Summary
Three distinct RNA polymerases (Pols) transcribe different classes of genes in the eukaryotic nucleus1. Pol III is the essential, evolutionarily conserved enzyme that generates short, non-coding RNAs, including transfer RNAs (tRNAs) and 5S ribosomal RNA (rRNA)2. Historical focus on transcription of protein-coding genes has left the roles of Pol III in organismal physiology relatively unexplored. The prominent regulator of Pol III activity, Target of Rapamycin kinase Complex 1 (TORC1), is an important longevity determinant3, raising the question of Pol III’s involvement in ageing. Here we show that Pol III limits lifespan downstream of TORC1. We find that a reduction in Pol III extends chronological lifespan in yeast and organismal lifespan in worms and flies. Inhibiting Pol III activity in the adult worm or fly gut is sufficient to extend lifespan, and in flies, longevity can be achieved by Pol III inhibition specifically in the intestinal stem cells (ISCs). The longevity phenotype is associated with amelioration of age-related gut pathology and functional decline, dampened protein synthesis and increased tolerance of proteostatic stress. Importantly, Pol III acts downstream of TORC1 for lifespan and limiting Pol III activity in the adult gut achieves the full longevity benefit of systemic TORC1 inhibition. Hence, Pol III is a pivotal output of this key nutrient signalling network for longevity; Pol III’s growth-promoting, anabolic activity mediates the acceleration of ageing by TORC1. The evolutionary conservation of Pol III affirms its potential as a therapeutic target.
The labour of transcription in the eukaryotic nucleus is divided amongst Pol I, II and III1,4. This specialisation is evident in the biogenesis of the translation machinery, a task for which all three Pols are co-ordinately required: Pol I generates the 45S pre-rRNA that is subsequently processed into mature rRNAs, Pol II transcribes various RNAs including messenger RNAs (mRNAs) encoding ribosomal proteins (RPs), while Pol III provides the tRNAs and 5S rRNA. To match the extrinsic conditions and the intrinsic need for protein synthesis, this costly process of generating protein synthetic capacity is tightly regulated by the key driver of cellular anabolism: TORC15,6. The central position of TORC1 in the control of fundamental cellular processes is mirrored by the striking impact of its activity on organismal physiology: following the initial discovery in worms7, the inhibition of TORC1 has been demonstrated to extend lifespan in all organisms tested, from yeast to mice8,9, with beneficial effects on a range of age-related diseases and dysfunctions3,10. TORC1 strongly activates Pol III transcription5,6 and this relationship suggests the possibility that inhibition of Pol III promotes longevity. Here, we test this hypothesis.
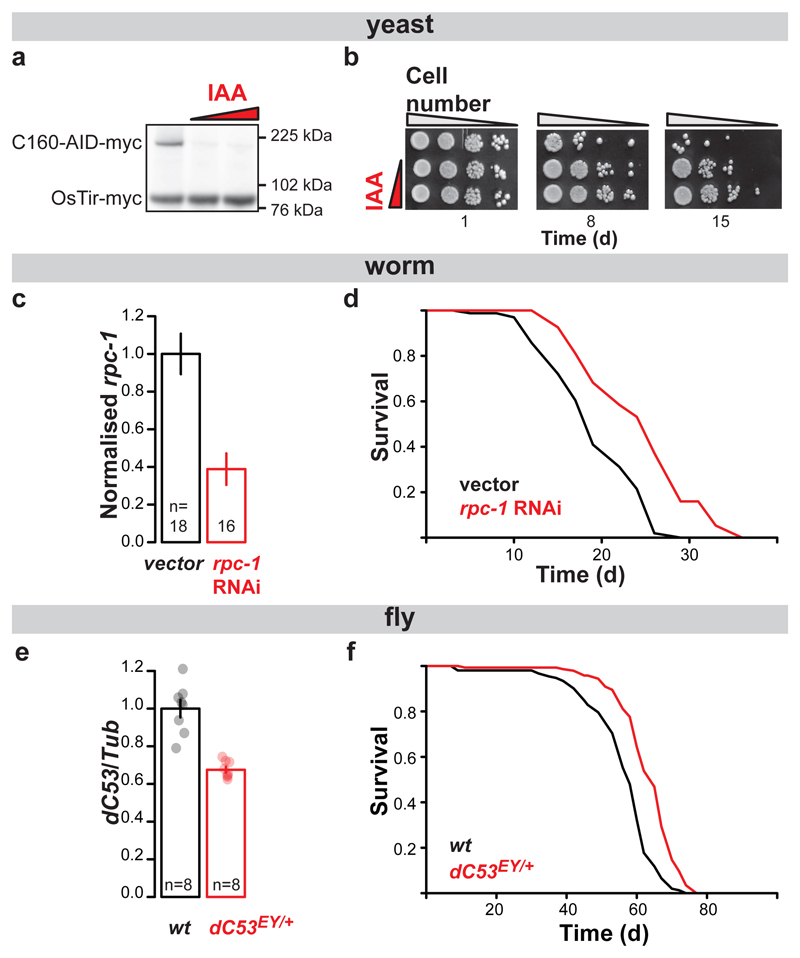
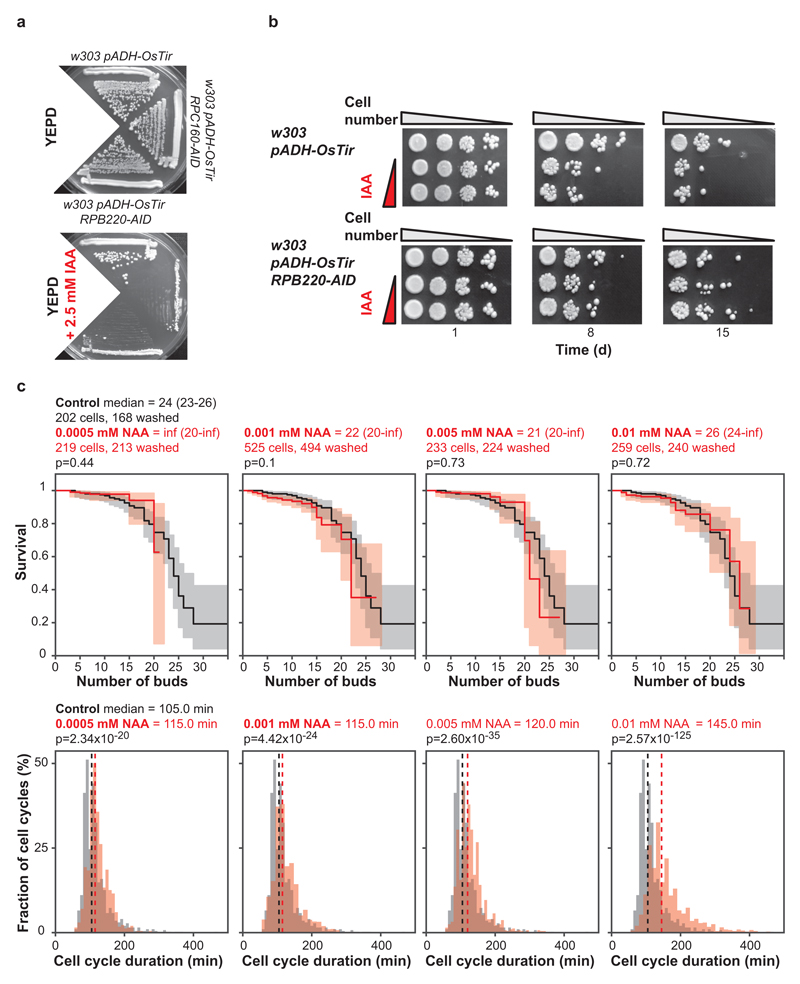
In S. cerevisiae, each of the 17 Pol III subunits is encoded by an essential gene. We generated a yeast strain in which its largest subunit (C160, encoded by RPC160/RPO314) is fused to the auxin-inducible degron (AID). The fusion protein can be targeted for degradation by the ectopically expressed E3 ubiquitin ligase (OsTir) in the presence of indole-3-acetic acid (IAA)11 to achieve conditional inhibition of Pol III (Extended Data Fig. 1a). We confirmed that IAA treatment triggered the degradation of the fusion protein (Fig. 1a) and observed IAA improve the survival of the RPC160-AID strain upon prolonged culture (Fig. 1b). In addition, IAA treatment of the control strain lacking the AID fusion reduced its survival, relative to same strain in absence of IAA and the RPC160-AID strain in presence of IAA (Extended Data Fig. 1b). Hence, Pol III depletion appears to extend yeast chronological lifespan. Note that IAA had no substantial effect on the survival of a strain carrying the AID domain fused to the largest subunit of Pol II (RPB220-AID), which appeared to survive better than the control strain in the presence of IAA (Extended Data Fig. 1a and b), indicating inhibition of Pol II may also extend chronological lifespan. Yeast chronological lifespan is a measure of survival in a nutritionally-limited, quiescent population. Replicative lifespan, on the other hand, measures the number of daughters produced by a single mother cell in its lifetime. We found no evidence that inhibition of Pol III causes an increase in yeast replicative lifespan (Extended Data Fig. 1c).
Figure 1. Inhibition of Pol III extends lifespan.
Treatment of the RPC160-AID-myc pADH-OsTir-myc budding yeast strain with 0, 0.125 and 0.25 mM IAA: a, triggers degradation of C160-AID-myc and b, extends its chronological lifespan, measured as colony formation after normalisation for optical density and 10-fold serial dilution ([a] and [b] show a representative of two experimental trials). Feeding N2 worms with E. coli expressing the rpc-1 RNAi construct from L4 stage: c, reduces the levels of rpc-1 mRNA (p<10-4, two-tailed t-test) and d, extends their lifespan relative to vector alone at 20°C in presence of FUDR (p=0.03, log-rank test, n= 86 control, 94 rpc-1 RNAi animals; representative of three trials). Female flies heterozygous for the dC53EY allele display: e, a reduction in dC53 transcript (p=10-4, two-tailed t-test, 95% confidence intervals [CI] = 0.89-1.1 wt, 0.64-0.71 dC53EY/+) and f, extended lifespan (p=6x10-13, log-rank test, n= 152 control, 144 dC53EY/+ animals; single trial). Bar charts show mean ± Standard Error of the Mean (SEM), with n= number of biologically independent samples indicated and overlay showing individual data points. For more detailed demography and summary of worm and fly lifespan trials see Extended Data Fig. 2c and 4a. For gel source data, see Sup. Fig. 1.
