Abstract
Paraburkholderia phymatum is a highly effective microsymbiont of Mimosa spp. and has also been shown to nodulate papilionoid legumes. P. phymatum was found to be highly competitive both in a natural environment as well as under controlled test conditions and is more competitive for nodulation over other α- and β-rhizobial strains in a variety of different plant hosts. In order to elucidate the factors that make this bacterium highly competitive for legume infection, we here characterized the type VI secretion system (T6SS) clusters of P. phymatum. T6SSs have been shown to function as a contact-dependent injection system for both bacterial and eukaryotic cells. We identified two T6SS clusters in the genome, created respective mutant strains and showed that they are defective in biofilm formation and in interbacterial competition in vitro. While the T6SS mutants were as efficient as the wild-type in nodulating the non-cognate host Vigna unguiculata, the mutants were less competitive in in planta competition assays, suggesting that the T6SS is one of the factors responsible for the success of P. phymatum in infecting legumes by directly inhibiting competitors.
Keywords: Rhizobium, legume, symbiosis, nodulation, competitiveness, T6SS, biofilm
Introduction
β-proteobacterial strains of the genus Burkholderia are found in a variety of environments and can be pathogenic (e.g., B. pseudomallei, B. thailandensis, B. cepacia, B. cenocepacia, B. glumae, and B. gladioli) or beneficial (e.g., B. phytofirmans, B. tuberum, and B. phymatum) to a wide range of eukaryotic hosts (Suarez-Moreno et al., 2012; Estrada-de los Santos et al., 2013; Eberl and Vandamme, 2016; Beukes et al., 2017). The discovery of β-proteobacteria (from the Burkholderia and Cupriavidus genera) inside root nodules of a leguminous plant in 2001 (Moulin et al., 2001) gave rise to a new class of rhizobia (the so called β-rhizobia) and highlighted the variety of bacteria present in the soil and in root nodules. Several studies that have followed the first description have rapidly increased the number of novel β-rhizobia, (Vandamme et al., 2002; Chen et al., 2003; Bontemps et al., 2010; Gyaneshwar et al., 2011). These new β-rhizobia are mainly found associated with Mimosa species from South America and Asia (Chen et al., 2005, 2006, 2007, 2008; Elliott et al., 2007b; dos Reis et al., 2010; Mishra et al., 2012); however, some species like B. phymatum and B. tuberum are able to enter symbiosis with papilionoid legumes such as those found in the South African fynbos, common bean (Phaseolus vulgaris) and siratro (Macroptilium atropurpureum) (Elliott et al., 2007a; Garau et al., 2009; Talbi et al., 2010; Angus et al., 2013; Beukes et al., 2013; Howieson et al., 2013; Lemaire et al., 2016). In the last few years, several plant-beneficial Burkholderia strains have been reclassified into the new genus, called Paraburkholderia (Sawana et al., 2014; Beukes et al., 2017). Most of the previously identified Burkholderia species that induce the formation of nodules and are able to fix nitrogen as symbionts are now referred to as Paraburkholderia.
Similarly to α-rhizobia, β-rhizobia have diverse host range and environmental preferences (Gyaneshwar et al., 2011). An important aspect of bacterial communities in the soil is the competition between the different species. Previous studies for nodule occupancy in leguminous plants have shown that β-rhizobia outcompete α-rhizobia for infection in different Mimosa species (Elliott et al., 2009; Melkonian et al., 2014). In addition, a recent study from our group identified Paraburkholderia phymatum LMG 21445T (=STM 815T) as a highly competitive symbiont that, when in competition with other β-rhizobia, was predominant in nodules of three out of four different legumes (Lardi et al., 2017). In that study, P. phymatum was also shown to produce higher amounts of exopolysaccharides (EPSs) and was the most competitive strain on plates, suggesting that inhibition of competitor growth was an important factor behind the successful competitiveness of P. phymatum (Lardi et al., 2017). Direct inhibition of bacteria growth can be achieved via secretion systems, which rely on structural components in the membrane to secrete effector proteins into the competitor cell (Stubbendieck and Straight, 2016; Bernal et al., 2017b). There are different types of such secretion systems in bacteria, including the newly described contact dependent type VI secretion systems (T6SSs), which are exclusively found in Gram-negative bacteria. T6SS can target either prokaryotic or eukaryotic cells (Schwarz et al., 2010; Angus et al., 2014; Ho et al., 2014). The structure and assembly of the T6SS complex has been determined for Vibrio cholerae, Pseudomonas aeruginosa, and Escherichia coli species (Basler et al., 2012, 2013; Shneider et al., 2013; Ho et al., 2014; Basler, 2015; Kudryashev et al., 2015; Vettiger and Basler, 2016; Vettiger et al., 2017). These studies showed that some structural elements of the T6SS share similarities with a phage tail spike (Records, 2011; Silverman et al., 2012; Ho et al., 2014) and also with the type IV secretion system T4SSb (Ma et al., 2009; Durand et al., 2012). A cluster of genes (tss – type six secretion) encodes the structural components of T6SS and functions as follow: the baseplate (TssA, E, F, G, K) and the membrane complex (TssJ, L, M), both anchored to the inner membrane, recruit the spike VgrG (tssI gene), which is attached to effectors; in the cytosol the sheath (TssB, C) polymerizes around the tube (Hcp, tssD gene); a contraction movement of the sheath expels the tube (Hcp) outside the cell, delivering the VgrG carrying effectors into the target cell. After the attack, ClpV (tssM gene), which is also attached to the membrane, uses ATP to reassemble the structure (Silverman et al., 2012; Ho et al., 2014; Alcoforado Diniz et al., 2015; Brunet et al., 2015; Gallique et al., 2017a). Phylogenetic reconstruction, using different genes of the T6SS cluster, revealed that most of the T6SSs are acquired by lateral gene transfer, and that small differences in regulation can lead to specificity and adaptation to different environments (Boyer et al., 2009).
Several plant-associated bacteria harbor genes coding for the T6SS in their genomes. The first description of a functional T6SS was in the symbiont Rhizobium leguminosarum (Bladergroen et al., 2003) where the system was shown to be important for the interaction with the plant host (Bladergroen et al., 2003; Nelson and Sadowsky, 2015; Ryu, 2015). On the other hand, in non-symbiotic plant-associated bacteria, T6SS was shown to be mainly involved in inter- or intra-bacterial competition. For instance, the biocontrol strain Pseudomonas putida KT2440 uses a T6SS for competition in soil and for protection of Nicotiana benthamiana plants from Xanthomonas campestris infection (Bernal et al., 2017a). In the plant pathogen Pantoea ananatis a T6SS mutant showed less virulence on onion plants and was affected in interbacterial competition against several bacteria (Shyntum et al., 2015). In Agrobacterium tumefaciens, competition against Pseudomonas aeruginosa inside a plant host (N. benthamiana) presented a T6SS-dependent advantage to A. tumefaciens (Ma et al., 2014).
In this work, we elucidated the role of two T6SS systems in the nitrogen-fixing and symbiotic strain P. phymatum. P. phymatum STM 815T was isolated by Moulin et al. (2001) and was described by Vandamme et al. (2002) under the accession number LMG 21445T. The genome of P. phymatum STM 815T has been sequenced (Moulin et al., 2014) and is available in NCBI under the following accession numbers for the replicons (NC_010622.1 for chromosome 1, NC_010623.1 for chromosome 2, NC_010625.1 for plasmid pBPHY01, and NC_010627.1 for the symbiotic plasmid pBPHY02). We show here that P. phymatum utilizes both T6SS systems to outcompete other β-rhizobial strains in vitro and to compete for legume root infection.
