Abstract
Hepsetidae is a small fish family with only the genus Hepsetus, with six described species distributed throughout the South, Central and Western regions of Africa, showing a close relationship with the Alestidae and some Neotropical fish families. However, no cytogenetic information is available for both Hepsetidae and Alestidae species, thus preventing any evolutionary comparative studies at the chromosomal level. In the present study, we are providing new cytogenetic data for Hepsetus odoe, including the standard karyotype, C-banding, repetitive DNAs mapping, comparative genomic hybridization (CGH) and whole chromosome painting (WCP), providing chromosomal patterns and subsidies for comparative cytogenetics with other characiform families. Both males and females H. odoe have 2n = 58 chromosomes (10m + 28sm + 20st/a), with most of the C-band positive heterochromatin localized in the centromeric and subtelomeric regions. Only one pair of chromosomes bears proximal 5S rDNA sites in the short arms, contrasting with the 18S rDNA sequences which are located in the terminal regions of four chromosome pairs. Clear interstitial hybridization signals are evidenced for the U1 and U2 snDNA probes, but in only one and two chromosome pairs, respectively. Microsatellite motifs are widely distributed in the karyotype, with exception for the (CGG)10, (GAA)10 and (GAG)10 probes, which highlight conspicuous interstitial signals on an unique pair of chromosomes. Comparative data from conventional and molecular cytogenetics, including CGH and WCP experiments, indicate that H. odoe and some Erythrinidae species, particularly Erythrinus erythrinus, share similar chromosomal sequences suggesting some relatedness among them, although bearing genomic specificities in view of their divergent evolutionary histories.
Keywords: fishes, molecular cytogenetics, chromosomal painting, comparative genomic hybridization (CGH), karyotype evolution
Introduction
Characiformes comprises 24 families and more than 2100 species (Eschmeyer and Fong, 2017), distributed in many Neotropical and Ethiopian rivers (Nelson et al., 2016). As they are exclusively freshwater fishes, their evolutionary history is related with continents fragmentations and settlement and, with the development of natural barriers during their dispersion throughout secondary habitats (Vari and Malabarba, 1998; Oliveira et al., 2007).
The most primitive characiforms are the African citharinoids (Arroyave et al., 2013), and the relationship between the Neotropical and Ethiopian species may be closely linked with the Gondwana break-up, with a fast diversification established in a new habitat free of competition (Calcagnotto et al., 2005). Despite significant efforts on morphological and molecular analyzes, phylogenetic relationships are currently still uncertain for several groups and even the monophyly of Characiformes is still debated (Arcila et al., 2017).
The wide diversification of the characiforms is highlighted by the high karyotype variability found within distinct Neotropical groups, showing the fast evolution of these fishes as expected by the high fragmentation observed in the South American rivers, in contrast with the African ones, which presents lower fragmentation and variability (Ortí and Meyer, 1997; Oliveira et al., 2007). One example of such scenario concerns the Erythrinidae, a small family widely distributed throughout South America, consisting of the genus Erythrinus, Hoplerythrinus, and Hoplias (Oyakawa, 2003). Cytogenetics of the Erythrinidae fishes have been quite investigated over years, especially for H. malabaricus and E. erythrinus, where a variety of chromosomal features occurs even within a same nominal species, thus supporting the presence of species complexes (Bertollo, 2007; Cioffi et al., 2012). In fact, erythrinids hold a variety of different karyomorphs, with diploid numbers (2n) varying from 39 in Hoplias malabaricus (karyomorph D) to 2n = 54 in Erythrinus erythrinus (karyomorph A), in addition to distinct sex chromosomes systems with independent origins and particular evolutionary trajectories (Cioffi et al., 2013). The diploid number found for most Erythrinus species (2n = 54) is also the common one observed for Characiformes, which possibly represents the ancestral condition for this order (Oliveira et al., 2007; Cioffi et al., 2012). However, the full comprehension of the evolutionary relationships of its families is not clear until now. A recent phylogeny based on 1,051 genetic markers showed that both African Hepsetidae and Alestidae families have closer relationship and, in a lower scale, to other Neotropical families, such as Erythrinidae, Cynodontidae and Hemiodotidae, but not with Lebiasinidae and Ctenoluciidae (Arcila et al., 2017). This result is not fully consensual with some previous phylogenetic proposals (Ortí and Meyer, 1997; Buckup, 1998; Calcagnotto et al., 2005), where some of above families were found to be related.
Notwithstanding, except for the Erythrinidae (see above), most of these families remain with kayotypes poorly analyzed, thus limiting any evolutionary comparative studies among them at the chromosomal level. In this sense, karyological data for Hemiodontidae are mainly restricted to chromosome numbers although all species presenting the same diploid number (2n = 54) and bi-armed chromosomes (Arefjev, 1990; Porto et al., 1992, 1993; Arai, 2011). Concerning Cynodontidae, the only species analyzed up to now (Rhaphiodon vulpinus) also presented the same 2n and karyotype structure (Pastori et al., 2009). In turn, the available chromosome data for Lebiasinidae are also mainly restricted to chromosome numbers (Scheel, 1973; Arai, 2011), with exception for a few species (Arefjev, 1990; Oliveira et al., 1991). Despite such largely limitation, a high diversity characterizes their diploid numbers, which ranges from 2n = 22 in Nannostomus unifasciatus, to 2n = 46 in N. trifasciatus (Oliveira et al., 2007; Arai, 2011). Occasional occurrence of large metacentric pairs, such as in N. unifasciatus (Arefjev, 1990) points to Robertsonian fusions in the karyotype differentiation. Pyrrhulina australis and Pyrrhulina aff. australis share 2n = 40 (4st + 36a), however, a significant genomic divergence was found between them, evidencing that they correspond to distinct evolutionary units (Moraes et al., 2017). As regards to Ctenoluciidae, four species of the Boulengerella genus from the Amazon River basin (Brazil), showed 2n = 36 and a very similar karyotype organization. A conspicuous chromosomal heteromorphism in male specimens point to a possible XX/XY sex chromosome system in such species (de Souza E Sousa et al., 2017). Besides, Ctenolucius hujeta (2n = 36) is the only additional species of Ctenoluciidae that has its chromosomal number already analyzed (Arefjev, 1990), coinciding with those found for the Boulengerella species.
The Hepsetidae family contains only a single genus (Hepsetus) and, for a long time, H. odoe was considered the only valid species. However, five additional species have been described by recent studies: H. kingsleyae, H. lineatus, H. occidentalis, H. cuvieri, and H. microlepis, distributed throughout the South, Central and Western regions of Africa (Decru et al., 2012, 2013a,b, 2015), where they have great significance for local economy (Kareem et al., 2016). Despite the economic and evolutionary importance of this group, no chromosome data are available for any Hepsetidae species.
In the present study, we provide, for the first time, cytogenetic data for Hepsetus odoe, including the standard karyotype, C-banding, repetitive DNAs mapping, comparative genomic hybridization (CGH) and whole chromosome painting (WCP), in order to investigate its chromosomal patterns and provide subsidies for comparative analyzes with some Neotropical fish families. In this sense, this study represents the first one of a series focusing on the cytogenetics and cytogenomics of the African species toward their karyoevolutionary processes.