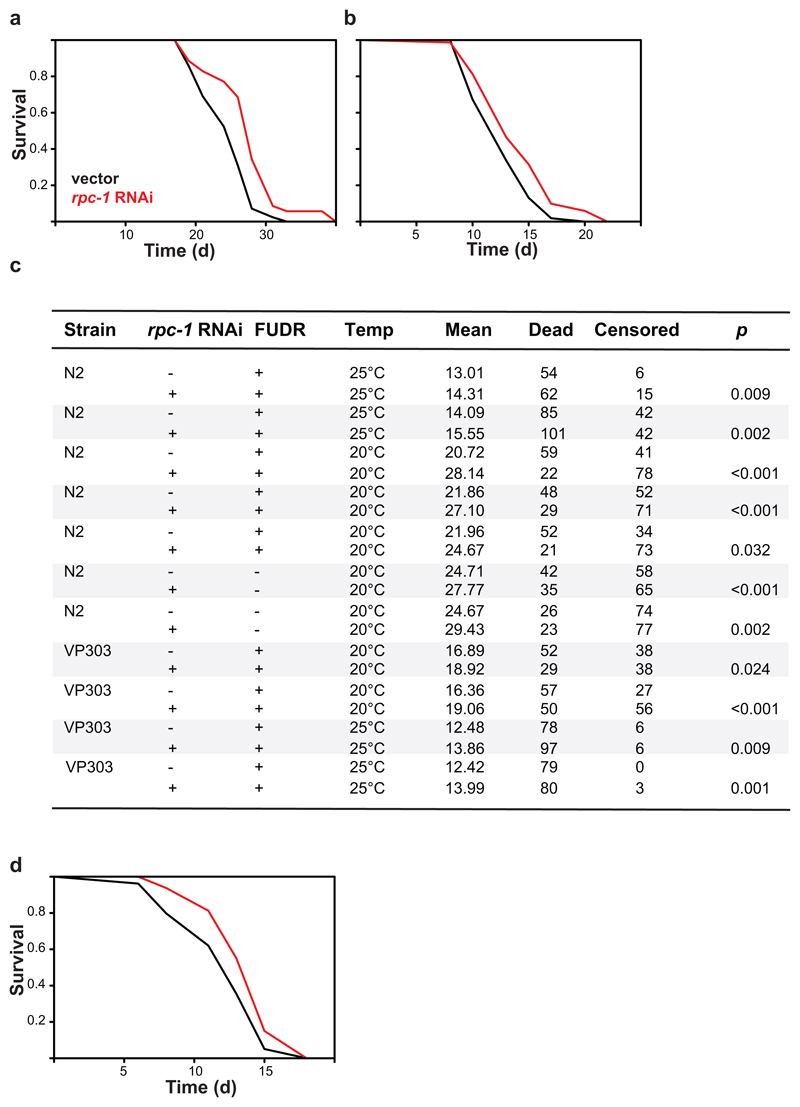
The observed increase in yeast chronological lifespan may simply be indicative of increased stress resistance and hence bear limited relevance to organismal ageing. To examine the role of Pol III in organismal ageing directly, we turned to animal models. We treated C. elegans from the L4 stage with RNAi against rpc-1, the worm orthologue of RPC160, achieving a partial knockdown of its mRNA (Fig. 1c). This consistently extended worm lifespan at both 20°C and 25°C (Fig. 1d; Extended Data Fig. 2a and b, see c for summary of worm lifespans). To reduce Pol III activity in the fruit fly, we backcrossed a P-element insertion deleting the transcriptional start site of the gene encoding the Pol III-specific subunit C53 (CG5147EY22749, henceforth called dC53EY, Extended Data Fig. 3) into a healthy, outbred D. melanogaster population. Homozygous dC53EY/EY mutants were not viable but heterozygous females had a partial reduction in dC53 mRNA and lived longer than controls (Fig. 1e and f; see Extended Data Fig. 4a for summary of fly lifespans). Taken together, our data strongly indicate that Pol III limits lifespan in multiple model organisms and, conversely, that partial inhibition of its activity is an evolutionarily conserved longevity intervention.
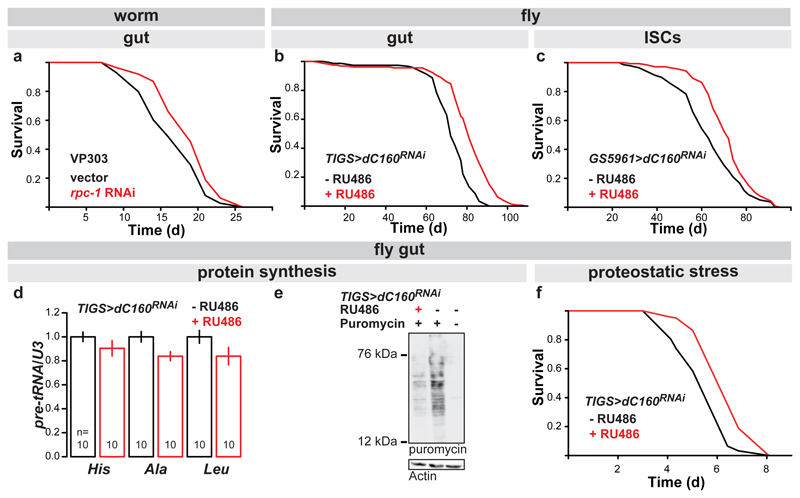
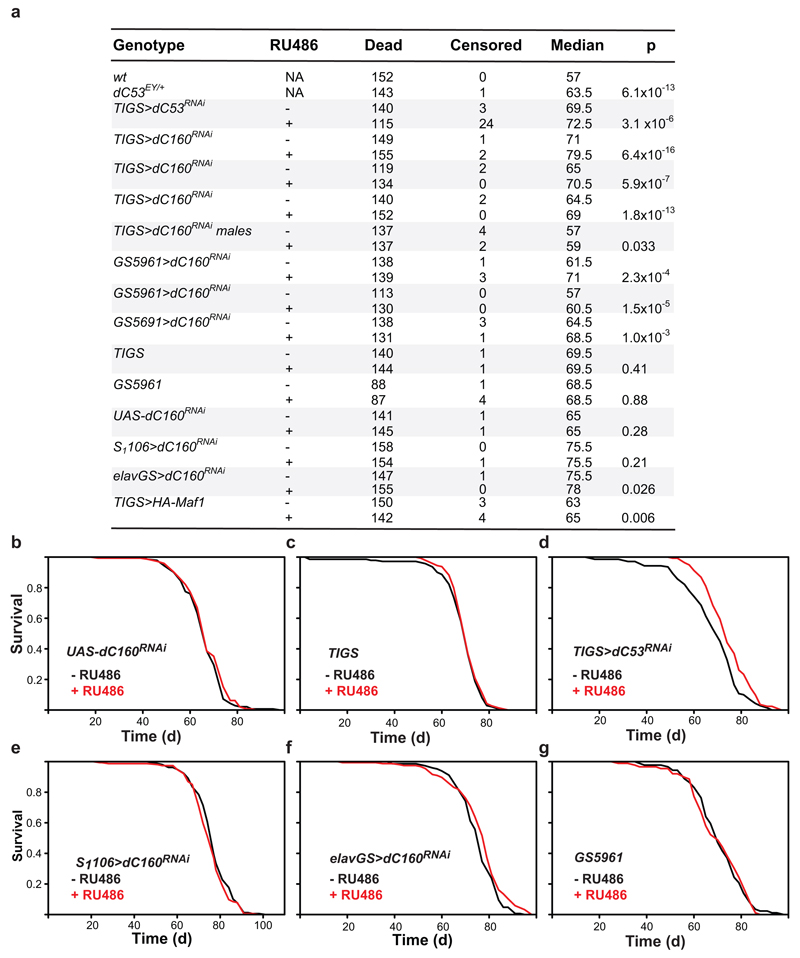
The longevity of an animal can be governed from a single organ. In the worm, this role is often played by the gut12,13. To restrict the rpc-1 knock-down to the gut, we used rde-1 null worms whose RNAi machinery deficiency is restored solely in the gut by gut-specific rde-1 rescue14. rpc-1 RNAi extended the lifespan of this strain, both at 20°C and 25°C (Fig. 2a, Extended Data Fig. 2d). Similarly, in the adult fly, driving an RNAi construct targeting the RPC160 orthologue (CG17209, henceforth called dC160, Extended Data Fig. 3) with the mid-gut-specific, RU486-inducible driver (TIGS) extended female lifespan (Fig. 2b), while the presence of the inducer (RU486) did not affect survival of the control lines (Extended Data Fig. 4b and c). The longevity phenotype could also be recapitulated with RNAi against another Pol III subunit (dC53, Extended Data Fig. 4d), indicating it is not due to off-target effects, or subunit-specific. The longevity phenotype appeared specific to the gut, since no significant lifespan extension was observed upon induction of dC160RNAi in the adult fly fat-body and only a modest, albeit significant extension resulted from neuronal induction (Extended Data Fig. 4e and f); fat-body and neurons being other two sites often associated with longevity13.
Figure 2. Gut-specific inhibition of Pol III extends lifespan, reduces protein synthesis and increases tolerance to proteostatic stress.
a, Activating RNAi against rpc-1 specifically in the worm gut, using the VP303 strain, extends worm lifespan at 20°C in presence of FUDR (p=0.02, log-rank test, n= 90 control, 67 rpc-1 RNAi animals; representative of two trials). b, Feeding RU486 to TIGS>dC160RNAi female fruit flies to induce dC160RNAi in the gut alone extends their lifespan (p=6x10-16, log-rank test, n= 150 -RU486, 157 +RU486 animals; representative of three trials). c, Feeding RU486 to GS5961>dC160RNAi female fruit flies to induce dC160RNAi in the ISCs alone extends their lifespan (p=2x10-4, log-rank test, n= 139 -RU486, 142 +RU486 animals; representative of three trials). Inducing dC160RNAi in the gut with RU486 feeding of TIGS>dC160RNAi females leads to: d, reduction in pre-tRNAs (mean ± SEM, p=0.04, Multivariate Analysis of Variance [MANOVA], n= 10 biologically independent samples per -/+RU486 condition, CI= 0.91-1.1, 0.76-1.0, 0.90-1.1, 0.75-0.92, 0.88-1.1, 0.68-1.0 left to right); e, reduction in gut protein synthesis, as quantified by ex-vivo puromycin incorporation and western blotting (representative of three biologically independent repeats; see Extended Data Fig. 6c); f, improved survival in response to tunicomycin challenge (p=3x10-15, log-rank test, n=185 animals per condition; representative of two trials). For more detailed demography and summary of lifespan trials see Extended Data Fig. 2c and 4a. For gel source data, see Sup. Fig. 1.