Results
Identification of Two Complete T6SS Clusters on the P. phymatum Megaplasmid pBPHY01
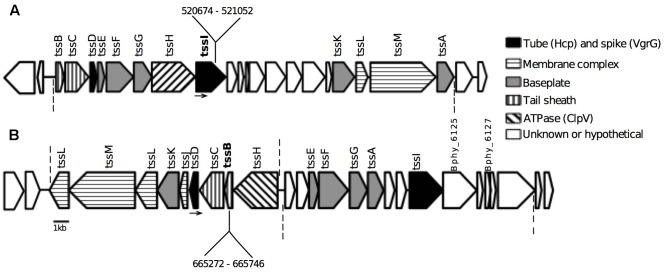
A BLAST search on the P. phymatum megaplasmid pBPHY01 sequence (GenBank sequence: CP001045.1) (Moulin et al., 2014) using the T6SS1 and T6SS2 of B. thailandensis (Schwarz et al., 2010) identified two complete T6SS clusters within pBPHY01. Table 1 lists the T6SS structural elements of both clusters and compares the amino acid sequences of the components. The low similarity between the components of both clusters suggests that the T6SS systems work independently of each other. The gene coding for the TssJ protein, which is part of the membrane complex, is present only in one of both clusters (Figure 1). Besides the cluster-encoded components, other T6SS related genes are found elsewhere in the P. phymatum genome, including six extra copies of the spike tssI-VgrG (Bphy_0023/BPHY_RS00115, Bphy_1932/BPHY_RS09825, Bphy_3640/BPHY_RS18400, Bphy_5197/BPHY_RS26040, Bphy_5744/BPHY_RS28740, and Bphy_7022/BPHY_RS34870). Both clusters contain, beside all structural type six secretion genes (tssA-M), several ORFs with unknown functions (Figure 1). According to a previous study by Angus et al. (2014), which presented a phylogenetic analysis based on proteins encoded by tss clusters of several Burkholderia strains, P. phymatum first tss cluster is a T6SS-b cluster (Figure 1A) and the second cluster a T6SS-3 (Figure 1B). An ORF (Bphy_5974/BPHY_RS29865), which is located upstream of the T6SS-b-type cluster, encodes a serine/threonine kinase that is involved in post-translational regulation of T6SS expression in P. aeruginosa (Casabona et al., 2013). The gene downstream of Bphy_5985 (BPHY_RS29920 – tssI), which codes for a VgrG effector, encodes a hypothetical protein (Bphy_5986/BPHY_RS29925) with an antitoxin domain (COG 2849) that may be required for immunity against self-killing. An ORF (Bphy_6117/BPHY_RS30535) coding for an accessory component, TagJ, is located within the T6SS-3 cluster. In P. aeruginosa TagJ, together with the product of tssH (ClpV), is involved in the recycling of the sheath (TssB, C) (Boyer et al., 2009; Lossi et al., 2012). The genomic region surrounding the P. phymatum T6SS-3 cluster contains several elements previously shown in other bacteria to be involved in the functioning of the system, such as a PAAR repeat-containing protein (Bphy_6127) (Shneider et al., 2013; Cianfanelli et al., 2016) and a putative peptidase M23 (Bphy_6125/BPHY_RS38880) (Shyntum et al., 2015; Weber et al., 2016). From the genomic organization and from our preliminary RNA-seq data on P. phymatum grown in free-living conditions, we suggest that both clusters are organized in operons (Figure 1) (Lardi et al., unpublished data).
Table 1.
Comparison between amino acid sequences of the components of both Paraburkholderia phymatum LMG 21445T type VI secretion systems (T6SSs) (T6SS-b and T6SS-3) (see Figure 1).
| T6SS component | Function | Cluster T6SS-b | Cluster T6SS-3 | % Identity (% similarity) |
|---|---|---|---|---|
| TssA | Baseplate | Bphy_5997 (BPHY_RS29980) | Bphy_6121 (BPHY_RS30555) | 22 (36) |
| TssB | Tail sheath | Bphy_5978 (BPHY_RS29885) | Bphy_6114 (BPHY_RS30520) | 49 (68) |
| TssC | Tail sheath | Bphy_5979 (BPHY_RS29890) | Bphy_6113 (BPHY_RS30515) | 48 (68) |
| TssD (Hcp) | Hcp Tube | Bphy_5980 (BPHY_RS29895) | Bphy_6112 (BPHY_RS30510) | 25 (46) |
| TssE | Baseplate | Bphy_5981 (BPHY_RS29900) | Bphy_6118 (BPHY_RS30540) | 32 (51) |
| TssF | Baseplate | Bphy_5982 (BPHY_RS29905) | Bphy_6119 (BPHY_RS30545) | 31 (47) |
| TssG | Baseplate | Bphy_5983 (BPHY_RS29910) | Bphy_6120 (BPHY_RS30550) | 28 (41) |
| TssH | ATPase | Bphy_5984 (BPHY_RS29915) | Bphy_6115 (BPHY_RS30525) | 46 (60) |
| TssI (VgrG) | VgrG Spike | Bphy_5985 (BPHY_RS29920) | Bphy_6124 (BPHY_RS30565) | 33 (48) |
| TssJ | Membrane complex | Not present | Bphy_6111 (BPHY_RS30505) | Not accessed |
| TssK | Membrane complex | Bphy_5994 (BPHY_RS29965) | Bphy_6110 (BPHY_RS30500) | 29 (47) |
| TssL | Membrane complex | Bphy_5995 (BPHY_RS29970) | Bphy_6109/6107 (BPHY_RS30495/BPHY_RS30485) | 25 (49)/24 (39) |
| TssM | Membrane complex | Bphy_5996 (BPHY_RS29975) | Bphy_6108 (BPHY_RS30490) | 21 (37) |
The genes mutated in this study (in bold) are indicated in Figure 1 (tssI from T6SS-b, and tssB from T6SS-3). The identification of the components is given with two Locus Tags (old and new NCBI Locus Tag in brackets) taken from the Burkholderia Genome Database (burkholderia.com).
FIGURE 1.
Type VI secretion system (T6SS) clusters on megaplasmid pBPHY01 of Paraburkholderia phymatum LMG 21445T. Nomenclature of the ORFs is according to the Burkholderia Genome Database (burkholderia.com). (A) T6SS-b cluster (position 510771–535444), structural elements coded between ORFs Bphy_5978 (BPHY_RS29885 – tssB) and Bphy_5997 (BPHY_RS29980 – tssA). (B) T6SS-3 cluster (position 654398–678783), structural elements coded between ORFs Bphy_6107 (BPHY_RS30485 – tssL) and Bphy_6124 (BPHY_RS30565 – tssI). The putative structural functions are marked in colors. Dashed lines indicate the beginning and the end of the putative operons based on preliminary RNA-seq data (Lardi et al., unpublished). Bphy_6125: (BPHY_RS38880) peptidase M23. Bphy_6127: PAAR repeat-containing protein. In bold are shown the genes mutated by insertion and the numbers indicate the position of the insertions (see “Materials and Methods”; Supplementary Table S1 and Table 1). Arrows indicate the external primers used for confirmation of the insertion (Bphy_5985For for the mutation in tssI-b gene and Bphy_6112Rev for the mutation in the tssB-3 gene – Supplementary Table S1).
According to Angus et al. (2014), cluster-types T6SS-b and T6SS-3 are not associated with the pathogenicity of Burkholderia species.
