Abstract
The pluripotency of embryonic stem cells (ESCs) relies on appropriate responsiveness to developmental cues. Promoter-proximal pausing of RNA polymerase II (Pol II) has been suggested to play a role in keeping genes poised for future activation. To identify the role of Pol II pausing in regulating ESC pluripotency, we have generated mouse ESCs carrying a mutation in the pause-inducing factor SPT5. Genomic studies reveal genome-wide reduction of paused Pol II caused by mutant SPT5 and further identify a tight correlation between pausing-mediated transcription effect and local chromatin environment. Functionally, this pausing-deficient SPT5 disrupts ESC differentiation upon removal of self-renewal signals. Thus, our study uncovers an important role of Pol II pausing in regulating ESC differentiation and suggests a model that Pol II pausing coordinates with epigenetic modification to influence transcription during mESC differentiation.
Keywords: Transcription pausing, Mouse embryonic stem cell, Global run-on sequencing (GRO-seq)
1. Introduction
Understanding the mechanisms governing self-renewal and differentiation of pluripotent embryonic stem cells (ESCs) is critical for designing efficient and safe protocols for their use in regenerative medicine. The hallmarks of pluripotency; i.e. maintaining unlimited self-renewal potential while being equipped to activate differentiation programs upon signals are regulated through a set of ESC transcription factors and epigenetic marks (Bernstein et al., 2006; Marks et al., 2012; Young, 2011). These together allow for continuous activation of stem cell factors and repression of developmental regulators. Many mechanisms have been proposed to regulate this unique ability of ESCs, including the deposition of “bivalent” histone marks and RNA polymerase II (Pol II) pausing at initial stages of transcription (Marks et al., 2012; Tee et al., 2014).
Pol II pausing occurs soon after transcription initiation and is mainly regulated by two pausing factors DSIF (DRB-Sensitivity Inducing Factor) and NELF (Negative Elongation Factor) (Rougvie and Lis, 1988; Yamaguchi et al., 1999). Release of Pol II to productive elongation is facilitated by recruitment of P-TEFb (positive transcription elongation factor b) that phosphorylates the C-terminal domain of Pol II as well as the pausing factors, leading to NELF dissociation and DSIF being converted to an elongation-stimulating factor (Cheng and Price, 2007; Liu et al., 2015; Peterlin and Price, 2006).
RNA Pol II pausing is suggested to be an important checkpoint in early transcription where signals can be integrated for rapid and synchronous gene activation (Adelman and Lis, 2012; Levine, 2011). Accordingly, genomic studies have found enrichment of paused Pol II at genes involved in signal transduction, developmental control and cell proliferation (Min et al., 2011). In mice, loss of the B subunit of NELF (NELF-B) leads to embryonic lethality at the inner cell mass stage (Amleh et al., 2009). In addition, deletion of Nelf-b in mESCs causes proliferation defects together with a blunted response to differentiation signals (Williams et al., 2015). Although these studies suggest crucial roles of NELF-mediated pausing in mouse embryonic development, the interpretation of these results could be complicated by other functions of NELF-B, such as the physical and functional interaction of NELF-B with the DNA repair protein BRCA1 (Aiyar et al., 2007; Nair et al., 2016; Ye et al., 2001). Thus, the direct role of Pol II pausing in mammalian embryonic development remains to be elucidated.
In our previous studies, we have reported an essential function of the pausing factor DSIF in the development of zebrafish hematopoietic stem cells by characterizing a missense mutation in the DSIF subunit gene spt5 (Yang et al., 2016). In vitro assays have shown that the mutation-caused single amino acid change in zebrafish SPT5 protein specifically disrupts the pausing function of DSIF without affecting its elongation stimulating activity (Guo et al., 2000). Here, based on the high conservation between zebrafish and mammalian SPT5, we incorporated the same mutation into mESCs by CRISPR/Cas9 genome editing. Using global run-on sequencing (GRO-seq), we verified the genome-wide reduction of Pol II pausing in Spt5 mutant mESCs. Although mutant mESCs can be maintained at the self-renewal culturing conditions, these cells show genome-wide transcriptional changes and have severe defects during differentiation. Importantly, we identified a tight correlation between pausing status and local chromatin environment. Our results suggest that genes with unfavorable chromatin environment may rely more on paused Pol II to make them permissive for future activation during differentiation.
