Supplemental Digital Content is available in the text.
Keywords: apoptosis, colorectal cancer, Kruppel like factor 6, over expression, proliferation
Abstract
Kruppel like factor 6 (KLF6), a member of KLF family, which has classic zinc finger structure, is broadly considered to have anticancer activity. The role of SV2 variant, one of KLF6 alternative splicing isoforms has not yet been definite in the colorectal cancer. This study aimed to detect the expression of the KLF6-SV2 in colorectal cancer and investigate its impact on cell proliferation and apoptosis. qRT-PCR was used to quantitatively determine KLF6-SV2 mRNA expression in colorectal cancer samples, corresponding normal tissue, normal colonic mucosal cell line FHC and seven colorectal cancer cell lines. SW480 and SW620 cell models with over-expressing KLF6-SV2 were constructed. Cell proliferation, cell cycle and apoptosis were measured respectively using MTT assay, DNA ploidy detection and Annexin V flow cytometry. Meanwhile, expression of p53, p21 and Bax were detected by qRT-PCR and western blot. The mRNA expression level of KLF6-SV2 in colorectal cancer tissues (0.783±0.409) was decreased than in corresponding normal tissues (1.086±0.449) (P<0.01), and expression in SW480 and SW620 were lower than in FHC, HCT116, LoVo, HT29, Caco-2 and RKO. In cell lines over-expressing KLF6-SV2, cell proliferation was markedly suppressed, cell cycle was blocked and cell apoptosis was significantly induced. Simultaneously, expression of p21 and Bax were remarkably up-regulated, while p53 remained unchanged. Decreased expression of KLF6-SV2 may be associated with the occurrence and development of colorectal cancer. KLF6-SV2 plays a role as tumor suppressor by efficiently blocking cell proliferation, arresting cell cycle and inducing apoptosis in colorectal cancer, which may be related to increased expression of p21 and Bax.
Introduction
Colorectal cancer (CRC) is one of the most prevalent neoplasms worldwide and is the fifth leading cause of male and female cancer-related death in China (Chen et al., 2016). Since CRC is highly malignant with an insidious onset and invasive fast-growing, most patients have reached the middle or advanced stages at the time of diagnosis. The mechanism of genesis and development of CRC is not well understood. It is consensus that more research on the cancer-related genes in CRC helps to reveal the mechanisms of CRC development and provide basis on finding more methods for early diagnosis and effective therapy.
Kruppel like factor 6 (KLF6) is considered as a tumor suppressor gene attracted attentions. KLF6 is located on human chromosome 10p15 (Fig. 1a). The full length is about 7.0 kb, containing four exons which encode protein that consists of 283 amino acids. Zinc finger domains of KLF6 protein are connected with promoter region elements like GC boxes of the target gene. KLF6 plays a role in regulating multiple biological processes, such as tissue growth and development (Fischer et al., 2001; Park et al., 2005), cell proliferation and differentiation(Inuzuka et al., 1999; Li et al., 2005) and angiogenesis (Botella et al., 2002). KLF6 may be associated with the development of tumors and is generally considered as a tumor suppressor. A study (Narla et al., 2001) found that single nucleotide polymorphisms of KLF6 could increase its alternative splicing, resulting into three kinds of splice variants named SV1, SV2 and SV3. The wild type KLF6 and SV3 variant localize in the nucleus because of a nuclear localization signal (NLS), whereas the SV1 and SV2 variants are predominantly cytoplasmic. KLF6-SV1 accelerates progression and metastasis of multiple cancers, such as prostate cancer (Narla et al., 2005), ovarian cancer (DiFeo et al., 2006) and lung adenocarcinoma (DiFeo et al., 2008).
Fig. 1.