Materials and Methods
Specimens, Chromosome Preparations, C-banding and DNA Samples
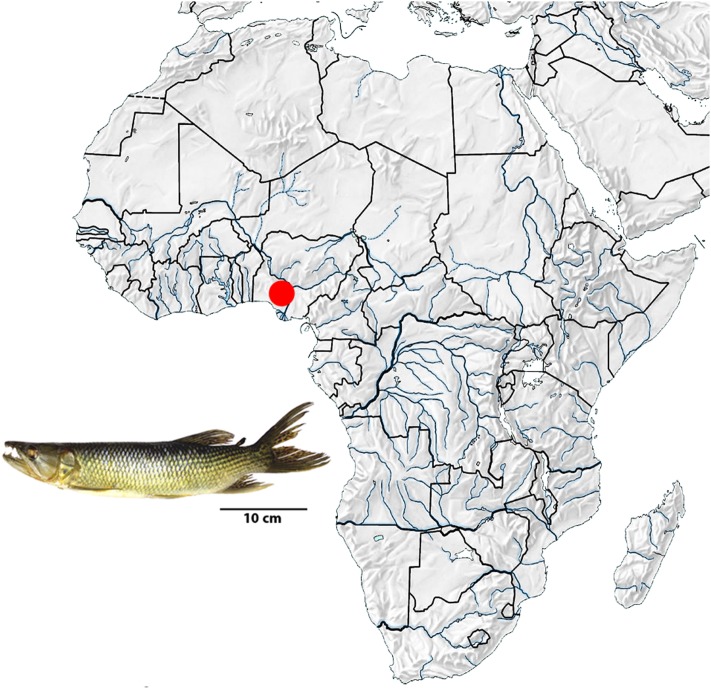
Eleven specimens of Hepsetus odoe (06 males and 05 females) from the Opa Reservoir, Obafemi Awolowo University, Nigeria (6°51′45″ N, 4°79′00″ E) were analyzed (Figure 1). The specimens were transferred to laboratory aquaria and kept under standard conditions for 1 day prior to the experiments. As H. odoe represent a non-CITES threatened species, no proper authorization was required for their sampling and/or transportation. All specimens were deposited in the Museu de Zoologia of the Universidade de São Paulo (MZUSP), under the accession No. 119844. Mitotic chromosomes were obtained by the protocols described in Bertollo et al. (2015) and experiments followed ethical conducts in accordance with the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (Process number CEUA 1853260315). The C-positive heterochromatin was detected using the Barium hydroxide protocol (Sumner, 1972). The genomic DNA was extracted according to standard phenol–chloroform procedures (Sambrook and Russell, 2001).
FIGURE 1.
Map of Africa showing the collect location of the Hepsetus odoe specimens (red point) in Nigeria, Oluwa River, Niger River basin. Map of Africa was adapted from http://geografiahistoriajodar.blogspot.com.br/.
Probes for Chromosome Hybridization
A total of 11 repetitive DNA sequences, including four multigene families (U1 and U2 snDNA, 5S and 18S rDNAs) and seven microsatellite repeat motifs (A)30, (CA)15, (GA)15, (CAC)10, (CGG)10, (GAA)10 and (GAG)10, were used as probes for FISH experiments. The oligonucleotide probes were directly labeled with Cy3 during synthesis according to Kubat et al. (2008). The other four tandemly arrayed DNA sequences were obtained via PCR from the nuclear DNA of H. odoe. The 5S rDNA repeat copy included 120 base pairs (bp) of the 5S rRNA transcribing gene and 200 bp of the non-transcribed spacer (NTS), produced according to Pendás et al. (1994). The second probe contained 1,400-bp repeats of the 18S rRNA gene, obtained according to Cioffi et al. (2009). Both rDNA probes were cloned into plasmid vectors and propagated in DH5α Escherichia coli competent cells (Invitrogen, San Diego, CA, United States). The U1 and U2 snDNA sequences were produced by PCR, according to Cross et al. (2005) and Silva et al. (2015), respectively. All these probes were directly labeled with Spectrum Orange-dUTP by nick translation, according to manufacturer’s recommendations (Roche, Mannheim, Germany), with the exception of 5S rDNA, which was directly labeled with Spectrum Green-dUTP, also by nick translation (Roche, Mannheim, Germany).
Fluorescence in Situ Hybridization (FISH) for Repetitive DNA Mapping
Fluorescence in situ hybridization (FISH) was performed under high stringency conditions on metaphase chromosome spreads, as described in Yano et al. (2017a). The chromosome slides were incubated with RNAse (10 μg/mL) for 1 h at 37°C in a wet chamber and then washed for 5 min in 1x PBS and incubated with pepsin 0,005% for 10 min at room temperature. It was followed a wash in 1x PBS, a fixation with 1% formaldehyde for 10 min at room temperature, and another 1x PBS wash. The slides were then set for an alcoholic series of 70, 85, and 100% 2 min each, followed by the DNA denaturation in 70% formamide/2x SSC for 3 min at 75°C. After denaturation, the chromosome spreads were dehydrated in an ethanol series of 70, 85, and 100% at room temperature, 2 min each. 20 μL of the hybridization mixture (100 ng probes, 50% deionized formamide, 10% dextran sulfate) were then dropped on the slides, and the hybridization was performed for 16–18 h at 37°C in a wet chamber containing 2x SSC. A post-hybridization wash was carried out with 2x SSC for 5 min followed by another wash in 1x SSC at 42°C, 5 min. A final washing series was then performed at room temperature, consisting of 1x PBS for 5 min, and ethanol 70, 85, and 100% for 2 min each. Finally, the chromosomes were counterstained with DAPI (1.2 μg/mL) and the slides mounted with an antifading solution (Vector, Burlingame, CA, United States).
Chromosomal Microdissection, Probe Preparation and Labeling
Fifteen copies of the following chromosomes were isolated by microdissection and amplified using the procedure described in Yang et al. (2009): (i) X chromosome of Hoplias malabaricus karyomorph B (HMB-X); (ii) Y1 chromosome of H. malabaricus karyomorph G (HMG-Y1) and (iii) Y chromosome of Erythrinus erythrinus karyomorph D (ERY-Y). These probes were labeled with Spectrum Orange-dUTP (ERY-Y) or Spectrum Green-dUTP (HMB-X and HMG-Y1) (Vysis, Downers Grove, IL, United States) in a secondary DOP PCR using 1 μL of the primarily amplified product as a template DNA, following Yang et al. (2009).
FISH of Whole Chromosome Specific Probes (W)
Chromosomal preparations of males and females of H. odoe were used for Zoo-FISH experiments with all the above mentioned probes. The hybridization procedures followed Yano et al. (2017a). To block the hybridization of high-copy repeat sequences 60 μg of C0t-1 DNA directly isolated from H. malabaricus (karyomorphs B and G) and E. erythrinus (karyomorph D) male genomes were prepared according to Zwick et al. (1997). Hybridization was performed for 144 h at 37°C in a moist chamber. The post-hybridization wash was carried out with 1x SSC for 5 min at 65°C, and in 4x SSC/Tween using a shaker at RT and then rinsed quickly in 1x PBS. Subsequently, the slides were dehydrated in an ethanol series (70, 85, and 100%), 2 min each. Finally, the chromosomes were counterstained with DAPI (1.2 μg/mL) and mounted in antifade solution (Vector).