Phenotypic Analysis of the P. phymatum T6SS Mutant Strains
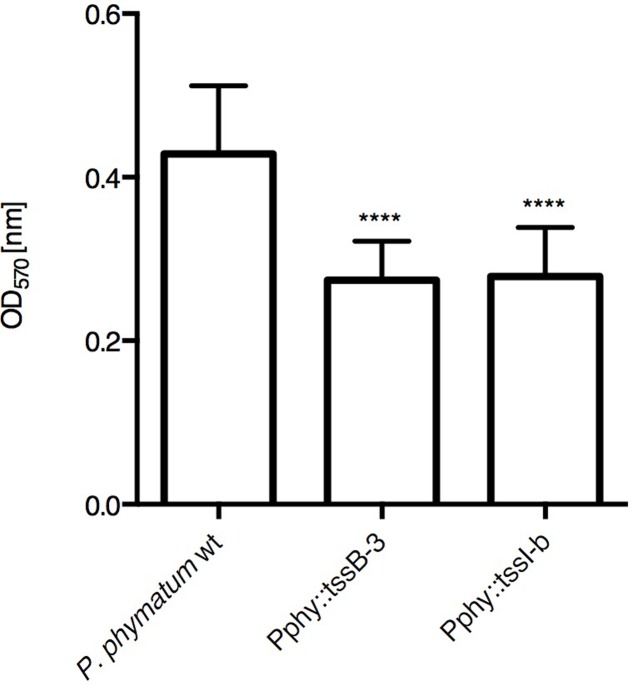
The gene tssI (Bphy_5985/BPHY_RS29920 position 519599–521524) from the cluster T6SS-b and tssB (Bphy_6114/BPHY_RS30520 position 665221–665754) from the cluster T6SS-3 were used as targets for the construction of insertional mutants, yielding the strains Pphy::tssI-b and Pphy::tssB-3, respectively. These genes were chosen because of their role as T6SS structural components and for being essential for the proper function of the secretion system (Boyer et al., 2009; Cianfanelli et al., 2016). The growth of the two P. phymatum T6SS mutant strains in rich media and minimal media (see “Materials and Methods”; Supplementary Figure S1) showed no difference compared to the wild-type, suggesting that the mutations do not affect the development of the bacteria. Phenotypes important for interaction with the plant host, such as EPS production and biofilm formation were tested in the T6SS mutant strains. Both phenotypes were previously shown to be dependent on a functional T6SS in Pseudomonas fluorescens (Decoin et al., 2015; Gallique et al., 2017b) in some Burkholderia species (Aubert et al., 2008) and in Ralstonia solanacearum (Zhang et al., 2014). In contrast, in P. phymatum EPS production was not influenced by the T6SSs (Supplementary Figure S2) as both mutant strains were able to produce as much EPS on YEM plates containing 0.06% yeast extract as the wild-type strain. On the other hand, biofilm formation was significantly decreased in the mutant strains, when compared to the wild-type (Figure 2).
FIGURE 2.
Biofilm formation in the P. phymatum strains. Biofilm formation was performed in 96-well plates. Each strain was measured in biological triplicates and eight wells per replica. ANOVA, Tukey’s test. ∗∗∗∗p ≤ 0.0001.
In order to verify differences in the symbiotic performance of mutant strains compared to the wild-type, the strains were inoculated on the legume cowpea (Vigna unguiculata). After 3 weeks, the nodulated roots were used to determine the number and dry weight of the nodules as well as the nitrogenase activity. The overall parameters analyzed did not show any significant differences between the mutant strains and the wild-type (Table 2). Although the mutant strain Pphy::tssB-3 presented higher values than the P. phymatum wild-type strain in all the parameters tested (nodule number, nodule dry weight and nitrogenase activity), these differences were not statistically significant.
Table 2.
Symbiotic properties of the T6SS mutant strains.
| Parameter | Bacterial strains |
||
|---|---|---|---|
| P. phymatum LMG21445T | Pphy::tssI-b | Pphy::tssB-3 | |
| Nitrogenase [% (C2H4 C2H2-1) g-1 min-1] | 0.46 ± 0.08 | 0.42 ± 0.12 | 0.55 ± 0.21 |
| Dry weight nodules [mg] | 0.17 ± 0.07 | 0.12 ± 0.06 | 0.19 ± 0.1 |
| Nodule number | 7.67 ± 3.79 | 8.75 ± 3.28 | 10.75 ± 3.92 |
Values ± SD (standard deviation, ANOVA, Tukey’s test with p ≤ 0.05). The average of the number and the dry weight of nodules in the inoculated cowpea plants is given as well as the specific nitrogenase activity calculated after the ARA (acetylene reduction assay), normalized by the dry weight of the nodules and acetylene incubation time.
With the purpose of verifying whether the T6SS mutant strains are impaired in competition for nodulation, a mixture containing equal amounts of the P. phymatum wild-type and of each of the mutant strains was tested on cowpea. For this, bacteria were mixed 1:1 (105 cells) and inoculated on germinated cowpea seeds. Nodules harvested after 3 weeks were surface sterilized and plated on selective media to quantify the proportion of mutant/wild-type in each plant. No significant difference in the amounts of nodules colonized by the wild-type or mutants were observed, suggesting that the mutations did not affect the competitiveness of P. phymatum (Supplementary Figure S3).
Competition with β-Rhizobial Strains for Legume Infection
To verify if the T6SS mutants are affected in interbacterial competition, we selected the legume host cowpea, which we previously showed to be exclusively nodulated by the wild-type strain of P. phymatum (Lardi et al., 2017). In fact, 100% of nodules recovered from cowpea plants inoculated with a mixture of β-rhizobia, containing P. diazotrophica, P. mimosarum, P. sabiae, and P. phymatum, were identified as P. phymatum. We therefore tested the same mixture of β-rhizobia against the T6SS mutants Pphy::tssB-3 and Pphy::tssI-b. The seeds were inoculated with two different cell densities (100 or 105 cells of each strain). The low density setting was the same as we used in our previous study (Lardi et al., 2017). The high density setting was thought to increase the contact between bacteria at the moment of infection.
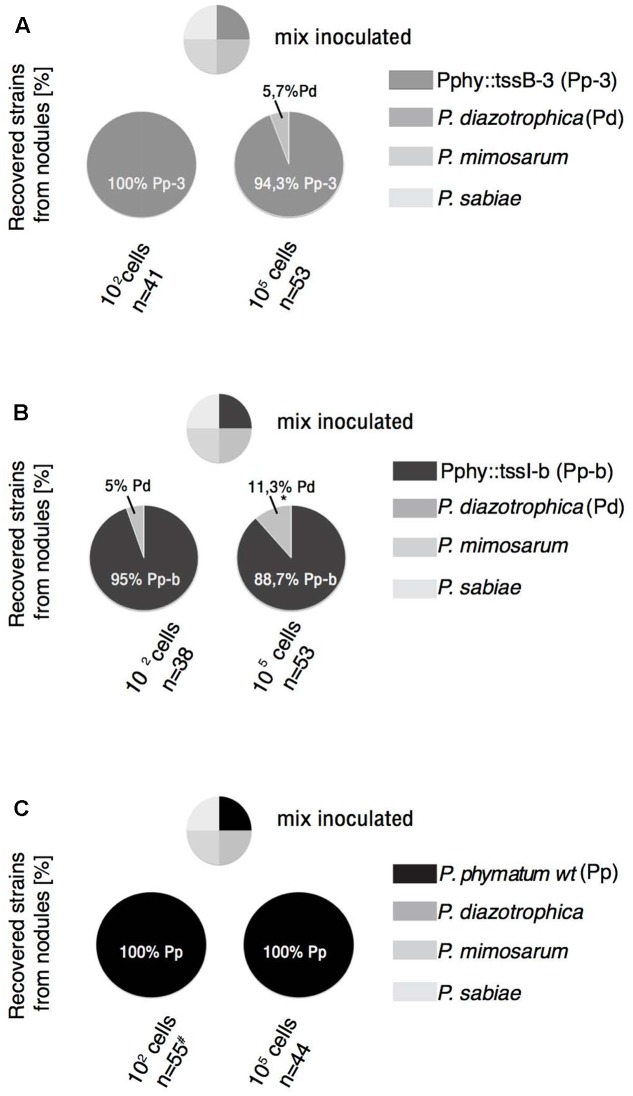
After 3 weeks of growth, all nodules from one plant were collected and subsequently analyzed for occupancy by plating on selective media (for the identification of P. phymatum mutant strains) and by using PCR with primers for the recA gene to identify the remaining strains (Mishra et al., 2012). While in the mixture containing 100 cells and the wild-type strain or the Pphy::tssB-3 mutant, 100% of the nodules were occupied by P. phymatum (Figures 3A,C), when the Pphy::tssI-b mutant was present in the mixture 5% of the nodules (2 out of 38) were occupied by P. diazotrophica (Figure 3B) and both nodules were located in the same plant root system. The plant assays with higher cell densities, where the P. phymatum wild-type was able to infect 100% of the nodules (Figure 3C), showed increased nodule colonization by strains other than the inoculated P. phymatum T6SS mutants. For the mixtures containing the Pphy::tssI-b or the Pphy::tssB-3 mutants, nodules infected by P. diazotrophica accounted for 11.3% (6 out of 53) and 5.7% (3 out of 53) of the total number of nodules analyzed, respectively (Figure 3). The nodules occupied by P. diazotrophica were found in the same root system of the nodules occupied by the T6SSs mutants, and in more than one plant. The other two strains of β-rhizobia tested (P. mimosarum and P. sabiae) were not identified in any of the experiments. These results suggest that T6SS is one of the factors responsible for the success of P. phymatum in infecting cowpea.