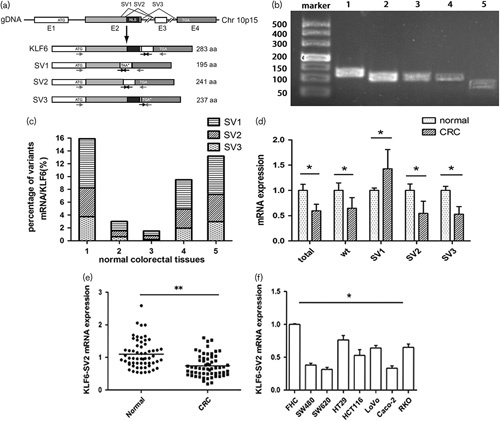
mRNA expression of KLF6-SV2 in CRC. Expression level was detected by qRT-PCR and calculated using the 2-ΔΔCt formula. (a) Gene structure of KLF6 and its splices variants. E1–E4 means exon 1–4. The arrows indicate the position of the primers used for PCR (Hanoun et al., 2010). (b) KLF6 and its splicing variants in normal colorectal tissue. PCR products were visualized after agarose gel electrophoresis. 1–5 stands for total KLF6, wild type KLF6, SV1, SV2, SV3. (c) Percentage of different variants mRNA expression in five normal colorectal tissues. (d) Different expression of KLF6 and alternative splicing in five pairs of normal and CRC tissues. Wt stands for wild type KLF6. (e) Different expression between CRC tumor tissue and adjacent tissue. (f) Different expression between FHC cell and seven CRC cell lines. **P<0.01, *P<0.05. CRC, colorectal cancer; KLF6, Kruppel like factor 6.
Hanoun et al. (2010)found that KLF6-SV2 was down-regulated in hepatocellular carcinoma and displayed antiproliferative and proapoptotic functions. However, another study (Zhenzhen et al., 2012) found KLF6-SV2 mRNA expression in hepatocellular carcinoma tissues was higher than in normal tissues. The different research results on KLF6-SV2 expression in hepatocellular carcinoma and its ambiguous function in CRC aroused our interest in its role in CRC. In this study we planned to investigate its impact on the biological characteristics of CRC and find out its possible mechanisms to provide new theoretical basis and pathogenesis of CRC.
Materials and methods
Materials and reagents
Sixty pairs of CRC tumor tissues and corresponding normal tissues more than 5 cm away from the neoplastic foci were obtained from patients at the Gastrointestinal Surgery Department of Fujian Provincial Hospital from October 2012 to December 2013. After separation, tissues were washed with saline and submerged in RNA preserving fluid, then stored in −80°C freezer. All specimens were diagnosed with colorectal cancer by pathological examination. All patients did not receive radiotherapy and chemotherapy before surgery. Written informed consents were obtained from the patients. The study was approved by Institutional Review Board (IRB) of Fujian Provincial Hospital (K2012-009-01). FHC (normal colonic mucosal cell line), Caco-2 and RKO (CRC cell lines) were purchased from ATCC (Manassas, Virginia, USA). SW480 (CRC cell line) was purchased from Institute of Biochemistry and Cell Biology, SIBS, CAS. SW620, HCT116, LoVo and HT29 (CRC cell lines) were obtained from our laboratory. Anti-p53 antibody was purchased from Abcam (Cambridge, Massachusetts, USA). Anti-p21 and anti-Bax antibody were purchased from Cell Signaling Technology (Danvers, Massachusetts, USA). Anti-GAPDH and second antibody were purchased from Beijing CWbiotech Co (Beijing, China).
Cell culture
FHC, Caco-2 and RKO were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum. SW480 and SW620 were grown in L-15 medium supplemented with 10% fetal bovine serum. HCT116, LoVo and HT29 were grown in DMEM medium supplemented with 10% fetal bovine serum. Penicillin and streptomycin were also added into medium.
Real-time fluorescent quantitative PCR
Total RNA was extracted from CRC tissues and cell lines using Trizol reagent. 1 μg RNA was reverse-transcribed to cDNA using Revert Aid cDNA Synthesis Kit (Invitrogen; Waltham, Massachusetts, USA). Quantitative PCR for five pairs of normal and CRC tissues was performed with primers (Supplementary Table S1, Supplemental digital content 1, http://links.lww.com/EJCP/A178) using SYBR Mix Plus (Takara; Japan). Real-time PCR was performed using GoldStar TaqMan Mixture (CWbiotech Co.) following manufacturer’s protocol. 1 µl of cDNA in 25 µl final volumes was subject to PCR amplification consisting of 95°C 10 min, 40 cycles (15 s at 95°C, 1 min at 60°C). The GAPDH mRNA expression was applied as an internal control. 2-ΔΔCt formula was used to calculate mRNA expression level. Primer and probe sequences are listed in Supplementary Table S2 (Supplemental digital content 1, http://links.lww.com/EJCP/A178).