Probes for Comparative Genomic Hybridization (CGH)
The gDNA of H. odoe was used for comparative analyzes with the gDNAs of several Erythrinidae species, namely E. erythrinus (karyomorph D), Hoplias lacerdae, H. malabaricus (karyomorph A) and Hoplerythrinus unitaeniatus (karyomorph D). The gDNA of H. odoe was labeled with biotin-16-dUTP using BIO-nick-translation Mix (Roche), while the male-derived gDNAs of E. erythrinus, H. malabaricus, H. unitaeniatus, and H. lacerdae were labeled with digoxigenin-11-dUTP using DIG-nick-translation Mix (Roche, Manheim, Germany). In all experiments it was utilized C0t-1 DNA (i.e., fraction of genomic DNA enriched for highly and moderately repetitive sequences), prepared according to Zwick et al. (1997), for blocking common genomic repetitive sequences. The final probe was composed of 500 ng of H. odoe gDNA plus 500 ng of the corresponding gDNA for each Erythrinidae species. The probe was ethanol-precipitated and the dry pellet dissolved in a hybridization buffer (20 μL per slide) containing 50% formamide + 2x SSC + 10% SDS+ 10% dextran sulfate and Denhardt’s buffer, pH 7.0).
Fluorescence in Situ Hybridization for CGH
CGH experiments were performed according to Symonová et al. (2013). Slides with the metaphase plates were stored overnight in a freezer, being submitted to an alcoholic series of 70, 85, and 100%, 3 min each, before and after the storage. After that, the slides were aged for 1–2 h at 60°C, washed in 2x SSC for 5 min, treated with RNAse (200 μg/mL) for 90 min at 37°C in a wet chamber and them washed in 2x SSC for 30 s. It was followed another alcoholic series treatment, a wash in 1x PBS for 5 min, a Pepsin (50 μg/mL) treatment, a wash in 1x PBS for 5 min and an additional alcoholic series treatment. Finally, the material was denaturated in 75% formamide/2x SSC at 74°C for 3 min, followed by an alcoholic series being the first 70% cold ethanol. 20 μL of the probes were spotted to the slides, which were them incubated at room temperature (37°C) in a dark humid chamber for 3 days, with rubber-sealed coverslips. The rubber cement and coverslips were removed in a solution of 4x SSC/0.1% Tween. The slides were then washed twice in 50% formamide/2x SSC for 10 min each, three times in 1x SSC, rinsed in 2x SSC at room temperature, and incubated 20 min. in a humid chamber with 500 μL of 3%BSA/4x SSC/Tween, with coverslips. The hybridization signal was detected with anti-digoxigenin-Rhodamin (Roche) diluted in 0.5% bovine serum albumin (BSA) in PBS, and avidin-FITC (Sigma) diluted in PBS containing 10% normal goat serum (NGS). Four final washes were performed at 44°C in 4x SSC/0.1% Tween, 7 min each. Finally, the chromosomes were counterstained with DAPI (1.2 μg/mL) and mounted in an antifade solution (Vector, Burlingame, CA, United States).
Microscopic Analyses
At least 30 metaphase spreads per individual were analyzed to confirm the diploid number, karyotype structure and FISH results. Images were captured using an Olympus BX50 microscope (Olympus Corporation, Ishikawa, Japan) with CoolSNAP and the images processed using Image Pro Plus 4.1 software (Media Cybernetics, Silver Spring, MD, United States). Chromosomes were classified as metacentric (m), submetacentric (sm), subtelocentric (st) or acrocentric (a), according to their arm ratios (Levan et al., 1964).
Results
Karyotype Composition and C-banding
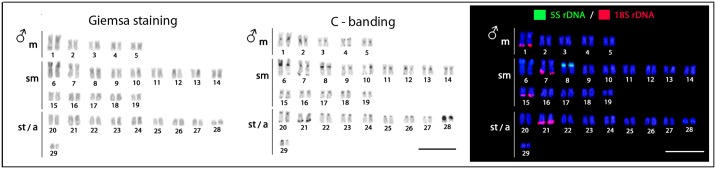
All specimens, both males and females, have 2n = 58 (10m + 28sm + 20st/a). The C-positive heterochromatic is most localized in the centromeric and subtelomeric regions, with a more conspicuous block present in the 28th chromosome pair of the karyotype (Figure 2).
FIGURE 2.
Hepsetus odoe male karyotypes under standard Giemsa staining, C-banding and double-FISH with 18S rDNA (red) and 5S rDNA (green) probes. Both males and females have the same karyotypes. Bar = 5 μm.
Chromosomal Mapping of Repetitive DNAs
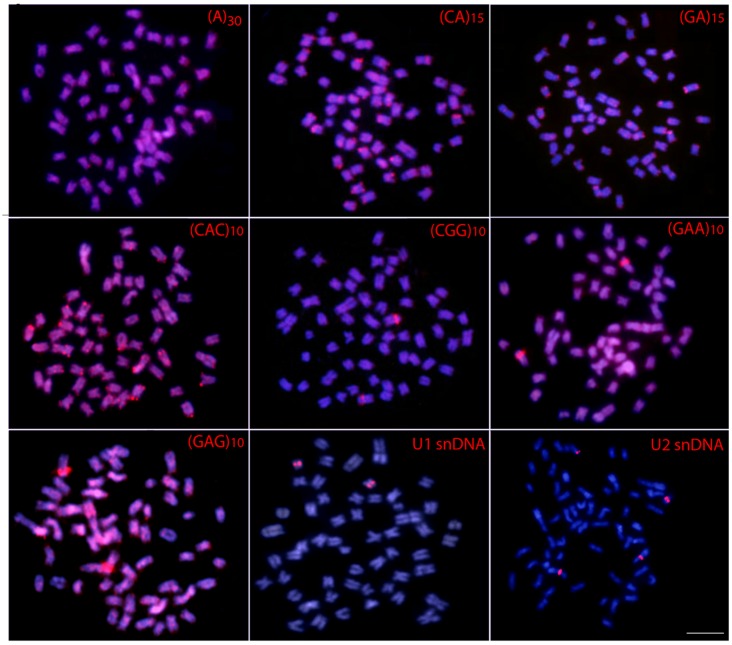
The 5S rDNA occurs in the proximal region of the short arms of only one sm chromosome pair, while the 18S rDNA is located in the terminal region of the long arms of four chromosome pairs (1m + 2sm + 1st) (Figure 2). Clear interstitial hybridization signals were observed in one pair of chromosomes for the U1 snDNA, and in two chromosome pairs for the U2 snDNA, being interstitial and telomeric located in each one of them, respectively (Figure 3). Widely distributed marks were evidenced by the microsatellite motifs. Signals were mainly telomeric, but some also interstitial, as for (A)30, (CA)15, (GA)15, (CAC)10 and (GAG)10 probes. Exceptions for these general patterns were presented by the (CGG)10, (GAA)10 and (GAG)10 probes, which highlighted a conspicuous interstitial signal on a unique pair of chromosomes (Figure 3).
FIGURE 3.
Metaphase plates of Hepsetus odoe hybridized with repetitive DNA sequences, including mono-, di- and trinucleotide microsatellites and the multigene families U1 and U2 snDNAs. Bar = 5 μm.