2. Material and methods
2.1. Cell culture and generation of ESC clones
ESCs were routinely cultured without feeders in mESC medium (DMEM+ 15% fetal bovine serum, 1mM sodium pyruvate, 50uM b-mercaptoethanol, MEM non-essential amino acids, 100 U/ml penicillin, 100 ug/ml streptomycin and 1000U/ml LIF) or in serum free N2B27 medium supplemented with 1uM MEK inhibitor (Cayman) and 3uM CHIR99021 (Cayman) (Ying et al., 2008) on 0.2% gelatin-coated plates. ES-E14TG2a (E14) ESCs were used for generation of Spt5V1008D clones. Cas9, sgRNA and donor DNA plasmids were introduced to E14 ESCs using the Neon transfection kit (Fisher), and integration of clones were confirmed using PCR followed by Sanger Sequencing. See supplemental information for further details.
2.2. Self-renewal assays and alkaline phosphatase staining
Single cells were plated 100cells/cm2 in 0.2% gelatin-coated 6-well plates and 24-well plates in triplicate. Every three days, cells in 6-well plates were dissociated with 0.25% Trypsin-EDTA and passaged to new plates at a concentration of 100cells/cm2. At the same time, the cells in 24-well plates were stained for alkaline phosphatase according to the manufacturer’s instructions (Stemgent). The number of positive colonies is counted. The cumulative number of colonies was calculated by multiplying the colony counts by the dilution factor used for passaging. Results are plotted as mean ± SEM of three wells.
2.3. RNA isolation and q-RT-PCR analysis
Total RNA was extracted using Trizol (Fisher) followed by DNAse treatment and reverse transcribed to cDNA with Superscript 3 cDNA synthesis kit (Fisher). qPCR was performed on Roche LightCycler 480 using the iQ SYBR Green Mastermix (BioRad). Gene expression was analyzed relative to Gapdh using the ΔΔCt method.
2.4. GRO-seq and data analysis
Nuclei isolation, run-on and preparation of libraries were performed as previously described (Franco et al., 2015). GRO-seq data were analyzed using the groHMM package described elsewhere (Chae et al., 2015). Additional details of GRO-seq library preparation and data analysis are described in Supplemental Information. ATAC-seq was performed as previously described (Buenrostro et al., 2013).
2.5. Differentiation assays
Serum-free differentiation was performed by removal of GSK3 and MEK inhibitors from ESCs in 2i media for indicated time lengths. EBs were prepared by titrating ESCs to single cell suspension in ESC media lacking LIF in hanging drops (300–500 cells/drop) and transferred to low-attachment plates in a shaking incubator after 3 days. EBs were collected at indicated time points for further analysis. Teratomas were prepared by intraperitoneal injection of 200,000 ESCs. Teratomas were collected after 21 days and stained with H&E for histological analysis (Histoserv Inc.).
2.6. Statistical Analyses
Data were analyzed by Student’s t-test. In all figures, data are represented by mean ± SEM from ≥3 independent experiments.
2.7. Accession numbers
GRO-seq and ATAC-seq data are available on GEO under accession numbers GSE99760.
