Abstract
In Chinese cabbage, heading type is a key agricultural trait of significant economic importance. Using a natural microspore-derived doubled haploid plant, we generated self-crossed progeny with overlapping or outward curling head morphotypes. Sequencing-based bulked segregant analysis (Seq-BSA) revealed a candidate region of 0.52 Mb (A06: 1,824,886~2,347,097 bp) containing genes enriched for plant hormone signal transduction. RNA Sequencing (RNA-Seq) analysis supported the hormone pathway enrichment leading to the identification of two key candidate genes, BrGH3.12 and BrABF1. The regulated homologous genes and the relationship between genes in this pathway were also revealed. Expression of BrGH3.12 varied significantly in the apical portion of the leaf, consistent with the morphological differences between overlapping and outward curling leaves. Transcript levels of BrABF1 in the top, middle and basal segments of the leaf were significantly different between the two types. The two morphotypes contained different concentrations of IAA in the apical portion of their leaves while levels of ABA differed significantly between plant types in the top, middle, and basal leaf segments. Results from Seq-BSA, RNA-Seq and metabolite analyses all support a role for IAA and ABA in heading type formation. These findings increase our understanding of the molecular basis for pattern formation of the leafy head in Chinese cabbage and will contribute to future work developing more desirable leafy head patterns.
Keywords: Chinese cabbage, heading type, plant hormone, BrGH3.12, BrABF1, Seq-BSA, RNA-Seq
Introduction
Chinese cabbage (Brassica rapa ssp. pekinensis) is widely cultivated in Asia and is becoming increasingly more popular in other countries. As a leafy vegetable crop, leaves at the seedling and rosette stage function primarily in photosynthesis and respiration. At the heading stage, a tight, leafy head is formed and serves as a storage organ and an edible product. Chinese cabbage heading type is an important commercial trait indicating how the top of leaves form the head. The patterns include overlapping, outward-curling, inward-curling without overlap, and spiral (Xu et al., 2004). The most commonly cultivated is the overlapping type, where the heading leaves curl inward at the top with the curling length exceeding the vertical central axis of the leafy head. This is the preferred type by both growers and consumers because it presents a closed top, cleaner inner leaves, a higher net-to-head ratio, and facilitates easier packaging and transportation. In contrast, outward-curling types produce leaves that curl outward and appear open at the top. The molecular regulatory mechanism responsible for heading type in Chinese cabbage remains elusive and has not to our knowledge been previously described.
The molecular mechanism of leafy head formation is also unclear (Wang et al., 2014), although some progress has been made. Changes in concentration of the plant hormone auxin (indole-3-acetic acid, or IAA) between the adaxial and abaxial sides were shown to cause leaves to curl inward to form a leafy head (Li, 1984). He et al. (2000) introduced IAA-related genes into Chinese cabbage and found that the transgenic plants showed an earlier heading date, produced more leaves, and developed heavier heads. Other studies have focused on head weight, diameter, and height, as well as heading time (Yu et al., 2013; Inoue et al., 2015); or the genes responsible for leaf curvature (Xiao et al., 2014). Based on results from adaxial-abaxial (ad-ab) polarity research in Arabidopsis thaliana (Fukushima and Hasebe, 2014), Brassica rapa ssp. pekinensis TEOSINTE BRANCHED1, cycloidea, and PCF transcription factor 4 (BrpTCP4) was shown to affect the leafy head size and shape of Chinese cabbage (Mao et al., 2014) while BrpSPL9 (B. rapa ssp. pekinensis SQUAMOSA PROMOTER BINDING-LIKE 9-2) was found to influence the timing of leafy head formation (Wang et al., 2014). The roles of BrARF3.1 (AUXIN RESPONSE FACTOR 3) and BrKAN2.1 (KANADA2) in determining the formation of leafy heads has also been examined through comparative genomic analysis (Cheng et al., 2016; Liang et al., 2016), while Genome-Wide Association Studies (GWAS) have been used to investigate the effect of WRKY and F-box genes on head diameter (Lim et al., 2015). Finally, through RNA-seq techniques, genes that are differentially expressed between the rosette and heading stages have been classified into four groups: transcription factors, protein kinases, calcium signaling, and auxin-related genes (Wang et al., 2012).
In this study, we obtained a natural microspore-derived doubled haploid plant, B3-29, from an introgression line containing chromosome segments of cabbage (B. oleracea var. capitata) in a Chinese cabbage background (Qin, 2013). Although the morphological phenotypes of individual field-grown plants were nearly identical during the seedling and rosette stages, two phenotypes of heading type were observed during the heading stage, overlapping and outward-curling (Figure 1; Dong, 2015). This provided a new opportunity for studying the heading type phenomenon. The availability of the Chinese cabbage genome (Wang, X. et al., 2011) facilitates the investigation of the molecular mechanisms responsible for the heading type trait. To identify the pathways and genes related to these two heading type phenotypes we applied a Sequencing-based Bulked Segregant Analysis (Seq-BSA) combined with RNA-Seq. The results were further validated at the metabolite level. Our findings provide the first molecular evidence of heading type formation using whole genome analysis in Chinese cabbage.
Figure 1.

Schematic representation of origins for experimental materials.
Materials and methods
Plant materials
The natural microspore-derived doubled haploid plant B3-29 used in this experiment was obtained by isolating microspore culture from M50-12 (Qin, 2013), which was derived from self-crossed progenies of a Brassica rapa-Brassica oleracea monosomic alien addition line AC4 (Gu et al., 2015). After nine generations of subculturing, seeds of B3-29 were planted in the field in January of 2013. In August of 2014, 125 seeds of selfed B3-29 were sown. The surviving plants displayed two contrasting heading type phenotypes: overlapping (82 plants) and outward-curling (31 plants) (Figure 1). From these, we selected 25 plants of each morphotype and used them to establish R01 and R02 bulks for Seq-BSA analysis. RNA-Seq was performed on three plants from each bulk.
Methods
Trait observations
Leaf growth rates were monitored from the rosette stage to the early heading stage of Chinese cabbage using 10 plants each of the overlapping and outward-curling types. The maximum length and width of leaves was recorded in 2-day intervals to track leaf growth. At the heading stage (65 days after transplanting), we examined the maximum width and length of leaves as they were naturally displayed as well as when individual leaves were spread out and pressed until almost completely flat. Total weight and the number of leaves were determined from three plants for each morphotype. For vasculature observations, the largest head-forming leaves were cleared of chlorophyll in 95% alcohol overnight and treated with distilled, deionized water: glycerol: phenol: lactic acid (1:1:1:1) for 20 min at 90°C.