FIGURE 3.
Effect of T6SS mutations in the competition of P. phymatum with other Paraburkholderia species in planta. The mixture containing 100 or 105 cells of each of the four strains, P. diazotrophica, P. mimosarum, P. sabiae and Pphy::tssB-3 (A), Pphy::tssI-b (B) or P. phymatum LMG 21445T wild-type (C) was inoculated on seedlings of cowpea (Vigna unguiculata) and each nodule of the plants were harvested after 21 days and analyzed. Two independent replicates for each experiment and at least four plants were analyzed. The percentage of recovered strains is given. n, number of nodules. Two-tailed z-test comparing the proportion of P. phymatum obtained with the mixture containing each mutant against the one containing the wild-type. ∗p ≤ 0.05. # result from Lardi et al. (2017).
Interbacterial in Vitro Competition Assays
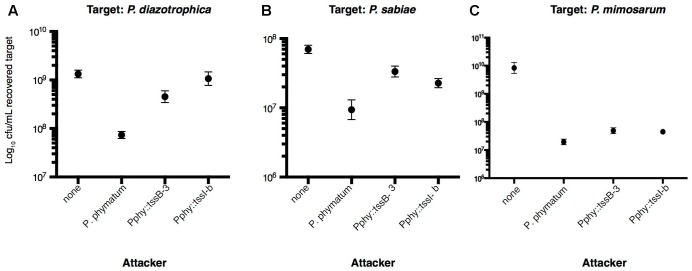
To better understand the interaction between the bacterial strains tested in plant infection experiments, we set up interbacterial competition assays between P. phymatum (dsRed-T6SS+ and mutants) as attacker and the other β-rhizobia strains used in the plant experiments (P. diazotrophica-gfp, P. mimosarum-gfp, and P. sabiae) as targets strains (see “Materials and Methods” for details of strains used). The bacteria were mixed 1:1 (attacker:target) and co-cultured on a LB without salt plate for 24 h. The spot containing the competing bacteria was collected and the bacterial colony forming units (CFUs) were determined on selective plates. The survival of target bacteria is shown in Figure 4. When P. phymatum was tested in competition against P. diazotrophica, the P. phymatum T6SS system was revealed to play an important role. In competition against the P. phymatum dsRed T6SS+, 18.2-fold fewer cells of P. diazotrophica were recovered, compared to the strain grown alone (p ≤ 0.01) (Figure 4A). However, when the Pphy::tssI-b mutant was tested as attacker, P. diazotrophica survived as well as in the control without attacker (1.2-fold less from the CFU recovered compared to when P. diazotrophica was grown without attacker). The same tendency could be observed for competition of the Pphy::tssB-3 mutant against P. diazotrophica (2.9-fold decreased number of cells recovered compared to when P. diazotrophica was grown without attacker). Our results suggest that the tssI gene of T6SS-b and the tssB of the T6SS-3 are involved in the competition of P. phymatum against P. diazotrophica (Figure 4A). Likewise, the competition of P. phymatum against P. sabiae was also T6SS-dependent: in competition against the mutants, the survival of P. sabiae was significantly higher (Pphy::tssB-3 showing only 1.6-fold and Pphy::tssI-b 2.3-fold fewer recovered CFU compared to the P. sabiae strain grown without attacker) than what was observed for the assay including the P. phymatum wild-type, with 5.7-fold fewer cells of P. sabiae than when the strain grown alone (p ≤ 0.05) (Figure 4B). In contrast, the competition between P. phymatum and P. mimosarum showed that both the wild-type and the mutant strains significantly affected (p ≤ 0.05) the survival of P. mimosarum (171.4-fold, 186.7-fold, and 430.8-fold fewer cells against Pphy::tssB-3, Pphy::tssI-b and P. phymatum dsRed T6SS+, respectively, when compared to the CFU recovered by the strain alone). This result indicates that for competition between these two bacteria factors other than T6SSs are involved (Figure 4C). Taken together, these data show that both T6SSs of P. phymatum play an important role in the competition against other bacteria such as P. diazotrophica and P. sabiae.
FIGURE 4.
Interbacterial competition assay with P. phymatum (dsRed T6SS+) and T6SS mutants as attackers. The graphs show the recovered colony forming units (CFU) of the target bacteria: P. diazotrophica gfp (A), P. sabiae (B), and P. mimosarum gfp (C). The dots represent the average of three independent experiments (+/– SEM). None: no attacker strain. Two-tailed t-test. (A) p = 0.0092 in the survival of targets against P. phymatum as attacker, p = 0.0445 against Pphy::tssB-3 and p = 0.6119 against Pphy::tssI-b. (B) p = 0.028 in the survival of the target against P. phymatum, p = 0.0282 against Pphy::tssB-3 and p = 0.0286 against Pphy::tssI-b. (C) p = 0.0139 in the survival of the target against P. phymatum, p = 0.1161 against Pphy::tssB-3 and p = 0.037 against Pphy::tssI-b.
Discussion
In this work we describe the role of T6SSs in a symbiotic and nitrogen fixing β-rhizobia that interacts with a variety of leguminous plants. We have previously shown that P. phymatum LMG 21445T outcompetes other β-rhizobia in plant infection of papilionoid legumes and in vitro on plates (Lardi et al., 2017). Here, we investigate the genetic mechanisms underlying the high level of competitiveness of P. phymatum by characterizing the importance of the T6SSs in this strain. T6SSs have been described in various Gram-negative bacteria and were shown to act against both prokaryotes and eukaryotes (Bingle et al., 2008; Filloux et al., 2008; Boyer et al., 2009; Basler et al., 2012, 2013; Alcoforado Diniz et al., 2015; Basler, 2015). Interestingly, the first report of a T6SS was in Rhizobium leguminosarum, where the wild-type strain was shown to be impaired in pea nodulation (Roest et al., 1997), while a mutation in the imp gene cluster (which was later identified as a T6SS) (Bladergroen et al., 2003) caused a nodulation phenotype. The authors could show that the impairment in pea nodulation was due to the secretion of proteins into the plant host cells (Bladergroen et al., 2003). Most of the research on T6SSs is done in pathogenic bacteria and the role of several effectors secreted through the system consists in the degradation of the cell wall of the attacked cell (Alcoforado Diniz et al., 2015). In pathogenic Burkholderia, T6SSs are used to attack both eukaryotic and prokaryotic target cells (Aubert et al., 2008; Schwarz et al., 2010, 2014; Burtnick et al., 2011; Angus et al., 2014). In the P. phymatum genome (Moulin et al., 2014) we identified two complete T6SS clusters. These T6SS clusters, according to the classification of Angus et al. (2014), are not associated with the pathogenicity of Burkholderia species (Schwarz et al., 2010, 2014; Burtnick et al., 2011). Moreover, the P. phymatum genome encodes eight VgrG proteins and only two of them are located inside a T6SS cluster.
Besides secreting effectors, T6SS are known to affect biofilm formation. For instance, the plant pathogen Ralstonia solanacearum possesses a T6SS gene cluster, which is not only important for pathogenicity in tomato plants but also has a strong effect on motility and biofilm formation (Zhang et al., 2014). In Pseudomonas fluorescens MFE01, a deletion mutant in the tssC gene, coding for the tail sheath component, was also impaired in biofilm formation (Gallique et al., 2017b). We found that both P. phymatum T6SS mutants showed a significantly decreased ability to form biofilms (Figure 2) while plant infection efficiency, at least for cowpea plants, was not affected (Table 2).