3. Results
3.1. Generation of Spt5 mutant mESCs
To identify the targets of Pol II pausing in ESCs, we aimed to generate pausing mutant mouse ESCs. Previous studies using mouse ESCs with a conditional deletion of the NELF complex subunit gene Nelf-b have shown a severe proliferation defect (Williams et al., 2015). Therefore, we opted for an alternative strategy to target the pausing complex DSIF subunit SPT5 (encoded by Supt5h). In vitro studies have shown that a single amino acid change (V1012D) in the C-terminal of zebrafish SPT5 protein specifically disrupted the pausing function of DSIF without affecting its elongation stimulation function (Guo et al., 2000). Because the residue mutated in zebrafish SPT5 is conserved in mouse (Figure S1A), we introduced the same mutation (V1008D) in mouse SPT5 using a CRISPR/Cas9-mediated knockin approach (Figure 1A). Multiple homozygous mESC clones carrying the V1008D mutation were successfully generated and confirmed by Sanger sequencing (Figure 1B, S1B). All clones show comparable ESC characters to wild-type cells based on colony morphology, growth rate and alkaline phosphatase staining (Figure 1C and data not shown), and can be maintained under both serum containing and serum-free ESC culture conditions.
Figure 1. Generation and characterization of Spt5V1008D mutant ESCs.
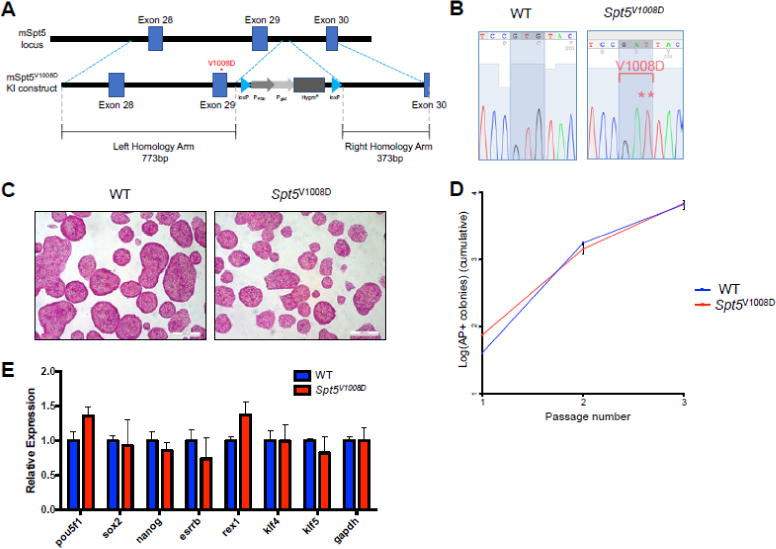
(A) Schematic of the murine Supt5h loci (exons 28 through 30) showing the wild-type and targeted allele that contains the V1008D mutation. The targeted allele contains a hygromycin resistance (HygroR) cassette flanked by LoxP sites.
(B) Sanger sequencing results of the wild-type and a homozygous clone. The mutated codon for V1008D is highlighted.
(C) Representative images of alkaline phosphatase staining of wild-type and Spt5V1008D homozygous ESCs.
(D) Clonal self-renewal assay for wild-type and Spt5V1008D ESCs (mean ± SEM, n=3)
(E) Quantitative RT-PCR comparing mRNA levels of ESC markers in wild-type and Spt5V1008D ESCs. Gene expression is normalized to Gapdh and presented as fold change relative to wild-type levels (n=3, mean ± SEM).
To quantify the clonal self-renewal ability of Spt5V1008D ESCs, we monitored the expansion of cells that are able to establish alkaline-phosphatase (AP) positive colonies at clonal density over three passages. Spt5V1008D ESCs behaved at similar levels compared to control ESCs (Figure 1D). In line with this, quantitative RT-PCR revealed no significant changes of expression levels of stem cell markers in mutant ESCs (Figure 1E).
To test the self-renewal ability of Spt5V1008D ESCs in long term culturing, we grew wild-type and Spt5V1008D ESCs for over 15 passages in the serum-free culture condition (Figure S2). Similar to wild-type ESCs, Spt5V1008D ESCs were able to maintain normal ESC morphology and positive AP staining at late passages (Figure S2A). Furthermore, mutant ESCs showed comparable cell cycle profiles and expression levels of pluripotency markers (Figure S2B–D). We conclude that the V1008D mutation in SPT5 does not cause significant defects in self-renewing ESCs.