Worm gut is composed of only post-mitotic cells. In flies, like in mammals, the adult gut epithelium contains the mitotically active ISCs15. ISC homeostasis is important for longevity16 and the TIGS gut driver appears active in at least some ISCs (Extended Data Fig. 5), prompting us to further restrict dC160RNAi induction to solely this cell type. ISC-specific dC160RNAi, achieved with the GS5961 driver, was sufficient to promote longevity (Fig. 2c, see Extended Data Fig. 4b and g for controls). In summary, Pol III activity in the gut limits survival in worms and flies, and in the fly, Pol III can drive ageing specifically from the gut stem cell compartment.
We assessed the consequences of Pol III inhibition in the fly gut. Pol III acts to generate precursor-tRNAs (pre-tRNAs) that are rapidly processed to mature tRNAs. Due to their short half-lives, pre-tRNA are suitable readouts of in-vivo Pol III activity. Profiling the levels of pre-tRNAHis, pre-tRNAAla and pre-tRNALeu, relative to the levels of U3, a small nucleolar RNA transcribed by Pol II17, revealed a moderate but significant reduction in Pol III activity upon gut-specific induction of dC160RNAi (Fig. 2d). The three Pols can be directly coordinated for the generation of translation machinery18. Indeed, Pol III inhibition had knock-on effects on Pol I but not Pol II-generated transcripts, revealing a partial cross-talk (Extended Data Fig. 6a and b). Consistent with reduced Pol III activity, dC160RNAi reduced protein synthesis in the gut (Fig. 2e, Extended Data Fig. 6c). These effects (reduction in pre-tRNAs or protein synthesis) were not observed after feeding RU486 to the driver-alone control (Extended Data Fig. 6d - f). The reduction in protein synthesis was not pathological: total protein content of the gut was unaltered; fecundity, a sensitive readout of a female’s nutritional status, was unaffected; and the flies’ weight, triacylglycerol and protein levels also remained unchanged (Extended Data Fig. 6g - i). Reduced protein synthesis can liberate protein-folding machinery from protein production and increase homeostatic capacity19. Indeed, inducing dC160RNAi in the gut increased the resistance of adult flies to a proteostatic challenge with tunicamycin (Fig. 2f, and Extended Data Fig. 6j for TIGS-alone control). Hence, Pol III can fine-tune the rate of protein synthesis in the adult fly gut, without obvious detrimental outcomes, while increasing resistance to proteotoxic stress.
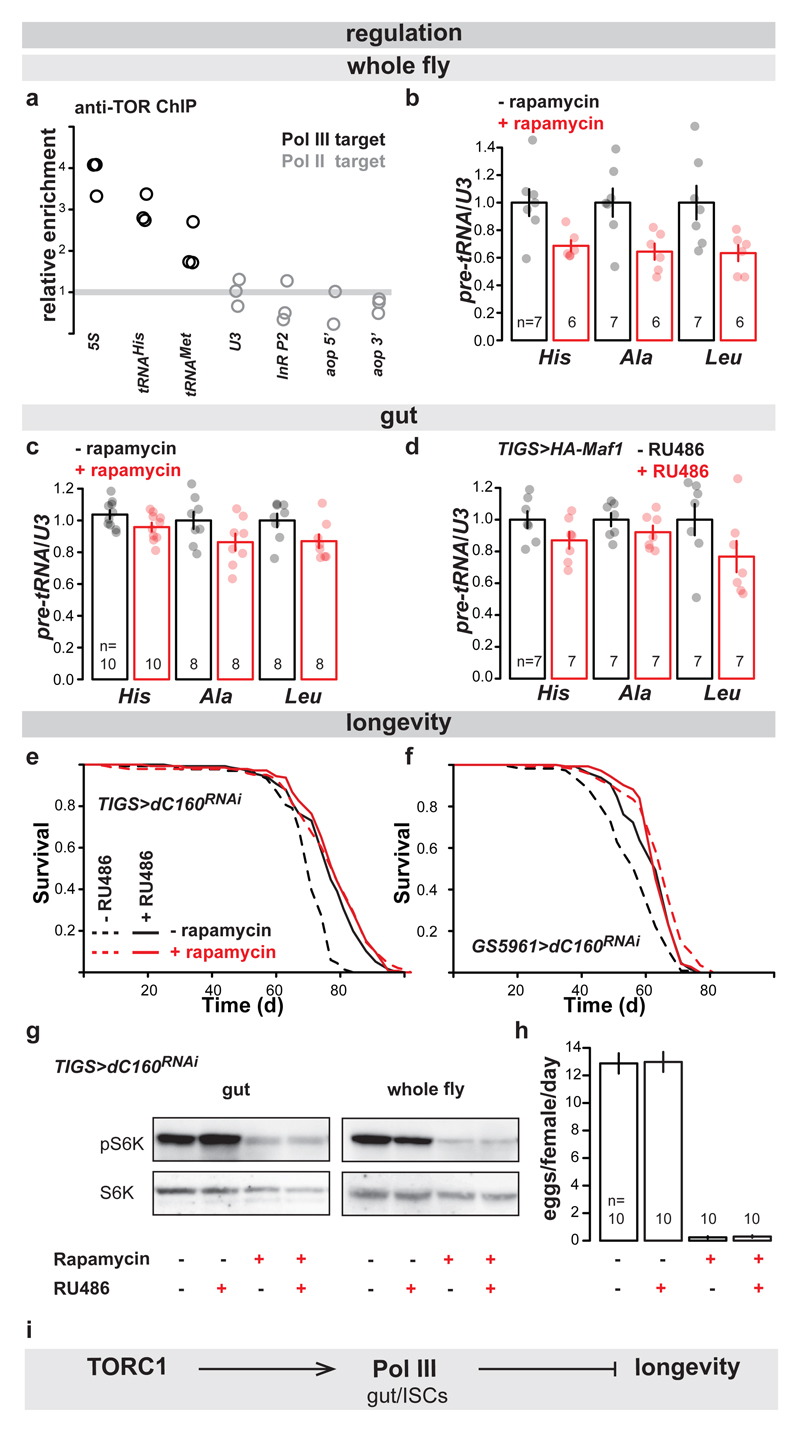
Having demonstrated the relevance of Pol III for ageing, we examined whether it acts downstream of TORC1 for lifespan using the fruit fly. Numerous observations in several organisms support the model where TORC1 localises on Pol III-transcribed loci and promotes the phosphorylation of the components of Pol III transcriptional machinery to activate transcription, in part by inhibition of the Pol III repressor, Maf15. Using chromatin immunoprecipitation (ChIP) with two independently generated antibodies against Drosophila TOR20,21, we observed the kinase enriched on Pol III-target genes in the adult fly, relative to Pol II targets (Fig. 3a; Extended Data Fig. 7a to d, and e for mock ChIP). Inhibition of TORC1 by feeding flies rapamycin reduced the levels of pre-tRNAs in whole flies (Fig. 3b). Rapamycin also reduced pre-tRNA levels specifically in the gut relative to U3 (Fig 3c). Since rapamycin results in re-scaling of the gut, evidenced by the reduction in the organ’s total RNA content (Extended Data Fig. 7f), we also confirmed that the drug reduced pre-tRNA levels relative to total RNA (Extended Data Fig. 7g). Interestingly, rapamycin did not cause a decrease in 45S pre-rRNA in the gut (Extended Data Fig. 7h and i), suggesting a lack of sustained Pol I inhibition. Additionally, gut-specific over-expression of Maf1 reduced the levels of pre-tRNAs and extended lifespan (Fig 3d, Extended Data Fig. 7j), confirming this Pol III repressor acts on Pol III in the adult gut. Our data are consistent with TORC1 driving systemic and gut-specific Pol III activity in the adult fruit fly.
Figure 3. Pol III acts downstream of TORC1 for lifespan.
a, Relative enrichment of Pol III-transcribed genes is higher than that of Pol II-transcribed genes after ChIP against TOR (p<10-4, Linear Model [LM] with an a priori contrast, n= 3 biologically independent samples). Rapamycin feeding causes a decrease in pre-tRNAs relative to U3 in: b, whole female flies (p=0.01, MANOVA, CI= 0.76-1.2, 0.58-0.79, 0.75-1.3, 0.49-0.79, 0.70-1.3, 0.48-0.78 left to right); c, their guts (p=0.01, MANOVA, CI= 0.98-1.1, 0.90-1.0, 0.87-1.1, 0.74-0.99, 0.90-1.1, 0.77-0.97 left to right). d, Induction of Maf1 in the guts of TIGS>HA-Maf1 females by RU486 feeding reduces the levels of pre-tRNAs relative to U3 (p=4x10-3, MANOVA, CI= 0.87-1.1, 0.74-1.0, 0.90-1.1, 0.82-1.0, 0.76-1.2, 0.53-1.0 left to right). e, Induction of dC160RNAi in the adult guts by RU486 feeding of TIGS>dC160RNAi females and rapamycin feeding both extend lifespan and are not additive (effect of rapamycin p=2x10-14, effect of RU486 p<2x10-16, interaction p=7x10-9, Cox Proportional Hazards [CPH], n= 135 control, 144 +RU486, 141 +rapamycin and 146 +RU486 and rapamycin animals; single trial). f, Induction of dC160RNAi in the ISCs by RU486 in GS5961>dC160RNAi females and rapamycin both extend lifespan and are not additive (effect of rapamycin p=8x10-14, effect of RU486 p=2x10-5, interaction p=3x10-7, CPH, n= 113 control, 130 +RU486, 145 +rapamycin and 144 +RU486 and rapamycin animals; single trial). Rapamycin but not dC160RNAi induction in the gut by RU468 feeding of TIGS>dC160RNAi females leads to: g, reduction in S6K phosphorylation in the gut or the whole fly (representative of four biologically independent repeats; see Extended Data Fig. 8c-f); h, reduction in egg laying (effect of rapamycin p<10-4, RU486 p=0.87 and interaction p=0.96, LM). i, Model of the relationship between TORC1, Pol III and lifespan. Bar charts show mean ± SEM, with n= number of biologically independent samples indicated and overlay showing individual data points. For more detailed demography, statistics and summary of lifespan trials see Extended Data Fig. 8a. For gel source data see Sup. Fig.1.
