Abstract
The proportion of starch, protein and oil in legume seeds is species dependent. The model legume, Medicago truncatula, has predominantly oil and protein stores. To investigate the regulation of seed oil production we compared M. truncatula with M. orbicularis, which has less oil and protein. The types of protein and fatty acids are similar between the two species. Electron microscopy indicated that the size and distribution of the oil bodies in M. orbicularis, is consistent with reduced oil production. M. orbicularis has more extruded endosperm mucilage compared to M. truncatula. The cotyledons have a greater cell wall content, visualized as thicker cell walls. The reduced oil content in M. orbicularis is associated with increased expression of the MtGLABRA2-like (MtGL2) transcription factor, linked to an inverse relationship between mucilage and oil content in Arabidopsis. The expression of the pectin biosynthesis GALACTURONOSYLTRANSFERASE (GAUT) genes, is also increased in M. orbicularis. These increases in extruded mucilage and cell wall storage components in M. orbicularis are accompanied by reduced expression of transcriptional regulators of oil biosynthesis, MtLEAFY COTYLEDON1-LIKE (MtL1L), MtABSCISIC ACID-INSENSITIVE3 (MtABI3), and MtWRINKLED-like (MtWRI), in M. orbicularis. The reduced oil in M. orbicularis, is consistent with increased synthesis of cell wall polysaccharides and decreased expression of master transcription factors regulating oil biosynthesis and embryo maturation. Comparative investigations between these two Medicago species is a useful system to investigate the regulation of oil content and carbon partitioning in legumes.
Keywords: GLABRA2, legumes, Medicago orbicularis, Medicago truncatula, mucilage, oil production, pectin, cell wall storage polysaccharides
Introduction
Legumes are the third largest family of flowering plants (Doyle and Luckow, 2003; Gepts et al., 2005) and the second most important crop family with approximately 20,000 species (Doyle and Luckow, 2003; Gepts et al., 2005), representing a significant source of protein and oil for the human diet, animal feedstock and industrial use. Legume seeds are rich in protein with globulin vicilins (7S) and legumins (11S) the most abundant (Shewry et al., 1995) but poor in sulfur-containing amino acids and contain nutritionally undesirable compounds such as protease inhibitors, which limits their nutritional value. Therefore, there has been a substantive focus on the improvement of seed protein quality (Molvig et al., 1997; Wang et al., 2003; Krishnan, 2005). There has also been a focus on sugar metabolism in non-oil storage legumes such as Vicia (Weber et al., 2005), the genetic modification of fatty acids and strategies to increase seed oil content in soybean (Clemente and Cahoon, 2009; Roesler et al., 2016). Soybean supplies nearly a third of world vegetable oil production (Graham and Vance, 2003). There is a need for more information on the cellular and genetic basis that underlies oil body formation during legume seed development (Repetto and Gallardo, 2012; Wang et al., 2012; Song et al., 2017).
Lipid storage starts from de novo fatty acids synthesis, followed by the synthesis of triacylglycerols (TAGs) and oil body biogenesis. The fatty acids are synthesized in the plastid, which relies on imported sucrose from maternal tissues to provide important precursors from glycolysis (Hills, 2004). TAGs are synthesized in the smooth ER by the esterification of fatty acids with the glycerol backbone, and eventually stored as individual oil bodies in the cytosol with oleosin as packaging protein. Recent studies suggest seipens are involved in early oil body assembly at specific ER domains (Cai et al., 2015). As a consequence, seed oil storage requires co-ordination of many steps. Master transcription factors identified to regulate oil storage in Arabidopsis, are LEAFY COTYLEDON1 (LEC1)/LEAFY COTYLEDON1-LIKE (L1L), LEAFY COTYLEDON2 (LEC2), FUSCA3 (FUS3) and ABSCISIC ACID INSENSITIVE3 (ABI3) (Crowe et al., 2000; Mendoza et al., 2005; Wang et al., 2007; Mu et al., 2008; Tan et al., 2011). For example, LEC1/L1L are key regulators of fatty acid synthesis, targeting multiple enzymes in the fatty acid pathway (Mu et al., 2008); FUS3 can induce de novo fatty acid synthesis in transgenic seedlings (Wang et al., 2007).
In addition to being used as a legume model for nodule development and the rhizobial-legume symbioses (Barker et al., 1990; Cook, 1999; Rose, 2008; Young et al., 2011) Medicago truncatula has been increasingly recognized as a model to study seed development and storage reserves in legumes (Gallardo et al., 2003; Wang and Grusak, 2005; Thompson et al., 2009; Wang et al., 2012; Verdier et al., 2013). We have studied embryogenesis from early development to cotyledon oil body formation (Wang et al., 2012) and profiled transcriptional regulation of early embryogenesis in preparation for seed storage (Kurdyukov et al., 2014).
An early study of Medicago seed composition (Tonnet and Snudden, 1974) showed that M. orbicularis seed contained approximately 3% oil compared to 9% in M. truncatula. As one approach to the regulation of Medicago seed oil production we carried out a comparative study, comparing M. truncatula with M. orbicularis. The comparative seed structure, cotyledon cell structure and storage composition between the two species was examined. M. orbicularis had more extruded endosperm mucilage compared to M. truncatula, thicker cotyledon cell walls and very small oil bodies. Investigations of endosperm mucilage, GALACTURONOSYLTRANSFERASES (GAUTs) involved in cell wall pectin biosynthesis and the transcriptional regulators of seed oil storage were consistent with more carbon being switched to polysaccharide biosynthesis, with less carbon available for oil biosynthesis in the M. orbicularis cotyledons.
Materials and Methods
Plant Materials
Medicago truncatula and M. orbicularis (South Australia Agriculture Research and Development Institute, SARDI, accessions SA1619 and 2554, respectively) plants were glasshouse grown with a 14 h photoperiod and 23/19°C day/night temperature. The visual characteristics of plant canopy, pods and seeds for both species are shown in Figure 1. Plants were grown in potting mix in 15 cm pots supplemented with 5 g Osmocote Exact Standard slow release fertilizer and with automated watering. Seeds from both species were isolated from pods at 10, 14, 18, 22, and 26 days after anthesis (DAA) and stored at -80°C. Embryos from seeds of both species reach maturity at approximately the same time with similar timing for the different developmental stages (see Wang et al., 2012, for M. truncatula embryo development stages).
FIGURE 1.
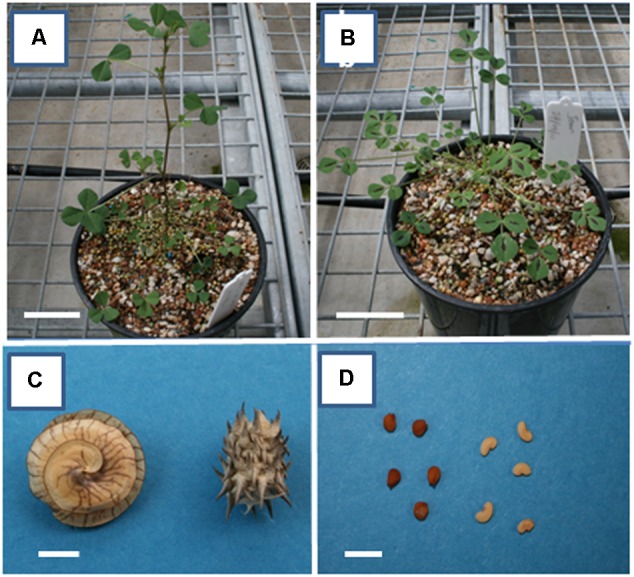
Vegetative canopy, pods and seeds of Medicago truncatula and M. orbicularis. (A) M. orbicularis and (B) M. truncatula plants 4 weeks after sowing. (C) Pods of M. orbicularis (left) and M. truncatula (right). (D) Seeds of M. orbicularis (left) and M. truncatula (right). Bars (A,B) 5 cm, (C,D) 5 mm.
Assays for Seed Protein, Oil and Starch
The protein assay was fully described by Wang et al. (2012). For each of three replicates, four seeds were ground in 500 μL of extraction buffer, sonicated and centrifuged at 16,000 × g at 4°C for 12 min, and the supernatant used to measure soluble protein by the Bradford Assay (Bradford, 1976), at a wavelength of 595 nm. The remaining insoluble protein was extracted by grinding in 500 μL of 1M NaOH and vortexing for 30 min. After centrifugation the protein in the supernatant was measured. A bovine serum albumin standard curve was used to derive protein concentrations.