3.2. Genome-wide reduction of Pol II pausing in Spt5V1008D ESCs
Previous in vitro studies have shown that the same mutation in zebrafish SPT5 specifically disrupts the pausing function of DSIF (Guo et al., 2000). To understand the genome-wide impact of this mutation on Pol II pausing, we performed GRO-seq to measure the distribution of transcriptionally engaged Pol II in wild-type and Spt5V1008D ESCs. We first validated our GRO-seq approaches in wild-type ESCs grown under serum/LIF vs. 2i conditions. A much higher occupancy of promoter-proximal Pol II in 2i condition than serum/LIF condition was observed, indicating strong pausing in cells grown in 2i (Figure S3A). This is consistent with previous studies showing increased pausing in 2i by Pol II ChIP-seq (Marks et al., 2012). Accordingly, calculation of pausing index (PI) by taking the ratio between Pol II around the promoter and the gene body revealed a much higher PI in 2i condition (Figure S3B). Although genome-wide pause was much higher in 2i condition, gene ontology (GO) analysis of paused genes (PI > 2) revealed similar ontology terms between 2i and serum conditions (Figure S3C), suggesting that the high level of Pol II pausing in 2i reflects a global effect rather than specific regulation for certain gene sets.
After confirming that ESCs cultured in 2i media exhibit more prominent Pol II pausing, we performed GRO-seq on wild-type and Spt5V1008D ESCs cultured in 2i media. Metagene analyses revealed that Spt5V1008D ESCs have a significant reduction of promoter-proximal Pol II but little change of Pol II levels within gene bodies (Figure 2A), suggesting a genome-wide reduction of transcription pausing. In support of reduced Pol II pausing, the average PI is significantly decreased in mutant ESCs (Figure 2B), with >70% genes showing lower PI than that in wild-type ESCs (Figure 2C).
Figure 2. Spt5V1008D ESCs show global reduction of Pol II pausing.
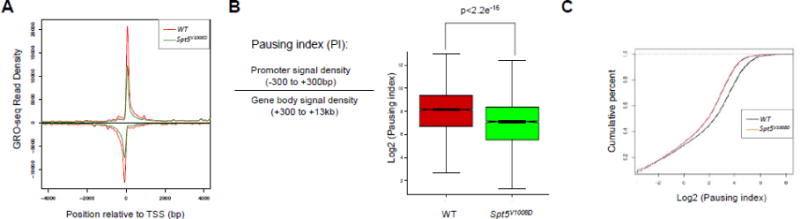
(A) Metagene analysis showing transcriptionally engaged Pol II occupancy centered at the transcription start site (TSS) measured by GRO-seq on sense and anti-sense strands in wild-type (red) and Spt5V1008D (green) ESCs.
(B) Left: depiction of pausing index (PI) calculation by ratio of Pol II signal around the promoter vs. the gene body. Right: boxplot analysis comparing PI between wild-type (red) and Spt5V1008D (green) ESCs.
(C) Cumulative distribution function analysis comparing PI distribution in wild-type (black) and Spt5V1008D (red) ESCs.
To identify gene classes most affected by pausing disruption, we carried out GO analysis on genes with at least 1.5-fold reduction of PI (n=4246). This identified high enrichment in GO terms related to transcription, cell cycle, kinase activity and signaling pathways such as MAPK and Wnt (Table S1). Together these results demonstrate a global reduction of transcription pausing in Spt5V1008D ESCs, providing the first genomic evidence demonstrating the in vivo effect of V1008D mutation of SPT5 on Pol II pausing.