Comparative Genomic Hybridization (CGH)
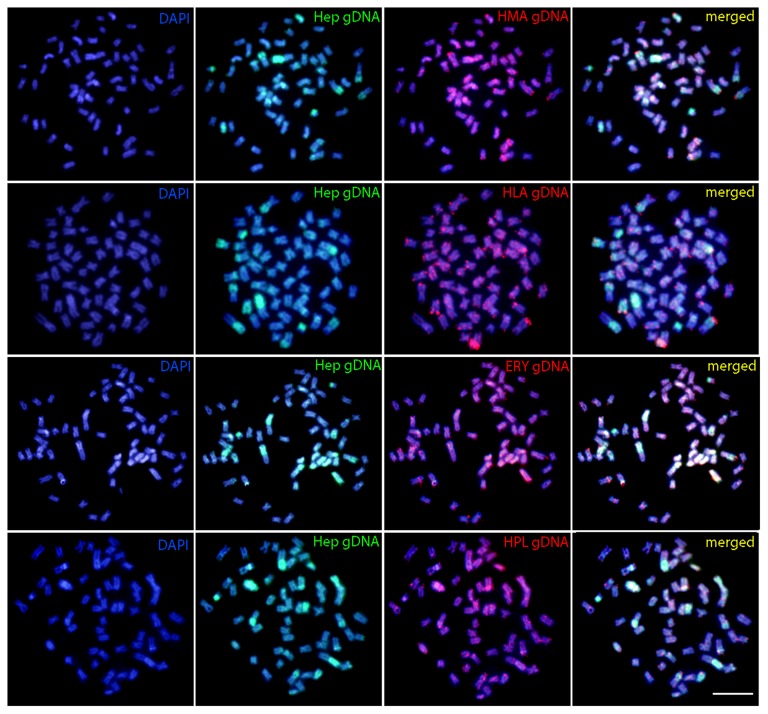
The comparative genomic hybridization showed that the gDNA of H. odoe shares some homologies with those of the Erythrinidae species analyzed. Despite some scattered hybridization, labeled telomeric and pericentromeric regions were evidenced according to each species. However, it stands out the hybridization pattern with E. erythrinus, where some whole chromosome pairs were labeled, in addition to telomeric overlaps in other ones. An exclusive acrocentric chromosome of H. odoe, that presented hybridization signals only with the gDNA of H. unitaeniatus, was also highlighted (Figure 4).
FIGURE 4.
Comparative genomic hybridization (CGH) in metaphase plates of Hepsetus odoe. First column: DAPI images (blue); Second column: hybridization pattern with Hepsetus odoe (Hep) gDNA probe; Third column: Hybridization patterns with Hoplias malabaricus (HMA) gDNA, Hoplias lacerdae (HLA) gDNA, Erythrinus erythrinus (ERY) gDNA and Hoplerythrinus unitaeniatus (HPL) gDNA probes; Fourth column: merged images of each genomic probes and DAPI staining. The common genomic regions are depicted in yellow. Bar = 5 μm.
Detection of Chromosomal Homeologies by Zoo-FISH Experiments
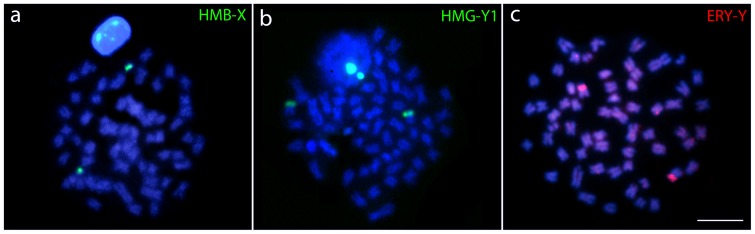
Hybridization performed with HMB-X (X chromosome from Hoplias malabaricus karyomorph B) probe highly painted one small st/a chromosome of H. odoe (Figure 5a). Both HMG-Y1 (Y1 chromosome from H. malabaricus karyomorph G) and ERY-Y (Y chromosome from Erythrinus erythrinus karyomorph D) probes painted the p arms of medium-sized st/a chromosomes (Figures 5b,c) of H. odoe. Besides, ERY-Y probe also produced faint scattered hybridization pattern on several other chromosomes of H. odoe (Figure 5c).
FIGURE 5.
Whole chromosome painting (WCP) in metaphase plates of Hepsetus odoe showing the chromosomes hybridized with (a) the X chromosome of Hoplias malabaricus karyomorph B (HMB-X), (b) the Y1 chromosome of Hoplias malabaricus karyomorph G (HMG-Y1) and (c) the Y chromosome of Erythrinus erythrinus karyomorph D (ERY-Y).
Discussion
General Chromosome Features of Hepsetus odoe
The lack of karyotype data for several fish groups impairs comparative analyzes on their evolutionary trends and chromosomal relationships. This is the case for the African Hepsetidae family for which chromosomal characteristics are completely unknown. In this sense, this study is the first one providing classical and molecular cytogenetic data for one of its representative species, H. odoe.
Both male and female specimens of H. odoe have the same karyotype structure, with 2n = 58 (5m + 14sm + 10st/a), with no evidence of differentiated sex chromosomes. The heterochromatin distribution follows the general pattern usually found in many other fish species, with preferential centromeric localization. Only one chromosome pair bears proximal 5S rDNA sites in their short arms, in contrast to the 18S rDNA sequences that are located in the telomeric regions of four different pairs in the karyotype. The distribution of these multigene families is shared among many fish groups (Pendás et al., 1994; Gornung, 2013) where the clustering of the 5S and 18S rDNAs in different chromosomes may avoid unwanted chromosomal changes between them (Martins and Galetti, 1999). In addition, this differential clustering is also true for the U2 snDNA sequences, since the cytogenetic mapping for different genes that composed this multigene family, although scarce among fishes, shows a preferential distribution among distinct chromosomes (reviewed in Yano et al., 2017b), as also observed in H. odoe.
With respect to microsatellites, although the scattered distribution of some of them is not so useful for comparative approaches, the conspicuous interstitial bands that (GAG)10, (CGG)10 and (GAA)10 probes highlighted in the genome of H. odoe, constitute important markers for comparative evolutionary analyzes with other Hepsetidae and also close related species. In fact, the clustering of microsatellites represents important evolutionary stages by composing non-coding genome regions, as well as relevant steps in the sex chromosome’s differentiation process (Bergero and Charlesworth, 2009).
DNA sequence analysis strongly suggests that Hepsetidae and Alestidae are phylogenetic close related families (Oliveira et al., 2011; Arcila et al., 2017). In this sense, the present data set for H. odoe are useful tools for complementary investigations covering other Hepsetidae and Alestidae species. In fact, this study represents the first one of a series focusing on the cytogenetics and cytogenomics of such African families, toward the investigation of their karyoevolutionary processes and relatedness.