Lipid extraction was based on the method of Folch et al. (1957). For each of three replicates, whole seed was homogenized with chloroform/methanol (2:1 v: v) in a final volume 20 times the volume of the sample. The mixture was agitated for 15–20 min in an orbital shaker at room temperature. The homogenate was centrifuged to recover the liquid phase. The solvent was washed with 0.2 volumes of a 0.9% NaCl solution, vortexed and then centrifuged at 3000 × g. The upper phase was discarded and the lower phase containing the lipid evaporated under a nitrogen stream. The fatty acid concentration was analyzed by direct transesterification of lipid using a gas chromatograph as described by Wang et al. (2012).
Total starch was extracted by following the instructions for the Abcam Starch Assay Kit (Cambridge, MA, United States). The method was slightly modified. For each of three replicates, tissue (5–10 mg) was ground using a plastic pestle in a 1.5 mL Eppendorf tube, followed by washing off of any free glucose and small oligosaccharides with 1 mL 90% ethanol at 60°C for 5 min with occasional vortexing. After removal of the supernatant, soluble starch in the pellet was extracted with 1 mL H2O and incubated in a boiling water bath for 60 min. After having extracted soluble starch, the water insoluble pellet was dissolved with 0.4 mL DMSO in a boiling water bath for 60 min. A standard curve was used to measure total starch concentration at OD 570 nm.
Cell Wall Isolation
The whole seed cell wall content was determined for 6 seeds with three replicates while embryo cell wall content was determined for 10 seeds with three replicates. Dry seed was pierced using a needle or sliced with a scalpel blade and soaked in water at 60°C (in a heating block) for 10–15 min. Embryo cell wall isolation was obtained after removing the seed coat, including any adhering mucilage, after soaking in water. The details for the cell wall isolation are described in the Foster et al. (2010) protocol and outlined here. The tissue was first ground, followed by adding 1.5 ml of 70% aqueous ethanol, and the mixture vortexed. The mixture was centrifuged at 10,000 × g for 10 min to pellet insoluble residue and the supernatant removed. The pellet was resuspended in 1.5 mL of chloroform/methanol (1:1 v: v), vortexed, centrifuged again and the supernatant discarded. The pellet was then resuspended in 500 μL of acetone and evaporated in a fume hood. Starch was removed from the sample as described in the Foster et al. (2010) protocol. The remaining pellet was washed three times by adding 1.5 mL water. The pellet was resuspended in 500 μL of acetone and evaporated until dry. The dried material represents the lignocellulosics (cellulose, various hemicelluloses, and the polyphenol lignin) of the isolated cell walls.
Major Seed Components
Dry weight was determined from mature seeds (three replicates of 100) after drying in an 80°C oven over 2 days. M. truncatula had a dry weight of 298.1 ± 25.2 mg and M. orbicularis 369.2 ± 3.05 mg per 100 seeds. Ten seeds with three replicates were used to determine the proportion of seed coat, embryo and extruded endosperm mucilage. The seed was first pierced, as described above, and imbibed in water at 60°C for 10 min. The seed coat, cotyledon and extruded endosperm mucilage were then separated and the dry weight determined.
Mucilage Staining
Mucilage was isolated as in the previous section and stained by Ruthenium Red (Western et al., 2000; Verdier et al., 2013). Mucilage still adhering to the inner side of the seed coat was also stained with Ruthenium Red. The isolated swollen endosperm layer was prepared for Ruthenium Red staining by a shorter incubation at 60°C and removal of seed coat and embryo.
Histology of Thin Sections
Dry seed was imbibed at 4°C for 5 days then fixed and embedded in LR White resin as previously described (Rose et al., 2006; Wang et al., 2012). Sections of 1 μm were stained in Toluidine Blue and Azur II and viewed and photographed with a Zeiss Axiophot microscope.
Transmission Electron Microscopy
The dry seed was first imbibed in water for 5 days at 4°C and then fixed using 4% (w:v) glutaraldehyde and 1.5% (w:v) paraformaldehyde in 25 mM phosphate buffer (pH 7) at 4°C for 4 h, with at least one change of buffer. Tissue was transferred to phosphate buffer, washed once and incubated overnight at 4°C. The tissue was then postfixed in 1% aqueous osmium tetroxide overnight at 4°C and washed twice with distilled water, then chilled to 4°C. Dehydration steps and infiltration into LR White resin has been described (Wang et al., 2012). Ultrathin sections were cut with a diamond knife and transferred to 200-mesh grids. The sections were stained with uranyl acetate and lead citrate and viewed using a JEOL JEM 1200-EXII electron microscope (JEOL Ltd., Tokyo, Japan).
Identification of Genes from the M. truncatula Genome Sequence
Homology-based methods using genes from other plant families have been successfully employed to identify specific classes of legume transcription factors (Udvardi et al., 2007). The phylogenetic analyses were based on protein or peptide sequences (e.g., Supplementary Figure S1a) from Arabidopsis, M. truncatula and soybean from phytozome v9.1 (M. truncatula genomic sequence 3.5). Candidate genes were checked against information from the NCBI database. Supplementary Table S1 provides information about the genes using the improved genome release M. truncatula version Mt4.0 (Tang et al., 2014).
Sequencing of Candidate Genes for M. orbicularis
Primers for sequencing were designed based on M. truncatula genes using Primer 3. The cDNA template from M. orbicularis was used to amplify the fragment, and a single band with similar size to M. truncatula was produced. The product was purified with Promega cleanup (Promega, Sydney, NSW, Australia) prior to sequencing at the Australian Genome Research Facility, Brisbane (AGRF1). Alignment of sequences between M. truncatula and M. orbicularis showed high similarity with only a few nucleotides varying (e.g., Supplementary Figure S1b). The same primers for gene expression comparisons between the two species could be used.
RNA Extraction, cDNA Synthesis and Real Time Quantitative PCR
RNA was extracted from young developing seed (before 18 DAA) using the RNAqueous-4PCR kit (Ambion, Sydney, NSW, Australia) and from older developing seed (after 18 DAA) using a phenol-chloroform method (Li and Trick, 2005) due to high phenolics or carbohydrate in these seeds. Genomic DNA was removed using DNase treatment. cDNA synthesis was performed with a SuperScript III first-strand synthesis system (Invitrogen, Melbourne, VIC, Australia) using 1 μg of total RNA and oligo(dT) primers.
The cDNA was diluted 1:25 for quantitative PCR (qPCR) reactions. All qPCR reactions were prepared using a CAS1200 robot (Qiagen, Melbourne, VIC, Australia) and run on a Rotor-Gene Q (Qiagen). Primers were designed using Primer3 (Supplementary Table S2). Reactions were performed with a 15 μL sample volume using Platinum Taq PCR polymerase and 2 μM SYTO9 fluorescent dye (Invitrogen). PCR cycling conditions were 94°C for 2 min, followed by 40 cycles of 94°C for 15 s, 60°C for 30 s and 72°C for 30 s. Dissociation analysis was performed in each run to verify the amplification of a specific product. Gene expression was normalized to the expression of GAPDH. GAPDH has been used as a suitable reference gene for M. truncatula embryogenesis and seed development investigations (Wang et al., 2012; Kurdyukov et al., 2014) and verified experimentally by other types of gene analysis (Nolan et al., 2003, 2009), microarray and RT-qPCR studies (Mantiri et al., 2008). Using geNORM software, Verdier et al. (2008) have also shown GAPDH to be a suitable reference gene.
PCR efficiency of each run was calculated using the LinRegPCR program of Ramakers et al. (2003). Relative expression was calculated using the Pfaffl (2001) method. Gene expression was relative to 10 days after anthesis. Results shown are means ± SE of three biological repeats.