3.3. Differentially regulated genes in Spt5V1008D mESCs have distinct chromatin states
Reduction of promoter-proximal Pol II in Spt5V1008D mESCs could lead to more release of paused Pol II into the gene body thus upregulated transcription, or less recruitment of Pol II to the promoter due to nucleosome reassembly that leads to transcription downregulation (Gilchrist et al., 2008). To identify genes that are differentially regulated in mutant ESCs, wild-type and mutant ESCs were compared for Pol II occupancy within gene bodies, which reflects the transcription activity of genes (Figure 3A, B). This analysis identified 416 genes with increased Pol II and 827 genes with reduced Pol II in gene bodies (≥1.5-fold, p<0.0001). We defined these genes as “upregulated” (up) vs. “downregulated” (down), respectively (Figure 3B). Interestingly, genes in the “up” group not only exhibited more gene-body associated Pol II but also have increased promoter-proximal Pol II in mutant ESCs, suggesting an overall increase of Pol II occupancy (Figure 3A). By contrast, genes in the “down” group have an overall reduction of Pol II occupancy from both promoters and gene bodies in mutant ESCs (Figure 3A). Moreover, in wild-type mESCs, “up” genes have significant lower PI than “down” genes (Figure 3C), suggesting that these genes are less paused in normal condition.
Figure 3. Differentially regulated genes in Spt5V1008D mESCs have distinct chromatin states.
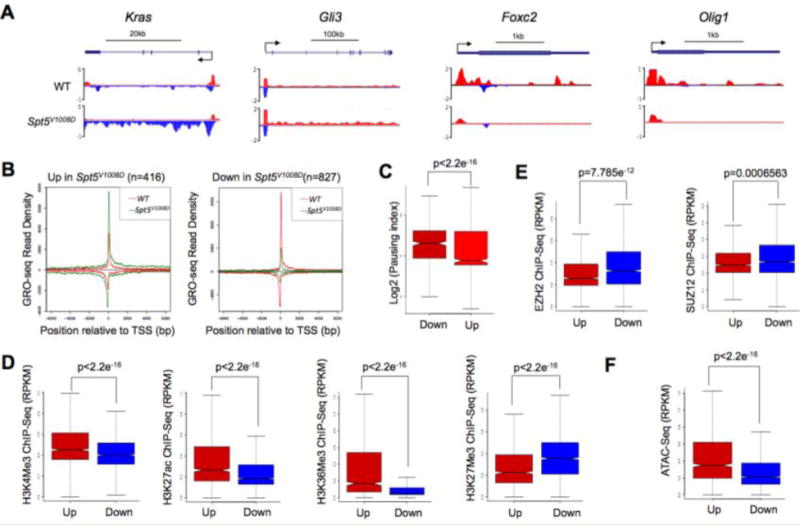
(A) Genome browser tracks of GRO-seq for upregulated genes (Kras and Gli3) and downregulated genes (Foxc2 and Olig1) in Spt5V1008D ESCs compared to wild-type (WT) cells. Scale bar and gene diagram are depicted above the browser tracks.
(B) Metagene analysis showing Pol II occupancy measured by GRO-seq on genes that show increased (Up) and decreased (Down) gene body reads in Spt5V1008D ESCs compared to wild-type (WT) cells.
(C) Boxplot analyses comparing the pausing index between the downregulated (Down) and upregulated (Up) genes in wild-type ESCs.
(D–F) Boxplot analysis comparing the occupancy of histone modification marks (D), enrichment of Polycomb proteins EZH2 and SUZ12 (E) and chromatin accessibility by ATAC-seq (F), between the upregulated (Up) genes and downregulated (Down) in wild-type ESCs.
To investigate what causes the different degree of pausing between “up” and “down” genes in wild-type ESCs, we compared the chromatin environment of these two gene sets in ESCs using published ChIP-seq data of histone modification marks from the same ES cell line grown in 2i condition (Joshi et al., 2015; Marks et al., 2012). Remarkably, these analyses revealed a higher occupancy on “up” genes of active histone marks such as H3K4me3, H3K27ac and H3K36me3 along with a lower level of the repressive mark H3K27me3 (Figure 3D), suggesting a relatively open chromatin state around these genes in normal ESCs. In contrast, genes in the “down” group are occupied by lower levels of active histone marks but higher levels of H3K27me3 and polycomb proteins EZH2 and SUZ12 (Figure 3D, E), suggesting a less permissive chromatin state. To further validate these results, we performed ATAC-seq (Buenrostro et al., 2013) to measure chromatin accessibility in wild-type ESCs. Consistent with ChIP-seq results, ATAC-seq showed a more accessible chromatin state for the “up” genes than “down” genes (Figure 3F).