Comparative Cytogenetics of Hepsetus odoe with Other Characiformes Species
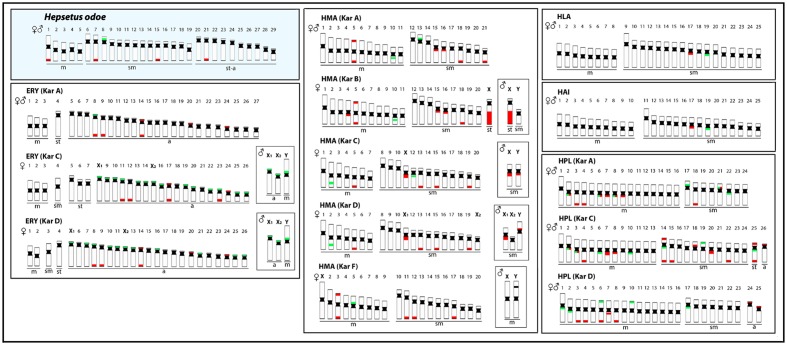
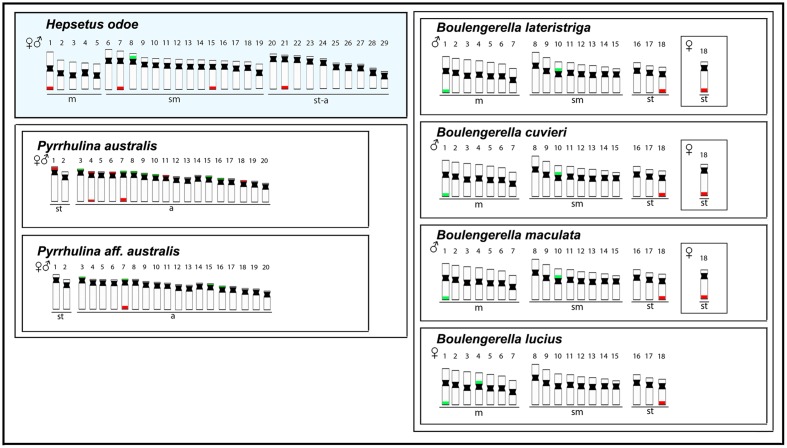
Some previous phylogenetic studies (Ortí and Meyer, 1997; Buckup, 1998; Calcagnotto et al., 2005) have suggested a relationship between Hepsetidae and some other Neotropical groups, such as the Erythrinidae, Ctenoluciidae and Lebiasinidae, although without a full consensus among them. Using new sequencing technology together with phylogenetic reconstructions, a new scenario was evidenced, discarding relationships of Hepsetidae with Lebiasinidae and Cnetoluciidae and, instead off, placing Hepsetidae and Alestidae in a closer clade which has a near position in the phylogenetic tree to some other Neotropical families, such as Erythrinidae, Cynodontidae and Hemiodontidae (Oliveira et al., 2011; Arcila et al., 2017). In this way, as the cytogenetic studies among Cynodontidae and Hemiodontidae families are until now restricted to 2n descriptions in few species, and Alestidae species are still unavailable in spite of recent collecting efforts, we performed a comparative analysis among H. odoe and Erythrinidae, Ctenoluciidae and Lebiasinidae representatives. In this sense, Figures 6, 7 depict some data, including chromosome number, karyotype organization, sex chromosome systems and distribution of the major and minor rDNA sequences in some Erytrinidae, Lebiasinidae, and Ctneoluciidae species. A general overview clearly indicates that Erythrinidae retains the highest amount of characters resembling those of H. odoe than Lebiasinidae and Ctenoluciidae species. Indeed, Erythrinus erythrinus (2n = 54/52), Hoplias lacerdae and H. aimara (2n = 50) and Hoplerythrinus unitaeniatus (2n = 48/52) show diploid numbers closer to that of H. odoe (2n = 58) then Pyrrhulina (2n = 40; Lebiasinadae) and Boulengerella (2n = 36; Ctenoluciidae) species.
FIGURE 6.
Representative idiograms of Hepsetus odoe and Erythrinidae species: Erythinus erythrinus (ERY) karyomorphs (Kar) A, C, D; Hoplias malabaricus (HMA) karyomorphs A. B, C, D, F; Hoplias lacerdae (HLA); Hoplias aimara (HAI) and Hoplerythrinus unitaeniatus (HPL) karyomorphs A.C, D. The distribution of the 18S and the 5S rDNAs for each species are highlighted in red and green, respectively. The sex chromosomes are boxed. Data from Cioffi et al. (2009), Martins et al. (2013), Martinez et al. (2015), and Oliveira et al. (2015).
FIGURE 7.
Representative idiograms of Hepsetus odoe and Pyrrhulina (Lebiasinidae) (data from Moraes et al., 2017) and Boulengerella (Ctenoluciidae) (data from de Souza E Sousa et al., 2017) species. The distribution of the 18S and the 5S rDNAs for each species are highlighted in red and green, respectively.
Particularly, inside Erythrinidae, E. erythrinus stand out as having more chromosomal similarities with H. odoe than the other ones, taking into account the broad organization of the karyotype and the amount of mono-armed chromosomes that they have. In fact, E. erythrinus karyomorph A shows the most basal karyotype inside this genus, considering that the other Erythrinus karyomorphs highlight clearly derived features, such as the differentiation of a multiple X1X1X2X2/X1X2Y sex chromosome system (Bertollo et al., 2004) and the huge dispersion of the 5S rDNA sequences in the genome (Cioffi et al., 2010; Martins et al., 2013). In addition, like H. odoe, E. erythrinus karyomorph A presents only one chromosome pair bearing 5S rDNA sequences at a similar position on the chromosomes, as well as a number of exclusive telomeric 18S rDNA sites. However, whereas in H. odoe the major rDNA sequences are only distributed in the long arms of the chromosomes, in E. erythrinus they are found both in the short as well as in the long arms (Cioffi et al., 2010). This is not an unexpected condition in view of differential distributions that can be set up along the evolutionary history of the species. In fact, repetitive DNAs have played a particular role on fish karyotyping (Cioffi and Bertollo, 2012), and variations in amount and types of several classes of repetitive DNAs are expected considering the inherent dynamism of these sequences during the evolutionary history of different taxa (Kubat et al., 2008; Cioffi et al., 2010, 2012; Pokorná et al., 2011; Yano et al., 2016). In spite of this, the distribution pattern of the (GAG)10 microsatellites in H. odoe also shows a significant accumulation on the E. erythrinus chromosomes (Yano et al., 2014).
Considering the above correlations between Hepsetus and Erythrinidae, comparative genomic hybridization (CGH) and whole chromosome painting (WCP) were also performed to obtain additional informative markers for comparative cytogenetics. Among fishes, CGH has been already applied for several purposes, such as to compare genomes of closely related species (Zhu and Gui, 2007; Knytl et al., 2013; Majtánová et al., 2016; Moraes et al., 2017), to detect parental genomes in hybrids (Symonová et al., 2013; Pereira et al., 2014), and to elucidate the origin and evolution of B and sex chromosomes (Fantinatti et al., 2011; Freitas et al., 2017; Yano et al., 2017c), among others. In our present case, CGH with four Erythrinidae species evidenced the co-localization of scattered signals in almost all chromosomes of H. odoe, together with the preferential signals in the terminal parts of some chromosomes, thus indicating the shared repetitive content of such regions. However, it stands out the hybridization pattern with E. erythrinus, where some whole chromosome pairs were painted, in addition to telomeric overlaps in other ones. Furthermore, the hybridization with H. odoe gDNA revealed the occurrence of conspicuous species-specific regions, very likely as a result of its particular evolutionary history, given that the resolution of the CGH method predominantly relies on the presence of species-specific (or sex-specific) repetitive DNA sequences and the evolutionary distance of the compared genomes.
Besides CGH, WCP experiments were also performed using microdissected sex chromosomes from H. malabaricus karyomorphs B (HMB-X) and G (HMG-Y1) and E. erythrinus karyomorph D (ERY-Y) as probes, in order to verify the occurrence of putative sex chromosomes in H. odoe. As a control experiment, all probes were previously hybridized in male chromosomal preparations of H. malabaricus (karyomorphs B and G) and E. erythrinus (karyomorph D), clearly demonstrating the hybridization signals on the sex chromosomes of these karyomorphs, thus corroborating previous data (Cioffi et al., 2013; Oliveira et al., 2017). When these probes were hybridized to chromosomal preparations of H. odoe, HMB-X highly painted one small st/a chromosome, while HMG-Y1 and ERY-Y probes painted the p arms of medium-sized st/a chromosomes. This way, these results highlight that such linkage groups are shared by H. odoe and Erythrinidae species, corroborating the CGH experiments which also demonstrated the sharing of a considerable genomic fraction among such groups. The maintenance of such linkage groups is somehow surprising considering the phylogenetic distance between these clades. However, chromosome homology across widely phylogenetically distributed clades have been also detected in several mammals (Balmus et al., 2007; Dementyeva et al., 2010; Kulemzina et al., 2011), birds (Oliveira et al., 2008, 2010; Tagliarini et al., 2011) and lizard (Pokorná et al., 2011) species. In the later, Zoo-FISH experiments using a Z-derived probe from Gallus gallus showed that the fraction of the reptile genome that is homologous to the avian Z chromosome exhibits a conserved synteny, despite the very ancient times (∼275 Mya) of their divergence (Pokorná et al., 2011).