Proteomics
Protein was extracted using a Plant Total Protein Extraction Kit (Sigma–Aldrich). Seeds from M. truncatula and M. orbicularis 12, 14, 18, and 22 DAA were used to profile protein composition. The protein was quantified using the Bradford (1976) assay. Protein was precipitated using methanol/chloroform, digested with sequencing grade trypsin (Promega) and protein digestion was stopped with a final concentration of 0.1% trifluoroacetic acid. The samples were then centrifuged twice at 16,000 × g for 30 min to pellet undigested protein. The supernatant was transferred to vials for nanoflow reversed phase liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.
Liquid chromatography tandem mass spectrometry analysis was similar to Baker et al. (2010) with the following deviations: peptides were sequenced by nanoflow reverse phase Liquid Chromatography (nanoACQUITY Ultra Performance LC, Waters, Milford, MA, United States) coupled directly to an ESI Q-ToF Mass Spectrometer (MicrOtof QII, Bruker GmbH, Preston, VIC, Australia) operating in positive ion MS/MS mode. Peptides were loaded at 5 μl/min onto a C18 trap column (Waters, nanoACQUITY UPLC Symmetry C18 Trap, 5 μm, 180 μm × 20 mm) for desalting and pre-concentration. Peptide separation was then performed at 300 nl/min over an nanoACQUITY UPLC BEH C18 Column, 1.7 μm, 75 μm × 150 mm (Waters), using a linear gradient of 0-60% buffer (84% ACN, 0.1% formic acid) over 200 min. The peptides were eluted directly into the nanoflow ESI ion source of the MS system for MS/MS analysis. The Q-ToF system was set to perform MS/MS on the top 5 ions present in each MS scan with an ion exclusion time of 30 s. Raw MS Files were converted into MASCOT Generic Format using Data Analysis 4.0 and imported into ProteinScape 2.1 platform (both Bruker, Bremen, Germany) for database searching. Searches were performed against the annotated M. truncatula protein database (in FASTA format; IMGAG 3.5 release was obtained using the Phytozome site2) and SwissProt (Green Plant) databases using an in-house licensed MASCOT server (version 2.3, Matrix Science). The number of allowed trypsin missed cleavages was set to 2. Deamidation of asparagine and glutamine, oxidation of methionine and phosphorylation of serine, threonine and tyrosine were set as variable modifications. The parent peptide ion tolerance was set to 1.2 Da with MS/MS fragment ion tolerance set to 0.7 Da. Peptide thresholds were set requiring False Positive Rate less than 0.05% with a low stringency MASCOT score greater than 35. Those spectra meeting these criteria were validated by manual inspection to ensure accurate y- and b-ion detection with overlapping sequence coverage. Each M. truncatula protein returning a positive peptide ID (significance value of p < 0.05) was interrogated against the non-redundant protein database at the NCBI3 and at Phytozome4 using BLAST-P to confirm protein identity and to classify the protein’s function.
Statistical Tests
Standard errors are indicated and Student’s two-tailed t-test was used for experimental comparisons. A single asterisk (∗) indicates that the difference is statistically significant at P < 0.05 while double asterisks (∗∗) indicate that the difference is statistically significant at P < 0.01. Treatments with different letters are significantly different at the 0.05 probability level, with the statistical analysis performed by comparing means in the SPSS 18.0 statistical package.
Results
In our initial analyses we characterized the seed composition of the M. truncatula and M. orbicularis accessions (Figure 1) used in this study.
Seed Oil and Fatty Acid Content
Seed oil content (oil weight/seed dry weight) and fatty acid composition of seeds in M. truncatula and M. orbicularis are shown in Figures 2A,B. The mature seed accumulated approximately 9% oil in M. truncatula and 3% in M. orbicularis Figure 2A. The most abundant fatty acids in Figure 2B for both species were C16:0 (palmitic acid), C18 1n-9 (oleic acid), C18:2n-6 (linoleic acid), and C18:3n-3 (alpha-linolenic acid, an omega-3 fatty acid). M. orbicularis was lower in C16:0, C18:1n-9, and C18:2n-6. Alpha-linolenic acid (C18:3n-3) was higher in M. orbicularis than in M. truncatula. Two minor fatty acids, C18:0 (stearic acid) and C18:3n-6 (gamma-linolenic acid, an omega-6 fatty acid), were also higher in M. orbicularis.
FIGURE 2.

Oil and fatty acid content of M. truncatula and M. orbicularis as a percentage of seed dry weight. (A) Oil and (B) Fatty acid content. SE indicated and ∗∗p < 0.01 (n = 3).
Seed Protein Content
The seed storage protein as a proportion of seed dry weight was compared between M. truncatula and M. orbicularis (Figure 3A). M. truncatula seed accumulated approximately 30% storage protein while M. orbicularis seed accumulated only 10% storage protein. We subsequently carried out proteomics using LC-MS/MS to identify the major storage proteins in both species. These are shown in Table 1 and Supplementary Table S3. The storage proteins identified in both Medicagos were very similar at both 18 and 22 days. The proteins identified were synthesized by multiple genes. As is known in M. truncatula the major storage proteins are vicilins/convicilins, legumins, and conglutins (Gallardo et al., 2003). The gene redundancy presumably reflects the importance of the storage proteins for survival in the next generation. To check that the M. truncatula and M. orbicularis storage protein profiles showed similar trends during development, data were obtained at 12, 14, 18, and 22 days after anthesis (DAA). The data again are consistent with similar protein profiles between the two species (Table 1 and Supplementary Table S3). In M. truncatula only the convicilin proteins are present at 12 DAA when SSP (seed storage protein) gene expression is initiated and the cotyledons have just started to develop (Supplementary Table S3 and Wang et al., 2012). A wider spectrum of SSPs are present at 14 DAA (Supplementary Table S3). In the case of M. orbicularis convicilin proteins are present at 14 DAA plus other SSPs, reflecting a slight difference (approximately 1 day) in cotyledon development timing. It was of interest that lipoxygenase is a major seed protein. This seed protein is important in germination to lyse oil bodies and is bound to the oil bodies at that time (Feussner et al., 2001). Proteome studies indicate it can also be bound to seed oil bodies (Jolivet et al., 2013).
FIGURE 3.

Protein and starch content of M. truncatula and M. orbicularis as a percentage of seed dry weight. (A) Total protein content. (B) Starch content. SE indicated and ∗∗p < 0.01 (n = 3).
Table 1.
Major proteins of Medicago truncatula and M. orbicularis, days after anthesis (DAA) indicated.
| Gene identifier (Supplementary Table S3) | Protein | M. truncatula 18 days | M. truncatula 22 days | M. orbicularis 18 days | M. orbicularis 22 days |
|---|---|---|---|---|---|
| Medtr7g079820 | Convicilin∗ | + | + | + | + |
| Medtr7g079780 | Convicilin∗ | + | + | + | + |
| Medtr7g079740 | Convicilin∗ | + | + | – | – |
| Medtr7g079730 | Convicilin∗ | + | + | + | + |
| Medtr7g079770 | Provicilin | + | + | + | + |
| VCLC_PEA | Vicilin Precursor | + | + | + | + |
| Medtr5g019780 | Vicilin-like antimicrobial | + | + | + | + |
| Medtr1g072630 | Legumin B | + | + | + | + |
| Medtr1g072610.2 | Legumin B | + | + | + | + |
| Medtr1g072600.1 | Legumin B | + | + | + | + |
| LEG_CICAR | Legumin | + | + | + | + |
| LEG_PEA | Legumin A | + | + | – | + |
| LEG_JPEA | Legumin J | – | – | + | + |
| LEGU_CANEN | Legumin | – | – | + | – |
| Medtr2g083160 | Conglutin | + | + | + | + |
| Medtr2g099570 | Seed lipoxygenase | + | + | + | + |
| Medtr2g099560 | Lipoxygenase | + | + | + | + |
The more detailed data relating to individual genes can be found in Supplementary Table S3. ∗Also annotated as vicilin (LegumeIP Database, Li et al., 2012).
Seed Starch Content
The content of total seed starch in M. truncatula and M. orbicularis is shown in Figure 3B. Seeds from both species had very low total starch content (with only 1–2% of seed dry weight) compared to protein or oil content. M. truncatula seed accumulated higher total starch than M. orbicularis mainly because of higher insoluble starch in M. truncatula. It was clear that the starch content in M. orbicularis was not increased despite there being less oil and protein.