Construction of cell lines over-expressing KLF6-SV2
KLF6-SV2 coding sequence were obtained by PCR amplification and inserted into Ubi-MCS-3FLAG-CMV-EGFP vector (GV365; Genechem; Nanjing, China). GV365, pHelper 1.0 and pHelper 2.0 cotransfected 293T cell. Lentivirus particles were obtained from supernatant after concentration and purification. SW480 and SW620 cells were incubated with lentivirus vector (MOI=10) in six-well plates. After 12 h, culture medium containing virus was aspirated from the well and fresh complete medium was added. Expression intensity of green fluorescent protein was observed 3 days later and the effect of over-expression of KLF6-SV2 was detected. Negative control group (infected with negative control lentivirus) and blank control group (uninfected) were set up simultaneously.
Cell proliferation assay
After transfection, cells in the logarithmic phase from each group were collected and seeded at 3000 cells/well of a 96-well plate. 10 μl 5 mg/ml of MTT solution was added to each well at the first, second, third, fourth and fifth day after cell seeding. After incubation for 4 h at 37°C, solution was discarded and 150 μl DMSO was added. The plate was taken for colorimetric analysis on microplate reader at 490 nm with 630 nm as reference wavelength.
Measurement of cell apoptosis
Cells were washed twice with cold PBS and then adjusted to 1×106/ml of cell concentration. Cell suspension was incubated with Annexin V-APC and PI in dark condition. Apoptotic cells were detected by flow cytometry in 1 h.
Detection of cell cycle
Cells collected from each group were washed with cold PBS and then adjusted to 1×106/ml of cell concentration. After fixation with 70% ethyl alcohol overnight, cells were incubated with RNaseA in 37°C water bath for 30 min followed by PI dye liquor at 4°C for 30 min. DNA content was measured by flow cytometry.
Western blot
Protein was extracted using RIPA lysis buffer and quantified by BCA method. 15 μg protein sample were subjected to SDS-PAGE and transferred onto PVDF membrane and blocked in 5% skimmed milk powder for 2 h and incubated with primary antibody overnight at 4°C. The membranes were washed with TBST and incubated with HRP conjugated goat anti-rabbit IgG for 2 h. Protein expression was detected by chemiluminescent system. GAPDH protein was used as the loading control.
Statistical analysis
SPSS 17.0 software (Armonk, New York, USA) was used to analyze data. Results were expressed as mean±SD. Significant difference of two groups was compared using t-test, and the correlation of KLF6 expression with clinicopathologic parameters was analyzed by χ2-test, with P value of less than 0.05 as statistically significance.
Results
KLF6-SV2 mRNA expression in colorectal cancer tissues and cell lines and its relationship with clinicopathologic parameters of colorectal cancer
Wild type, splice variants and total KLF6 mRNA expression in colorectal sample were showed by agarose gel electrophoresis in Fig. 1b. The splice variants represented 1.5–15.9% compared to wild type KLF6, and SV1 mRNA expression was higher than SV2 and SV3 (Fig. 1c).The total, wild type, SV2 and SV3 of KLF6 were decreased except for SV1 expression increased in CRC samples (Fig. 1d). The mRNA expression level of KLF6-SV2 in CRC tissues was 0.783±0.409 (in the value of 2-ΔΔCt, the same below), which was lower than in corresponding normal tissues at 1.086±0.449 (P<0.01) (Fig. 1e). Reduced expression of KLF6-SV2 was found in 46/60 (76.7%) samples, while increased expression was found in 14 cases. There was no significant correlation between KLF6-SV2 expression and sex, ages, tumor location, depth of invasion, lymph node metastasis and pathological grading (P>0.05, Table 1). KLF6-SV2 expression in SW480 and SW620 were lower than in FHC, HCT116, LoVo, HT29, Caco-2 and RKO (P<0.05) (Fig. 1f).
Table 1.