Conclusion
This study, focusing on standard and molecular cytogenetic approaches of H. odoe, represents the first data set for an Hepsetidae species. Our data supports the likely proximity between African and Neotropical families, such as Hepsetidae and Erythrinidae. In fact, our experiments, including CGH and WCP, indicate that H. odoe and some Erythrinidae species, in special from the genus Erythrinus, share similar chromosomal sequences, thus reflecting some degree of relationship among them. In fact, Erythrinus seems to carry the most basal karyotype organization within Erythrinidae, and likely the most proximal to that highlighted by H. odoe. This study represents the first one of a series of further investigations focusing on the African Characiformes chromosomal and genomic characteristics, allowing a broader and more detailed view on the evolutionary history of this group through a cytogenetic approach. Such additional data will securely improve our knowledge about the relatedness of the African and the Neotropical characiform families.
Author Contributions
PC carried out the cytogenetic analysis and drafted the manuscript. EdO, CY, AA-R, and TH helped in the cytogenetic analysis, drafted and revised the manuscript. CO, ED, OJ, and TL drafted and revised the manuscript. MC and LB coordinated the study, drafted and revised the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (Proc. nos. 304992/2015-1 and 401575/2016-0) and Fundação de Amparo à Pesquisa do Estado de São Paulo – FAPESP (Proc. nos. 2016/21411-7, 2016/17556-0 and 2017/08471-3) (FAPESP 2014/26508-3 and CNPq 306054/2006-0 to CO).
References
- Arai R. (2011). Fish Karyotypes: A Check List. Tokyo: Springer; 10.1007/978-4-431-53877-6 [DOI] [Google Scholar]
- Arcila D., Ortí G., Vari R., Armbruster J. W., Stiassny M. L., Ko K. D., et al. (2017). Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat. Ecol. Evolut. 1:0020. 10.1038/s41559-016-0020 [DOI] [PubMed] [Google Scholar]
- Arefjev V. A. (1990). Problems of karyotypic variability in the family Characidae (Pisces, Characiformes) with the description of somatic karyotypes for six species of tetras. Caryologia 43 305–319. 10.1080/00087114.1990.10797009 [DOI] [Google Scholar]
- Arroyave J., Denton J. S., Stiassny M. L. (2013). Are characiform fishes Gondwanan in origin? Insights from a time-scaled molecular phylogeny of the Citharinoidei (Ostariophysi: Characiformes). PLOS ONE 8:e77269. 10.1371/journal.pone.0077269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmus G., Trifonov V. A., Biltueva L. S., O’Brien P. C. M., Alkalaeva E. S., Fu B., et al. (2007). Cross species painting among camel, cattle, pig and human: further insights into the putative Cetartiodactyla ancestral karyotype. Chromosome Res. 15 499–514. 10.1007/s10577-007-1154-x [DOI] [PubMed] [Google Scholar]
- Bergero R., Charlesworth D. (2009). The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evolut. 24 94–102. 10.1016/j.tree.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Bertollo L. A. C. (2007). “Chromosome evolution in the Neotropical Erythrinidae fish family: an overview,” in Fish Cytogenetics, eds Pisano E., Ozouf-Costaz C., Foresti F., Kapoor B. G. (Enfield, NH: Science Publishers; ), 195–211. [Google Scholar]
- Bertollo L. A. C., Cioffi M. B., Moreira-Filho O. (2015). “Direct chromosome preparation from freshwater teleost fishes,” in Fish Cytogenetic Techniques, eds Ozouf-Costaz C., Pisano E., Foresti F., Almeida Toledo L. F. (Enfield, CT: CRC Press; ), 21–26. [Google Scholar]
- Bertollo L. A. C., Oliveira C., Molina W. F., Margarido V. P., Fontes M. S., Pastori M. C., et al. (2004). Chromosome evolution in the erythrinid fish, Erythrinus erythrinus (Teleostei: Characiformes). Heredity 93 228–233. 10.1038/sj.hdy.6800511 [DOI] [PubMed] [Google Scholar]
- Buckup P. A. (1998). “Relationships of the Characidiinae and phylogeny of characiform fishes (Teleostei: Characiformes),” in Phylogeny and Classification of Neotropical Fishes, eds Malabarba L. R., Reis R. E., Vari R. P., Lucena Z. M. S., Lucena C. A. S. (Porto Alegre: EDIPUCRS; ), 193–234. [Google Scholar]
- Calcagnotto D., Schaefer S. A., DeSalle R. (2005). Relationships among characiform fishes inferred from analysis of nuclear and mitochondrial gene sequences. Mol. Phylogenet. Evol. 36 135–153. 10.1016/j.ympev.2005.01.004 [DOI] [PubMed] [Google Scholar]
- Cioffi M. B., Bertollo L. A. C. (2012). “Chromosomal distribution and evolution of repetitive DNAs in fish,” in Repetitive DNA, ed. Garrido-Ramos M. A. (Basel: Karger; ), 197–221. 10.1159/000337950 [DOI] [PubMed] [Google Scholar]
- Cioffi M. B., Liehr T., Trifonov V., Molina W. F., Bertollo L. A. C. (2013). Independent sex chromosome evolution in lower vertebrates: a molecular cytogenetic overview in the Erythrinidae fish family. Cytogenet. Genome Res. 141 186–194. 10.1159/000354039 [DOI] [PubMed] [Google Scholar]
- Cioffi M. B., Martins C., Bertollo L. A. (2010). Chromosome spreading of associated transposable elements and ribosomal DNA in the fish Erythrinus erythrinus. Implications for genome change and karyoevolution in fish. BMC Evolut. Biol. 10:271. 10.1186/1471-2148-10-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi M. B., Martins C., Bertollo L. A. C. (2009). Comparative chromosome mapping of repetitive sequences. Implications for genomic evolution in the fish, Hoplias malabaricus. BMC Genetics 10:34. 10.1186/1471-2156-10-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi M. B., Molina W. F., Artoni R. F., Bertollo L. A. C. (2012). “Chromosomes as tools for discovering biodiversity – the case of Erythrinidae fish family,” in Recent Trends in Cytogenetic Studies – Methodologies and Applications, ed. Tirunilai P. (Rijeka: InTech Publisher; ), 125–146. [Google Scholar]
- Cross I., Díaz E., Sánchez I., Rebordinos L. (2005). Molecular and cytogenetic characterization of Crassostrea angulata chromosomes. Aquaculture 247 135–144. 10.1016/j.aquaculture.2005.02.039 [DOI] [Google Scholar]
- de Souza E Sousa J. S., Viana P. F., Bertollo L. A. C., Cioffi M. B., Feldberg E. (2017). Evolutionary relationships among Boulengerella species (Ctenoluciidae, Characiformes): genomic organization of repetitive DNAs and highly conserved karyotypes. Cytogenet. Genome Res. 152 194–203. 10.1159/000480141 [DOI] [PubMed] [Google Scholar]
- Decru E., Snoeks J., Vreven E. (2013a). The true identity of the holotype of Hepsetus odoe and the names of the two west african species of Hepsetus (Teleostei: Hepsetidae). Ichthyol. Explor. Freshw. 24 187–192. [Google Scholar]