Ultrastructure of Mature Cotyledons in M. truncatula and M. orbicularis
To obtain more information on the storage components, transmission electron micrographs of cotyledons of mature seeds in M. truncatula and M. orbicularis were obtained (Figure 4). The cotyledons are the major site of storage components. The cotyledon cells of M. truncatula are shown in Figures 4A,C and of M. orbicularis in Figures 4B,D. At higher magnification oil bodies could be readily visualized as white spherical to ovoid structures in both species (Figures 4E,F), but were very much smaller in M. orbicularis. As previously observed, the M. truncatula oil bodies surrounded the protein bodies and also aligned along the plasma membrane. The M. orbicularis oil bodies also surrounded the protein bodies but not in concentric layers and not as uniformly. The small M. orbicularis oil bodies were also randomly spread throughout the cytoplasm.
FIGURE 4.
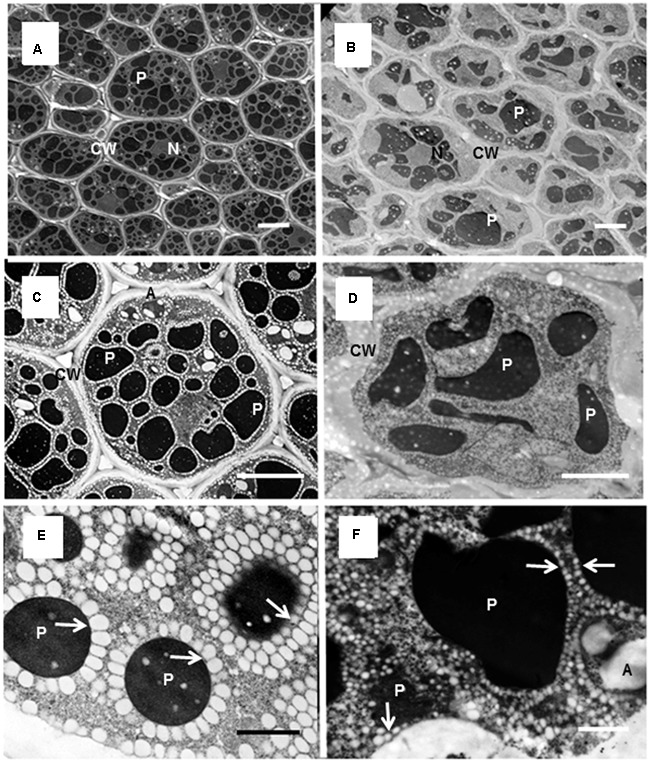
Electron micrographs of cells of M. truncatula and M. orbicularis cotyledons. View of multiple cells of M. truncatula (A) and M. orbicularis (B); single cells of M. truncatula (C) and M. orbicularis (D) and subcellular structure of M. truncatula (E) and M. orbicularis (F). Amyloplasts (A), cell walls (CW), nucleus (N), oil bodies (arrows), protein bodies (P). Bars (A–D) 5 μm, (E,F) 1 μm.
We considered whether oleosin might be limiting in M. orbicularis but we couldn’t obtain any evidence for that, consistent with the small size of the oil bodies. The transcription of a highly expressed oleosin gene (Medtr3g109190) we had previously identified as MtOLEOSIN2 (Wang et al., 2012) and present in both M. truncatula and M. orbicularis (Supplementary Table S3) was examined. There was clearly greater early expression in M. truncatula than in M. orbicularis. Then expression in M. orbicularis rapidly increased to M. truncatula levels or higher (Figure 5) and had not plateaued at the last measurement. Small oil bodies are usually associated with a low ratio of oil to oleosin (Shimada and Hara-Nishimura, 2010).
FIGURE 5.

Relative expression (relative to 10 DAA) of Mt-OLEOSIN2 (Medtr3g109190) during M. truncatula and M. orbicularis seed development. SE indicated and treatments with different letters are significantly different at the 0.05 probability level (n = 3).
Starch containing amyloplasts were observed in both species but more commonly in M. truncatula, reflecting starch levels (Figure 3B).
Cell Wall and Gross Seed Components Including Mucilage
The data obtained for the common storage products did not provide an explanation for the differences in the storage components of the two different Medicagos. Cell wall material is also a potential storage component (Daveby and Aman, 1993; Stombaugh et al., 2000, 2004) and the M. orbicularis cotyledon storage cells had a notably thicker cell wall (Figures 4A–D). The cell wall content of whole seeds (cell wall dry weight/seed dry weight) was 45.4% in M. truncatula and 50.0% in M. orbicularis while the cell wall content of the cotyledons alone was 41.7% in M. truncatula and 44.9% in M. orbicularis. In both cases, the cell wall content in M. truncatula was significantly lower compared to M. orbicularis (p < 0.05). This prompted an examination of the gross seed structure.
In legumes, the mature seed consists of an embryo, a reduced endosperm and a seed coat (Figures 6A,B). The seed coat, with several layers of cells is developed from the integuments of the ovule (Beeckman et al., 2000; Wang et al., 2012). In order to separate the seed coat from the cotyledons the seed was soaked in water. What we were surprised to observe was the extrusion of a gelatinous mass of mucilaginous material between the seed coat and the cotyledons, essentially enveloping the embryo. The mucilage (Figures 6C,D) characteristically stained with Ruthenium Red (Western et al., 2000; Verdier et al., 2013). The mucilage could be isolated as a gel-like sheet in which fibrillar material could be visualized (Figure 6C). After removal of most of the mucilage some traces of it still adhered to the inner seed coat in both species (Figure 6D). This differs from Arabidopsis with mucilage coming from the epidermal cell walls (Beeckman et al., 2000) which when hydrated forms a sphere around the seed. In Medicago the remnant endosperm forms a layer of cells surrounding the embryo (Wang and Grusak, 2005). The cell walls of these cells contain galactomannans (Reid, 1985; Edwards et al., 1999; Naoumkina et al., 2008), that releases as the mucilage gel forms between the embryo and the seed coat on hydration. This remnant endosperm layer can be isolated as a discrete envelope from around the embryo, staining with Ruthenium Red (Figures 6E,F).
FIGURE 6.
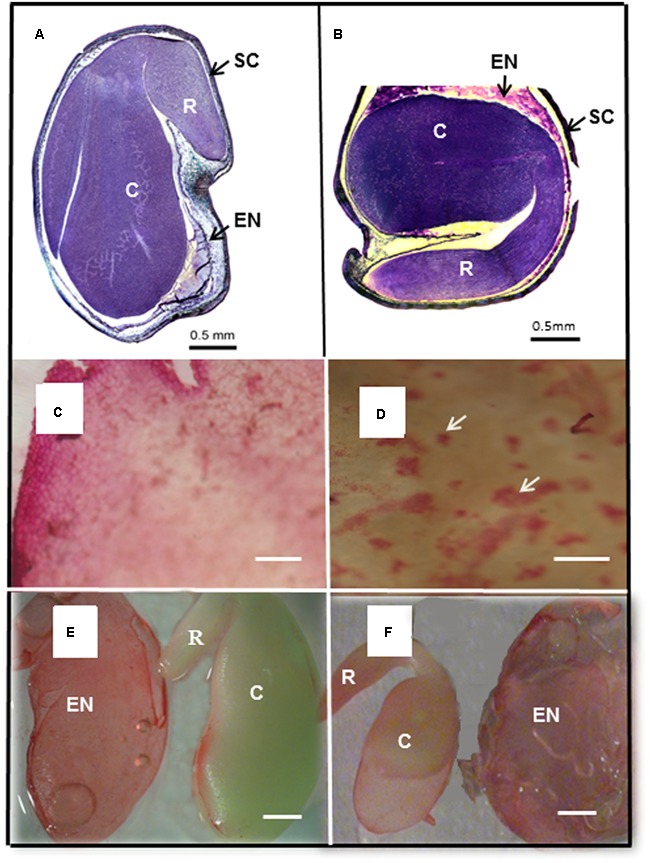
Seed morphology of M. truncatula and M. orbicularis and extruded endosperm mucilage. (A) Section (1 μm) through M. truncatula and (B) M. orbicularis seed stained with Toluidine Blue and Azur II. Seed coat (SC), endosperm (EN), embryonic cotyledons (C) and radicle (R). (C) Extruded endosperm mucilage with fine fibrous material visible and (D) mucilage (arrows) adhering to the inner side of the seed coat, stained with Ruthenium Red. (E) Isolated endosperm (EN) and embryo with cotyledons (C) and radicle (R) of M. truncatula and (F) M. orbicularis stained with Ruthenium Red. Bars (A,B,E,F) 0.5 mm, (C,D) 20 μm.