These findings suggest that the effect of pausing on transcription may be determined by the local chromatin environment. For genes that have a relatively open chromatin state, paused Pol II may negatively influence their transcription by inhibiting productive elongation. By contrast, genes with unfavorable chromatin environment may rely on paused Pol II to maintain chromatin accessibility for basal transcription and future activation as suggested previously (Gilchrist et al., 2008), therefore are more likely to be downregulated when Pol II pausing is disrupted. Indeed, GO analysis revealed that the “down” group is enriched with genes functioning in multicellular organism development and cellular differentiation processes (Table S2).
3.4. Spt5V1008D mESCs have differentiation defects
Our GO analyses of genes with reduced pausing index in mutant ESCs identified signaling pathways MAPK and Wnt (Table S1), which are known to play important roles in regulating the balance between ESC self-renewal and differentiation (Ying et al., 2008). To understand whether the reduction of pausing in those gene sets have a functional consequence, we tested whether mutant ESCs behave normally in response to loss of self-renewal signals by removing GSK3 and MEK inhibitors in 2i media. While withdraw of inhibitors induced an immediate upregulation of ERK signaling and a graduate reduction of Wnt signaling in wild-type ESCs, mutant ESCs failed to show these responses (Figure 4A, S4), indicating a blunted response to differentiation cues by pausing disruption.
Figure 4. Spt5V1008D ESCs show defects in differentiation.
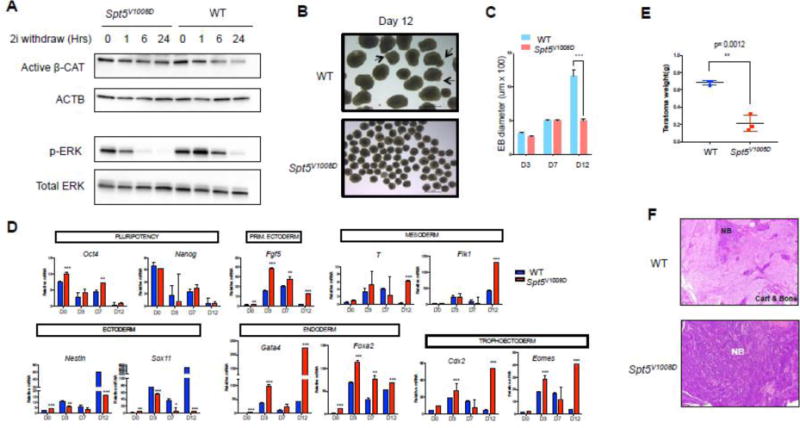
(A) Western blots comparing the activity of Wnt (active β-Catenin) and ERK signaling (p-ERK) between wild-type and mutant ESCs after removal of GSK3 and ERK inhibitors (2i) from the serum free media for different time length (Hrs: hours) as indicated. β-actin (ACTB) and total ERK serve as loading controls.
(B) Bright field imaging of day-12 embryoid bodies (EBs) from wild-type and mutant ESCs. Arrows depict cystic structures seen in wild-type day-12 EBs.
(C) Comparison of mean diameters (uM) of 20 EBs at indicated days of differentiation (n=3, mean ± SEM, ***p<0.001).
(D) Quantitative RT-PCR comparing mRNA levels of pluripotency and differentiation marker genes between wild-type and mutant EBs. Gene expression is normalized to Gapdh and presented as fold change relative to day0 (n=3, mean ± SEM, * p<0.05, **p<0.01, ***p<0.001).
(E) Teratoma weight comparison at time of collection. Data represents the average from three tumors in each group.
(F) Representative histology images of H&E stained teratoma sections. NB: undifferentiated neuroblastic tissue. Cart: cartilage. Note the teratoma derived from mutant ESCs (bottom) is full of NB with no signs of mature tissues (bone and cartilage).