To examine whether Pol III is downstream of TORC1 for lifespan, we combined adult-onset Pol III inhibition with rapamycin treatment. Rapamycin feeding or gut-specific dC160RNAi resulted in the same magnitude of lifespan extension (Fig. 3e). The two treatments were not additive (see Extended Data Fig. 8a for summary and statistical analyses), consistent with their acting in the same longevity pathway. The same was observed with RNAi against dC53 in the gut (Extended Data Fig. 8b), as well as when dC160RNAi expression was restricted to the ISCs (Fig. 3f). Importantly, rapamycin feeding also inhibited the phosphorylation of TORC1 substrate, S6 kinase3 (S6K), in both the gut and the whole fly, and decreased fecundity, whilst gut-specific induction of C160RNAi did not (Fig. 3g and h, Extended Data Fig. 8c - f). This confirms that Pol III inhibition does not impact TORC1 activity, neither locally nor systemically, and hence, Pol III acts down-stream of TORC1 in ageing (Fig. 3i).
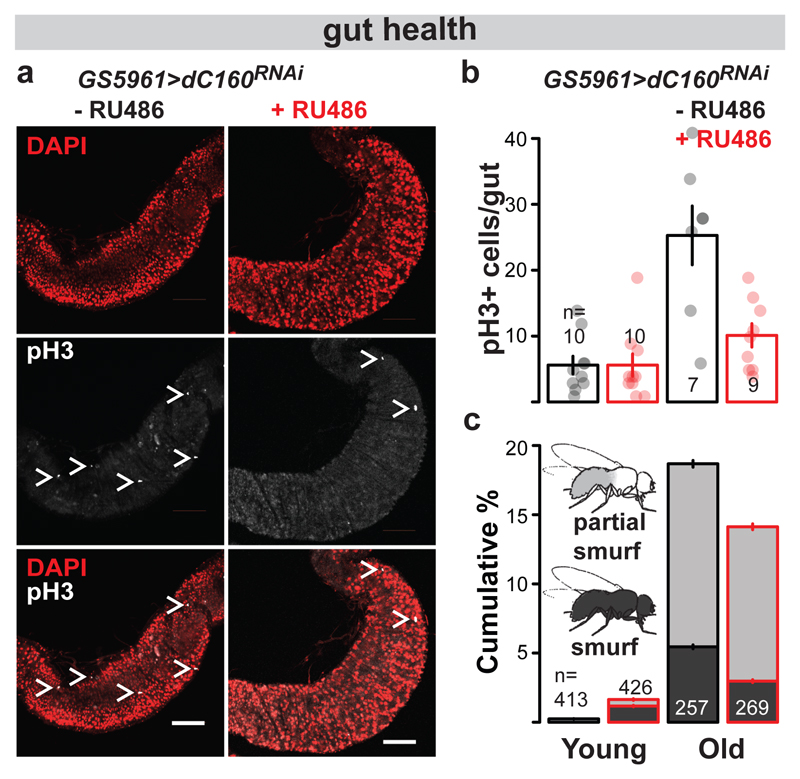
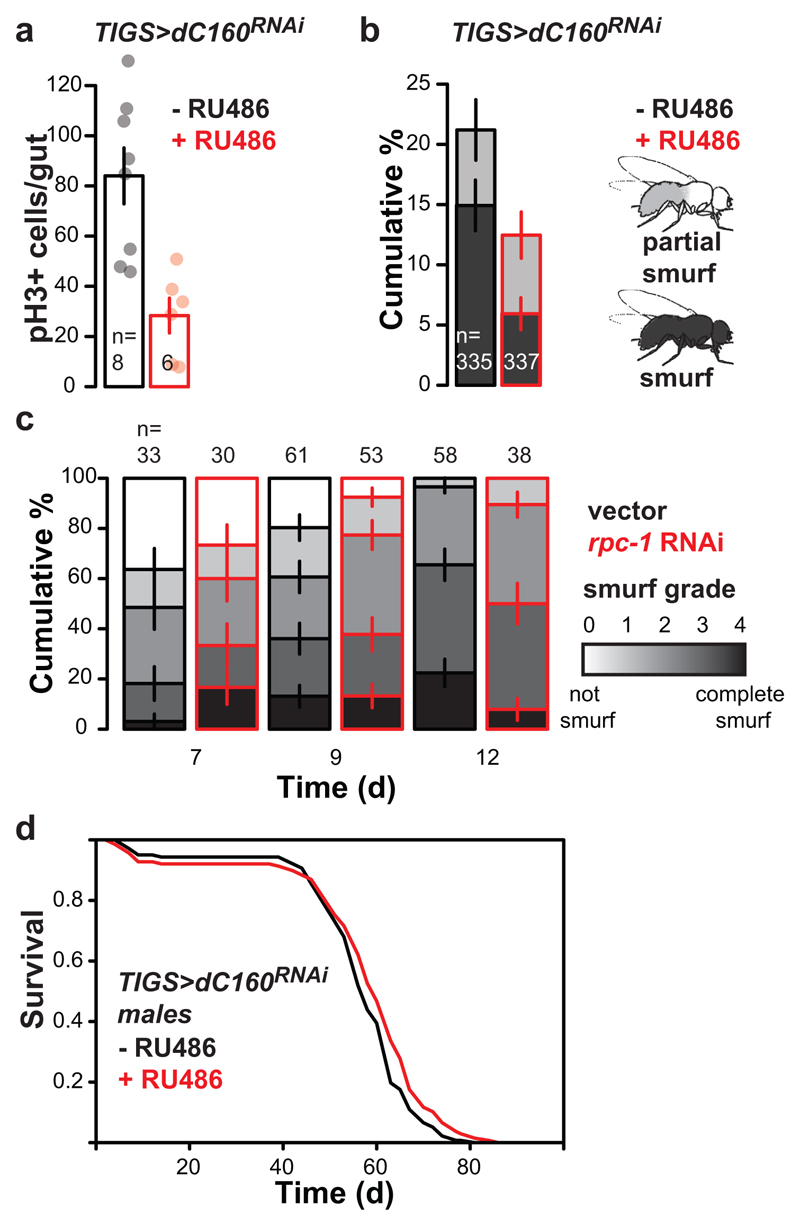
TORC1 inhibition is known to ameliorate age-related pathology and functional decline of the gut22. We examined whether inhibition of Pol III was sufficient to block the dysplasia resulting from ISC hyperproliferation and mis-differentiation by assessing the characteristic, age-dependent increase in dividing, phospho-histone H3 positive (pH3+) cells16. Inducing dC160RNAi in the fly gut or solely in the ISCs ameliorated this pathology (Extended Data Fig. 9a, Fig. 4a and b). The treatment was also sufficient to counteract the age-related loss of gut barrier function, decreasing the number of flies displaying extra-intestinal accumulation of a blue food dye (“smurf” phenotype23, Extended Data Fig. 9b, Fig. 4c). Interestingly, we also found that rpc-1 RNAi feeding reduced the severity of age-related loss of gut-barrier function in worms (Extended Data Fig. 9c). In Drosophila, gut health24 and TORC1 inhibition25 are specifically linked to female survival. Indeed, induction of dC160RNAi in the gut had a sexually dimorphic effect on lifespan, as the effect on males, albeit significant, was reduced in magnitude relative to females (Extended Data Fig. 9d). Overall, our data show that gut/ISC-restricted inhibition of Pol III, which extends lifespan, is sufficient to ameliorate age-related impairments in gut health, which may be causative or correlate with this longevity.
Figure 4. Stem cell-restricted Pol III inhibition improves age-related dysplasia and gut barrier function.
dC160RNAi induction in the ISCs of adult GS5961>dC160RNAi females by RU486 feeding supresses age-related accumulation of pH3 positive cells: a, images of pH3 staining in the posterior mid-gut at 70 d of age (white bar = 100 μm, white “>” marks pH3+ cells, representative of seven -RU486 and nine +RU486 animals); b, the number of pH3+ cells per gut (effect of age p<10-4, RU486 p=2x10-3, interaction p=2x10-3, LM, young: 7-9 d, old: 56-70 d, CI= 2.6-8.6, 1.8-9.4, 14-36, 6.0-14 left to right). c, dC160RNAi induction in the ISCs of adult GS5961>dC160RNAi females by RU486 feeding reduces the age-related increase in the number of flies with a leaky gut (effect of age p<10-4, RU486 p=0.09, interaction p=0.01, Ordinal Logistic Regression, young: 21 d, old: 58 d, CI= 0.19-0.28, 1.5-1.8, 18-19, 14-15 % smurf left to right). Bar charts show mean ± SEM, with n= number of biologically independent animals indicated and overlay showing individual data points.
Our study demonstrates that adult-onset decrease in the growth-promoting, anabolic function mediated by Pol III in the gut, and specifically in the stem cell compartment, is sufficient to recapitulate the longevity benefits of rapamycin treatment. Pol III activity is essential for growth6; its detrimental effects on ageing suggest an antagonistic pleiotropy26 where wild-type levels of Pol III activity are optimised for growth and reproductive fitness in early life but prove detrimental for later health. We reveal a fundamental role for Pol III in adult physiology, implicating wild-type Pol III activity in age-related stem cell dysfunction, declining gut health and organismal survival, downstream of nutrient signalling pathways. The longevity resulting from partial Pol III inhibition in adulthood likely stems from the reduced provision of protein synthetic machinery, however, differential regulation of tRNA genes or Pol III-mediated changes to chromatin organisation may also be involved, as has been suggested in other contexts2. The strong structural and functional conservation of Pol III in eukaryotes suggests that studies of its influence on mammalian ageing are warranted and may lead to important therapies.
Methods
Yeast stocks, chronological lifespans and microfluidics assessment
pMK43-based cassette was integrated into w303 MATa leu2-3,112 trp1-1 can1-100 ura3-1 pADH-OsTir-9Myc::ADE2::ade2-1 his3-11,15 to produce RPC160 or RPB220 C-terminal AID fusions as described11, confirmed by PCR and absence of growth in presence of 2.5 mM IAA.