After soaking the seed in water the seed was separated into the seed coat, extruded mucilage and cotyledons and the dry weights determined. M. truncatula seed had 14.0% seed coat, 15.3% extruded seed mucilage and 70.7% embryo while M. orbicularis had 34.4% seed coat, 23.6% extruded seed mucilage and 42.0% embryo (Figure 7). M. orbicularis seed had a higher content of seed coat and mucilage but lower embryo dry weight compared to M. truncatula. Measurement of mucilage by chemical analysis in M. orbicularis, in an older study, reported 18.6% seed mucilage rich in galactomannans (Tookey et al., 1962). The endosperm of M. orbicularis also stains more pink with Toluidine Blue and AzurII (Figures 6A,B). A pink colouration is consistent with increased pectinaceous material, such as mucilage (Verdier et al., 2013). Given the thicker cotyledon storage cell walls and extruded mucilage derived from the endosperm cell walls of M. orbicularis (Figures 4A–D, 6, 7); we investigated whether the increased mucilage and cell wall material could be related to changes in the expression of genes potentially involved in the regulation of mucilage biosynthesis and cell wall pectin biosynthesis. Cell wall polysaccharides can have an important storage role in addition to their function in the primary cell wall (Buckeridge et al., 2000).
FIGURE 7.
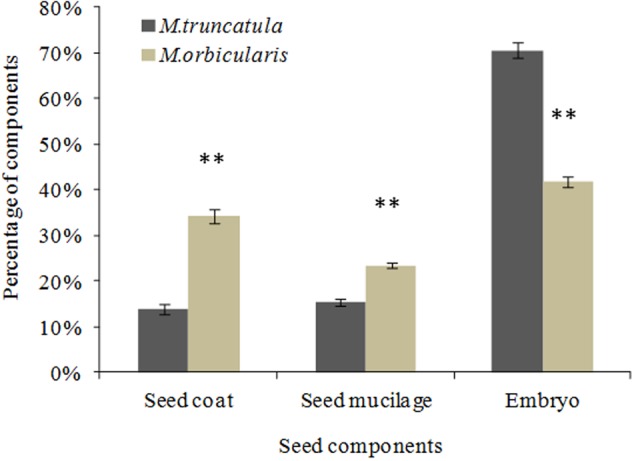
Proportion of seed dry weight of seed coat, extruded endosperm mucilage and embryo for M. truncatula and M. orbicularis. SE indicated and ∗∗p < 0.01 (n = 3).
Expression of GLABRA2-Like and GALACTURONOSYLTRANSFERASE Genes
There is an inverse relationship between mucilage synthesis and oil production when M. orbicularis and M. truncatula are compared. Similarly, Arabidopsis mutants for the GL2 transcription factor (Shi et al., 2012) have reduced mucilage and increased oil production. Could the expression of a GL2 transcription factor be a contributor to the differences in oil production between the two Medicago species?
We identified a GL2 homolog in M. truncatula (Supplementary Figures S1a,b and Table S1) and gene expression of MtGL2-like was compared between M. truncatula and M. orbicularis (Figure 8). Mt-GL2-like expression decreased from 10 DAA onward in both species. From 10 to 18 DAA, expression of MtGL2-like in M. truncatula was markedly lower compared to M. orbicularis while from 18 DAA onward, the expression was at a low level in both species. This suggests that the MtGL2-like transcription factor is potentially a negative regulator of oil production, as in Arabidopsis.
FIGURE 8.

Relative expression (relative to 10 DAA) of MtGLABRA2- like (Medtr2g101720) during M. truncatula and M. orbicularis seed development. SE indicated and treatments with different letters are significantly different at the 0.05 probability level (n = 3).
Given the increased cell wall material and the thicker cell walls in the M. orbicularis cotyledons, we also investigated the expression of GALACTURONOSYLTRANSFERASES (GAUTs) involved in the biosynthesis of the homogalacturonan pectin polysaccharide (Doong and Mohnen, 1998; Atmodjo et al., 2011), potentially a cell wall storage polysaccharide. We located seven GAUT genes (Supplementary Table S1) and studied their expression in the two Medicagos. Four of these genes showed enhanced expression in M. orbicularis (Figure 9) while the other three showed low expression with no difference between the two Medicagos. These data suggest cell wall pectins also contribute to the drain on carbon available for oil biosynthesis in M. orbicularis. In the case of M. orbicularis expression of both GL2 and GAUTs is high early in seed development, just prior to the laying down of oil bodies (Wang et al., 2012).
FIGURE 9.

Relative expression (relative to 10 DAA) of four different GAUT genes during M. truncatula and M. orbicularis seed development. (A) Mt-GAUT1 (Medtr7g075840). (B) Mt-GAUT3 (Medtr3g107930). (C) Mt-GAUT4 (Medtr 2g027740) (D) Mt-GAUT7 (Medtr7g055600). SE indicated and treatments with different letters are significantly different at the 0.05 probability level (n = 3).
Gene Expression of Transcriptional Regulators of Seed Oil Storage
If in M. orbicularis there was more carbon being directed into mucilage and pectin polysaccharides, and less into oil, then it would be expected that transcriptional regulators of oil biosynthesis and storage would subsequently be down-regulated. Expression of homologs of the master regulators MtLEC1-LIKE, MtFUS3 and MtABI3 in Medicago was therefore carried out. MtFUS3, was expressed at a lower level in both species (Figure 10C). The expression patterns of MtL1L and Mt-ABI3 are shown in Figures 10A,B. MtL1L expression from 10 to 18 DAA in M. orbicularis was lower than in M. truncatula, but similar after 18 DAA. Mt-ABI3 expression increased from 10 DAA and reached a plateau at 18 DAA in M. truncatula and at 14 DAA in M. orbicularis. ABI3 expression in M. orbicularis from 14 DAA onward was much lower than that in M. truncatula. The comparisons indicated that lower expression of MtL1L and MtABI3 was associated with the lower seed protein and oil in M. orbicularis, with lower MtL1L expression in the early to middle stage and lower ABI3 expression in the middle and late stage of seed development. The expression of the transcription factor MtWRI-like, which regulates fatty acid biosynthesis in Arabidopsis seed (To et al., 2012) was also reduced in M. orbicularis (Figure 10D).
FIGURE 10.

Relative expression (relative to 10 DAA) of four different transcription factors, linked to regulation of oil biosynthesis, during M. truncatula and M. orbicularis seed development. (A) MtLEC1-LIKE (Medtr4g133952), (B) MtABI3 (Medtr7g059330), (C) MtFUS3 (Medtr7g083700) and (D) MtWRINKLE-like (Medtr8g044070). SE indicated and treatments with different letters are significantly different at the 0.05 probability level (n = 3).
Discussion
The distinctive disparity of storage protein and oil content between M. truncatula and M. orbicularis, with their similar genetic background, provides a natural system for comparative studies of the regulation of seed storage accumulation (Tesfaye et al., 2006). The significant difference in oil between M. truncatula and M. orbicularis, shown in this study, is consistent with the report by Tonnet and Snudden (1974). This served as the basis of this investigation, following our earlier studies with M. truncatula on cellular and gene expression aspects of embryogenesis and storage filling (Wang et al., 2012; Kurdyukov et al., 2014).
The composition of major fatty acids was similar between M. truncatula and M. orbicularis (mainly palmitic, linolenic, α-linolenic, and oleic acids) but the quantity of individual fatty acids varied. Similar fatty acids were found by Djemel et al. (2005) in M. truncatula Jemalong 5. Lotus japonicus has higher stearate and linoleate levels than Medicago (Dam et al., 2009). Soybean seed oil is also primarily composed of five fatty acids (Kinney and Clemente, 2005) but with different levels, i.e., palmitic acid (∼13%), stearic acid (∼4%), oleic acid (∼18%), linoleic acid (∼55%), and linolenic acid (∼10%).