We previously showed that P. phymatum is highly competitive for nodulation of cowpea, where all nodules formed on the roots were infected by P. phymatum. In this study, we showed that using the same mixture of strains and the same growth conditions we described previously (Lardi et al., 2017) but with the P. phymatum T6SS insertional mutants instead of the wild-type, we have a different outcome in our nodulation competition assay (Figure 3). With the P. phymatum T6SS mutants in the bacterial mixture and a higher cell density inoculum (105 cells), we were able to recover, besides P. phymatum, P. diazotrophica from the nodules. The latter is a β-rhizobial strain isolated from nodules of different Mimosa species from several geographical locations (Sheu et al., 2013) and its genome has recently been sequenced (Beukes et al., 2017). The results on the plant competition could be explained either by the impairment of the P. phymatum T6SS mutants to establish an efficient infection with the plant host (cowpea) or by a disadvantage to compete with other soil bacteria. Inspection of the recently sequenced P. diazotrophica and P. mimosarum genomes revealed the presence of a complete T6SS system also in these strains. The role of the T6SSs in these strains is currently unknown. Since the P. phymatum mutants constructed in our study were not impaired in the infection of cowpea (Table 2), we suggest that the P. phymatum T6SS plays a role in β-rhizobial competition for plant infection.
Even though the T6SS in bacterial pathogens was firstly described as usually important for the interaction with eukaryotic host cells, the role in interbacterial competition was shown to be the most common feature of T6SSs. Many reports, on different bacteria, have shown that the system is successfully used to kill bacteria. Plant-associated bacteria belonging to the genera Agrobacterium, Pantoea, and Pseudomonas (Ma et al., 2009, 2014; Shyntum et al., 2015; Bernal et al., 2017a) were shown to utilize T6SSs to outcompete other members of the soil community (Bernal et al., 2017a). Here we show that the T6SS clusters present in P. phymatum are important for interbacterial competition of the strain in vitro and in vivo. In contrast to what has been shown previously with R. leguminosarum (Bladergroen et al., 2003; Ryu, 2015), P. phymatum T6SSs do not affect the establishment of a symbiosis with the plant host. In term of nodule dry weight as well as nodule number and nitrogenase activity, cowpea plants infected with the T6SS mutant strains behaved similar to the ones inoculated with the wild-type. Preliminary tests suggested similar results using common bean as host legume (data not shown). We show here that P. phymatum uses T6SSs to outcompete P. diazotrophica and P. sabiae. The survival rate of these two strains (shown in Figure 4 as the recovered CFU) when attacked by a P. phymatum T6SS mutant was significantly higher relative to survival against the wild-type and comparable with the survival rate without attacker. We cannot rule out that the slight differences in the phenotype observed with both mutants are due to the gene disrupted (tssB in cluster T6SS-3 and tssI-b in cluster T6SS-b) or to a polar effect caused by the insertion of the plasmid. Taken together, the results of the in vitro assays are in good agreement with the competition experiments for legume nodulation. The reason why P. diazotrophica is outcompeting the other β-rhizobia when the P. phymatum wild-type was replaced with the T6SS mutants is unclear.
Interestingly, killing of P. mimosarum by P. phymatum seemed independent of the T6SSs (Figure 4C). Maybe other P. phymatum competition traits, such as the contact-dependent inhibition system (CDI) or antibiotic production (Aoki et al., 2010; Stubbendieck and Straight, 2016) could be responsible for the competitive advantage of P. phymatum over P. mimosarum. It is noteworthy that P. mimosarum was shown to be more effective for N2-fixation and nodulation than P. phymatum on the cognate β-rhizobial legume host Mimosa pudica (Lardi et al., 2017). Therefore, T6SS-mediated competition prior to infection may be less important for β-rhizobial competition in the case of M. pudica infection.
This study reports on the initial characterization of the role of the T6SSs in the β-rhizobial strain P. phymatum. We demonstrate that in this bacterium the T6SS is involved in biofilm formation and plays an important role in competition against other β-rhizobial strains in vitro and in vivo on the papilionoid legume cowpea as host. Our results support a scenario where T6SS-mediated competition is needed for selective killing of competitors, which in turn will have a strong impact on which of the present soil bacteria will infect the roots of a given plant host. Further work will be required to elucidate the contribution of each components of the systems and to figure out how P. phymatum T6SSs are activated in a natural environment.
Materials and Methods
Bacterial Strains and Growth Conditions
Paraburkholderia strains (described in the Supplementary Table S1) were cultivated under aerobic conditions at 30°C in LB medium without salt (10 g/L tryptone and 5 g/L yeast extract), and supplemented with appropriate antibiotics. E. coli strains (Supplementary Table S1) for cloning and helper strain for tri-parental mating were multiplied in LB (10 g/L tryptone, 5 g/L yeast extract, and 4 g/L NaCl). For plant nodulation tests, for tri-parental mating and biofilm formation the cultures were washed in defined buffered AB-minimal medium (Clark and Maaloe, 1967) without nitrogen and with 10 mM sodium citrate as the carbon source while for EPS and interbacterial competition assays the cells were washed in LB medium without salt.
Construction of Mutant Strains
A fragment of 378 bp from the gene Bphy_5985 (BPHY_RS29920) (tssI-b, 519599–521524) and of 456 bp from the gene Bphy_6114 (BPHY_RS30520) (tssB-3, 665221665754) was amplified from the genomic DNA of P. phymatum with primers that introduced EcoRI sites on the extremities (tssI-bFor and tssI-bRev; tssB-3For and tssB-3Rev – Supplementary Table S1). The PCR products were purified with the Qiagen® PCR purification kit, and digested with EcoRI (NEB). The suicide vector pSHAFT2 (Shastri et al., 2017) was also digested with EcoRI, and used for a ligation with the digested PCR fragments using T4 DNA Ligase from Roche®. The ligation reaction was transformed in chemically competent E. coli CC118pir cells, and the insertion of the fragments was confirmed by sequencing with the primer pSHAFTseqFor (Supplementary Table S1). The constructs were introduced in P. phymatum by tri-parental mating using E. coli DH5α pRK2013 as helper. The transconjugants were purified twice in ABC minimal media with chloramphenicol 80 μg/ml. The correct insertion was confirmed by amplification with external primers annealing in the genome of P. phymatum (Bphy_5985For position 519929 in the case of tssI-b and Bphy_6112Rev position 663149 in the case of tssB-3, Figure 1) with the pSHAFTseqFor primer (Supplementary Table S1). The resulting strains are called Pphy::tssI-b and Pphy::tssB-3.
For the generation of the tagged P. diazotrophica, P. mimosarum, and P. phymatum strains, tri-parental mating was performed, using E. coli DH5α containing pRK2013 as helper, and E. coli with the plasmids pBAH8 (Huber et al., 2002) and pIN62 (Vergunst et al., 2010) as donors. The resulting strains were used in the in vitro competition assays.
Phenotypic Characterization of the T6SS Mutant Strains
In order to verify the phenotype of the P. phymatum mutants, different aspects of the physiology were determined and compared to the wild-type strain. The growth of the strains was analyzed in rich media (LB without salt) and minimal media (ABC). For the assessment of EPS production, a cotton swab was plunged in a bacterial suspension with an OD600 0.5 and streaked on modified YEM plates (1% mannitol, 0.06% yeast extract) (Richau et al., 2000). The plates were kept for 72 h at 30°C. Biofilm formation was detected in cell culture plate, 96-well, polystyrene (Sarstedt), inoculated with AB-minimal medium (Clark and Maaloe, 1967) and 10 mM of sodium citrate as carbon source, as described previously (Huber et al., 2002; Pessi et al., 2013; Lardi et al., 2015) with a longer incubation time (72 h).