Primers for strain construction:
C160 Fw
|
TGTCTATTTGAAAGTCTCTCAAATGAGGCAGCTTTAAAAGCGAACCGTACGCTGCAGGTCGAC
|
C160 Rv
|
AGAAAAATAATACAAATGCTATAAAAAAGTTTAAAAACGACTACTATCGATGAATTCGAGCTCG
|
B220 Fw
|
CCAAAGCAAGACGAACAAAAGCATAATGAAAATGAAAATTCCAGACGTACGCTGCAGGTCGAC
|
B220 Rv
|
ATATATAATGTAATAACGTCAAATACGTAAGGATGATATACTATAATCGATGAATTCGAGCTCG
|
Primers for verification:
C160 Fw
|
TTGGGTCAAACGATGTCG
|
B220 Fw
|
CGCCTTCATACTCTCCAAC
|
C160/B220 Rv
|
TGCCCATCATGGTACCTG
|
For chronological lifespans, the strains were grown to exponential phase (OD600~0.4) in Synthetic Complete medium (2% glucose, 0.5% ammonium sulphate, 0.17% yeast nitrogen base, 0.001% adenine, uracil, tryptophan, histidine, arginine, methionine, 0.0025% phenylalanine, 0.003% tyrosine, lysine, 0.004% isoleucine, 0.005% glutamate, aspartate, 0.0075% valine, 0.01% threonine, 0.02% serine and leucine [w/v]), the culture split and treated with IAA in acetone or acetone alone (0.1%, day 0) and kept with aeration and shaking at 30°C. Cell were harvested for protein extraction after 30 min. Cultures essentially reached stationary phase after 24h. The viability was measured on the indicated days by plating 5 μl of 10-fold serial dilutions starting from initial concentration corresponding to OD600=0.5 on YEPD plates and growth for 2 d at 30°C.
For replicative lifespan, cells from single colonies were inoculated in 10 mL of minimal medium27 with 1% glucose, 0.02% leucine and 0.001% tryptophan, arginine, histidine and uracil (w/v) and pH 5 maintained with K-Phthalate-KOH buffer. The 10 mL cultures were cultivated overnight in 100 mL shake flasks at 30°C and 300 rpm. Still exponential, they were diluted next morning to OD600 of 0.005-0.01 and cultivated for several hours to OD600 of 0.045-0.09 when they were loaded into the microfluidics device as described28,29. The growth medium was aerated in advance by shaking for at least two hours. Trapped in the device, the cells were constantly provided with fresh medium containing the synthetic auxin hormone 1-naphthaleneacetic acid (NAA) at the concentrations of 0.0005, 0.001, 0.005 or 0.01 mM; the control did not contain NAA. These concentrations of the hormone span through the dynamic range of the auxin-based degron system where the degree of protein depletion can be efficiently modulated in the set-up used30. Temperature of 30°C was maintained throughout the experiment.
Microscopic imaging in the bright field channel was performed for up to 5 days with the time interval of 5 minutes, using a Nikon Ti-E inverted microscope equipped with a 40x Nikon Super Fluor Apochromat objective, and halogen lamp with additional UV-blocking filter. For each cell, the time points of (1) the budding events, (2) the eventual cell losses due to wash away, or (3) cell death were recorded by visual inspection of the movies with the help of ImageJ and a custom written macro. For assessment of cell division times, the first six cell cycles of each cell were used.
Worm husbandry, lifespans and gut integrity assay
Prior to experiments animals were maintained at 20°C and grown for at least three generations with ample OP50 Escherichia coli food to assure full viability. The rpc-1 RNAi clone, gene code C42D4.8, was obtained from the Ahringer library. Lifespan assays were performed on HT115 E. coli expressing either the rpc-1 RNAi plasmid or pL4440 empty vector control. Experiments were carried out at both 20°C and 25°C. Worms were scored as dead or alive at intervals and worms that crawled off the plate or died from explosion or bagging phenotypes were censored. rpc-1 RNAi treatment from L4 stage increased the incidence of a vulval explosion phenotype (noted in Extended Data Fig. 2c). However, we found that at 25°C this phenotype was greatly reduced (Extended Data Fig. 2c). For gut-restricted RNAi, the VP303 strain was used14. The “smurf” assay for gut integrity was carried out essentially as described31. Worms were aged from the L4 stage at 25°C and on the appropriate day soaked in blue dye for 3 hours. The dye was removed by allowing the worms to crawl around on a bacterial lawn for 30 minutes prior to microscopy analysis. Individual worms were scored from 0-4 for their degree of “smurfness” with 0 being no blue beyond the gut barrier and 4 being completely blue.
Fly husbandry, lifespan, tunicamycin survival, smurf and fecundity assays
The outbred, wild-type stock was collected in 1970 in Dahomey (now Benin) and has been kept in population cages to maintain lifespan and fecundity at levels similar to wild-caught flies. w1118 mutation was backcrossed into the stock and Wolbachia cleared by tetracycline treatment. TIGS32 (a.k.a. TIGS-2), GS596133, S110634, elavGS35, UAS-HA-Maf136, UAS-dC160RNAi and UAS-dC53RNAi (v30512 and v103810 from Vienna Drosophila Resource Centre), and dC53EY22749 (CG5147EY22749 from Bloomington Stock Centre) were all backcrossed at least 6 times into the w1118 Dahomey background.
Stocks were maintained and experiments conducted at 25°C on a 12L:12D cycle at 60% humidity, on SYA food37 containing 10% brewer’s yeast, 5% sucrose, and 1.5% agar (all w/v; with propionic acid and Nipagin as preservatives). RU486 (Sigma) and Rapamycin (LC Laboratories, both dissolved in ethanol) were added to 200μM final concentration as required. Tunicamycin (Cell Signaling, DMSO stock) was used in food with only sugar and agar at concentration of 10mg/l. For control treatments, equivalent volumes of the vehicle alone were added.
Flies were reared at standardised larval density and adults collected over 12 h, allowed to mate for 48 h and sorted into experimental vials at a density of 15 flies per vial (10 flies per vial for RNA extractions). For lifespan experiments, flies were transferred to fresh vials and their survival scored three times a week. Flies were transferred onto food containing tunicamycin on day 9 and survival scored once or twice daily. For smurf assays, at the indicated age the flies were placed on SYA food containing 2.5% (w/v) blue dye (FD&C blue dye no. 1, Fastcolors) for 48 h and scored as full smurfs if completely blue or partial smurfs if the dye had leaked out of the gut but not reached the head. Eggs layed over ~24h were counted on day 10. Other phenotypic tests were performed essentially as described38.
RNA extraction, qPCR and RNA-Seq
Synchronised populations of worms were placed on control or rpc-1 RNAi at the L4 stage, grown at 20°C and harvested after 4 days. Ten whole adult flies, or ten dissected mid-guts, were harvested on day 7-9. Total RNA was isolated using TRIZOL (Invitrogen). RNA isolation was quantitative – the amount obtained was proportional to the starting amount. RNA was converted to cDNA using random hexamers and Superscript II (Invitrogen). Quantitative PCR was performed using Power SYBR Green PCR Master Mix (ABI), with the relative standard curve method. For worms, rpc-1 transcript levels were normalised to the geometric mean of three stably expressed reference genes cdc-42, pmp-3 and Y45F10D.4 as described39. For flies, primers specific for pre-tRNAs were designed based on previous biochemical characterisation40 or predicted intronic sequences41.
Primer sequences:
Flies:
Gene of Interest |
Forward Primer |
Reverse Primer |
dC53 (CG5147)
|
GGGTGACCCAGAGTCCCT |
GGCGAGCTCAGCGAAGAG |
pre-rRNA ITS
|
TTAGTGTGGGGCTTGGCAACCT
|
CGCCGTTGTTGTAAGTACTCGCC
|
pre-rRNA ETS
|
GTTGCCGACCTCGCATTGTTCG
|
CGGAGCCAAGTCCCGTGTTCAA
|
pre-tRNAHIS
|
CGTGATCGTCTAGTGGTTAG |
CCCAACTCCGTGACAATG |
pre-tRNAALA
|
CGCACGGTACTTATAATCAG |
CCAGGTGAGGCTCGAACTC |
pre-tRNALEU
|
GCGCCAGACTCAAGATTG |
TGTCAGAAGTGGGATTCG |
Tub
|
TGGGCCCGTCTGGACCACAA |
TCGCCGTCACCGGAGTCCAT |
U3
|
CACACTAGCTGAAAGCCAAG |
CGAAGCCCTGCGTCCCGAAC |
Worms:
Gene of Interest |
Forward Primer |
Reverse Primer |
rpc-1
|
ACGATGGATCACTTGTTTGAAGC |
GTTCCGACAGTCATTGGGGT |
cdc-42
|
CTGCTGGACAGGAAGATTACG |
CTCGGACATTCTCGAATGAAG |
pmp-3
|
GTTCCCGTGTTCATCACTCAT |
ACACCGTCGAGAAGCTGTAGA |
Y45F10D.4
|
GTCGCTTCAAATCAGTTCAGC |
GTTCTTGTCAAGTGATCCGACA |
For RNA-Seq, RNA was further cleaned up with Qiagen RNeasy (Qiagen), ribo-depleted (Ribo-Zero Gold; Illumina) and sequenced on the Illumina platform at Glasgow Polyomics (75 bp, pair-end reads). Transcript abundance was estimated with Salmon42 (https://combine-lab.github.io/salmon/) in quazi-mapping mode onto all Drosophila cDNA sequences (BDGP6), imported with tximport (https://bioconductor.org/packages/release/bioc/html/tximport.html) into R (https://www.r-project.org/) and differential expression determined with DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) using dissection batch as covariate at 10% false discovery rate43.
Western blots, immunoprecipitation (IP), S2 dsRNA treatment, translation assays and ChIP
Proteins were extracted from yeast (10 ml culture), S2 cells (0.1-2 ml culture; S2 cells were obtained from L. Partridge), 10 flies or ten dissected mid-guts with trichloroacetic acid, washed and re-suspended in SDS-PAGE loading buffer, separated by SDS-PAGE and transferred to nitrocellulose. Western blots were performed with anti-puromycin 12D10 antibody (Millipore), anti-Actin (Abcam, ab1801 or ab8224), anti-Myc (Sigma), anti-FLAG (Sigma), anti-phospho-T398-S6K (Cell Signaling, 9209), anti-S6K antibody44 or anti-TOR antibody20.
IPs were performed on ~2mg of total protein extracted from 2-5 ml of S2 cell culture (transfected with pAFW-dTOR, treated with dsRNA or untreated) into 50mM HEPES-KOH pH 8, 100 mM KCl, 5 mM EDTA, 10% glycerol, 0.5% NP-40 and protease inhibitors with 0.5 μl of anti-dTOR serum20, washed five times with the same buffer and eluted into SDS-PAGE sample buffer. dsRNA against dTOR corresponds to fragment 3694-4208 bp of the dTOR open reading frame (this is DRSC02811 from DRSC/TRiP) and was generated with Megascript RNAi Kit (Thermo Fisher Scientific).