Medicago orbicularis has a much lower protein content compared to M. truncatula, but with a similar spectrum of storage proteins (also including lipoxygenases), though the contribution of the different genes differs (Table 1 and Supplementary Table S3). The starch content is minor in both M. truncatula and M. orbicularis, but is higher in M. truncatula. Why are all the common storage components – oil, protein and starch, lower in M. orbicularis?
Medicago orbicularis has a higher seed dry weight compared to M. truncatula. Nevertheless, M. truncatula has three times the protein and oil relative to M. orbicularis. Further investigation indicated that M. orbicularis has a much higher seed coat dry weight than M. truncatula (Figure 7). The seed coat does not store significant storage protein and oil which is characteristically stored in the cotyledons as shown in the ultrastructure images (Figure 4). If the seed coat is deducted, M. orbicularis has a lower embryo dry weight compared to M. truncatula (Figure 8). This contributes to the protein and oil differences between the embryos of both species. However, taking this into account, M. truncatula embryos still have approximately 80% more protein and oil relative to M. orbicularis. So what is happening in the M. orbicularis embryo to reduce seed oil?
What is of special interest is the mucilage which on hydration forms a layer between the cotyledons and seed coat. Commonly when a seed has substantive mucilage it forms a gel-like capsule surrounding the seed (Western, 2012) where it could mediate seed dispersal through adhesion or possibly facilitate seed hydration (Deng et al., 2012; Western, 2012). There is little recent information on legume mucilage in the literature (Moise et al., 2005; Western, 2012), including M. truncatula (Wang and Grusak, 2005; Verdier et al., 2013). However, a much older publication identified mucilage in M. orbicularis at a similar level to the accession used here (18.6% on a protein free basis, Tookey et al., 1962). Subsequent investigation by Tookey and Jones (1965) of 300 legume species found a negative correlation between oil plus protein and mucilage. Mucilage is carbon rich [a galactomannan in legumes (Tookey et al., 1962; Edwards et al., 1999; Naoumkina et al., 2008)] and the increased mucilage in M. orbicularis correlates with the decreased oil production relative to M. truncatula. Recently there has been more attention given to mucilage biochemistry and formation in Arabidopsis (e.g., Saez-Aguayo et al., 2013; Voiniciuc et al., 2013). Of particular interest is the demonstration by Shi et al. (2012) showing that the glabra2 mutant deficient in mucilage produced more oil. Consistent with this, M. truncatula with low expression of a GL2 homolog, has more oil and less mucilage than M. orbicularis. In M. orbicularis there is a surge of GL2 expression just prior to oil accumulation starting (Figures 5, 8). This is consistent with reducing the carbon flow into oil in M. orbicularis. It has to be noted that in Arabidopsis mucilage is rich in rhamnogalacturonan I (Macquet et al., 2007) while it is rich in galactomannans in legumes (Tookey et al., 1962; Edwards et al., 1999; Naoumkina et al., 2008). However, it is feasible that glycotransferases have similar regulation strategies in cell wall – linked polysaccharide assembly. It also seems likely that the increased biosynthesis of cell wall pectins (homogalacturonans) contributes to lower oil and protein in M. orbicularis cotyledons, based on the expression of GAUT genes. This is also consistent with total cell wall measurements, electron micrographs of cotyledon cells (Figure 4) and the storage role of cell wall polysaccharides (Buckeridge et al., 2000; Buckeridge, 2010).
We have focused on the embryo and the relationship to the extruded endosperm mucilage and cotyledon cell walls, but carbon also flows to the seed coat. In M. orbicularis the seed coat represents a greater proportion of the seed and as discussed above there are smaller embryos in M. orbicularis (Figure 7). There is also another aspect of the seed coat which differs between the two Medicagos. M. orbicularis has a very hard seed coat which is reddish brown, whereas M. truncatula has a light brown seed coat which is not as hard. This may reflect differences in phenylpropanoids which are negatively correlated with omega-3 fatty acids in flax (Ramsay et al., 2014). In Arabidopsis there is a negative correlation between proanthocyanidins and the amount of fatty acids (Wang et al., 2014). Further comparative metabolomic investigations between the Medicago seed coats is warranted, in conjunction with further studies on the cotyledons, as a precursor to further gene regulation studies.
The final storage outcome depends on the efficiency of the biosynthetic pathways once carbon is uploaded into the embryo. The master transcription factors AtLEC1/L1L, AtLEC2, AtFUS3, and AtABI3 have been shown to regulate seed protein and oil accumulation in Arabidopsis and homologs have been identified in M. truncatula (Verdier et al., 2008; Thompson et al., 2009; Tang et al., 2014). In situ hybridisation of MtABI3 in Kurdyukov et al. (2014) showed expression throughout the whole M. truncatula cotyledon, after the torpedo stage, supporting the role of the MtABI3 gene in M. truncatula. In M. orbicularis, MtFUS3 expression was relatively low and similar to M. truncatula. MtL1L and MtABI3 have reduced expression in M. orbicularis, consistent with lower accumulation of seed protein and oil in M. orbicularis (Kagaya et al., 2005; Tan et al., 2011). These data also support the role of these two transcription factors in seed storage in M. truncatula (Verdier et al., 2013). The expression of the MtWRI-like transcription factor, regulating fatty acid biosynthesis (To et al., 2012), also supports lower oil production in M. orbicularis.
The differences between M. truncatula and M. orbicularis reflects differences in ecology with M. orbicularis having a very thick seed coat (Patanè and Gresta, 2006), thick walled cells and increased mucilage which would facilitate hydration (Deng et al., 2012). These characteristics would be useful in more arid conditions.
The differences highlighted between M. orbicularis and M. truncatula provide additional approaches into carbon partitioning and the modification of oil content in legumes (Song et al., 2017). The direct testing of the transcription factor GL2 and the GAUT biosynthesis genes can be carried out in M. truncatula, using overexpression or gene knockout using CRISPR/Cas9 (Luo et al., 2016), where there are robust transformation protocols (Song et al., 2013).
Author Contributions
All authors contributed to the drafting/writing and interpretation of the work, and approve and are accountable for the final version of the manuscript. In addition, YS and RR designed experimentation. LH did the statistics. YS did the majority of experiments. X-DW did the cytology. YS, NS, and SW did the proteomics and MG supervised and interpreted the fatty acid analysis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to Melissa Fry for assistance in oil and fatty acid measurement and Joseph Enright for caring for plants in the glasshouse.