Seq-BSA analysis
Illumina library construction and sequencing
A total of 50 plants (25 overlapping, 25 outward-curling) were used for sequencing. Genomic DNA was isolated from young leaves with an extraction kit (Aidlab Biotechnologies Co. Ltd., China), and two DNA pools from R01 and R02 bulks were prepared by mixing equimolar concentrations of DNA samples from each morphotype. The Illumina libraries for these two pools were prepared using a NEBNext® DNA Library Prep Reagent Set for Illumina® (E6000L; Illumina Inc., San Diego, CA, USA). A total of 5 μg of DNA from each sample was sheared via Ultrasonic cleaning (KQ-50E; Kunshan Ultrasonic Instrument Co. Ltd.,) followed by end-repair and adapter-ligation. Size selection of libraries was performed using a 2% agarose gel to obtain a target insert size of 500 bp. The purified libraries were amplified using adaptor-compatible PCR primers and the size distribution was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The DNA libraries were sequenced on an Illumina HiSeq 2500 platform with an FC-401-4002_HiSeq SBS Kit V4 (50 cycles) and a PE-401-4001_HiSeq PE Cluster Kit V4 cBot (Illumina Inc.,) to generate 125 bp paired-end reads.
Sequence alignment and variant calling
A custom-built program in C++ was used to filter out low-quality reads and duplicates were discarded using SAMtools (Li et al., 2009). Filtered reads were aligned to the reference Chinese cabbage genome (http://brassicadb.org/brad/datasets/pub/Genomes/Brassica_rapa/V1.5/) using BWA (Li and Durbin, 2009). The GATK Toolkit (McKenna et al., 2010) was used to correct for alignment errors due to local rearrangements around insertions and deletions as well as recalibration of read-base quality and variant calling. The variants were removed if the distance was less than 10 bp between two indels, if more than two SNPs occurred in a 5 bp window, or SNPs were within 5 bp of an indel.
Mapping of candidate genomic regions
Candidate regions were selected based on Euclidean distance (ED) associations (Hill et al., 2013). After the loci for multiple mutations, undetected sites in any single bulk, or homozygous and consistent loci in both bulks were filtered out, the ED was calculated at each SNP location using the following equation:
Where A, C, G, and T represented their corresponding bases. The ED was then raised to a power (default = 5), without masking repetitive regions (default = not masked). These data were subsequently passed to R for signal processing and peak identification. The ED values across the genome were fitted and sliding-window averages of 1 Mb were plotted. Peak regions were defined as candidate regions where the Distance-fitted values were greater than three standard deviations above the genome-wide median. From this, R could be used to plot Distance fits.
RNA-Seq analysis
At the early heading stage (68 days after sowing), we sampled the ninth leaf in from the exterior of the developing head at three distinct positions: apical (S), middle (Z, corresponding to the bend position on the overlapping morphotype), and basal (X) (Figure S2). The morphotypes were referred to as DB for overlapping and SX for outward-curling. Each sample was divided into two sets: one for RNA-Seq, the other for determining IAA and ABA concentrations.
RNA extraction and quality test
Total RNA was isolated with an RNA extraction kit (Huayueyang, China), and quantified with a Qubit® RNA Assay Kit on the Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). The RNA integrity was assessed on the Bioanalyzer 2100 System using the RNA Nano 6000 Assay Kit (Agilent).
RNA-Seq library construction and sequencing
A total of 3 μg RNA per sample was used as input material for the RNA sample preparations. Our RNA-Seq libraries were constructed with a NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA). Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was performed using divalent cations at an elevated temperature in the NEB proprietary fragmentation buffer (5X). The RNA-Seq libraries were sequenced on an Illumina HiSeq 4000 platform to generate 150 bp paired-end reads.
Bioinformatics analysis of RNA-Seq data
Raw reads were pre-processed to remove adapter, poly-N sequences and low quality reads. Processed reads were mapped to the Chinese cabbage reference genome using TopHat (Trapnell et al., 2009, 2012) v. 2.0.12. Read counts for each gene were summarized by HTSeq (Anders, 2010) v. 0.6.1. and FPKM values were calculated based gene length and read counts mapping to the gene. Differential expression analysis of the two groups (three biological replicates per group) was performed using the DESeq package (Anders and Huber, 2010, 2012; Wang et al., 2010) in R (1.18.0). This software provides statistical routines for determining differential expression using a negative binomial distribution model. The resulting P-values were adjusted using the Benjamini and Hochberg method to control for false discovery rates. Genes with an adjusted P < 0.05 were considered differentially expressed.
Real-time quantitative RT-PCR
The qRT-PCR verifications were conducted as previously described (Gu et al., 2016), using a Thunderbird SYBR qPCR Mix (Toyobo, Shanghai, China) and a LightCycler® 96 (Roche). All primers are listed in Table S1.
Quantification of IAA and ABA
Concentrations of IAA and ABA were determined using a liquid chromatography–tandem mass spectrometry (LC–MS/MS) system [MS API 4000 (Applied Biosystems, USA) and LC 1200 (Agilent)]. Leaf samples were frozen in liquid nitrogen and 200-mg portions were ground, placed in ultrasonic pots and extracted in 3 mL of solution (n-propyl alcohol/water/hydrochloric acid; v/v/v = 200.0/100.0/0.2) for 30 min in an ice bath in the dark. Following incubation, 2 mL of dichloromethane was added to the extraction and incubated for 30 min. The substratum liquid was collected following centrifugation at 4,500 rpm for 10 min, dried under nitrogen and re-dissolved in 1 mL of 80% (v/v) methanol. The supernatants were collected after centrifugation at 12,000 rpm for 10 min and passed through a 0.22-μm filter membrane. After 200 μL of this supernatant was loaded onto the LC-MS/MS system, the procedure continued on a 2.1 mm × 50 mm XBridgeTM C18 2.5 μm column (Waters, USA), at a column temperature of 30°C. The mobile phase comprised solvent A (0.05% HCOOH, v/v) (Waters) and solvent B (0.05% HCOOH-CH3CN, v/v) in a gradient mode (time/concentration of A/concentration of B) of 0.01/0/10, 2.0/90/10, 12.0/50/50, 12.1/90/10, and 15/90/10, at a flow rate of 0.2 mL min−1. The MS settings were as follows: capillary voltage, −4.5 kV; source temperature, 350°C; scan type, multiple reaction monitoring (MRM); dwell time, 200 ms; settling time, 700 ms; and MR pause, 5.007 ms. For IAA, the MRM was set at: Q1/Q3 174/130, CE −13, DP −43, CXP −21, and EP −10; whereas for ABA, the MRM was set at: Q1/Q3 137/93, CE −22, DP −37, CXP −16, and EP −10.
Statistical analysis
Significant differences in phenotypic traits and hormone levels between overlapping and outward-curling plants was assessed by Fisher's exact test using a P < 0.05.
Genes in the candidate region from the Seq-BSA analysis were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.ad.jp/kegg/; Kanehisa et al., 2004) to classify genes based on biological function. Pathway enrichment for genes in the candidate region was determined using the entire genome as the background set. Statistical significance was assessed by Fisher's exact test using a P<0.05.
We used KOBAS software (Mao et al., 2005) to test the statistical enrichment of differentially expressed genes in KEGG pathways.