- Decru E., Snoeks J., Vreven E. (2015). Taxonomic evaluation of the Hepsetus from the Congo basin with the revalidation of H. microlepis (Teleostei: Hepsetidae). Ichthyol. Explor. Freshw. 26 273–287. [Google Scholar]
- Decru E., Vreven E., Snoeks J. (2012). A revision of the West African Hepsetus (Characiformes: Hepsetidae) with a description of Hepsetus akawo sp. nov. and a redescription of Hepsetus odoe (Bloch, 1794). J. Nat. Hist. 46 1–23. 10.1080/00222933.2011.622055 [DOI] [Google Scholar]
- Decru E., Vreven E., Snoeks J. (2013b). A revision of the Lower Guinean Hepsetus species (Characiformes; Hepsetidae) with the description of Hepsetus kingsleyae sp. nov. J. Fish Biol. 82 1351–1375. 10.1111/jfb.12079 [DOI] [PubMed] [Google Scholar]
- Dementyeva P. V., Trifonov V. A., Kulemzina A. I., Graphodatsky A. S. (2010). Reconstruction of the putative Cervidae ancestral karyotype by chromosome painting of Siberian roe deer (Capreolus pygargus) with dromedary probes. Cytogenet. Genome Res. 128 228–235. 10.1159/000298878 [DOI] [PubMed] [Google Scholar]
- Eschmeyer W. N., Fong J. D. (2017). Species by Family/Subfamily. Available at: http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp [accessed March 30, 2017]. [Google Scholar]
- Fantinatti B. E., Mazzuchelli J., Valente G. T., Cabral-de-Mello D. C., Martins C. (2011). Genomic content and new insights on the origin of the B chromosome of the cichlid fish Astatotilapia latifasciata. Genetica 139 1273–1282. 10.1007/s10709-012-9629-x [DOI] [PubMed] [Google Scholar]
- Freitas N. L., Al-Rikabi A. B. H., Bertollo L. A. C., Ezaz T., Yano C. F., Oliveira E. A., et al. (2017). Early stages of XY sex chromosomes differentiation in the fish Hoplias malabaricus (Characiformes, Erythrinidae) revealed by DNA repeats accumulation. Curr. Genomics 18 10.2174/1389202918666170711160528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornung E. (2013). Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: a review of research. Cytogenet. Genome Res. 141 90–102. 10.1159/000354832 [DOI] [PubMed] [Google Scholar]
- Kareem O. K., Olanrewaju A. N., Osho E. F., Orisasona O., Akintunde M. A. (2016). Growth patterns and condition factor of Hepsetus odoe (Bloch, 1794) captured in eleyele lake, Southwest Nigeria. Fish. Aquacult. J. 7:178 10.4172/2150-3508.1000178 [DOI] [Google Scholar]
- Knytl M., Kalous L., Symonová R., Rylková K., Ráb P. (2013). Chromosome studies of european cyprinid fishes: cross-species painting reveals natural allotetraploid origin of a Carassius female with 206 chromosomes. Cytogenet. Genome Res. 139 276–283. 10.1159/000350689 [DOI] [PubMed] [Google Scholar]
- Kubat Z., Hobza R., Vyskot B., Kejnovsky E. (2008). Microsatellite accumulation on the Y chromosome in Silene latifolia. Genome 51 350–356. 10.1139/G08-024 [DOI] [PubMed] [Google Scholar]
- Kulemzina A. I., Yang F., Trifonov V. A., Ryder O. A., Ferguson-Smith M. A., Graphodatsky A. S. (2011). Chromosome painting in Tragulidae facilitates the reconstruction of Ruminantia ancestral karyotype. Chromosome Res. 19 531–539. 10.1007/s10577-011-9201-z [DOI] [PubMed] [Google Scholar]
- Levan A., Fredga K., Sandberg A. A. (1964). Nomenclature for centromeric position on chromosomes. Hereditas 52 201–220. 10.1111/j.1601-5223.1964.tb01953.x [DOI] [Google Scholar]
- Majtánová Z., Choleva L., Symonová R., Ráb P., Kotusz J., Pekárik L., et al. (2016). Asexual reproduction does not apparently increase the rate of chromosomal evolution: karyotype stability in diploid and triploid clonal hybrid fish (Cobitis, Cypriniformes, Teleostei). PLOS ONE 11:e0146872. 10.1371/journal.pone.0146872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins C., Galetti P. M. (1999). Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Res. 7 363–367. 10.1023/A:1009216030316 [DOI] [PubMed] [Google Scholar]
- Martins N. F., Bertollo L. A. C., Troy W. P., Feldberg E., de Souza Valentin F. C., Cioffi M. B. (2013). Differentiation and evolutionary relationships in Erythrinus erythrinus (Characiformes, Erythrinidae): comparative chromosome mapping of repetitive sequences. Rev. Fish Biol. Fish. 23 261–269. 10.1007/s11160-012-9292-4 [DOI] [Google Scholar]
- Martinez J. D. F., Lui R. L., Traldi J. B., Blanco D. R., Moreira-Filho O. (2015). Occurrence of natural hybrids among sympatric karyomorphs in Hoplerythrinus unitaeniatus (Characiformes, Erythrinidae). Zebrafish 12 281–287. 10.1089/zeb.2015.1083 [DOI] [PubMed] [Google Scholar]
- Moraes R. L. R., Bertollo L. A. C., Marinho M. M. F., Yano C. F., Hatanaka T., Barby F. F., et al. (2017). Evolutionary relationships and cytotaxonomy considerations in the genus Pyrrhulina (Characiformes, Lebiasinidae). Zebrafish 10.1089/zeb.2017.1465 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Nelson J. S., Grande T. C., Wilson M. V. (2016). Fishes of the World. Somerset, NJ: John Wiley & Sons Inc. 10.1002/9781119174844 [DOI] [Google Scholar]
- Oliveira C., Almeida-Toledo L. F., Foresti F. (2007). “Karyotypic evolution in Neotropical fishes,” in Fish Cytogenetics, eds Pisano E., Ozouf-Costaz C., Foresti F., Kapoor B. G. (Enfield, CT: Science Publishers; ), 111–164. [Google Scholar]
- Oliveira C., Andreata A. A., Toledo L. F. A., Toledo S. A. (1991). Karyotype and nucleolus organizer regions of Pyrrhulina cf australis (Pisces, Characiformes, Lebiasinidae). Brazilian Journal of Genetics 14 685–690. [Google Scholar]
- Oliveira C., Avelino G. S., Abe K. T., Mariguela T. C., Benine R. C., Ortí G., et al. (2011). Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol. Biol. 11:275. 10.1186/1471-2148-11-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira E. A., Bertollo L. A. C., Yano C. F., Cioffi M. B. (2015). Comparative cytogenetics in the genus Hoplias (Characiformes, Erythrinidae) highlights contrasting karyotype evolution among congeneric species. Mol. Cytogenet. 8:56. 10.1186/s13039-015-0161-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira E. A., Sember A., Bertollo L. A. C., Yano C. F., Ezaz T., Moreira-Filho O., et al. (2017). Tracking the evolutionary pathway of sex chromosomes among fishes: characterizing the unique XX/XY1Y2 system in Hoplias malabaricus (Teleostei, Characiformes). Chromosoma 1–14. 10.1007/s00412-017-0648-3 [DOI] [PubMed] [Google Scholar]
