung adenocarcinomas presenting with GGN lesion tissue. The sensitivity and specificity of miRNA panel were 86.4% and 60.6%, respectively, while CEA, NSE and CYFRA21-1 showed sensitivities of 5%, 4% and 17% in stage I and 47%, 21% and 56% in stage IV, respectively.
MiR-210 has consistently been found to be induced by hypoxia and implicated in cancer progression.30–32 Consistent with our data, it is upregulated in multiple tumors and associated with poor patient prognosis. Ding et al31 high-lighted the importance of miR-210 in ovarian cancer progression by demonstrating that it promoted cancer cell mobility by modulating epithelial–mesenchymal transition. Furthermore, one study recently indicated that miR-210 could be considered as a poor prognostic biomarker for lung adenocarcinoma.32 Thus, previous studies support our findings that miR-210 could be a biomarker for the early diagnosis of lung adenocarcinoma.
Altered miR-378 expression has been shown to contribute to human cancers, including breast, prostate, gastric and hepatocellular carcinoma.33,34 Peng et al33 found that miRNA-378 is a biomarker for early detection and diagnosis of colorectal cancer. Decreased expression of miR-378 has been reported in prostate cancer tissue; its overexpression results in inhibition of cancer cell migration and invasion and promotes cell apoptosis.34 These other findings of altered miR-378 in cancer corroborates our finding that miR-378 could be helpful in the diagnosis of early lung adenocarcinoma presenting with GGNs.
Our finding that miR-138 is downregulated in adenocarcinoma presenting with GGNs concurs with mounting evidence presented in other studies that miR-138 is dysregulated in numerous human cancers including lung and may play an important role in tumorigenesis.35,36
Consistent with our findings for lower miR-199a-3p expression in tumor samples, other studies have noted its altered expression in cancer. Kinose et al37 reported that miR-199a-3p dramatically suppressed the progression of ovarian cancer by downregulating expression of c-Met. Wang et al38 found that MiR-199a inhibited proliferation and migration by regulating CD44-ezrin signaling in cutaneous cancer.
We have found that miR-148a expression level was significantly elevated in lung adenocarcinoma patients presenting with GGNs. Several studies have revealed differential expression of miR-148a in tumor versus non-tumor tissue in liver and colorectal cancer.39,40 Li et al41 found that the expression levels of circulating miR-148 family miRNAs might serve as biomarkers for distinguishing NSCLC from benign pulmonary diseases.
Finally, pathway enrichment analysis for the predicted target genes of our validated differentially expressed miRNA results show strong evidence that these miRNAs were linked to cancer pathways. Similar targeting patterns and pathways have been observed in many cancer types including colorectal, pancreatic, bladder and chronic myeloid leukemia, which supports our findings. Others including Kim et al42 have identified some of the same miRNA targets such as the MAPK family as well.
Our work contributes novel data to the literature of miRNAs in cancer because we detected miRNAs via NGS, enabling us to better identify comprehensive potential diagnostic markers. Moreover, we analyzed lung adenocarcinoma patients presenting with GGNs, which are known to be correlated to the early stages of lung adenocarcinoma. In addition, the differential miRNA expression identified in our study was validated in both tumor and matched adjacent non-tumor tissue. We also used TarBase v7.0 to identify target genes of significantly differentially expressed miRNAs.
There are several limitations in our study. Circulating miRNAs in the blood should be analyzed for their potential use as the markers for minimally invasive lung cancer diagnostics, prediction of treatment efficacy and disease prognosis. It has been reported that large numbers of miRNAs are released into peripheral blood when cell damage occurs.43–46 In upcoming research, we plan to compare miRNA levels in the blood of adenocarcinoma patients to healthy individuals with the goal of identifying and verifying a miRNA signature to detect the cancer non-invasively.
This study was aimed to identify characteristic, statistically significantly different levels of miRNA expression in lung adenocarcinoma presenting with GGNs compared to non-cancerous tissue. We found seven miRNAs whose differential expression could potentially illuminate or even initiate lung adenocarcinoma presenting with GGNs, though additional research would be necessary to draw any definitive conclusions. However, this miRNA panel has more immediate potential as a tool for early diagnosis of lung cancer.
Data sharing statement
Our NGS data were uploaded to the website Zenodo: https://zenodo.org/record/50498/export/xm#.Wblpgvl71gk.
Acknowledgments
This study was supported in part by a grant from Shanghai Pujiang Program (17PJD036); 3-year action plan for promoting major disease clinical skills, Shanghai Shenkang Hospital Development Center (16CR1001A); the fundamental research funds for the central universities.