Relationship between KLF6-SV2 expression and clinicopathologic parameters of colorectal cancer
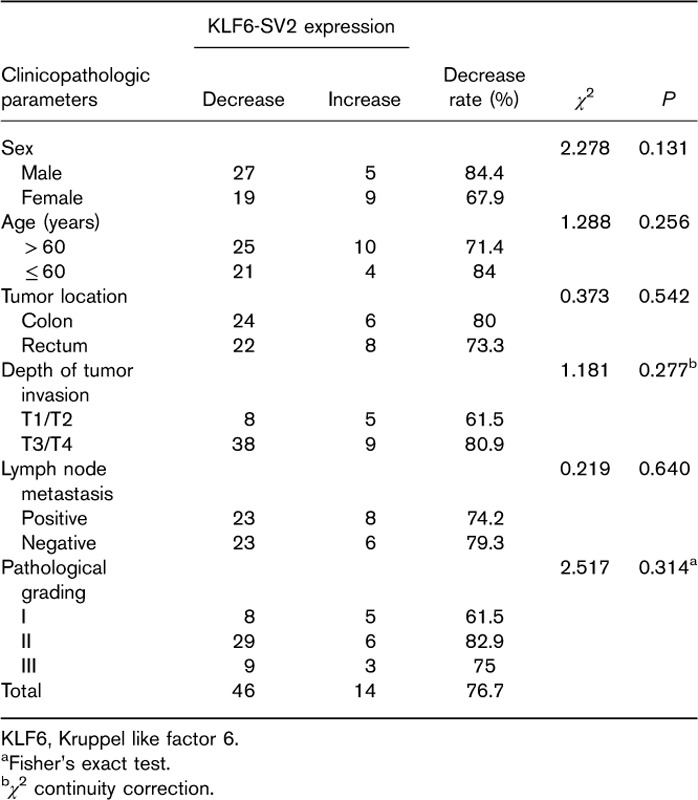
Detection of KLF6-SV2 over-expression
After transfection, green fluorescent protein expression was firstly observed under fluorescent microscope. The result showed transfection efficiency was above 80% (Fig. 2a). Quantitative PCR results revealed that KLF6-SV2 mRNA expression in over-expressing group of SW480 and SW620 cell lines were significantly increased compared with negative control group (Fig. 2b), which indicated KLF6-SV2 was over-expressed successfully.
Fig. 2.
Construction of SW480 and SW620 stable cell lines over-expressing KLF6-SV2. (a) Expression intensity of GFP under fluorescent microscope. (b) Expression of the KLF6-SV2 mRNA in every group. BC, blank control group; GFP, green fluorescent protein; KLF6, Kruppel like factor 6; NC, negative control group; OE, over-expressing group. **P<0.01, #P>0.05.
Impact of KLF6-SV2 over-expression on proliferation of SW480 and SW620 cells
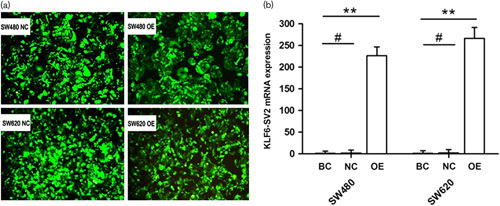
MTT assay demonstrated that as culture time went on, proliferative activity of over-expressing group in SW480 and SW620 on third to fifth day was remarkably lower than negative control group and cell growth was significantly inhibited (Fig. 3a). Proliferation inhibition rates of SW480 were 19.8, 22.0 and 25.8%, and those of SW620 were 23.3, 25.3 and 32.4%.
Fig. 3.
Impact of over-expressing SV2 on proliferation, apoptosis and cell cycle. (a) Cell proliferation of SW480 and SW620 was detected by MTT assay at different time. (b) Apoptotic difference of groups in SW480 and SW620. (c) Cell proportion in different phases of the cell cycle measured by flow cytometry. BC, blank control group; NC, negative control group; OE, over-expressing group. *P<0.05.
Impact of KLF6-SV2 over-expression on apoptosis of SW480 and SW620 cells
The apoptotic rates of over-expressing group in SW480 and SW620 were 35.87±3.52 and 11.69±0.85%, respectively, while the rates of negative control groups were 5.45±1.83 and 0.46±0.25%. The results suggested that apoptotic cells were increased in over-expressing group (P<0.05) (Fig. 3b).
Impact of KLF6-SV2 over-expression on cell cycle of SW480 and SW620 cells
DNA ploidy assay manifested that the percentage of G0/G1 phase cells in SW480 over-expressing group was 66.79±3.42%, which was dramatically higher than negative control group 55.97±3.18% (P<0.05). Similarly, the result also appeared on SW620, with 62.46±3.87% in over-expressing group and 41.17±2.56% in control group (Fig. 3c) (P<0.05). The percentage of S phase and G2/M phase cells were reduced.
mRNA and protein expression of p53, p21 and Bax
PCR and western blot results showed, compared with negative control group, mRNA and protein expression levels of p21 and Bax were significantly up-regulated (P<0.05), whereas p53 expression appeared no different (P>0.05) (Fig. 4a and b).
Fig. 4.