To further test the differentiation ability of mutant cells, we differentiated ESCs into embryonic bodies (EBs). Mutant ESCs initiate differentiation but start to show significant growth retardation after day 7 and fail to form cystic structures at late stages of differentiation (Figure 4B, C). We examined the expression of self-renewal genes and differentiation markers throughout the time course of differentiation by q-RT-PCR (Figure 4D). Mutant EBs exhibited increased levels of self-renewal gene Oct4, and showed abnormal expression of multiple differentiation markers. The ectoderm markers Nestin and Sox11 were induced less efficiently in mutant EBs at most time points of EB differentiation. Although markers of other primitive and definitive germ layers were induced at similar (T, Flk1, Cdx2) or even higher levels (Fgf5, Gata4, Foxa2, Eomes) at early stages of differentiation in mutant EBs compared to those of the wild-type counterparts, most of these germ-layer markers failed to undergo downregulation at later stages of differentiation in mutant EBs. Furthermore, unlike wild-type EBs that express terminal differentiation markers for the cardiomyocyte lineage and start beating at day14 of EB differentiation, mutant EBs had severe defects in expressing these terminal lineage markers and most of them failed to show any sign of beating (Figure S3).
To test the effect of Spt5V1008D on longer-term differentiation, we performed teratoma assays by injecting control or mutant ESCs into NOD/SCID mice. Examination of teratomas at day 21 revealed that the mutant ESCs gave rise to much smaller teratomas with an overrepresentation of undifferentiated neuroblastic tissue and diminished terminally differentiated mesoderm-derived tissues such as bone and cartilage (Figure 4E, F). Together with the gene expression data, these results suggest that the Spt5V1008D mutation causes general defects in mESC differentiation with particular blockage of terminal lineage differentiation.
4. Discussion
Transcriptional regulation of gene expression is crucial for embryonic development. It has been shown that transcription can be regulated at the early elongation stage of transcription by Pol II pausing and elongation factors (Adelman and Lis, 2012). Previous attempts to elucidate the function of pausing in mammalian embryonic development have made use of targeted deletion of Nelf-b in mESCs (Williams et al., 2015). However, the proliferation defects of Nelf-b depleted mESCs prevents its use in functional study of Pol II pausing in long-term ESC differentiation. To circumvent these issues, here we attempted to generate a pausing mutant mESCs by targeting the pausing factor DSIF. We have incorporated a single amino acid substitution V1008D in the DSIF subunit SPT5 that has been shown to specifically disrupt the pausing function of SPT5 by in vitro studies (Guo et al., 2000). GRO-seq analyses of transcriptionally engaged Pol II revealed a genome-wide reduction of paused Pol II in promoter-proximal regions in mutant ESCs, validating previous in vitro studies. The fact that we did not see a complete abolishment of paused Pol II peak could be due to the remaining pausing mediated by NELF. Alternatively, V1008D substitution could be a hypomorphic mutant and other residues may contribute to the pausing function of SPT5.
RNA Pol II pausing is suggested to affect gene transcription in both positive and negative manner (Aida et al., 2006; Gilchrist et al., 2010; Gilchrist et al., 2008). Our work supports this conclusion and further suggests that the influence of pausing on transcription may depend on local chromatin environment. We observed that genes with a more permissive chromatin environment tend to have a less degree of pausing and are more likely to be upregulated in pausing-deficient ESCs, suggesting an inhibitive role of Pol II pausing on these genes. By contrast, genes occupied by higher levels of repressive histone marks and Polycomb proteins tend to require paused Pol II for their transcription. Accordingly, these genes tend to bear a higher level of paused Pol II, which may be more competent to overcome the unfavorable chromatin environment to keep promoter accessible for the basal transcriptional machinery, as suggested by previous studies (Gilchrist et al., 2010; Gilchrist et al., 2008), thereby making them more malleable for future activation. This may partially explain why the genome-wide reduction of pausing in Spt5V1008D ESCs did not cause significant phenotypes under self-renewing condition but severely disrupted differentiation. Compared to the self-renewal state, differentiating ESCs have a great increase of inaccessible chromatin due to chromatin condensation and heterochromatin formation. Such an increase of unfavorable chromatin environment may render genes to rely more on paused Pol II to keep promoter accessible for the transcriptional machinery as cells differentiate.