Results
Characteristics of overlapping and outward-curling morphotypes
From seedling to the early heading stage, we did not observe noticeable differences in phenotype (Figures 2A,B,D,E) or leaf growth over time (Figure S1). However, their respective heading type showed visible distinctions during head formation between overlapping (Figures 2C,I,J) vs. outward-curling types (Figures 2F,K,L). When measurements were taken of heads in their natural state, leaves were longer in the outward-curling group, and differences were not significant (P = 0.28). However, when completely spread out, the leaves were significantly longer in the overlapping group (P = 0.02). In both measurements, the leaf width was similar between types (P = 0.52, natural state; P = 0.78, completely spread out). On average, the outward-curling plants were slightly heavier (P = 0.55) and produced significantly more leaves than the overlapping type (P = 0.01). The overlapping plants showed more branching of the leaf veins, with at least three secondary veins produced for each primary vein resulting in a greater number of small secondary veins (Figure 2G). By comparison, the primary veins on the outward-curling leaves were spread more widely toward the leaf edge with barely visible secondary veins (Figure 2H).
Figure 2.
Two types of head leaf patterns in top region during the formation of heads (red arrows) for self-crossed progenies from line B3-29. Overlapping type: (A) seedling stage; (B) rosette stage; (C) heading stage. Outward-curling type: (D) seedling stage; (E) rosette stage; (F) heading stage. Leaf veins: (G) overlapping type; (H) outward-curling type. Appearance after head-forming leaves were removed in individual layers (left to right, outer to innermost layer): (I,J), overlapping type; (K,L), outward-curling type.
Seq-BSA analysis
Construction and sequencing of R01 and R02 bulks
Two Illumina libraries (R01 and R02) were constructed and subjected to whole-genome re-sequencing using Illumina HiSeq2500. In total, 123.18 million paired-end (PE) reads (60.88 million and 62.31 million reads for R01 and R02, respectively) were generated. Mapping of those reads to the reference genome resulted in 23X and 28X coverage with 87.40 and 86.31% mapping efficiency for R01 and R02 bulks, respectively (Table 1).
Table 1.
Sequencing and mapping of sequence reads.
| Bulk | Clean reads | Data generated (Gb) | Q30 (%) | Genome coverage (%) | Average depth (X) | SNP number |
|---|---|---|---|---|---|---|
| R01 | 121763612 | 15.34 | 85.73 | 87.40 | 23 | 1328929 |
| R02 | 124615198 | 15.70 | 86.86 | 86.31 | 28 | 1336057 |
Selection of candidate regions
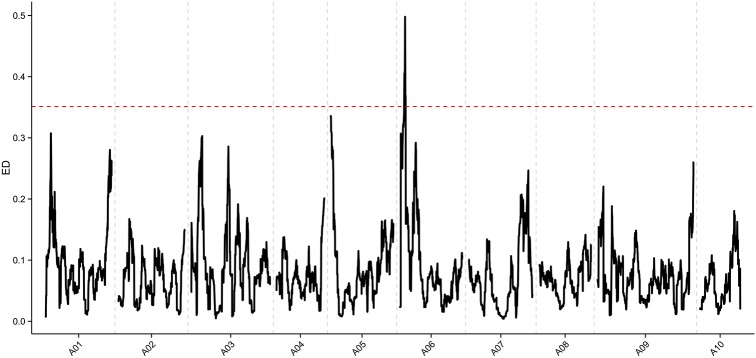
We used 72,962 SNPs between R01 and R02 for association analysis. Values for Euclidean distance (ED) were fit across the genome and sliding-window averages of 1 Mb were plotted. The ED threshold was 0.35 when three standard deviations above the genome-wide median were chosen. The candidate regions responsible for heading type in overlapping and outward-curling types were selected from peaks with an ED above the threshold. From this, one candidate region at chromosome A06 was detected, with a total length of 0.52 Mb (1,824,886 ~ 2,347,097 bp) (Figure 3) containing 90 genes (Table S2).
Figure 3.
Values calculated for ED on chromosomes, based on Seq-BSA analysis.
Analysis of genes in the candidate region
To identify the biological pathways that are active in the formation of heading type, we mapped 90 candidate genes to KEGG database and compared their enrichment against the whole genome. The proportion of genes that were enriched for plant hormone signal transduction (ko04075) was significant (P = 0.02) and contained the greatest representation by unique genes. Of these, four were enriched for plant hormone signal transduction including eight parallel pathways (auxin, cytokinin, gibberellins, ABA, ethylene, brassinosteroid, jasmonic acid, and salicylic acid), with three (Bra018749, Bra018750, and Bra018751) assigned to the auxin pathway and one (Bra018800) to the ABA pathway. One of the genes in the auxin pathway was BrGH3.12, an ortholog of the early-auxin-response gene GRETCHEN HAGEN3 (GH3) (Hagen and Guilfoyle, 2002; Staswick et al., 2005; Bitto et al., 2009; Westfall et al., 2010, 2012; Peat et al., 2012). In the ABA pathway, we identified BrABF1 (Bra018800), an ortholog of ABSCISIC ACID RESPONSIVE ELEMENT BINDING FACTOR1 (ABF1) (Yoshida et al., 2010, 2015).
Swissport annotation and results from our literature search (Table 2) revealed six additional auxin-related genes involved in auxin biosynthesis, transport, and binding proteins, as well as an additional gene involved in the ABA response. Gene Ontology enrichment also identified the four genes mentioned above for the hormone signal transduction pathway, two (Bra018787 and Bra018772) for the auxin response, and two (Bra018787 and Bra018745) for the ABA response. The consensus results between the two database enrichment tests suggests that genes related to auxin and ABA pathways are important candidates for heading type formation in Chinese cabbage.
Table 2.
Functions of auxin- and ABA-related genes, based on Swissport annotation.
| Hormone | Function | Gene name | Gene ID |
|---|---|---|---|
| Auxin | Biosynthesis | YUCCA (Mashiguchi et al., 2011; Won et al., 2011; Zhao, 2012) | Bra018763, Bra018766, Bra018761 |
| Receptor system | Arabidopsis SKP1-like 1 (ASK1) Grones and Friml, 2015 | Bra018756, Bra018757 | |
| Transporter | WALLS ARE THIN1 (WAT1) (Ranocha et al., 2013) | Bra018748 | |
| ABA | Response | RESPONSIVE TO DEHYDRATION 22 (RD22) (Zhao, Y. et al., 2013; Harshavardhan et al., 2014) | Bra018785 |
RNA-Seq analysis
Comparison of gene expression between morphotypes
The RNA-Seq analysis generated 42.27–55.95 million reads for each sample. Three biological replicates were performed for each line. After removal of adaptor sequences, duplicated sequences, ambiguous reads, and low-quality reads, 30.60–41.11 million reads were mapped to the Brassica rapa genome (Table 3). Expression was normalized and differentially expressed genes (DEGs) were detected between the morphotypes. The following codes were used to describe the formation patterns and locations along the leaf where samples were taken: DB (overlapping) vs. SX (outward-curling); S, Z, and X to represent the top, middle, and basal portion of the leaf, respectively. From this analysis, we found 6034 DEGs between DB_S and SX_S, with 2453 genes significantly up-regulated and 3581 significantly down-regulated (Figure 4A). Among the 2227 DEGs between DB_Z and SX_Z, 756 were significantly up-regulated and 1471 were significantly down-regulated (Figure 4B). Finally, 1445 DEGs were detected between DB_X and SX_X, with 535 significantly up-regulated and 910 significantly down-regulated (Figure 4C). Thus, with regard to leaf position, when morphological differences were more significant, more DEGs were detected.