Funding. This research was supported by an Australian Research Council (ARC) Grant CEO348212 to RR.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02070/full#supplementary-material
References
- Atmodjo M. A., Sakuragi Y., Zhu X., Burrell A. J., Mohanty S. S., Atwood J. A., et al. (2011). Galacturonosyltransferase (GAUT) 1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan: galacturonosyltransferase complex. Proc. Natl. Acad. Sci. U.S.A. 108 20225–20230. 10.1073/pnas.1112816108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. A., Reeves G., Hetherington L., Aitken R. J. (2010). Analysis of proteomic changes associated with sperm capacitation through the combined use of IPG-strip pre-fractionation followed by RP chromatography LC-MS/MS analysis. Proteomics 10 482–495. 10.1002/pmic.200900574 [DOI] [PubMed] [Google Scholar]
- Barker D. G., Bianchi S., Blondon F., Dattée Y., Duc G., Essad S., et al. (1990). Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol. Biol. Rep. 8 40–49. 10.1007/BF02668879 22475377 [DOI] [Google Scholar]
- Beeckman T., De Rycke R., Viane R., Inzé D. (2000). Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 113 139–148. 10.1007/PL00013924 [DOI] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Buckeridge M. S. (2010). Seed cell wall storage polysaccharides: models to understand cell wall biosynthesis and degradation. Plant Physiol. 154 1017–1023. 10.1104/pp.110.158642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckeridge M. S., dos Santos H. P., Tiné M. A. S. (2000). Mobilisation of storage cell wall polysaccharides in seeds. Plant Physiol. Biochem. 38 141–156. 10.1016/S0981-9428(00)00162-5 [DOI] [Google Scholar]
- Cai Y., Goodman J. M., Pyc M., Mullen R. T., Dyer J. M., Chapman K. D. (2015). Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27 2616–2636. 10.1105/tpc.15.00588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente T. E., Cahoon E. B. (2009). Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol. 151 1030–1040. 10.1104/pp.109.146282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. R. (1999). Medicago truncatula — a model in the making! Commentary. Curr. Opin. Plant Biol. 2 301–304. 10.1016/S1369-5266(99)80053-3 [DOI] [PubMed] [Google Scholar]
- Crowe A. J., Abenes M., Plant A., Moloney M. M. (2000). The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci. 151 171–181. 10.1016/S0168-9452(99)00214-9 [DOI] [PubMed] [Google Scholar]
- Dam S., Laursen B. S., Ørnfelt J. H., Jochimsen B., Stærfeldt H. H., Friis C., et al. (2009). The proteome of seed development in the model legume Lotus japonicus. Plant Physiol. 149 1325–1340. 10.1104/pp.108.133405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daveby Y. D., Aman P. (1993). Chemical composition of certain dehulled legume seeds their hulls with special reference to carbohydrates. Swedish J. Agric. Res. 23 133–139. [Google Scholar]
- Deng W., Jeng D.-S., Toorop P. E., Squire G. R., Iannetta P. P. M. (2012). A mathematical model of mucilage expansion in myxospermous seeds of Capsella bursa-pastoris (shepherd’s purse). Ann. Bot. 109 419–427. 10.1093/aob/mcr296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djemel N., Guedon D., Lechevalier A., Salon C., Miquel M., Prosperi J. M., et al. (2005). Development and composition of the seeds of nine genotypes of the Medicago truncatula species complex. Plant Physiol. Biochem. 43 557–566. 10.1016/j.plaphy.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Doong R. L., Mohnen D. (1998). Solubilization and characterization of a galacturonosyltransferase that synthesizes the pectic polysaccharide homogalacturonan. Plant J. 13 363–374. 10.1046/j.1365-313X.1998.00042.x [DOI] [Google Scholar]
- Doyle J. J., Luckow M. A. (2003). The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 131 900–910. 10.1104/pp.102.018150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M. E., Dickson C. A., Chengappa S., Sidebottom C., Gidley M. J., Reid J. S. G. (1999). Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 19 691–697. 10.1046/j.1365-313x.1999.00566.x [DOI] [PubMed] [Google Scholar]
- Feussner I., Kühn H., Wasternack C. (2001). Lipoxygenase-dependent degradation of storage lipids. Trends Plant Sci. 6 268–273. 10.1016/S1360-1385(01)01950-1 [DOI] [PubMed] [Google Scholar]
- Folch J., Lees M., Sloane-Stanley G. H. (1957). A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226 497–509. [PubMed] [Google Scholar]
- Foster C. E., Martin T. M., Pauly M. (2010). Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. J. Visual. Exp. 37:1837. 10.3791/1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K., Le Signor C., Vandekerckhove J., Thompson R. D., Burstin J. (2003). Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol. 133 664–682. 10.1104/pp.103.025254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P., Beavis W. D., Brummer E. C., Shoemaker R. C., Stalker H. T., Weeden N. F., et al. (2005). Legumes as a model plant family. Genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol. 137 1228–1235. 10.1104/pp.105.060871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham P. H., Vance C. P. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131 872–877. 10.1104/pp.017004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills M. J. (2004). Control of storage-product synthesis in seeds. Curr. Opin. Plant Biol. 7 302–308. 10.1016/j.pbi.2004.03.003 [DOI] [PubMed] [Google Scholar]
- Jolivet P., Acevedo F., Boulard C., d’Andréa S., Faure J. D., Kohli A., et al. (2013). Crop seed oil bodies: from challenges in protein identification to an emerging picture of the oil body proteome. Proteomics 13 1836–1849. 10.1002/pmic.201200431 [DOI] [PubMed] [Google Scholar]
- Kagaya Y., Toyoshima R., Okuda R., Usui H., Yamamoto A., Hattori T. (2005). LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46 399–406. 10.1093/pcp/pci048 [DOI] [PubMed] [Google Scholar]
- Kinney A. J., Clemente T. E. (2005). Modifying soybean oil for enhanced performance in biodiesel blends. Fuel Process. Technol. 86 1137–1147. 10.1016/j.fuproc.2004.11.008 [DOI] [Google Scholar]
- Krishnan H. B. (2005). Engineering soybean for enhanced sulfur amino acid content. Crop Sci. 45 454–461. 10.2135/cropsci2005.0454 [DOI] [Google Scholar]
- Kurdyukov S., Song Y., Sheahan M. B., Rose R. J. (2014). Transcriptional regulation of early embryo development in the model legume Medicago truncatula. Plant Cell Rep. 33 349–362. 10.1007/s00299-013-1535-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Dai X., Liu T., Zhao P. X. (2012). LegumeIP: an integrative database for comparative genomics and transcriptomics of model legumes. Nucleic Acids Res. 40 D1221–D1229. 10.1093/nar/gkr939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Trick H. N. (2005). Rapid method for high-quality RNA isolation from seed endosperm containing high levels of starch. BioTechniques 38 872–876. 10.2144/05386BM05 [DOI] [PubMed] [Google Scholar]
- Luo M., Gilbert B., Ayliffe M. (2016). Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants. Plant Cell Rep. 35 1439–1450. 10.1007/s00299-016-1989-8 [DOI] [PubMed] [Google Scholar]
- Macquet A., Ralet M. C., Kronenberger J., Marion-Poll A., North H. M. (2007). In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol. 48 984–999. 10.1093/pcp/pcm068 [DOI] [PubMed] [Google Scholar]
- Mantiri F. R., Kurdyukov S., Lohar D. P., Sharopova N., Saeed N. A., Wang X. D., et al. (2008). The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol. 146 1622–1636. 10.1104/pp.107.110379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M. S., Dubreucq B., Miquel M., Caboche M., Lepiniec L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579 4666–4670. 10.1016/j.febslet.2005.07.037 [DOI] [PubMed] [Google Scholar]
- Moise J. A., Han S., Gudynaite-Savitch L., Johnson D. A., Miki B. L. (2005). Seed coats: structure, development, composition, and biotechnology. In Vitro Cell Dev. Biol. Plant 41 620–644. 10.1079/IVP2005686 [DOI] [Google Scholar]
- Molvig L., Tabe L. M., Eggum B. O., Moore A. E., Craig S., Spencer D., et al. (1997). Enhanced methionine levels and increased nutritive value of seeds of transgenic lupins (Lupinus angustifolius L.) expressing a sunflower seed albumin gene. Proc. Natl. Acad. Sci. U.S.A. 94 8393–8398. 10.1073/pnas.94.16.8393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Tan H., Zheng Q., Fu F., Liang Y., Zhang J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148 1042–1054. 10.1104/pp108.126342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina M., Vaghchhipawala S., Tang Y., Ben Y., Powell R. J., Dixon R. A. (2008). Metabolic and genetic perturbations accompany the modification of galactomannan in seeds of Medicago truncatula expressing mannan synthase from guar (Cyamopsis tetragonoloba L.). Plant Biotechnol. J. 6 619–631. 10.1111/j.1467-7652.2008.00345.x [DOI] [PubMed] [Google Scholar]