In conclusion, by generating stable mutants for the pausing factor SPT5, our work reveals a crucial role of RNA Pol II pausing in ESC differentiation. Notably, we identify a tight correlation between pausing-mediated transcription regulation and local chromatin environment. Future studies to identify chromatin remodelers and histone modifiers that regulates Pol II pausing will be important to deepen our understanding on how to maintain adequate levels of gene transcription in cells to prevent developmental abnormalities and malignant diseases.
Supplementary Material
Highlights.
Pausing in ES cells occurs primarily at cell cycle control and signaling genes
Disruption of DSIF-mediated RNA Pol II pausing impairs mESC differentiation
Pol II pausing coordinates with chromatin environment to regulate transcription
Acknowledgments
This work was supported by the National Institutes of Health (R00DK088963 and R01DK105287) (X. B.), the Cancer Prevention Research Institute of Texas (R1115 for X. B. and RR140042 for L. A. B.) and the Cecil H. and Ida Green Center Training Program in Reproductive Biology Sciences Research at UTSW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
M.T. performed experiments and analyzed results; A.A.G. and V.S.M. performed genomic analyses. C.M., K.N. and L.A.B. contributed to some experiments; X.B. conceived the study and supervised all experiments. M.T. and X.B. wrote the manuscript.
The authors declare no competing financial interests.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Molecular and cellular biology. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar SE, Cho H, Lee J, Li R. Concerted transcriptional regulation by BRCA1 and COBRA1 in breast cancer cells. International journal of biological sciences. 2007;3:486–492. doi: 10.7150/ijbs.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amleh A, Nair SJ, Sun J, Sutherland A, Hasty P, Li R. Mouse cofactor of BRCA1 (Cobra1) is required for early embryogenesis. PloS one. 2009;4:e5034. doi: 10.1371/journal.pone.0005034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae M, Danko CG, Kraus WL. groHMM: a computational tool for identifying unannotated and cell type-specific transcription units from global run-on sequencing data. BMC bioinformatics. 2015;16:222. doi: 10.1186/s12859-015-0656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. The Journal of biological chemistry. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- Franco HL, Nagari A, Kraus WL. TNFalpha signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Molecular cell. 2015;58:21–34. doi: 10.1016/j.molcel.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes & development. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Yamaguchi Y, Schilbach S, Wada T, Lee J, Goddard A, French D, Handa H, Rosenthal A. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- Joshi O, Wang SY, Kuznetsova T, Atlasi Y, Peng T, Fabre PJ, Habibi E, Shaik J, Saeed S, Handoko L, et al. Dynamic Reorganization of Extremely Long-Range Promoter-Promoter Interactions between Two States of Pluripotency. Cell stem cell. 2015;17:748–757. doi: 10.1016/j.stem.2015.11.010. [DOI] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kraus WL, Bai X. Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends in biochemical sciences. 2015;40:516–525. doi: 10.1016/j.tibs.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes & development. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SJ, Zhang X, Chiang HC, Jahid MJ, Wang Y, Garza P, April C, Salathia N, Banerjee T, Alenazi FS, et al. Genetic suppression reveals DNA repair-independent antagonism between BRCA1 and COBRA1 in mammary gland development. Nature communications. 2016;7:10913. doi: 10.1038/ncomms10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, Adelman K. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Molecular cell. 2015;58:311–322. doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yang Q, Liu X, Zhou T, Cook J, Nguyen K, Bai X. RNA polymerase II pausing modulates hematopoietic stem cell emergence in zebrafish. Blood. 2016;128:1701–1710. doi: 10.1182/blood-2016-02-697847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Hu YF, Zhong H, Nye AC, Belmont AS, Li R. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J Cell Biol. 2001;155:911–921. doi: 10.1083/jcb.200108049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