Table 3.
Summary of transcriptome sequencing data.
| Sample | Clean reads | Q30 (%) | Total mapped | Uniquely mapped |
|---|---|---|---|---|
| DB_S | 43377979 | 96.85 | 31981991 73.73% |
31402433 72.39% |
| DB_Z | 55948586 | 97.00 | 41112704 73.48% |
40214398 71.84% |
| DB_X | 44958501 | 96.85 | 32878413 73.14% |
32123772 71.45% |
| SX_S | 42791156 | 96.52 | 31444702 73.49% |
30836394 72.07% |
| SX_Z | 48400741 | 95.78 | 34768856 71.88% |
34123332 70.54% |
| SX_X | 42270630 | 96.78 | 30598448 72.45% |
30002739 71.04% |
Figure 4.
Volcano plots of DEGs between overlapping and outward-curling types. (A) DB_S vs. SX_S; (B) DB_Z vs. SX_Z; (C) DB_S vs. SX_S.
Analysis of the biological pathways involved in heading type formation in Chinese cabbage
We mapped the DEGs to the reference canonical pathways in KEGG. Up-regulated DEGs with roles in plant hormone signal transduction were significantly enriched in the three leaf positions between each heading type: DB_S vs. Sx_S, DB_Z vs. Sx_Z and DB_X vs. Sx_X (Figure 5). The corrected p-values (q-values) indicated that these enrichments were statistically significant at 0.02, 0.04, and 0.00, respectively. Furthermore, the candidate genes overlapped with the genes identified from our Seq-BSA analysis, providing additional support for the role of plant hormone signal transduction in specifying heading types in Chinese cabbage. Among the eight parallel pathways, the most significantly enriched were auxin and ABA (Table S3). The proportions of up-regulated DEGs in the auxin and ABA pathways compared to the overall pathway were 17.78 and 24.44%, respectively, when DB_S was compared with SX_S. For DB_Z vs. SX_Z, the proportions were 43.75 and 31.25%, respectively; while those proportions were 30.00 and 55.00% for DB_X vs. SX_X. Based on the Seq-BSA analysis, genes in the candidate region related to plant hormone signal transduction were entirely committed to the auxin and ABA pathways. Therefore, the results from the Seq-BSA and RNA-Seq analysis are consistent.
Figure 5.
Enriched KEGG pathway scatterplots for upregulated DEGs. (A) DB_S vs. SX_S; (B) DB_Z vs. SX_Z; (C) DB_S vs. SX_S.
Identification of candidate genes
Among the candidates related to auxin and ABA signaling detected in the Seq-BSA analysis, two (BrGH3.12 and BrABF1) exhibited significantly different expression in the RNA-Seq data. Transcript levels of BrGH3.12 and BrABF1 were significantly different between types when measured at the apical region of the leaf. Furthermore, BrABF1 expression was significantly different between types when detected at the middle and basal positions. Therefore, the combination of Seq-BSA and RNA-Seq analyses indicated that BrGH3.12 and BrABF1 are strong candidate genes for heading type.
Expression changes of auxin and ABA-related genes
The DEGs assigned to the auxin pathway (Table S4) when DB_S was compared to Sx_S showed that one orthologs of TIR1 (TRANSPORT INHIBITOR RESPONSE 1) was up-regulated; six orthologs of early auxin-responsive AUXIN/INDOLEACETIC ACID-INDUCED PROTEINs (Aux/IAAs) were up-regulated including IAA2, IAA7, IAA9, IAA12, IAA16 and IAA28 while three IAA17 and one IAA15 were down-regulated. According to the auxin pathway description in the KEGG database, Aux/IAAs are regulated at multiple levels, consistent with their critical role in maintaining proper auxin response (Figure S3). In addition to the Aux/IAAs, three orthologs of AUXIN RESPONSE FACTORs (ARFs) were up-regulated including ARF6, ARF10 and ARF16. Except for one up-regulated ortholog of SMALL AUXIN UP-RNA 20 (SAUR20), 15 orthologs of SAURs were down-regulated including one SAUR19, SAUR23, SAUR24, SAUR36, SAUR61, SAUR62, two SAUR66, four SAUR20s, and three SAUR21s (Figure S3). In addition, significant differences in expression of NIT2, TSB1, TSB2, and YUCCA genes that are involved in regulating the IAA biosynthetic pathway were detected in the DB_S to Sx_S comparison (Table S4).
Finally, significant expression differences were found for the ABA related genes PYL/PYR, PP2C, SRK2, and ABF between DB and SX (Table S5) suggesting some involvement of ABA signaling.
Validation of RNA-Seq data by quantitative RT-PCR (qRT-PCR)
To verify the RNA-Seq data, we performed qRT-PCR on 10 unigenes at three leaf positions in overlapping and outward-curling plant types. In addition to BrGH3.12 and BrABF1, eight unigenes were randomly selected. As shown in Figure 6, all 10 genes displayed the same expression patterns in the qRT-PCR assays as in the RNA-Seq analysis. The Pearson correlation coefficient between the two methods was 0.9542, confirming the quality of the RNA-seq data.
Figure 6.
Comparison of results obtained via RNA-Seq and qRT-PCR for 10 genes expressed at different locations along the leaf sampled from overlapping and outward-curling morphotypes. 1t, 1m, and 1l: BrGH3.12 expression at top, middle, and basal position, respectively; 2t, 2m, and 2l: BrABF1 expression at top, middle, and basal position, respectively.
Concentrations of IAA and ABA
At the early heading stage, the IAA concentration was significantly higher for the outward-curling type than for the overlapping type when measured at the apical region of the leaf (P = 0.03). In contrast, the differences in IAA levels were not significant between morphotypes when measured at the middle (P = 0.28) and basal (P = 0.27) segments of the leaf (Figure 7). This trend in IAA concentrations across leaf positions and between overlapping and outward-curling types is consistent with the variation in expression of BrGH3.12 detected under the same parameters. In the outward-curling type, IAA concentrations were 1.27-fold higher in the apical region than in the middle while the basal portion contained less IAA than the apical region (Figure 7). However, in the overlapping type, the IAA concentration was 0.86-fold lower at the apical than in the middle region and greater at the basal region compared to the apical region (Figure 7).
Figure 7.