- Nolan K. E., Irwanto R. R., Rose R. J. (2003). Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol. 133 218–230. 10.1104/pp.103.020917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan K. E., Kurdyukov S., Rose R. J. (2009). Expression of the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) gene is associated with developmental change in the life cycle of the model legume Medicago truncatula. J. Exp. Bot. 60 1759–1771. 10.1093/jxb/erp046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patanè C., Gresta F. (2006). Germination of Astragalus hamosus and Medicago orbicularis as affected by seed-coat dormancy breaking techniques. J. Arid Environ. 67 165–173. 10.1016/j.jaridenv.2006.02.001 [DOI] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in realtime RT-PCR. Nucleic Acids Res. 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J. M., Deprez R. H., Moorman A. F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339 62–66. 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- Ramsay A., Fliniaux O., Fang J., Molinie R., Roscher A., Grand E., et al. (2014). Development of an NMR metabolomics-based tool for selection of flaxseed varieties. Metabolomics 10 1258–1267. 10.1007/s11306-014-0664-8 [DOI] [Google Scholar]
- Reid J. S. G. (1985). Cell wall storage carbohydrates in seeds – biochemistry of the seed “gums” and “hemicelluloses”. Adv. Bot. Res. 11 125–155. 10.1016/S0065-2296(08)60170-6 [DOI] [Google Scholar]
- Repetto O., Gallardo K. (2012). “Organelle proteomics of developing seeds: comparison with other plant tissue organelles,” in Seed Development: OMICS Technologies Toward Improvement of Seed Quality and Crop Yield eds Agrawal G. K., Rakwal R. (Dordrecht: Springer; ) 213–246. [Google Scholar]
- Roesler K., Shen B., Bermudez E., Li C., Hunt J., Damude H. G., et al. (2016). An improved variant of soybean type 1 diacylglycerol acyltransferase increases the oil content and decreases the soluble carbohydrate content of soybeans. Plant Physiol. 171 878–893. 10.1104/pp.16.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R. J. (2008). Medicago truncatula as a model for understanding plant interactions with other organisms, plant development and stress biology: past, present and future. Funct. Plant Biol. 35 253–264. 10.1071/FP07297 [DOI] [PubMed] [Google Scholar]
- Rose R. J., Wang X. D., Nolan K. E., Rolfe B. G. (2006). Root meristems in Medicago truncatula tissue culture arise from vascular-derived procambial-like cells in a process regulated by ethylene. J. Exp. Bot. 57 2227–2235. 10.1093/jxb/erj187 [DOI] [PubMed] [Google Scholar]
- Saez-Aguayo S., Ralet M. C., Berger A., Botran L., Ropartz D., Marion-Poll A., et al. (2013). PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25 308–323. 10.1105/tpc.112.106575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry P. R., Napier J. A., Tatham A. S. (1995). Seed storage proteins: structures and biosynthesis. Plant Cell 7 945–956. 10.1105/tpc.7.7.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Katavic V., Yu Y., Kunst L., Haughn G. (2012). Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J. 69 37–46. 10.1111/j.1365-313X.2011.04768.x [DOI] [PubMed] [Google Scholar]
- Shimada T. L., Hara-Nishimura I. (2010). Oil-body-membrane proteins and their physiological functions in plants. Biol. Pharm. Bull. 33 360–363. 10.1248/bpb.33.360 [DOI] [PubMed] [Google Scholar]
- Song Y., Nolan K. E., Rose R. J. (2013). “Stable transformation of Medicago truncatula cv. Jemalong for gene analysis,,” in Legume Genomics. Methods and Protocols, Methods in Molecular Biology Vol. 1069 ed. Rose R. J. (New York, NY: Springer; ) 203–214. [DOI] [PubMed] [Google Scholar]
- Song Y., Wang X.-D., Rose R. J. (2017). Oil body biogenesis and biotechnology in legume seeds. Plant Cell Rep. 36 1519–1532. 10.1007/s00299-017-2201-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stombaugh S. K., Jung H. G., Orf J. H., Somers D. A. (2000). Genotypic and environmental variation in soybean seed cell wall polysaccharides. Crop Sci. 40 408–412. 10.2135/cropsci2000.402408x [DOI] [Google Scholar]
- Stombaugh S. K., Orf J. H., Jung H. G., Chase K., Lark K. G., Somers D. A. (2004). Quantitative trait loci associated with cell wall polysaccharides in soybean seed. Crop Sci. 44 2101–2106. 10.2135/cropsci2004.2101 [DOI] [Google Scholar]
- Tan H., Yang X., Zhang F., Zheng X., Qu C., Mu J., et al. (2011). Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 156 1577–1588. 10.1104/pp.111.175000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Krishnakumar V., Bidwell S., Rosen B., Chan A., Zhou S., et al. (2014). An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15:312. 10.1186/1471-2164-15-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye M., Silverstein K. A., Bucciarelli B., Samac D. A., Vance C. P. (2006). The affymetrix Medicago gene chip array is applicable for transcript analysis of alfalfa (Medicago sativa). Funct. Plant Biol. 33 783–788. 10.1071/FP06065 [DOI] [PubMed] [Google Scholar]
- Thompson R., Burstin J., Gallardo K. (2009). Post-genomics studies of developmental processes in legume seeds. Plant Physiol. 151 1023–1029. 10.1104/pp.109.143966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A., Joubès J., Barthole G., Lécureuil A., Scagnelli A., Jasinski S., et al. (2012). WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24 5007–5023. 10.1105/tpc.112.106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnet M., Snudden P. (1974). Oil and protein content of the seeds of some pasture legumes. Aust. J. Agric. Res. 25 767–774. 10.1071/AR9740767 [DOI] [Google Scholar]
- Tookey H. L., Jones Q. (1965). New sources of water-soluble seed gums. Econ. Bot. 19 165–174. 10.1007/BF02862828 [DOI] [Google Scholar]
- Tookey H. L., Lohmar R. L., Wolff I. A., Jones Q. (1962). Seed polysaccharides, new sources of seed mucilages. J. Agric. Food Chem. 10 131–133. 10.1021/jf60120a014 [DOI] [Google Scholar]
- Udvardi M. K., Kakar K., Wandrey M., Montanari O., Murray J., Andriankaja A., et al. (2007). Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol. 144 538–549. 10.1104/pp.107.098061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J., Dessaint F., Schneider C., Abirached-Darmency M. A. (2013). A combined histology and transcriptome analysis unravels novel questions on Medicago truncatula seed coat. J. Exp. Bot. 64 459–470. 10.1093/jxb/ers304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J., Kakar K., Gallardo K., Le Signor C., Aubert G., Schlereth A., et al. (2008). Gene expression profiling of M. truncatula transcription factors identifies putative regulators of grain legume seed filling. Plant Mol. Biol. 67 567–580. 10.1007/s11103-008-9320-x [DOI] [PubMed] [Google Scholar]
- Voiniciuc C., Dean G. H., Griffiths J. S., Kirchsteiger K., Hwang Y. T., Gillett A., et al. (2013). FLYING SAUCER 1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in ARABIDOPSIS seed mucilage. Plant Cell 25 944–959. 10.1105/tpc.112.107888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Guo J., Lambert K. N., Lin Y. (2007). Developmental control of Arabidopsis seed oil biosynthesis. Planta 226 773–783. 10.1007/s00425-007-0524-0 [DOI] [PubMed] [Google Scholar]
- Wang H. L., Grusak M. A. (2005). Structure and development of Medicago truncatula pod wall and seed coat. Ann. Bot. 95 737–747. 10.1093/aob/mci080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. L., Domoney C., Hedley C. L., Casey R., Grusak M. A. (2003). Can we improve the nutritional quality of legume seeds? Plant Physiol. 131 886–891. 10.1104/pp.102.017665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-D., Song Y., Sheahan M. B., Garg M. L., Rose R. J. (2012). From embryo sac to oil and protein bodies: embryo development in the model legume Medicago truncatula. New Phytol. 193 327–338. 10.1111/j.1469-8137.2011.03925.x [DOI] [PubMed] [Google Scholar]
- Wang Z., Chen M., Chen T., Xuan L., Li Z., Du X., et al. (2014). TRANSPARENT TESTA2 regulates embryonic fatty acid biosynthesis by targeting FUSCA3 during the early developmental stage of Arabidopsis seeds. Plant J. 77 757–769. 10.1111/tpj.12426 [DOI] [PubMed] [Google Scholar]
- Weber H., Borisjuk L., Wobus U. (2005). Molecular physiology of legume seed development. Annu. Rev. Plant Biol. 56 253–279. 10.1146/annurev.arplant.56.032604.144201 [DOI] [PubMed] [Google Scholar]
- Western T. L. (2012). The sticky tale of seed coat mucilages: production, genetics, and role in seed germination and dispersal. Seed Sci. Res. 22 1–25. 10.1017/S0960258511000249 [DOI] [Google Scholar]
- Western T. L., Skinner D. J., Haughn G. W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122 345–355. 10.1104/pp.122.2.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. D., Debellé F., Oldroyd G. E., Geurts R., Cannon S. B., Udvardi M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480 520–524. 10.1038/nature10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