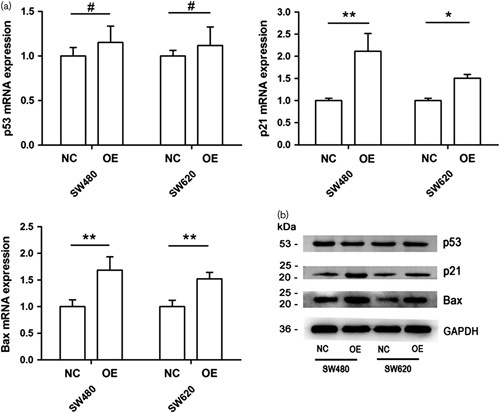
Impact of KLF6-SV2 over-expression on p53, p21 and Bax genes. (a) Expression of p53, p21 and Bax mRNA by qRT-PCR in SW480 and SW620. (b) Expression of p53, p21 and Bax protein by western blot in SW480 and SW620. NC, negative control group; OE, over-expressing group.**P<0.01, *P<0.05, #P>0.05. KLF6, Kruppel like factor 6.
Discussion
Proliferation of tumor cells is a continuous process, which can be regulated by activation of oncogenes or inactivation of tumor suppressors. Rapid cell proliferation would further enhance the ability of invasion and migration, leading the tumor to easily diffuse to other tissues and organs. Apoptosis refers to spontaneous programmed cell death, which can maintain the stability of internal environment in the tissue. Disturbance of apoptosis mechanism will result in reduction of tumor cell death, rapid increase of tumor cell number and fast growth of tumor tissue. Bax is the most critical proapoptotic protein in Bcl-2 family. Two monomer molecules of Bax combine together to form a functional homodimer. Higher content of Bax monomer will produce more homodimers to induce cells to get into the program of apoptosis.
Many studies have demonstrated that KLF6 is related to various cell biological functions such as proliferation and apoptosis in multiple tumors. It was documented that 73% of hepatocellular carcinoma showed decreased expression of KLF6 mRNA, and the extent of decline was much more significant during the transfer stages of advanced liver cancer (Kremer-Tal et al., 2007). Camacho-Vanegas et al. (2007) found that cell proliferation dropped nearly to a half of the original level when KLF6 was over-expressed in glioblastoma study. Conversely, after knocking out KLF6 gene, cell proliferation was obviously increased to about 4.5 times. A similar finding that interfering the expression of KLF6 accelerated cell proliferation was also reported in ovarian cancer (DiFeo et al, 2006). In non-small-cell lung cancer with silenced p53 gene and over-expressed KLF6, apoptotic cells showed an obvious upward trend (Ito et al., 2004). One study indicated that KLF6 could also up-regulate cyclin-dependent kinase inhibitor p27 to inhibit cell proliferation (Tahara et al., 2009). Huang et al. (2008)reported that knockout of AFT3 through siRNA interference could block apoptosis induced by KLF6.
In the present study, we firstly determined the mRNA expression of KLF6 in five pairs of normal and CRC tissues. The results showed that total, wild type, SV2 and SV3 of KLF6 were decreased except for SV1 expression increased in CRC samples. The phenomenon of total KLF6 decrease and SV1 expression increase were also observed in the previously reported in other tumor tissues (Ito et al., 2004; DiFeo et al., 2006). In this study we focused on KLF6-SV2 expression and function in the CRC. We further examined its expression levels in 60 pairs of CRC tissues and eight CRC cell lines. We found its expression in CRC tumor tissue on the whole was reduced in 46 out of 60 samples and in the cell lines were showed various levels. We chose two cell lines with lowest expression levels, SW480 and SW620, to construct cell models over-expressing KLF6-SV2 by lentivirus transfection for biological characteristics experiments. We observed that over-expression of KLF6-SV2 could inhibit CRC cell proliferation, block cell cycle and induce cell apoptosis. These results were consistent with Hanoun’s research on hepatocellular carcinoma. Our study on the mechanism of KLF6-SV2 on the CRC cells proliferative inhibition and apoptosis induction demonstrated that after over-expression of KLF6-SV2, expression of cyclin-dependent kinase inhibitor p21 and proapoptotic gene Bax were significantly up-regulated, while no noticeable change of p53 was found. This result suggested that KLF6-SV2 plays a role as tumor suppressor in CRC by up-regulating expression of p21 and Bax with p53-independent manner and through arresting cell cycle at G1 phase and inducing apoptosis.