Plant Growth Conditions and Nitrogenase Activity
Cowpea (Vigna unguiculata (L.) Walp. cv. Red Caloona, kindly provided by Prof. Hans-Martin Fischer –ETH Zurich) surface sterilized seeds (Klonowska et al., 2012) were placed on 0.8% agar plates and incubated in the dark at 30°C for 24–36 h. Germinated seedlings were introduced into autoclaved yoghurt-jar containing vermiculite (VTT-Group, Muttenz, Switzerland) and 170 ml diluted Jensen medium (Hahn and Hennecke, 1984). Direct seed inoculation with the desired bacterial strains (105 cells) or with the bacterial mixtures (102 and 105 cells) was done. The plants were grown as follows in a growth chamber: 22°C (night) and 27°C (day); 12 h light (200 μMol intensity); humidity 60%. The plants were harvested 21 days post-infection (dpi), and the determination of symbiotic properties (nodule number, nodule dry weight and determination of nitrogenase activity) was done according to Göttfert et al. (1990). ARA (acetylene reduction assay) was done to determine the specific nitrogenase activity of each plant. For that, 1 ml of acetylene (PanGas, Zürich, Switzerland) was injected in 50 ml tubes (Infochroma AG, Zug, Switzerland) containing the root of the plant to be analyzed. Gas chromatography (Agilent Technologies, 6850 Network GC System) (Koch et al., 2010) was performed by injecting 25 μl of the incubated gas. The specific nitrogenase activity was obtained by taking the relative area of ethylene produced, normalized by nodule number, dry weight and incubation time. At least three plants per experiment were tested for single infection. Two independent experiment were performed.
Competition Assays in Planta
The strains for inoculation were grown until stationary phase in LB medium without salt. After washing the cultures twice in minimal medium (AB) without nitrogen, the OD600 of each culture was adjusted to 0.01. The CFU was verified by plating on LB without salt plates. For the first test, the cultures were diluted to 102 cells, and for the second test to 105 cells. The bacterial mixture containing P. phymatum (wild-type, Pphy::tssI-b or Pphy::tssB-3), P. diazotrophica, P. mimosarum, P. sabiae in equal amounts was immediately applied on cowpea germinated seeds (1 ml/seedling). The mixture containing the P. phymatum wild-type was tested only with an inoculum of 105 cells. Two independent experiments were performed per test and per experiment a minimum of four plants of cowpea were analyzed. After 3 weeks of incubation, the roots were surface sterilized by plunging for 10 s in 100% ethanol, then 3 min in 2.5% sodium hypochloride and finally washed five times with sterile deionized water (Vincent, 1970). All the nodules present on the roots of the plants were collected individually and placed on an Eppendorf tube containing 100 μl LB without salt with 50% glycerol. The nodules were then individually crushed and plated onto two solid LB medium without salt, one of them containing chloramphenicol for the selection of the mutant P. phymatum strains. Single colonies grown on the plate without antibiotic were tested via colony PCR with degenerated recA primers (recABurk1_F and recABurk1_R; Supplementary Table S1), which generate a product with all the four Paraburkholderia strains used in this study (Mishra et al., 2012). The obtained PCR product was purified and sequenced (at Microsynth AG, Switzerland).
Competition Assays in Vitro
Interbacterial competition was tested by killing assays on plate, as described by Shyntum et al. (2015), with the following modifications. P. phymatum strains (wild type-dsRed tagged T6SS+ and T6SS mutants) were used as attackers, while P. diazotrophica (gfp tagged), P. mimosarum (gfp tagged), and P. sabiae were target bacteria. Bacteria were grown until stationary phase in LB medium without salt, washed once with the same media and normalized to OD600 0.1 (Supplementary Figure S4). The inoculum corresponded to a CFU of 108 for each strain. The strains were mixed 1:1 (attacker:target) and 5 μl were spotted on LB plates without salt and incubated 24 h at 30°C. After incubation, the bacteria were recovered from the plate, resuspended in 1 ml of the same media and serial dilution (10-3 until 10-8) was plated. CFU were counted on LB without salt plates and on selective plates, i.e., LB medium without salt containing chloramphenicol (for P. phymatum dsRed and for the P. phymatum mutant strains), and containing gentamycin (for P. mimosarum and P. diazotrophica gfp tagged strains). P. sabiae recovery was counted by subtracting the number of attacker colonies grown in chloramphenicol with the ones grown without antibiotic.
Statistical Analyses
Prism software version 6.0c was used for all the statistical analyses. Biofilm formation and plant test phenotypes (nodule number, nodule dry weight and specific nitrogenase activity) were analyzed using one-way ANOVA with p ≤ 0.05. Interbacterial competition experiments were analyzed with two-tailed t-test with p ≤ 0.05, while the proportions of bacteria after competition on cowpea plants were analyzed by z-test (chi-square) with p ≤ 0.05.
Author Contributions
SdC: Conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper. ML: Performed the experiments, Analyzed the data. AG: Performed the experiments. LE: Analyzed the data. GP: Conceived and designed the experiments, analyzed the data, wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Prof. Hans-Martin Fischer for the use the gas chromatograph at the Institute of Microbiology at the ETH Zürich and for providing the cowpea seeds. They are grateful to Yilei Liu for her precious support in the laboratory and Felipe Fenselau de Felippes for his feedback on the manuscript.
Footnotes
Funding. This work was supported by University of Zurich Forschungskredit to SdC (FK-16-096) and by the Swiss National Science Foundation (31003A_153374) to GP.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02473/full#supplementary-material
References
- Alcoforado Diniz J., Liu Y. C., Coulthurst S. J. (2015). Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell. Microbiol. 17 1742–1751. 10.1111/cmi.12532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus A. A., Agapakis C. M., Fong S., Yerrapragada S., Estrada-de los Santos P., Yang P., et al. (2014). Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLOS ONE 9:e83779. 10.1371/journal.pone.0083779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus A. A., Lee A., Lum M. R., Shehayeb M., Hessabi R., Fujishige N. A., et al. (2013). Nodulation and effective nitrogen fixation of Macroptilium atropurpureum (siratro) by Burkholderia tuberum, a nodulating and plant growth promoting beta-proteobacterium, are influenced by environmental factors. Plant Soil 369 543–562. 10.1007/s11104-013-1590-7 [DOI] [Google Scholar]
- Aoki S. K., Diner E. J., De Roodenbeke C. T., Burgess B. R., Poole S. J., Braaten B. A., et al. (2010). A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468 439–442. 10.1038/nature09490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert D. F., Flannagan R. S., Valvano M. A. (2008). A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect. Immun. 76 1979–1991. 10.1128/IAI.01338-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M. (2015). Type VI secretion system: secretion by a contractile nanomachine. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:20150021. 10.1098/rstb.2015.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M., Ho B. T., Mekalanos J. J. (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152 884–894. 10.1016/j.cell.2013.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M., Pilhofer M., Henderson G. P., Jensen G. J., Mekalanos J. J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483 182–186. 10.1038/nature10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal P., Allsopp L. P., Filloux A., Llamas M. A. (2017a). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11 972–987. 10.1038/ismej.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal P., Llamas M. A., Filloux A. (2017b). Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 10.1111/1462-2920.13956 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukes C. W., Palmer M., Manyaka P., Chan W. Y., Avontuur J. R., Van Zyl E., et al. (2017). Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 8:1154. 10.3389/fmicb.2017.01154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukes C. W., Venter S. N., Law I. J., Phalane F. L., Steenkamp E. T. (2013). South African papilionoid legumes are nodulated by diverse Burkholderia with unique nodulation and nitrogen-fixation loci. PLOS ONE 8:e68406. 10.1371/journal.pone.0068406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle L. E., Bailey C. M., Pallen M. J. (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol. 11 3–8. 10.1016/j.mib.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Bladergroen M. R., Badelt K., Spaink H. P. (2003). Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Interact. 16 53–64. 10.1094/MPMI.2003.16.1.53 [DOI] [PubMed] [Google Scholar]