Relative translation rates were determined with the SUnSET assay45: 10 mid-guts of 7 day old flies per sample were dissected in ice-cold PBS and kept in 200 μl of ice-cold Schneider’s medium followed by incubation in 1 ml of Schneider’s medium with 10 μg/ml puromycin for 30 min at 25°C. 333 μl of 50 % trichloroacetic acid were added to stop the reaction. Level of puromycin incorporation was determined by western blots.
ChIP was performed as described46 on chromatin prepared from 7-day old, wild-type females using either the anti-TOR antibody raised against a recombinant TOR protein fragment20, or anti-TOR raised against a peptide21. The mock control included no antibody. Enrichment after IP was measured relative to input with qPCR. Primers for 5’ and 3’ end of aop and the P2 InR promoter have been described46,47.
Further primers used:
Gene of Interest |
Forward Primer |
Reverse Primer |
5S rRNA
|
GCCAACGACCATACCACGCTG
|
AGTACTAACCGCGCCCGACG
|
tRNAMET
|
CGCAGTTGGCAGCGCGTAAG
|
CCCCGGGTGAGGCTCGAACT
|
pH3, Prospero and anti-HRP staining
Guts were dissected at indicated ages in ice-cold PBS and immediately fixed in 4% formaldehyde for 30 minutes. The staining was performed essentially as described38 with anti-phospho-H3 antibody (Cell Signaling, 9701), anti-Prospero (Developmental Studies Hybridoma Bank) or anti-HRP48. Guts were mounted in mounting medium with DAPI (Vectastain). pH3 positive cells per midgut were counted on a fluorescence microscope. Representative images were acquired with the Zeiss LSM700 confocal microscope.
Statistical Analysis
Fly and worm: Survival assays were analysed with log-rank test in Excel (Microsoft) or JMP (SAS), or with CPH in R using the survival package (https://cran.r-project.org/web/packages/survival/index.html). All other tests were performed in JMP. Data obtained from dissections, ChIP or westerns were scaled to dissection/chromatin/replicate batch, except for pH3 counts, to account for batching effects. MANOVA was used to test for overall effect of RU486 or rapamycin feeding. For ChIP analysis, “gene” was used as covariate in a LM with an a priory contrast comparing Pol III- to Pol II-transcribed genes. All regression models had a fully factorial design.
Yeast microfluidics platform: The data, including the number of buds produced by each cell and its final event (death or washout), were used for Kaplan-Meier estimation of survival curves with the Lifelines module (Davidson-Pilon, C., Lifelines, (2016), Github repository, https://github.com/CamDavidsonPilon/lifelines) in Python. Plotting and statistical analysis were done in Python.
Extended Data
Extended Data Figure 1. Inhibition of Pol III in yeast.
a, The growth of strains carrying pADH-OsTir with RPC160-AID, RPB220-AID or the control lacking any AID fusion in the presence or absence of 2.5 mM IAA (single trial). b, Chronological lifespans of the control and RPB220-AID strains treated with 0, 0.125 and 0.25 mM IAA. Top panels show a representative of two experiments, performed in parallel with RPC160-AID shown in Fig. 1b. The bottom panels show a single experiment; the improved survival of RPB220-AID was also observed at a higher IAA concentration in a second experiment. c, Yeast replicative lifespan (top panels) is not altered by 1-naphthaleneacetic acid (NAA; analogue of IAA) while cell cycle duration (bottom panels) is. Both were assessed in the pADH-OsTir RPC160-AID strain on a microfluidics dissection platform. The concentrations of NAA span the dynamic range where the degree of protein depletion can be efficiently modulated in this set-up30. The same control data are shown in each panel for comparison. For each NAA concentration one experiment was performed. For replicative lifespans, 95% CIs are indicated by shading, or in brackets for median lifespan, together with log-rank p value. One-sided Mann-Whitney U test was used to test for significant differences in cell cycle duration. No adjustments were made for multiple comparisons. Dashed lines on bottom panels represent medians.
Extended Data Figure 2. Inhibition of Pol III extends worm lifespan.
a, Lifespan is extended by feeding N2 worms with rpc-1 RNAi at 20°C in absence of FUDR (p<10-3 log-rank test, n= 100 control and rpc-1 RNAi treated animals). b, It is also extended at 25°C in presence of FUDR (p=9x10-3, log-rank test, n= 60 control, 77 rpc-1 RNAi animals). c, Summary of each worm lifespan experiment performed including the representative trials presented in the figures. Log-rank test p value is reported. The total number of animals in the trial = dead + censored. In general, fewer worms were censored in control vs rpc-1 RNAi conditions (average of N2 at 25°C 25% vs 38%; average of N2 at 20°C 53% vs 73%; average of VP303 at 25°C 3% vs 4% and average of VP303 at 20°C 37% vs 54%) which is likely to be due to an increased number of gut explosions in the rpc-1 RNAi treated worms. On average 84.9% of control and 85.6% of rpc-1 RNAi-treated censored events occurred before the 25th percentile of the survival curve. Overall, increasing the temperature to 25°C reduced censoring without altering our findings. d, Lifespan is extended when the RNAi against rpc-1 is restricted to the gut using VP303 strain, at 25°C in presence of FUDR (p=9x10-3, log-rank test, n= 84 control, 103 rpc-1 RNAi treated animals). In (a), (b) and (d), a representative of two trials is shown.
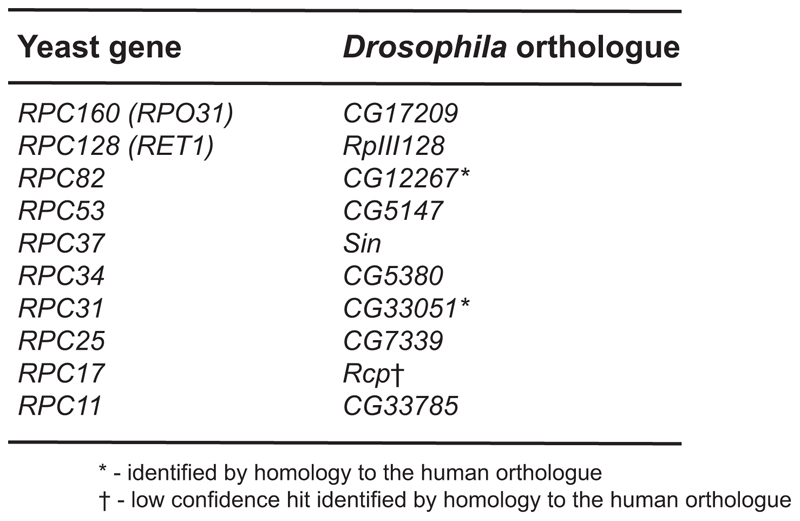
Extended Data Figure 3. Genes corresponding to unique Pol III subunits in Drosophila.
The genes encoding the unique Pol III subunits were identified in fruit flies based on their homology to the yeast genes (BLAST, followed by reverse BLAST), or to the human orthologue.
Extended Data Figure 4. Inhibition of Pol III extends fly lifespan.
a, Summary of each fly lifespan experiment performed including the representative trials presented in the figures but excluding the ones with rapamycin (see Extended Data Fig. 8a). Experiments were performed on females unless otherwise noted. The total number of animals in the trial = dead + censored. Log-rank test p value is reported. RU486 feeding does not have an effect on the lifespans of: b, UAS-dC160RNAi alone (p=0.28, log-rank test, n= 142 -RU486, 146 +RU486 animals); or c, TIGS alone controls (p=0.41, log-rank test, n= 141 -RU486, 145 +RU486 animals). d, Inducing dC53RNAi in the gut of TIGS>dC53RNAi females by RU486 extends their lifespan (p=3x10-6, log-rank test, n= 143 -RU486, 139 +RU486 animals). e, Inducing dC160RNAi predominantly in the fat body of S1106>dC160RNAi females by RU486 has no effect on their lifespan (p=0.21, log-rank test, n= 158 -RU486, 155 +RU486 animals). f, Inducing dC160RNAi in neurons of elavGS>dC160RNAi females by RU486 has a modest effect on their lifespan (p=0.03, log-rank test, n= 148 -RU486, 155 +RU486 animals). g, RU486 feeding does not have an effect on the lifespan of the GS5961 alone control (p=0.88, log-rank test, n= 89 -RU486, 91 +RU486 animals). In (b) to (g) the single trial performed is shown.
Extended Data Figure 5. TIGS is active in ISCs.
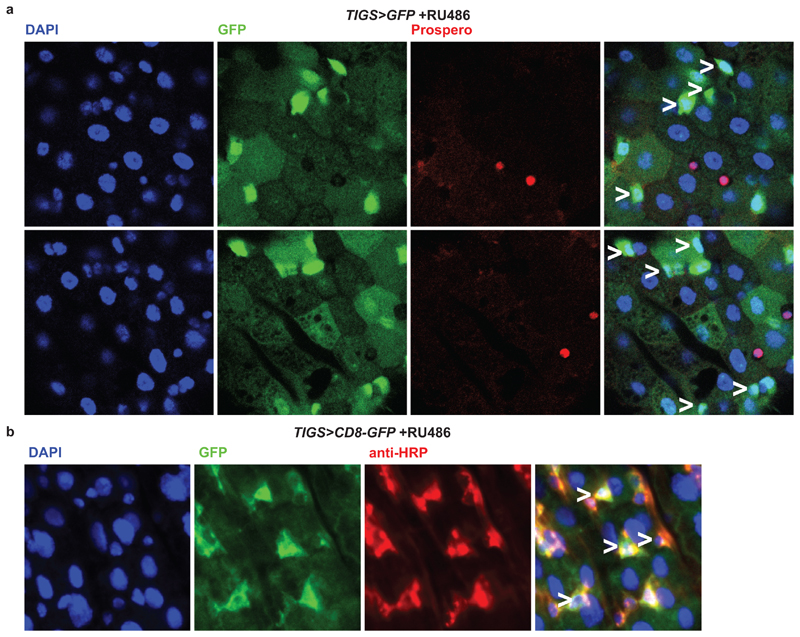
Images from the posterior region of the midgut showing GFP expression driven by TIGS in the presence of RU486 and stained with: a, anti-Prospero; b, anti-HRP. GFP expression can be observed in cells with small nuclei that are Prospero-negative in (a) and those that stain with anti-HRP in (b). Examples of both types are indicated with a white “>” on the merged images. GFP-positive cells can be observed whose morphology and staining pattern correspond closely to that of the ISCs (small nucleus, small cell size, Prospero-negative, anti-HRP-positive [see ref. 48 regarding anti-HRP]). TIGS has a complex expression pattern, showing variation between neighbouring cells of the same type and between gut regions. TIGS appears active in at least some ISCs. Images are representative of two animals.