The abstract of this paper was presented at the World Conference on Lung Cancer (WCLC) as an oral presentation with interim findings. The oral abstract was published in “Oral Abstracts” on the website of the International Association for the Study of Lung Cancer (IASLC).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NL, Remy J. Fleischner Society: glossary of terms for thoracic imaging. Radiology. 2008;246(3):697–722. doi: 10.1148/radiol.2462070712. [DOI] [PubMed] [Google Scholar]
- 4.Matsuguma H, Mori K, Nakahara R, et al. Characteristics of subsolid pulmonary nodules showing growth during follow-up with CT scanning. Chest. 2013;143(2):436–443. doi: 10.1378/chest.11-3306. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12(12):580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353(17):1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 10.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Rikke BA, Wynes MW, Rozeboom LM, Barón AE, Hirsch FR. Independent validation test of the vote-counting strategy used to rank biomarkers from published studies. Biomark Med. 2015;9(8):751–761. doi: 10.2217/BMM.15.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 15.Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 16.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 18.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 21.Garzon R, Pichiorri F, Palumbo T, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103(13):5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6(3):482–488. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 23.Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, Harris CC. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer. 2012;130(6):1378–1386. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J, Todd NW, Zhang H, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91(4):579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng D, Haddadin S, Wang Y, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4(6):575–586. [PMC free article] [PubMed] [Google Scholar]
- 26.Võsa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microRNA expression in lung cancer. Int J Cancer. 2013;132(12):2884–2893. doi: 10.1002/ijc.27981. [DOI] [PubMed] [Google Scholar]
- 27.Guan P, Yin Z, Li X, Wu W, Zhou B. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res. 2012;31:54. doi: 10.1186/1756-9966-31-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen J, Liu Z, Todd NW, et al. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011;11:374. doi: 10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boeri M, Sestini S, Fortunato O, et al. Recent advances of microRNA-based molecular diagnostics to reduce false-positive lung cancer imaging. Expert Rev Mol Diagn. 2015;15(6):801–813. doi: 10.1586/14737159.2015.1041377. [DOI] [PubMed] [Google Scholar]
- 30.Noman MZ, Janji B, Hu S, et al. Tumor-promoting effects of myeloid-derived suppressor cells are potentiated by hypoxia-induced expression of miR-210. Cancer Res. 2015;75(18):3771–3787. doi: 10.1158/0008-5472.CAN-15-0405. [DOI] [PubMed] [Google Scholar]
- 31.Ding L, Zhao L, Chen W, Liu T, Li Z, Li X. miR-210, a modulator of hypoxia-induced epithelial-mesenchymal transition in ovarian cancer cell. Int J Clin Exp Med. 2015;8(2):2299–2307. eCollection 2015. [PMC free article] [PubMed] [Google Scholar]
- 32.Osugi J, Kimura Y, Owada Y, et al. Prognostic impact of hypoxia-inducible miRNA-210 in patients with lung adenocarcinoma. J Oncol. 2015;2015:316745. doi: 10.1155/2015/316745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng J, Xie Z, Cheng L, et al. Paired design study by real-time PCR: miR-378* and miR-145 are potent early diagnostic biomarkers of human colorectal cancer. BMC Cancer. 2015;15:158. doi: 10.1186/s12885-015-1123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen QG, Zhou W, Han T, et al. MiR-378 suppresses prostate cancer cell growth through downregulation of MAPK1 in vitro and in vivo. Tumour Biol. 2016;37(2):2095–2103. doi: 10.1007/s13277-015-3996-8. [DOI] [PubMed] [Google Scholar]
- 35.Yu C, Wang M, Li Z, et al. MicroRNA-138-5p regulates pancreatic cancer cell growth through targeting FOXC1. Cell Oncol (Dordr) 2015;38(3):173–181. doi: 10.1007/s13402-014-0200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu R, Zeng G, Gao J, et al. miR-138 suppresses the proliferation of oral squamous cell carcinoma cells by targeting Yes-associated protein 1. Oncol Rep. 2015;34(4):2171–2178. doi: 10.3892/or.2015.4144. [DOI] [PubMed] [Google Scholar]
- 37.Kinose Y, Sawada K, Nakamura K, et al. The hypoxia-related microRNA miR-199a-3p displays tumor suppressor functions in ovarian carcinoma. Oncotarget. 2015;6(13):11342–11356. doi: 10.18632/oncotarget.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang SH, Zhou JD, He QY, Yin ZQ, Cao K, Luo CQ. MiR-199a inhibits the ability of proliferation and migration by regulating CD44-Ezrin signaling in cutaneous squamous cell carcinoma cells. Int J Clin Exp Pathol. 2014;7(10):7131–7141. eCollection 2014. [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Jia HL, Zhou HJ, et al. Identification of metastasis-related microRNAs of hepatocellular carcinoma in hepatocellular carcinoma cell lines by quantitative real time PCR. Zhonghua Gan Zang Bing Za Zhi. 2009;17(7):526–530. Chinese. [PubMed] [Google Scholar]
- 40.Song Y, Xu Y, Wang Z, et al. MicroRNA-148b suppresses cell growth by targeting cholecystokinin-2 receptor in colorectal cancer. Int J Cancer. 2012;131(5):1042–1051. doi: 10.1002/ijc.26485. [DOI] [PubMed] [Google Scholar]
- 41.Li L, Chen YY, Li SQ, Huang C, Qin YZ. Expression of miR-148/152 family as potential biomarkers in non-small-cell lung cancer. Med Sci Monit. 2015;21:1155–1161. doi: 10.12659/MSM.892940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S, Lee UJ, Kim MN, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283(26):18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J, Yu L, Gao X, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29(36):4781–4788. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 44.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol. 2014;32(8):768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fortunato O, Boeri M, Verri C, et al. Assessment of circulating microRNAs in plasma of lung cancer patients. Molecules. 2014;19(3):3038–3054. doi: 10.3390/molecules19033038. [DOI] [PMC free article] [PubMed] [Google Scholar]