- Bontemps C., Elliott G. N., Simon M. F., dos Reis Junior F. B., Gross E., Lawton R. C., et al. (2010). Burkholderia species are ancient symbionts of legumes. Mol. Ecol. 19 44–52. 10.1111/j.1365-294X.2009.04458.x [DOI] [PubMed] [Google Scholar]
- Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. 10.1186/1471-2164-10-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet Y. R., Zoued A., Boyer F., Douzi B., Cascales E. (2015). The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLOS Genet. 11:e1005545. 10.1371/journal.pgen.1005545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtnick M. N., Brett P. J., Harding S. V., Ngugi S. A., Ribot W. J., Chantratita N., et al. (2011). The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79 1512–1525. 10.1128/IAI.01218-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabona M. G., Silverman J. M., Sall K. M., Boyer F., Coute Y., Poirel J., et al. (2013). An ABC transporter and an outer membrane lipoprotein participate in posttranslational activation of type VI secretion in Pseudomonas aeruginosa. Environ. Microbiol. 15 471–486. 10.1111/j.1462-2920.2012.02816.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. M., De Faria S. M., Chou J. H., James E. K., Elliott G. N., Sprent J. I., et al. (2008). Burkholderia sabiae sp. nov., isolated from root nodules of Mimosa caesalpiniifolia. Int. J. Syst. Evol. Microbiol. 58 2174–2179. 10.1099/ijs.0.65816-0 [DOI] [PubMed] [Google Scholar]
- Chen W. M., De Faria S. M., James E. K., Elliott G. N., Lin K. Y., Chou J. H., et al. (2007). Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int. J. Syst. Evol. Microbiol. 57 1055–1059. 10.1099/ijs.0.64873-0 [DOI] [PubMed] [Google Scholar]
- Chen W. M., De Faria S. M., Straliotto R., Pitard R. M., Simões-Araùjo J. L., Chou J. H., et al. (2005). Proof that Burkholderia strains form effective symbioses with legumes: a study of novel Mimosa-nodulating strains from South America. Appl. Environ. Microbiol. 71 7461–7471. 10.1128/AEM.71.11.7461-7471.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. M., James E. K., Coenye T., Chou J. H., Barrios E., De Faria S. M., et al. (2006). Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 56 1847–1851. 10.1099/ijs.0.64325-0 [DOI] [PubMed] [Google Scholar]
- Chen W. M., Moulin L., Bontemps C., Vandamme P., Bena G., Boivin-Masson C. (2003). Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J. Bacteriol. 185 7266–7272. 10.1128/JB.185.24.7266-7272.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli F. R., Alcoforado Diniz J., Guo M., De Cesare V., Trost M., Coulthurst S. J. (2016). VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLOS Pathog. 12:e1005735. 10.1371/journal.ppat.1005735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J., Maaloe O. (1967). DNA replication and division cycle in Escherichia coli. J. Mol. Biol. 23 99–112. 10.1016/S0022-2836(67)80070-6 4578860 [DOI] [Google Scholar]
- Decoin V., Gallique M., Barbey C., Le Mauff F., Poc C. D., Feuilloley M. G., et al. (2015). A Pseudomonas fluorescens type 6 secretion system is related to mucoidy, motility and bacterial competition. BMC Microbiol. 15:72. 10.1186/s12866-015-0405-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis F. B., Jr., Simon M. F., Gross E., Boddey R. M., Elliott G. N., Neto N. E., et al. (2010). Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol. 186 934–946. 10.1111/j.1469-8137.2010.03267.x [DOI] [PubMed] [Google Scholar]
- Durand E., Zoued A., Spinelli S., Watson P. J., Aschtgen M. S., Journet L., et al. (2012). Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. J. Biol. Chem. 287 14157–14168. 10.1074/jbc.M111.338731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl L., Vandamme P. (2016). Members of the genus Burkholderia: good and bad guys. F1000Res. 5 1–10. 10.12688/f1000research.8221.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G. N., Chen W. M., Bontemps C., Chou J. H., Young J. P., Sprent J. I., et al. (2007a). Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann. Bot. 100 1403–1411. 10.1093/aob/mcm227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G. N., Chen W. M., Chou J. H., Wang H. C., Sheu S. Y., Perin L., et al. (2007b). Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 173 168–180. 10.1111/j.1469-8137.2006.01894.x [DOI] [PubMed] [Google Scholar]
- Elliott G. N., Chou J. H., Chen W. M., Bloemberg G. V., Bontemps C., Martinez-Romero E., et al. (2009). Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ. Microbiol. 11 762–778. 10.1111/j.1462-2920.2008.01799.x [DOI] [PubMed] [Google Scholar]
- Estrada-de los Santos P., Vinuesa P., Martinez-Aguilar L., Hirsch A. M., Caballero-Mellado J. (2013). Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr. Microbiol. 67 51–60. 10.1007/s00284-013-0330-9 [DOI] [PubMed] [Google Scholar]
- Filloux A., Hachani A., Bleves S. (2008). The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154 1570–1583. 10.1099/mic.0.2008/016840-0 [DOI] [PubMed] [Google Scholar]
- Gallique M., Bouteiller M., Merieau A. (2017a). The type VI secretion system: a dynamic system for bacterial communication? Front. Microbiol. 8:1454. 10.3389/fmicb.2017.01454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallique M., Decoin V., Barbey C., Rosay T., Feuilloley M. G., Orange N., et al. (2017b). Contribution of the Pseudomonas fluorescens MFE01 type VI secretion system to biofilm formation. PLOS ONE 12:e0170770. 10.1371/journal.pone.0170770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garau G., Yates R. J., Deiana P., Howieson J. G. (2009). Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol. Biochem. 41 125–134. 10.1016/j.soilbio.2008.10.011 [DOI] [Google Scholar]
- Göttfert M., Hitz S., Hennecke H. (1990). Identification of nodS and nodU, two inducible genes inserted between the Bradyrhizobium japonicum nodYABC and nodIJ genes. Mol. Plant Microbe Interact. 3 308–316. [DOI] [PubMed] [Google Scholar]
- Gyaneshwar P., Hirsch A. M., Moulin L., Chen W. M., Elliott G. N., Bontemps C., et al. (2011). Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol. Plant Microbe Interact. 24 1276–1288. 10.1094/MPMI-06-11-0172 [DOI] [PubMed] [Google Scholar]
- Hahn M., Hennecke H. (1984). Localized mutagenesis in Rhizobium japonicum. Mol. Gen. Genet. 193 46–52. 10.1007/Bf00327412 [DOI] [Google Scholar]
- Ho B. T., Dong T. G., Mekalanos J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15 9–21. 10.1016/j.chom.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howieson J. G., De Meyer S. E., Vivas-Marfisi A., Ratnayake S., Ardley J. K., Yates R. J. (2013). Novel Burkholderia bacteria isolated from Lebeckia ambigua - a perennial suffrutescent legume of the fynbos. Soil Biol. Biochem. 60 55–64. 10.1016/j.soilbio.2013.01.009 [DOI] [Google Scholar]
- Huber B., Riedel K., Kothe M., Givskov M., Molin S., Eberl L. (2002). Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46 411–426. 10.1046/j.1365-2958.2002.03182.x [DOI] [PubMed] [Google Scholar]
- Klonowska A., Chaintreuil C., Tisseyre P., Miche L., Melkonian R., Ducousso M., et al. (2012). Biodiversity of Mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in New Caledonia and their adaptation to heavy metal-rich soils. FEMS Microbiol. Ecol. 81 618–635. 10.1111/j.1574-6941.2012.01393.x [DOI] [PubMed] [Google Scholar]
- Koch M., Delmotte N., Rehrauer H., Vorholt J. A., Pessi G., Hennecke H. (2010). Rhizobial adaptation to hosts, a new facet in the legume root-nodule symbiosis. Mol. Plant Microbe Interact. 23 784–790. 10.1094/Mpmi-23-6-0784 [DOI] [PubMed] [Google Scholar]