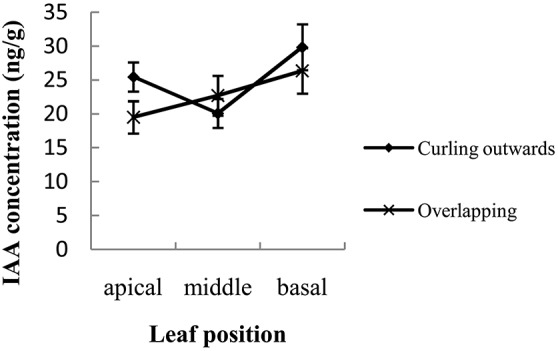
Concentrations of IAA measured at 3 positions within the leaf sampled from plants showing overlapping or outward-curling morphotypes of head formation.
At each corresponding position, the ABA concentration was always significantly higher in the outward-curling type than in the overlapping type (P < 0.01) (Figure 8). When comparisons were made at each leaf position, the apical also had consistently more ABA than either the middle or basal portions, regardless of morphotype (Figure 8). Within an individual type, the ABA concentration at the apical region of the leaf was 1.01-fold higher than the middle for the outward-curling type and 1.52-fold higher than the middle for the overlapping type (Figure 8). In the latter type, the apical region of the leaf tended to grow more slowly than the middle portion; this discrepancy was supported by the difference in levels of IAA and ABA measured at those positions. Based on the Gaussian curvature theory, leaves for which marginal regions grow slower than the central regions form a cup-like shape with positive Gaussian curvature (Nath et al., 2003). Therefore, one would expect to see this difference in growth rates manifested by leaves that curl inward and overlap when the head is forming. By comparison, faster growth at the apical region of the leaf rather than in the middle would naturally lead to an outward-curling pattern.
Figure 8.

Concentrations of ABA measured at 3 positions within the leaf sampled from plants showing overlapping or outward-curling morphotypes of head formation.
Discussion
The heading type is an important agricultural trait in Chinese cabbage. To investigate the mechanism of overlapping and outward curling heading type, the self-crossed progeny of one natural microspore-derived doubled haploid plant were examined. The candidate biological pathway contributing to this trait was identified by Seq-BSA and RNA-Seq analysis, highlighting the success of this joint approach. Furthermore, the concentrations of related metabolites in the pathway were detected. The results from the genomic, transcript and metabolite level analyses support a role for auxin and ABA signaling during heading type specification.
Merging divergent genomes into a single nucleus can trigger a “highly-programmed sequence of events within the cell that serves to cushion the effect of [genomic] shock” (McClintock, 1984). Genetic alterations have been reported for some synthetic polyploids, such as Arabidopsis thaliana (Comai et al., 2000; Madlung et al., 2002), Triticeae (Ozkan et al., 2001; Han et al., 2003; Ma et al., 2004), and Brassica (Song et al., 1995). The introgression of alien chromosome fragments can also severely affect the accepting genome, thereby triggering genetic and genomic changes (Liu et al., 2015). In this study, heading type showed segregation in self-crossed progenies from a single natural doubled haploid plant that integrated cabbage chromosome segments in the Chinese cabbage genome background. Similar segregations in self-crossed progenies have been derived from other Chinese cabbage introgression lines that integrated various segments from different chromosomes of cabbage (Kang et al., 2015; Li, 2015; Yan et al., 2015; Geng et al., 2016). These findings provide further evidence that trait segregation is caused by variations in receptor genomes that are induced by genomic shock and not, in the case of Chinese cabbage, by some genes added from a donor cabbage. In addition, using the Seq-BSA data, we mapped the remaining reads to the cabbage reference genome after mapping reads to the Chinese cabbage reference genome but did not obtain a candidate region. This led us to focus on the accepting Chinese cabbage genome in this study.
Auxin influences many processes in the plant including cell division, elongation, and differentiation and is active throughout the development of leaves (Ludwig-Muller, 2011; Garrett et al., 2012), flowers (Cheng et al., 2006; Tabata et al., 2009), fruits (Eklund et al., 2010; Shalom et al., 2014), and roots (Overvoorde et al., 2010; Jones and Ljung, 2012). Auxin also plays an important role during embryogenesis (Cheng et al., 2007; Braun et al., 2008; Wang, W. et al., 2011), hypocotyl elongation (Lilley et al., 2012), shoot regeneration (Qiao et al., 2012), branch point density (Esteve-Bruna et al., 2013), and plant responses involving phototropism, gravitropism, and apical dominance (Horiguchi et al., 2006; Cho et al., 2007).
During leaf development, auxin affects vein formation (Scarpella et al., 2006; Li et al., 2008), the number of palisade mesophyll cells, and air spaces within the spongy mesophyll (Esteve-Bruna et al., 2013). Auxin modulates the balance between adaxial and abaxial cell growth, giving rise to curly phenotypes (Bowman et al., 2002; Byrne, 2005; Qin et al., 2005; Izhaki and Bowman, 2007; Kidner and Timmermans, 2007; Wu et al., 2007, Braun et al., 2008; Wu et al., 2008; Husbands et al., 2009; Liu et al., 2011), epinastic phenotypes (Qin et al., 2005), and variations in leaf angle (Song et al., 2009; Bian et al., 2012). Cheng et al. (2016) reported that auxin-related genes are strongly associated with the leaf-curling trait (heading or non-heading) in Brassica rapa. Our data suggest that the significant differences in IAA concentrations and venation patterns between morphotypes at the apical region of the leaf is largely responsible for the determination of outward-curling vs. overlapping types of head formation. This is also supported by the Seq-BSA and RNA-Seq analyses that identified genes involved in the auxin pathway.
As an early-auxin-response gene, GH3 encodes indole-3-acetic acid-amido synthetase and functions in maintaining auxin homeostasis by conjugating excess IAA to various amino acids (Hagen and Guilfoyle, 1985; Staswick et al., 2002). The OsGH3 gene family member in rice (Oryza sativa) has a crucial role in controlling leaf inclination (Zhang, L. Y. et al., 2009; Zhang, S. W. et al., 2009; Du et al., 2012; Zhao, S. Q. et al., 2013), while an auxin-response factor, OsARF19, modulates rice leaf angle through positive regulation of OsGH3-5 and OsBRI1 (Zhang et al., 2015). In Brassica rapa, ARF3 is also associated with heading and non-heading (Cheng et al., 2016). Overexpression of OsGH3 family genes in rice results in a decrease in free-IAA and similar morphological phenotypes that include dwarfism, greater leaf angle, shorter leaves, smaller panicles, and fewer crown roots and root hairs (Ding et al., 2008; Domingo et al., 2009; Zhang, S. W. et al., 2009; Fu et al., 2011; Du et al., 2012; Zhao, S. Q. et al., 2013). Based on the results from the combined Seq-BSA and RNA-Seq analysis, we selected BrGH3.12 as a candidate gene. BrGH3.12 expression at the apical region of the leaf was significantly higher in the overlapping type than in the outward-curling type, a trend that was opposite to the levels of IAA. However, neither the expression of BrGH3.12 nor IAA concentrations differed between morphotypes when we examined the middle and basal portions of the leaf. The relationship between BrGH3.12 expression levels and IAA concentrations are consistent with previous studies.