Conclusion
This study showed that KLF6-SV2 expression was decreased in CRC, and indicated that KLF6-SV2 plays a role as tumor suppressor in CRC by efficiently blocking cell proliferation, arresting cell cycle and inducing apoptosis, which may be related to increased expression of p21 and Bax. Nevertheless, regulatory mechanism of KLF6-SV2 on CRC invasion and migration still needs further research.
Supplementary Material
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (www.eurjcancerprev.com).
Acknowledgements
This work was supported by the Medical Innovation Project of Fujian Province, China (2012-CXB-6).
Conflicts of interest
There are no conflicts of interest.
References
- Botella LM, Sanchez-Elsner T, Sanz-Rodriguez F, Kojima S, Shimada J, Guerrero-Esteo M, et al. (2002). Transcriptional activation of endoglin and transforming growth factor-beta signaling components by cooperative interaction between Sp1 and KLF6: their potential role in the response to vascular injury. Blood 100:4001–4010. [DOI] [PubMed] [Google Scholar]
- Camacho-Vanegas O, Narla G, Teixeira MS, DiFeo A, Misra A, Singh G, et al. (2007). Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int J Cancer 121:1390–1395. [DOI] [PubMed] [Google Scholar]
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. (2016). Cancer statistics in China, 2015. CA Cancer J Clin 66:115–132. [DOI] [PubMed] [Google Scholar]
- DiFeo A, Narla G, Hirshfeld J, Camacho-Vanegas O, Narla J, Rose SL, et al. (2006). Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res 12:3730–3739. [DOI] [PubMed] [Google Scholar]
- DiFeo A, Feld L, Rodriguez E, Wang C, Beer DG, Martignetti JA, et al. (2008). A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res 68:965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer EA, Verpont MC, Garrett-Sinha LA, Ronco PM, Rossert JA. (2001). Klf6 is a zinc finger protein expressed in a cell-specific manner during kidney development. J Am Soc Nephrol 12:726–735. [DOI] [PubMed] [Google Scholar]
- Hanoun N, Bureau C, Diab T, Gayet O, Dusetti N, Selves J, et al. (2010). The SV2 variant of KLF6 is down-regulated in hepatocellular carcinoma and displays anti-proliferative and pro-apoptotic functions. J Hepatol 53:880–888. [DOI] [PubMed] [Google Scholar]
- Huang X, Li X, Guo B. (2008). KLF6 induces apoptosis in prostate cancer cells through up-regulation of ATF3. J Biol Chem 283:29795–29801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Nanbu-Wakao R, Masuho Y, Muramatsu M, Tojo H, Wakao H. (1999). Differential regulation of immediate early gene expression in preadipocyte cells through multiple signaling pathways. Biochem Biophys Res Commun 265:664–668. [DOI] [PubMed] [Google Scholar]
- Ito G, Uchiyama M, Kondo M, Mori S, Usami N, Maeda O, et al. (2004). Kruppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res 64:3838–3843. [DOI] [PubMed] [Google Scholar]
- Kremer-Tal S, Narla G, Chen Y, Hod E, DiFeo A, Yea S, et al. (2007). Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. J Hepatol 46:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Yea S, Li S, Chen Z, Narla G, Banck M, et al. (2005). Kruppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J Biol Chem 280:26941–26952. [DOI] [PubMed] [Google Scholar]
- Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, et al. (2001). KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563–2566. [DOI] [PubMed] [Google Scholar]
- Narla G, DiFeo A, Helen LR, Schaid DJ, Hirshfeld J, Hod E, et al. (2005). A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res 65:1213–1222. [DOI] [PubMed] [Google Scholar]
- Park JH, Eliyahu E, Narla G, DiFeo A, Martignetti JA, Schuchman EH. (2005). KLF6 is one transcription factor involved in regulating acid ceramidase gene expression. Biochim Biophys Acta 1732:82–87. [DOI] [PubMed] [Google Scholar]
- Tahara E, Kadara H, Lacroix L, Lotan D, Lotan R. (2009). Activation of protein kinase C by phorbol 12-myristate 13-acetate suppresses the growth of lung cancer cells through KLF6 induction. Cancer Biol Ther 8:801–807. [DOI] [PubMed] [Google Scholar]
- Zhenzhen Z, De’an T, Limin X, Wei Y, Min L. (2012). New candidate tumor-suppressor gene KLF6 and its splice variant KLF6 SV2 counterbalancing expression in primary hepatocarcinoma. Hepatogastroenterology 59:473–476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (www.eurjcancerprev.com).