- Kudryashev M., Wang R. Y., Brackmann M., Scherer S., Maier T., Baker D., et al. (2015). Structure of the type VI secretion system contractile sheath. Cell 160 952–962. 10.1016/j.cell.2015.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardi M., Aguilar C., Pedrioli A., Omasits U., Suppiger A., Carcamo-Oyarce G., et al. (2015). σ54-dependent response to nitrogen limitation and virulence in Burkholderia cenocepacia strain H111. Appl. Environ. Microbiol. 81 4077–4089. 10.1128/AEM.00694-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardi M., De Campos S. B., Purtschert G., Eberl L., Pessi G. (2017). Competition experiments for legume infection identify Burkholderia phymatum as a highly competitive β-rhizobium. Front. Microbiol. 8:1527. 10.3389/fmicb.2017.01527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire B., Chimphango S. B., Stirton C., Rafudeen S., Honnay O., Smets E., et al. (2016). Biogeographical patterns of legume-nodulating Burkholderia spp.: from African fynbos to continental scales. Appl. Environ. Microbiol. 82 5099–5115. 10.1128/AEM.00591-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossi N. S., Manoli E., Simpson P., Jones C., Hui K., Dajani R., et al. (2012). The archetype Pseudomonas aeruginosa proteins TssB and TagJ form a novel subcomplex in the bacterial type VI secretion system. Mol. Microbiol. 86 437–456. 10.1111/j.1365-2958.2012.08204.x [DOI] [PubMed] [Google Scholar]
- Ma L. S., Hachani A., Lin J. S., Filloux A., Lai E. M. (2014). Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16 94–104. 10.1016/j.chom.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. S., Lin J. S., Lai E. M. (2009). An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191 4316–4329. 10.1128/JB.00029-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian R., Moulin L., Bena G., Tisseyre P., Chaintreuil C., Heulin K., et al. (2014). The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype. Environ. Microbiol. 16 2099–2111. 10.1111/1462-2920.12286 [DOI] [PubMed] [Google Scholar]
- Mishra R. P., Tisseyre P., Melkonian R., Chaintreuil C., Miche L., Klonowska A., et al. (2012). Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol. Ecol. 79 487–503. 10.1111/j.1574-6941.2011.01235.x [DOI] [PubMed] [Google Scholar]
- Moulin L., Klonowska A., Caroline B., Booth K., Vriezen J. A., Melkonian R., et al. (2014). Complete genome sequence of Burkholderia phymatum STM815(T), a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand. Genomic Sci. 9 763–774. 10.4056/sigs.4861021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin L., Munive A., Dreyfus B., Boivin-Masson C. (2001). Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 411 948–950. 10.1038/35082070 [DOI] [PubMed] [Google Scholar]
- Nelson M. S., Sadowsky M. J. (2015). Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 6:491. 10.3389/fpls.2015.00491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G., Braunwalder R., Grunau A., Omasits U., Ahrens C. H., Eberl L. (2013). Response of Burkholderia cenocepacia H111 to micro-oxia. PLOS ONE 8:e72939. 10.1371/journal.pone.0072939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Records A. R. (2011). The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol. Plant Microbe Interact. 24 751–757. 10.1094/MPMI-11-10-0262 [DOI] [PubMed] [Google Scholar]
- Richau J. A., Leitao J. H., Correia M., Lito L., Salgado M. J., Barreto C., et al. (2000). Molecular typing and exopolysaccharide biosynthesis of Burkholderia cepacia isolates from a Portuguese cystic fibrosis center. J. Clin. Microbiol. 38 1651–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roest H. P., Mulders I. H., Spaink H. P., Wijffelman C. A., Lugtenberg B. J. (1997). A Rhizobium leguminosarum biovar trifolii locus not localized on the sym plasmid hinders effective nodulation on plants of the pea cross-inoculation group. Mol. Plant Microbe Interact. 10 938–941. 10.1094/MPMI.1997.10.7.938 [DOI] [PubMed] [Google Scholar]
- Ryu C. M. (2015). Against friend and foe: type 6 effectors in plant-associated bacteria. J. Microbiol. 53 201–208. 10.1007/s12275-015-5055-y [DOI] [PubMed] [Google Scholar]
- Sawana A., Adeolu M., Gupta R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:429. 10.3389/fgene.2014.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S., Singh P., Robertson J. D., Leroux M., Skerrett S. J., Goodlett D. R., et al. (2014). VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82 1445–1452. 10.1128/IAI.01368-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S., West T. E., Boyer F., Chiang W. C., Carl M. A., Hood R. D., et al. (2010). Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLOS Pathog. 6:e1001068. 10.1371/journal.ppat.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastri S., Spiewak H. L., Sofoluwe A., Eidsvaag V. A., Asghar A. H., Pereira T., et al. (2017). An efficient system for the generation of marked genetic mutants in members of the genus Burkholderia. Plasmid 89 49–56. 10.1016/j.plasmid.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu S. Y., Chou J. H., Bontemps C., Elliott G. N., Gross E., dos Reis F. B., et al. (2013). Burkholderia diazotrophica sp. nov., isolated from root nodules of Mimosa spp. Int. J. Syst. Evol. Microbiol. 63 435–441. 10.1099/ijs.0.039859-0 [DOI] [PubMed] [Google Scholar]
- Shneider M. M., Buth S. A., Ho B. T., Basler M., Mekalanos J. J., Leiman P. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500 350–353. 10.1038/nature12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyntum D. Y., Theron J., Venter S. N., Moleleki L. N., Toth I. K., Coutinho T. A. (2015). Pantoea ananatis utilizes a type VI secretion system for pathogenesis and bacterial competition. Mol. Plant Microbe Interact. 28 420–431. 10.1094/MPMI-07-14-0219-R [DOI] [PubMed] [Google Scholar]
- Silverman J. M., Brunet Y. R., Cascales E., Mougous J. D. (2012). Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66 453–472. 10.1146/annurev-micro-121809-151619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbendieck R. M., Straight P. D. (2016). Multifaceted interfaces of bacterial competition. J. Bacteriol. 198 2145–2155. 10.1128/JB.00275-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Moreno Z. R., Caballero-Mellado J., Coutinho B. G., Mendonca-Previato L., James E. K., Venturi V. (2012). Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 63 249–266. 10.1007/s00248-011-9929-1 [DOI] [PubMed] [Google Scholar]
- Talbi C., Delgado M. J., Girard L., Ramirez-Trujillo A., Caballero-Mellado J., Bedmar E. J. (2010). Burkholderia phymatum strains capable of nodulating Phaseolus vulgaris are present in Moroccan soils. Appl. Environ. Microbiol. 76 4587–4591. 10.1128/AEM.02886-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Goris J., Chen W. M., De Vos P., Willems A. (2002). Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25 507–512. 10.1078/07232020260517634 [DOI] [PubMed] [Google Scholar]
- Vergunst A. C., Meijer A. H., Renshaw S. A., O’callaghan D. (2010). Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect. Immun. 78 1495–1508. 10.1128/IAI.00743-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettiger A., Basler M. (2016). Type VI secretion system substrates are transferred and reused among sister cells. Cell 167 99.e12–110.e12. 10.1016/j.cell.2016.08.023 [DOI] [PubMed] [Google Scholar]
- Vettiger A., Winter J., Lin L., Basler M. (2017). The type VI secretion system sheath assembles at the end distal from the membrane anchor. Nat. Commun. 8:16088. 10.1038/ncomms16088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J. M. (1970). A Manual for the Practical Study of Root Nodule Bacteria. Oxford: Blackwell Scientific Publications. [Google Scholar]
- Weber B. S., Hennon S. W., Wright M. S., Scott N. E., De Berardinis V., Foster L. J., et al. (2016). Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. mBio 7:e01253-16. 10.1128/mBio.01253-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xu J., Xu J., Zhang H., He L., Feng J. (2014). TssB is essential for virulence and required for type VI secretion system in Ralstonia solanacearum. Microb. Pathog. 74 1–7. 10.1016/j.micpath.2014.06.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.