The phytohormone ABA serves as an endogenous messenger that plays a key role in plant responses to environmental stress and several developmental processes, including root growth (McAdam et al., 2016), seed maturation, and dormancy (Yoshida et al., 2006; Miyakawa et al., 2013). Endogenous ABA has an obvious and ubiquitous inhibitory effect on shoot growth, which is thought to stem primarily from its ability to induce stomatal closure and, ultimately, decrease assimilation (Tardieu et al., 2010). The balance of IAA and ABA homeostasis plays a crucial role in plant development and diverse stress responses in rice (Du et al., 2012). In our study, the concentrations of auxin and ABA were significantly different between morphotypes. Therefore, both BrGH3.12, an early auxin-responsive gene, and BrABF1, a gene involved in ABA signaling, appear to be suitable candidates for regulating heading type in Chinese cabbage.
Ethics statement
The study was approved by Hebei Agricultural University, China. All provided written informed consent.
Author contributions
AG and SS conceived the original screening and research plans; AG and JZ supervised the experiments; AG, CM, HD, and YC performed most of the experiments; XC and YL provided technical assistance; AG, YW and LW designed the experiments and analyzed the data; AG conceived the project and wrote the article with contributions from all authors; JZ and SS supervised and reviewed the writing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. Kathleen Greenham at Dartmouth College in Hanover, NH, USA for critical reading of the manuscript.
Footnotes
Funding. Financial support for this work was provided by the National Key R&D of China (Grant no. 2016YFD0100204-17), the National Natural Science Foundation of China (Grant no. 31772324, 31572120, 31672151), the Science and Technology Research Project of Hebei Colleges and Universities (Grant no. ZD2017236), International Cooperation project in the Science and Technology Support Program of Hebei (Grant no. 17396315D), Hundred Innovative Talent Program of Hebei (Grant no. SLRC2017040).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2017.00176/full#supplementary-material
Leaf growth over time for plants showing overlapping or outward-curling morphotype.
Sampling segments for leaves analyzed via RNA-seq.
Difference genes expression in the auxin pathway.
References
- Anders S. (2010). HTSeq: Analysing High-Throughput Sequencing Data with Python. Available online at: http://htseq.readthedocs.io/ [DOI] [PMC free article] [PubMed]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. (2012). Differential Expression of RNA-Seq Data at the Gene Level-the DESeq Package. Heidelberg: Embl. [Google Scholar]
- Bian H., Xie Y., Guo F., Han N., Ma S., Zeng Z., et al. (2012). Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 196, 149–161. 10.1111/j.1469-8137.2012.04248.x [DOI] [PubMed] [Google Scholar]
- Bitto E., Bingman C. A., Bittova L., Houston N. L., Boston R. S., Fox B. G., et al. (2009). X-ray structure of ILL2, an auxin-conjugate amidohydrolase from Arabidopsis thaliana. Proteins 74, 61–71. 10.1002/prot.22124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N., Wyrzykowska J., Muller P., David K., Couch D., Perrot-Rechenmann C., et al. (2008). Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell 20, 2746–2762. 10.1105/tpc.108.059048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Eshed Y., Baum S. F. (2002). Establishment of polarity in angiosperm lateral organs. Trends. Genet. 18, 134–141. 10.1016/S0168-9525(01)02601-4 [DOI] [PubMed] [Google Scholar]
- Byrne M. E. (2005). Networks in leaf development. Curr. Opin. Plant. Biol. 8, 59–66. 10.1016/j.pbi.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Cheng F., Sun R. F., Hou X. L., Zheng H. K., Zhang F. L., Zhang Y. Y., et al. (2016). Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 48, 1218–1224. 10.1038/ng.3634 [DOI] [PubMed] [Google Scholar]
- Cheng L., Wang F., Shou H., Huang F., Zheng L., He F., et al. (2007). Mutation in nicotianamine aminotransferase stimulated the Fe (II) acquisition system and led to iron accumulation in rice. J. Plant. Physiol. 145, 1647–1657. 10.1104/pp.107.107912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Gene. Dev. 20, 1790–1799. 10.1101/gad.1415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K. H., Jun S. E., Jeong S. J., Lee Y. K., Kim G. T. (2007). Developmental processes of leaf morphogenesis in Arabidopsis. J. Integr. Plant. Biol. 50, 282–290. 10.1007/BF03030656 [DOI] [Google Scholar]
- Comai L., Tyagi A. P., Winter K., Holmes-Davis R., Reynolds S. H., Stevens Y., et al. (2000). Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant. Cell 12, 1551–1567. 10.1105/tpc.12.9.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Cao Y., Huang L., Zhao J., Xu C., Li X., et al. (2008). Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant. Cell 20, 228–240. 10.1105/tpc.107.055657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C., Andrés F., Tharreau D., Iglesias D. J., Talón M. (2009). Constitutive expression of OsGH3. 1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant. Microbe. Interact. 22, 201–210. 10.1094/MPMI-22-2-0201 [DOI] [PubMed] [Google Scholar]
- Dong H. (2015). Translocation Lines Identification in Progenies of Chinese cabbage-cabbage Alien Addition Line AC4 and Expression of Genes Related to Head Development. Dissertation, Baoding, IL: Hebei Agricultural University. [Google Scholar]
- Du H., Wu N., Fu J., Wang S., Li X., Xiao J., et al. (2012). A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 63, 6467–6480. 10.1093/jxb/ers300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund D. M., Ståldal V., Valsecchi I., Cierlik I., Eriksson C., Hiratsu K., et al. (2010). The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant. Cell 22, 349–363. 10.1105/tpc.108.064816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Bruna D., Pérez-Pérez J. M., Ponce M. R., Micol J. L. (2013). incurvata13, a novel allele of AUXIN RESISTANT6, reveals a specific role for auxin and the SCF complex in Arabidopsis embryogenesis, vascular specification, and leaf flatness. J. Plant. Physiol. 161, 1303–1320. 10.1104/pp.112.207779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Liu H., Li Y., Yu H., Li X., Xiao J., et al. (2011). Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. J. Plant. Physiol. 155, 589–602. 10.1104/pp.110.163774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima K., Hasebe M. (2014). Adaxial–abaxial polarity: the developmental basis of leaf shape diversity. Genesis 52, 1–18. 10.1002/dvg.22728 [DOI] [PubMed] [Google Scholar]
- Garrett J. J., Meents M. J., Blackshaw M. T., Blackshaw L. C., Hou H., Styranko D. M., et al. (2012). A novel, semi-dominant allele of MONOPTEROS provides insight into leaf initiation and vein pattern formation. Planta 236, 297–312. 10.1007/s00425-012-1607-0 [DOI] [PubMed] [Google Scholar]
- Geng Q. Q., Wang Y. H., Xuan S. X., Mao Q. Y., Zhao J. J., Shen S. X. (2016). Obtaining and genetic stability of Chinese cabbage - cabbage translocation lines with fragment of cabbage chromosome 2. Acta. Hortic. Sin. 43, 261–270. 10.16420/j.issn.0513-353x.2015-0858 [DOI] [Google Scholar]
- Gu A. X., Shen S. X., Wang Y. H., Zhao J. J., Xuan S. X., Chen X. P., et al. (2015). Generation and characterization of Brassica rapa ssp. pekinensis–B. oleracea var. capitata monosomic and disomic alien addition lines. J. Genet. 94, 435–444. [DOI] [PubMed] [Google Scholar]