- Oliveira E. H. C., De Moura S. P., Dos Anjos L. J. S., Nagamachi C. Y., Pieczarka J. C., Ferguson-Smith M. A. (2008). Comparative chromosome painting between chicken and spectacled owl (Pulsatrix perspicillata): implications for chromosomal evolution in the Strigidae (Aves, Strigiformes). Cytogenet. Genome Res. 122 157–162. 10.1159/000163093 [DOI] [PubMed] [Google Scholar]
- Oliveira E. H. C., Tagliarini M. M., Rissino J. D., Pieczarka J. C., Nagamachi C. Y., O’Brien P. C., et al. (2010). Reciprocal chromosome painting between white hawk (Leucopternis albicollis) and chicken reveals extensive fusions and fissions during karyotype evolution of Accipitridae (Aves, Falconiformes). Chromosome Res. 18 349–355. 10.1007/s10577-010-9117-z [DOI] [PubMed] [Google Scholar]
- Ortí G., Meyer A. (1997). The radiation of characiform fishes and the limits of resolution of mitochondrial ribosomal DNA sequences. Syst. Biol. 46 75–100. 10.1093/sysbio/46.1.75 [DOI] [PubMed] [Google Scholar]
- Oyakawa O. T. (2003). “Family erythrinidae,” in Check List of the Freshwater Fishes of South and Central America, eds Reis R. E., Kullander S. O., Ferraris C. J., Jr. (Porto Alegre: EDIPUCRS; ), 238–240. [Google Scholar]
- Pastori M. C., Roncati H. C., Aichino D. R., Ledesma M. A., Fenocchio A. S. (2009) Chromosome characterization and cytotaxonomic considerations on Characidae, Acestrorhynchidae and Cynodontidae (Pisces, Characiformes) from the Paraná River (Argentina). Caryologia 62 69–74. 10.1080/00087114.2004.10589668 [DOI] [Google Scholar]
- Pendás A. M., Moran P., Freije J. P., Garcia-Vazquez E. (1994). Chromosomal mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenet. Genome Res. 67 31–36. 10.1159/000133792 [DOI] [PubMed] [Google Scholar]
- Pereira C. S. A., Aboim M. A., Ráb P., Collares-Pereira M. J. (2014). Introgressive hybridization as a promoter of genome reshuffling in natural homoploid fish hybrids (Cyprinidae, Leuciscinae). Heredity 112 343–350. 10.1038/hdy.2013.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorná M., Kratochvíl L., Kejnovský E. (2011). Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox). BMC Genet. 12:90. 10.1186/1471-2156-12-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto J. I. R., Feldberg E., das Neves Falcao J., Nakayama C. M. (1993). Cytogenetic studies in hemiodidae (Ostariophysi, Characiformes) fishes from the central Amazon. Cytologia 58 397–402. 10.1508/cytologia.58.397 [DOI] [Google Scholar]
- Porto J. I. R., Feldberg E., Nakayama C. M., Falcão J. (1992). A checklist of chromosome numbers and karyotypes of Amazonian freshwater fishes. Rev. D’hydrobiol. Trop. 25 287–299. [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scheel J. J. (1973). Fish Chromosomes and Their Evolution. Internal Report of Danmarks Akvarium 22 NAID: 10006435147 Charlottenlund. [Google Scholar]
- Silva D. M., Utsunomia R., Pansonato-Alves J. C., Oliveira C., Foresti F. (2015). Chromosomal mapping of repetitive DNA sequences in five species of Astyanax (Characiformes, Characidae) reveals independent location of U1 and U2 snRNA sites and association of U1 snRNA and 5S rDNA. Cytogenet. Genome Res. 146 144–152. 10.1159/000438813 [DOI] [PubMed] [Google Scholar]
- Sumner A. T. (1972). A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75 304–306. 10.1016/0014-4827(72)90558-7 [DOI] [PubMed] [Google Scholar]
- Symonová R., Majtánová Z., Sember A., Staaks G. B. O., Bohlen J., Freyhof J., et al. (2013). Genome differentiation in a species pair of coregonine fishes: an extremely rapid speciation driven by stress-activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol. Biol. 13:42. 10.1186/1471-2148-13-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliarini M. M., O’Brien P., Ferguson-Smith M. A., de Oliveira E. H. (2011). Maintenance of syntenic groups between Cathartidae and Gallus gallus indicates symplesiomorphic karyotypes in new world vultures. Genet. Mol. Biol. 34 80–83. 10.1590/S1415-47572010005000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vari R. P., Malabarba L. R. (1998). “Neotropical ichthyology: an overview,” in Phylogeny and Classification of Neotropical Fishes, eds Malabarba L. R., Reis R. E., Vari R. P., Lucena Z. M. S., Lucena C. A. S. (Porto Alegre: EDIPUCRS; ), 1–12. [Google Scholar]
- Yang F., Trifonov V., Ng B. L., Kosyakova N., Carter N. P. (2009). “Generation of paint probes by flow-sorted and microdissected chromosomes,” in Fluorescence In Situ Hybridization (FISH)–Application Guide, ed. Liehr T. (Berlin: Springer; ), 35–52. 10.1007/978-3-540-70581-9_3 [DOI] [Google Scholar]
- Yano C. F., Bertollo L. A. C., Cioffi M. B. (2017a). “Fish-FISH: molecular cytogenetics in fish species,” in Fluorescence in situ Hybridization (FISH) - Application Guide, ed. Liehr T. (Berlin: Springer; ), 429–444. [Google Scholar]
- Yano C. F., Bertollo L. A. C., Ezaz T., Trifonov V., Sember A., Liehr T., et al. (2017c). Highly conserved Z and molecularly diverged W chromosomes in the fish genus Triportheus (Characiformes, Triportheidae). Heredity 118 276–283. 10.1038/hdy.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano C. F., Bertollo L. A. C., Liehr T., Troy W. P., Cioffi M. B. (2016). W chromosome dynamics in Triportheus species (Characiformes, Triportheidae). An ongoing process narrated by repetitive sequences. J. Heredity 107 342–348. 10.1093/jhered/esw021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano C. F., Bertollo L. A. C., Molina W. F., Liehr T., Cioffi M. B. (2014). Genomic organization of repetitive DNAs and its implications for male karyotype and the neo-Y chromosome differentiation in Erythrinus erythrinus (Characiformes, Erythrinidae). Comp. Cytogenet. 8 139–151. 10.3897/CompCytogen.v8i2.7597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano C. F., Bertollo L. A. C., Rebordinos L., Merlo M. A., Liehr T., Portela-Bens S., et al. (2017b). Evolutionary dynamics of rDNAs and U2 small nuclear DNAs in Triportheus (Characiformes, Triportheidae): high variability and particular syntenic organization. Zebrafish 14 146–154. 10.1089/zeb.2016.1351 [DOI] [PubMed] [Google Scholar]
- Zhu H. P., Gui J. F. (2007). Identification of genome organization in the unusual allotetraploid form of Carassius auratus gibelio. Aquaculture 265 109–117. 10.1016/j.aquaculture.2006.10.026 [DOI] [Google Scholar]
- Zwick M. S., Hanson R. E., McKnight T. D., Islam-Faridi M. N., Stelly D. M., Wing R. A., et al. (1997). A rapid procedure for the isolation of C0t-1 DNA from plants. Genome 40 138–142. 10.1139/g97-020 [DOI] [PubMed] [Google Scholar]