Extended Data Figure 6. Effects of dC160RNAi induction in Drosophila adult gut.
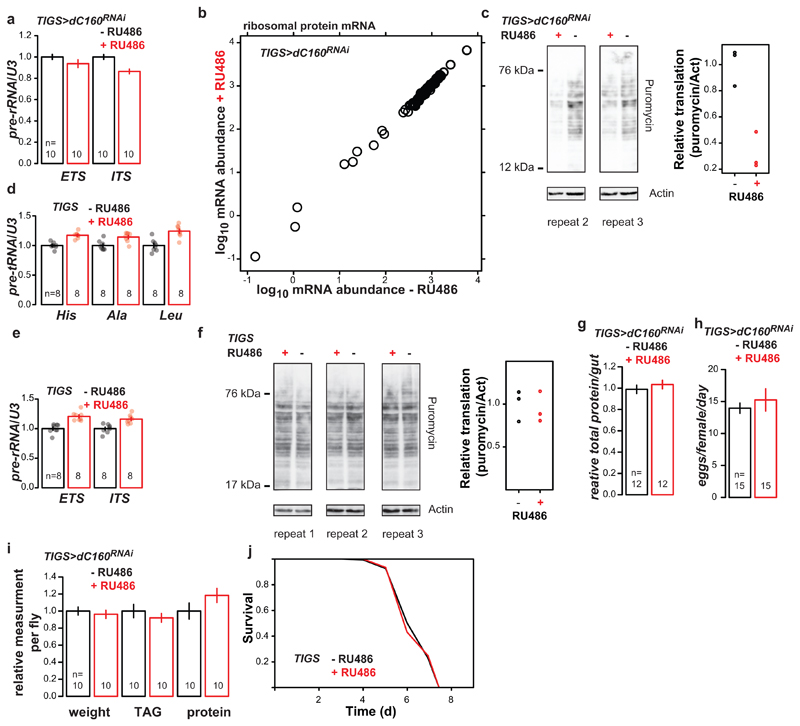
Induction of dC160RNAi in the guts of TIGS>dC160RNAi females results in: a, decreased levels of 45S pre-rRNA (p=4x10-3, MANOVA, CI= 0.95-1.1, 0.85-1.0, 0.95-1.1, 0.81-0.92 left to right; EST = 5’ external transcribed spacer, IST = internal transcribed spacer), indicating a reduction in Pol I activity as a result of Pol III – Pol I crosstalk; b, unaltered levels of mRNAs encoding ribosomal proteins (RNA-Seq data, no significant differences at 10% false discovery rate, DESeq2, n= 3 biologically independent samples), indicating no crosstalk between Pol III and Pol II; c, decreased protein synthesis (two further biological repeats and quantification related to Fig. 2e; p=4x10-3, two-sided t-test, n=3 biologically independent samples, CI= 0.65-1.4 -RU486, -0.033-0.68 +RU486). RU486 feeding of TIGS-alone control females does not result in a significant decrease in: d, levels of pre-tRNAs (p=1x10-4, MANOVA, CI= 0.96-1.0, 1.1-1.2, 0.93-1.1, 1.1-1.2, 0.91-1.1, 1.2-1.3 left to right); e, levels of 45S pre-rRNA (p=2x10-4, MANOVA, CI= 0.94-1.1, 1.1-1.3, 0.94-1.1, 1.1-1.2 left to right); f, protein synthesis (p=0.74, two-sided t-test, n=3 biologically independent samples). Induction of dC160RNAi in the guts of TIGS>dC160RNAi females does not result in significant changes to: g, total gut protein content (p=0.43, two-sided t-test); h, female fecundity (p=0.51, two-sided t-test); i, whole-fly body weight, triacylglycerol or protein content (p=0.58, 0.40, 0.16 respectively, two-sided t-test). j, RU486 feeding of TIGS-alone control females does not result in increased resistance to tunicamycin (p=0.89, log-rank test, n= 149 -RU486, 153 +RU486 animals; single trial). Bar charts show mean ± SEM, with n= number of biologically independent samples indicated and overlay showing individual data points. For gel source data see Sup. Fig. 1.
Extended Data Figure 7. Regulation of Pol III activity by TORC1 in Drosophila.
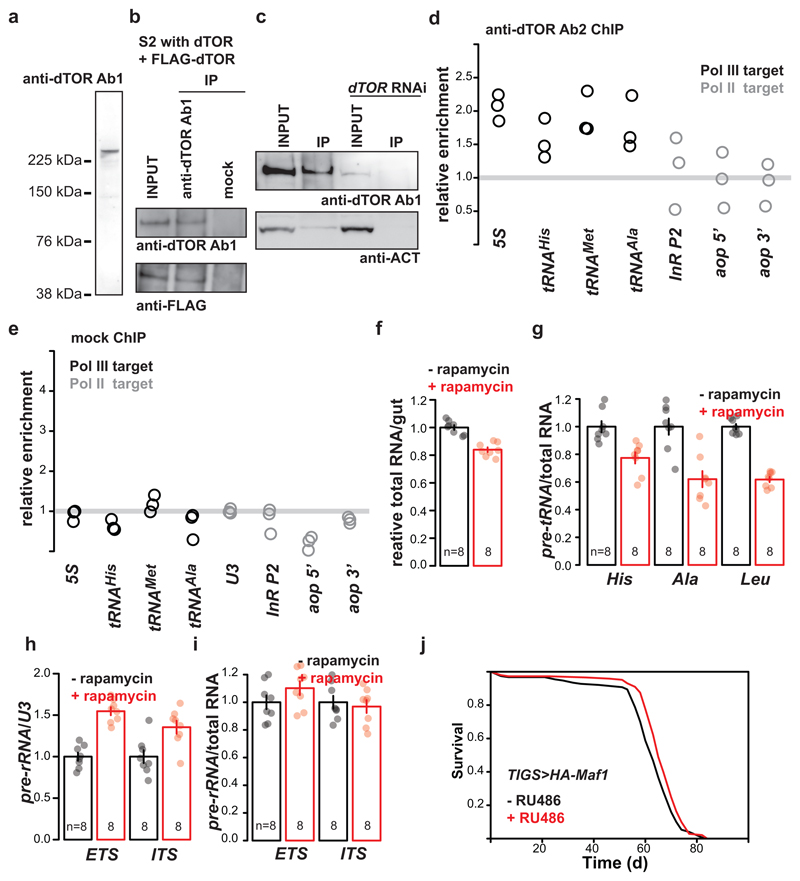
a, The antibody raised against a recombinant fragment of Drosophila TOR protein 20 and used for ChIP (Fig. 3a) recognises a single band of the expected size on western blots of S2 cell extracts. b, The same antibody can immunoprecipitate (IP) dTOR from S2 cells expressing the endogenous and FLAG-tagged dTOR. c, It can also IP endogenous dTOR and the intensity of this band is reduced upon treatment of S2 cells with dsRNA against dTOR. For (a) to (c), a single experiment was performed; the ability of the dTOR RNAi to reduce the intensity of the band was confirmed in an independent experiment. d, Relative enrichment of Pol III-transcribed genes is higher than that of Pol II-transcribed genes after ChIP using a second antibody against Drosophila TOR (raised against a peptide21, p=2x10-4; LM with an a priori contrast, n= 3 biologically independent samples, CI= 1.6-2.6, 0.81-2.3, 1.1-2.7, 0.77-2.8, -0.24-2.5, -0.065-2.0, 0.13-1.7 left to right). e, No enrichment for Pol III-transcribed genes over Pol II-transcribed genes is observed after mock ChIP with no antibody (p=0.09, LM with an a priori contrast, n=3 biologically independent samples). f, Rapamycin feeding results in a decrease in total RNA content of the adult gut (p<10-4, two-sided t-test). g, Rapamycin feeding results in reduction of pre-tRNAs relative to total RNA in the fly gut (p=10-4, MANOVA). Rapamycin feeding does not result in a reduction of pre-rRNA in the fly gut relative to:h, U3 (p<10-4, MANOVA); i, total RNA (p=0.57, MANOVA). j, Feeding RU486 to TIGS>HA-Maf1 female fruit flies to induce HA-Maf1 in the gut alone extends their lifespan (p=0.006, log-rank test, n= 153 -RU486, 146 +RU486 animals; single trial). Bar charts show mean ± SEM, with n= number of biologically independent samples indicated and overlay showing individual data points. For gel source data see Sup. Fig. 1.
Extended Data Figure 8. Relationship between TORC1 and Pol III.
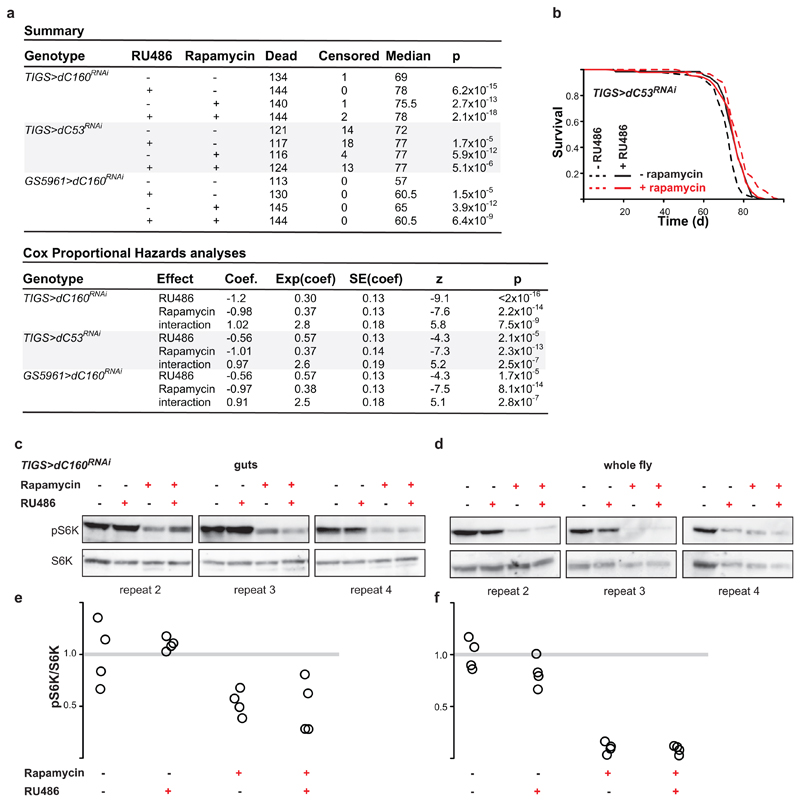
a, Summary of fly lifespans examining the epistasis between Pol III and TORC1 inhibition (top), including the CPH analyses results (bottom). In the summary (top), log-rank test p value, relative to -RU486 -rapamycin control, is reported and the total number of animals in the trial = dead + censored. b, Induction of dC53RNAi in the adult guts by RU486 feeding of TIGS>dC53RNAi females and rapamycin feeding both extend lifespan and are not additive (for statistical analysis see [a]; n= 135 control, 135 +RU486, 120 +rapamycin and 137 +RU486 and rapamycin animals; single trial). c - f, Rapamycin but not induction of dC160RNAi in the guts of TIGS>dC160RNAi females by RU486 reduces the phosphorylation of S6K in the gut (effect of rapamycin p=3x10-4, RU486 p=0.77 and interaction p=0.55, LM, CI= 0.51-1.5, 1.0-1.2, 0.33-0.73, 0.08-0.91 left to right) and whole flies (effect of rapamycin p<10-4, RU486 p=0.10 and interaction p=0.16, LM, CI=0.77-1.2, 0.60-1.0, 0.016-0.19, 0.019-0.15 left to right). Further biological repeats related to Fig. 3g are presented in (c) for the gut and in (d) for the whole fly. These are quantified in (e) and (f) respectively. In (c) to (f), data from four biologically independent samples are shown. For gel source data see Sup. Fig. 1.