- Gu A. X., Zhao J. J., Li L. M., Wang Y. H., Zhao Y. J., Hua F., et al. (2016). Analyses of phenotype and ARGOS and ASY1 expression in a ploidy Chinese cabbage series derived from one haploid. Breed. Sci. 66, 161–168. 10.1270/jsbbs.66.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grones P., Friml J. (2015). Auxin transporters and binding proteins at a glance. J. Cell. Sci. 128, 1–7. 10.1242/jcs.159418 [DOI] [PubMed] [Google Scholar]
- Hagen G., Guilfoyle T. J. (1985). Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 5, 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G., Guilfoyle T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant. Mol. Biol. 49, 373–385. 10.1023/A:1015207114117 [DOI] [PubMed] [Google Scholar]
- Han F. P., Fedak G., Ouellet T., Liu B. (2003). Rapid genomic changes in interspecific and intergeneric hybrids and allopolyploids of Triticeae. Genome. 46, 716–723. 10.1139/g03-049 [DOI] [PubMed] [Google Scholar]
- He Y. K., Xue W. X., Sun Y. D., Yu X. H., Liu P. L. (2000). Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell. Res. 10, 151–160. 10.1038/sj.cr.7290044 [DOI] [PubMed] [Google Scholar]
- Hill J. T., Demarest B. L., Bisgrove B. W., Gorsi B., Su Y. C., Yost H. J. (2013). MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome. Res. 23, 687–697. 10.1101/gr.146936.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Ferjani A., Fujikura U., Tsukaya H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant. Res. 119, 37–42. 10.1007/s10265-005-0232-4 [DOI] [PubMed] [Google Scholar]
- Husbands A. Y., Chitwood D. H., Plavskin Y., Timmermans M. C. (2009). Signals and prepatterns: new insights into organ polarity in plants. Genes Dev. 23, 1986–1997. 10.1101/gad.1819909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Kubo N., Kondo T., Hirai M. (2015). Detection of quantitative trait loci for heading traits in Brassica rapa using different heading types of Chinese cabbage. J. Hortic. Sci. Biotechnol. 90, 311–317. 10.1080/14620316.2015.11513188 [DOI] [Google Scholar]
- Izhaki A., Bowman J. L. (2007). KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant. Cell 19, 495–508. 10.1105/tpc.106.047472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B., Ljung K. (2012). Subterranean space exploration: the development of root system architecture. Curr. Opin. Plant. Biol. 15, 97–102. 10.1016/j.pbi.2011.10.003 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, 277–280. 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X.-H., Xuan S.-X., Li X.-F., Zhao J.-J., Shen S.-X., Wang Y.-H. (2015). Obtaining and genetic stability analysis of Chinese cabbage (Brassica oleracea var. capitata)-cabbage (Brassica compestris ssp. pekinensis) translocation lines with fragment of cabbage chromosome 7. J. Agric. Biot. 23, 1368–1376. 10.3969/j.issn.1674-7968.2015.10.013 [DOI] [Google Scholar]
- Kidner C. A., Timmermans M. C. (2007). Mixing and matching pathways in leaf polarity. Curr. Opin. Plant. Biol. 10, 13–20. 10.1016/j.pbi.2006.11.013 [DOI] [PubMed] [Google Scholar]
- Harshavardhan V. T., Seiler C., Junker A., Weigelt-Fischer K., Klukas C., Altmann T., et al. (2014). AtRD22 and AtUSPL1, members of the plant-specific BURP domain family involved in Arabidopsis thaliana drought tolerance. PLoS ONE 9:e110065. 10.1371/journal.pone.0110065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. BMC 25, 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The Sequence Alignment/Map (SAM) format and SAMtools. BMC 25, 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. C., Qin G. J., Tsuge T., Hou X. H., Ding M. Y., Aoyama T., et al. (2008). SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol. 179, 751–764. 10.1111/j.1469-8137.2008.02514.x [DOI] [PubMed] [Google Scholar]
- Li J. W. (1984). Chinese Cabbage. Beijing: Agricultural Press. [Google Scholar]
- Li Y. B. (2015). Genetic Analysis of Alien Chromosome in the Derivative Progenies from Chinese Cabbage-Head Cabbage Additional Line with Chromosome No. 1. Masters thesis, Baoding, IL: Hebei Agricultural University. [Google Scholar]
- Liang J., Liu B., Wu J., Cheng F., Wang X. (2016). Genetic variation and divergence of genes involved in leaf adaxial-abaxial polarity establishment in Brassica rapa. Front. Plant Sci. 7:94. 10.3389/fpls.2016.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley J. L., Gee C. W., Sairanen I., Ljung K., Nemhauser J. L. (2012). An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant. Physiol. 160, 2261–2270. 10.1104/pp.112.205575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y. P., Dhandapani V., Paul P., Pang W., Choi S., Li X. (2015). Genome-wide association studies of morphological related traits in chinese cabbage (B. rapa), in PAG (Singapore: ). [Google Scholar]
- Liu S., Li F., Kong L., Sun Y., Qin L. M., Chen S. Y., et al. (2015). Genetic and epigenetic changes in somatic hybrid introgression lines between wheat and tall wheatgrass. Genetics 199, 1035–1045. 10.1534/genetics.114.174094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Jia L., Wang H., He Y. (2011). HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J. Exp. Bot. 62, 4367–4381. 10.1093/jxb/err167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Muller J. (2011). Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773. 10.1093/jxb/erq412 [DOI] [PubMed] [Google Scholar]
- Ma X. F., Fang P., Gustafson J. P. (2004). Polyploidization induced genome variation in triticale. Genome 47, 839–848. 10.1139/g04-051 [DOI] [PubMed] [Google Scholar]
- Madlung A., Masuelli R. W., Watson B., Reynolds S. H., Davison J., Comai L. (2002). Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant. Physiol. 129, 733–746. 10.1104/pp.003095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J. G., Wei L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- Mao Y., Wu F., Yu X., Bai J., Zhong W., He Y. (2014). MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in Chinese cabbage by differential cell division arrest in leaf regions. Plant. Physiol. 164, 710–720. 10.1104/pp.113.228007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., et al. (2011).The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam S. A., Brodribb T. J., Ross J. J. (2016). Shoot-derived abscisic acid promotes root growth. Plant Cell Environ. 39, 652–659. 10.1111/pce.12669 [DOI] [PubMed] [Google Scholar]
- McClintock B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T., Fujita Y., Yamaguchi-Shinozaki K., Tanokura M. (2013). Structure and function of abscisic acid receptors. Trends. Plant. Sci. 5, 259–266. 10.1016/j.tplants.2012.11.002 [DOI] [PubMed] [Google Scholar]
- Nath U., Crawford B. C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. 10.1126/science.1079354 [DOI] [PubMed] [Google Scholar]
- Overvoorde P., Fukaki H., Beeckman T. (2010). Auxin control of root development. Cold. Spring. Harb. Perspect. Biol. 2:a001537. 10.1101/cshperspect.a001537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan H., Levy A. A., Feldman M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant. Cell. 13, 1735–1748. 10.1105/TPC.010082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peat T. S., Böttcher C., Newman J., Lucent D., Cowieson N., Davies C. (2012). Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant. Cell 24, 4525–4538. 10.1105/tpc.112.102921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M., Zhao Z. J., Xiang F. N. (2012). Arabidopsis thaliana in vitro shoot regeneration is impaired by silencing of TIR1. Biol. Plantarum 56, 409–414. 10.1007/s10535-011-0233-1 [DOI] [Google Scholar]
- Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., et al. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17, 2693–2704. 10.1105/tpc.105.034959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y. M. (2013). Analysis of Chinese Cabbage-Cabbage Alien Addition Lines with EST-SSRs and DH Population Creation of the Bolting Tolerance Alien Addition Line. Masters thesis, Baoding, IL: Hebei Agricultural University. [Google Scholar]
- Ranocha P., Dima O., Nagy R., Felten J., Corratgé-Faillie C., Novák O., et al. (2013). Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 4:2625. 10.1038/ncomms3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015–1027. 10.1101/gad.1402406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom L., Samuels S., Zur N., Shlizerman L., Doron-Faigenboim A., Blumwald E., et al. (2014). Fruit load induces changes in global gene expression and in abscisic acid (ABA) and indole acetic acid (IAA) homeostasis in citrus buds. J. Exp. Bot. 65, 3029–3044. 10.1093/jxb/eru148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Lu P. I. N. G., Tang K., Osborn T. C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. U.S.A. 92, 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., You J., Xiong L. (2009). Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant. Mol. Biol. 70, 297–309. 10.1007/s11103-009-9474-1 [DOI] [PubMed] [Google Scholar]
- Staswick P. E., Tiryaki I., Rowe M. L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. 10.1105/tpc.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E., Serban B., Rowe M., Tiryaki I., Maldonado M. T., Maldonado M. C., et al. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole−3–acetic acid. Plant Cell 17, 616–627. 10.1105/tpc.104.026690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R., Ikezaki M., Fujibe T., Aida M., Tian C. E., Ueno Y., et al. (2009). Arabidopsis auxin response factor 6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 51, 164–175. 10.1093/pcp/pcp176 [DOI] [PubMed] [Google Scholar]
- Tardieu F., Parent B., Simonneau T. (2010). Control of leaf growth by abscisic acid: hydraulic or non-hydraulic processes? Plant Cell Environ. 33, 636–647. 10.1111/j.1365-3040.2009.02091.x [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Kim D., Kelley D. R., Pimentel H., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Li L., Li H., Liu L., Zhang Y., Gao J., et al. (2012). Transcriptome analysis of rosette and folding leaves in Chinese cabbage using high-throughput RNA sequencing. Genomics 99, 299–307. 10.1016/j.ygeno.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Zhang X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–148. 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- Wang W., Xu B., Wang H., Li J., Huang H., Xu L. (2011). YUCCA genes are expressed in response to leaf adaxial-abaxial juxtaposition and are required for leaf margin development. Plant Physiol. 157, 1805–1819. 10.1104/pp.111.186395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- Wang Y. L., Wang F. J., Bai J. J., He Y. K. (2014). BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant. Biotechnol. J. 12, 312–321. 10.1111/pbi.12138 [DOI] [PubMed] [Google Scholar]
- Westfall C. S., Herrmann J., Chen Q., Wang S., Jez J. M. (2010). Modulating plant hormones by enzyme action: the GH3 family of acyl acid amido synthetases. Plant. Signal. Behav. 5, 1607–1612. 10.4161/psb.5.12.13941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall C. S., Zubieta C., Herrmann J., Kapp U., Nanao M. H., Jez J. M. (2012). Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 336, 1708–1711. 10.1126/science.1221863 [DOI] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523. 10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Yu L., Cao W., Mao Y., Liu Z., He Y. (2007). The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-micro RNA processing. Plant Cell 19, 914–925. 10.1105/tpc.106.048637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lin W. C., Huang T., Poethig R. S., Springer P. S., Kerstetter R. A. (2008). KANADI1 regulates adaxial–abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc. Natl. Acad. Sci. U.S.A. 105, 16392–16397. 10.1073/pnas.0803997105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D., Wang H., Basnet R. K., Zhao J., Lin K., Hou X., et al. (2014). Genetic dissection of leaf development in Brassica rapa using a genetical genomics approach. Plant Physiol. 164, 1309–1325. 10.1104/pp.113.227348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. B., Lv B., Zhang F. L., Chen G., Liu X. S., Wang J. Y., et al. (2004). Guidelines for the Conduct of Tests for Distinctness Uniformity and Stability-Chinese Cabbage. Beijing: The national standard of the People's Republic of China. GB/T 19557.5-2004 [Google Scholar]
- Yan Z., Wang Y., Xuan S., Zhao J., Shen S. (2015). Obtaining and genetic stability of Chinese cabbage—cabbage translocation lines with fragment of cabbage chromosome 8. Acta. Hortic. Sin. 43, 261–270. [Google Scholar]
- Yoshida T., Nishimura N., Kitahata N., Kuromori T., Ito T., Asami T., et al. (2006). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant. Physiol. 140, 115–126. 10.1104/pp.105.070128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant. J. 61, 672–685. 10.1111/j.1365-313X.2009.04092.x [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Maruyama K., Mogami J., Todaka D., Shinozaki K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic-acid signaling in response to osmotic stress. Plant Cell Environ. 38, 35–49. 10.1111/pce.12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wang H., Zhong W., Bai J., Liu P., He Y. (2013). QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS ONE 8:e76059. 10.1371/journal.pone.0076059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Y., Bai M. Y., Wu J., Zhu J. Y., Wang H., Zhang Z., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767–3780. 10.1105/tpc.109.070441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wang S., Xu Y., Yu C., Shen C., Qian Q., et al. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38, 638–654. 10.1111/pce.12397 [DOI] [PubMed] [Google Scholar]
- Zhang S. W., Li C. H., Cao J., Zhang Y. C., Zhang S. Q., Xia Y. F., et al. (2009). Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 151, 1889–1901. 10.1104/pp.109.146803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338. 10.1093/mp/ssr104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. Q., Xiang J. J., Xue H. W. (2013). Studies on the rice leaf inclination1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol. Plant. 6, 174–187. 10.1093/mp/sss064 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Chan Z., Xing L., Liu X., Hou Y. J., Chinnusamy V., et al. (2013). The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 23, 1380–1395. 10.1038/cr.2013.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Leaf growth over time for plants showing overlapping or outward-curling morphotype.
Sampling segments for leaves analyzed via RNA-seq.
Difference genes expression in the auxin pathway.