Extended Data Figure 9. Inhibition of Pol III in the gut preserves organ health.
a, dC160RNAi induction in the gut of adult TIGS>dC160RNAi females by RU486 feeding supresses accumulation of pH3 positive cells in old flies (p=1x10-3, 2-tailed t-test, CI= 58-110 -RU486, 10-46 +RU486). b, dC160RNAi induction in the gut of adult TIGS>dC160RNAi females by RU486 feeding supresses loss of gut barrier function (number of “smurfs”) in old flies (p=5x10-4, χ2-test, CI=16-26 -RU486, 8.7-16 +RU486 % smurf). c, rpc-1 RNAi suppresses the severity of the age-related loss of gut barrier function in worms (effect of age p<10-4, rpc-1 RNAi p=0.51, interaction p=0.01, Ordinal Logistic Regression, CI=5.0-31, 16-50, 24-48, 25-51, 53-78, 34-66 % smurf grades 3 and 4). Age-related loss of gut barrier function has been previously described for worms31. d, dC160RNAi induction in the gut of adult TIGS>dC160RNAi males by RU486 feeding results in a small but significant extension of lifespan (p=0.03, log-rank-test, n= 141 –RU486, 139 +RU486 animals; single trial). Bar charts show mean ± SEM with n= number of animals indicated and overlay showing individual data points.
Supplementary Material
Acknowledgments
The authors thank S. Grewal, B. Ohlstein, L. Partridge and S. Pletcher for fly lines; C. Bouchoux and F. Uhlmann for yeast reagents; G. Juhasz and A. Teleman for antibodies; E. Bolukbasi and L. Partridge for FLAG-tagged dTor construct and S2 cells; M. Hill and D. Ivanov for help with RNA-Seq analysis; L. Conder, A. Garaeva, D. Mostapha, G. Phillips and P. van der Poel for technical assistance and M. Piper, J. Bähler and the IHA members for support, comments and critical reading of the manuscript. Reagents were obtained from Developmental Studies Hybridoma Bank, Vienna Drosophila Resource Centre, Bloomington Stock Center and the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was funded in part by Biotechnology and Biological Sciences Research Council grant BB/M029093/1, Royal Society grant RG140694 and Medical Research Council grant MR/L018802/1 to NA, and Royal Society grant RG140122 to JMAT. MH and VT received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 642738. DF is a recipient of the UCL Impact PhD studentship.
Footnotes
Code Availability statement
A custom designed code was used to analyse the yeast replicative lifespan data and will be made available upon reasonable request.
Data Availability statement
The data that support the findings of this study are available within the paper and its supplementary information files, including source data for figures, or are available from the corresponding author upon reasonable request. RNA-Seq data have been deposited in ArrayExpress: accession code E-MTAB-5252.
Author contributions
NA conceived the study; DF and NA made the yeast strains and performed chronological lifespans; VT performed and analysed yeast replicative lifespans under supervision of MH; MAT and JWMG performed and analysed worm experiments under supervision of JMAT; DF, AJD, IK and NA performed and analysed fly experiments under supervision of NA; DF, MAT, JMAT and NA wrote the manuscript with contributions from AJD.
Author information
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Roeder RG, Rutter WJ. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 2.Arimbasseri AG, Maraia RJ. RNA Polymerase III Advances: Structural and tRNA Functional Views. Trends in biochemical sciences. 2016;41:546–559. doi: 10.1016/j.tibs.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy BK, Lamming DW. The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metab. 2016;23:990–1003. doi: 10.1016/j.cmet.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Molecular cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. S1097-2765(12)00089-5 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Moir RD, Willis IM. Regulation of pol III transcription by nutrient and stress signaling pathways. Bba-Gene Regul Mech. 2013;1829:361–375. doi: 10.1016/j.bbagrm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grewal SS. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Bba-Gene Regul Mech. 2015;1849:898–907. doi: 10.1016/j.bbagrm.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Vellai T, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 8.Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. 20/2/174 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitto A, et al. Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. eLife. 2016;5 doi: 10.7554/eLife.16351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. nmeth.1401 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 13.Piper MD, Selman C, McElwee JJ, Partridge L. Separating cause from effect: how does insulin/IGF signalling control lifespan in worms, flies and mice? J Intern Med. 2008;263:179–191. doi: 10.1111/j.1365-2796.2007.01906.x. [DOI] [PubMed] [Google Scholar]
- 14.Espelt MV, Estevez AY, Yin X, Strange K. Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-trisphosphate receptor and phospholipases C beta and gamma. The Journal of general physiology. 2005;126:379–392. doi: 10.1085/jgp.200509355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemaitre B, Miguel-Aliaga I. The digestive tract of Drosophila melanogaster. Annual review of genetics. 2013;47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- 16.Biteau B, et al. Lifespan Extension by Preserving Proliferative Homeostasis in Drosophila. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Laferte A, et al. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. 20/15/2030 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Molecular cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 20.Nagy P, et al. Atg17/FIP200 localizes to perilysosomal Ref(2) P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy. 2014;10:453–467. doi: 10.4161/auto.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsokanos FF, et al. eIF4A inactivates TORC1 in response to amino acid starvation. The EMBO journal. 2016;35:1058–1076. doi: 10.15252/embj.201593118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan XL, et al. Rapamycin preserves gut homeostasis during Drosophila aging. Oncotarget. 2015;6:35274–35283. doi: 10.18632/oncotarget.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci U S A. 2012;109:21528–21533. doi: 10.1073/pnas.1215849110. 1215849110 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regan JC, et al. Sex difference in pathology of the ageing gut mediates the greater response of female lifespan to dietary restriction. eLife. 2016;5:e10956. doi: 10.7554/eLife.10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjedov I, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams GC. Pleiotropy, Natural-Selection, and the Evolution of Senescence. Evolution. 1957;11:398–411. doi: 10.2307/2406060. [DOI] [Google Scholar]
Additional references for Methods
- 27.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 28.Lee SS, Avalos Vizcarra I, Huberts DH, Lee LP, Heinemann M. Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform. Proc Natl Acad Sci U S A. 2012;109:4916–4920. doi: 10.1073/pnas.1113505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huberts DH, et al. Construction and use of a microfluidic dissection platform for long-term imaging of cellular processes in budding yeast. Nature protocols. 2013;8:1019–1027. doi: 10.1038/nprot.2013.060. [DOI] [PubMed] [Google Scholar]
- 30.Papagiannakis A, Jonge J, Zhang Z, Heinemann M. Quantitative characterization of the auxin-inducible degron: a guide for dynamic protein depletion in single yeast cells. Scientific Reports. 2017 doi: 10.1038/s41598-017-04791-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gelino S, et al. Intestinal Autophagy Improves Healthspan and Longevity in C. elegans during Dietary Restriction. PLoS genetics. 2016;12:e1006135. doi: 10.1371/journal.pgen.1006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poirier L, Shane A, Zheng J, Seroude L. Characterization of the Drosophila gene-switch system in aging studies: a cautionary tale. Aging Cell. 2008;7:758–770. doi: 10.1111/j.1474-9726.2008.00421.x. [DOI] [PubMed] [Google Scholar]
- 33.Mathur D, Bost A, Driver I, Ohlstein B. A transient niche regulates the specification of Drosophila intestinal stem cells. Science. 2010;327:210–213. doi: 10.1126/science.1181958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giannakou ME, et al. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 35.Niccoli T, et al. Increased Glucose Transport into Neurons Rescues Abeta Toxicity in Drosophila. Current biology : CB. 2016;26:2291–2300. doi: 10.1016/j.cub.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rideout EJ, Marshall L, Grewal SS. Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc Natl Acad Sci U S A. 2012;109:1139–1144. doi: 10.1073/pnas.1113311109. 1113311109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bass TM, et al. Optimization of dietary restriction protocols in Drosophila. J Gerontol A Biol Sci Med Sci. 2007;62:1071–1081. doi: 10.1093/gerona/62.10.1071. 62/10/1071 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alic N, Hoddinott MP, Vinti G, Partridge L. Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging Cell. 2011;10:137–147. doi: 10.1111/j.1474-9726.2010.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC molecular biology. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frendewey D, Dingermann T, Cooley L, Soll D. Processing of Precursor Transfer-Rnas in Drosophila Processing of the 3' End Involves an Endonucleolytic Cleavage and Occurs after 5' End Maturation. Journal of Biological Chemistry. 1985;260:449–454. [PubMed] [Google Scholar]
- 41.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nature methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15 doi: 10.1186/S13059-014-0550-8. Artn 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hahn K, et al. PP2A regulatory subunit PP2A-B' counteracts S6K phosphorylation. Cell Metab. 2010;11:438–444. doi: 10.1016/j.cmet.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nature methods. 2009;6:275–277. doi: 10.1038/NMETH.1314. [DOI] [PubMed] [Google Scholar]
- 46.Alic N, et al. Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol Syst Biol. 2011;7:502. doi: 10.1038/msb.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alic N, et al. Interplay of dFOXO and two ETS-family transcription factors determines lifespan in Drosophila melanogaster. PLoS genetics. 2014;10:e1004619. doi: 10.1371/journal.pgen.1004619. PGENETICS-D-14-00538 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. S0092-8674(11)01081-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.