Abstract
Significant endocrine therapy-resistant tumor proliferation is present in ≥20% of estrogen receptor positive (ER+) primary breast cancers and is associated with disease recurrence and death. Here, we uncover a link between intrinsic endocrine therapy resistance and dysregulation of the MutL mismatch repair complex (MLH1/3, PMS1/2), and demonstrate a direct role for MutL complex loss in resistance to all classes of endocrine therapy. We find that MutL deficiency in ER+ breast cancer abrogates Chk2-mediated inhibition of CDK4, a prerequisite for endocrine therapy responsiveness. Consequently, CDK4/6 inhibitors (CDK4/6i) remain effective in MutL-defective ER+ breast cancer cells. These observations are supported by data from a clinical trial where a CDK4/6i was found to strongly inhibit AI-resistant proliferation of MutL-defective tumors. These data suggest that diagnostic markers of MutL deficiency could be used to direct adjuvant CDK4/6i to a population of breast cancer patients who exhibit marked resistance to the current standard of care.
Keywords: Mismatch repair, estrogen receptor, breast cancer, CDK4/6 inhibitor
Introduction
Resistance to endocrine therapy remains a significant cause of death for the ~175,000 women diagnosed each year with Estrogen Receptor positive (ER+) breast cancer(1). While some headway has been made in understanding underlying mechanisms, the majority of cases remain unexplained(1). Traditionally, growth factor receptor pathway activation has been implicated as one underlying mechanism of acquired resistance to endocrine therapy. Recently, studies of endocrine therapy resistance in metastatic breast cancer, where patients have been exposed to long periods of treatment, have identified the acquisition of mutations in the estrogen receptor gene (ESR1) causing ligand-independent activation and aromatase inhibitor (AI) resistance to constitute an alternative underlying mechanism for endocrine resistance in the advanced disease setting(2–4). However, ER+ primary breast cancer can also be endocrine therapy resistant at diagnosis (intrinsic resistance) where ESR1 mutations cannot be the sole explanation, as they are too rare. Intrinsic endocrine resistance is easily diagnosed, based on failure to fully suppress Ki67 (proliferation marker) in tumor biopsies after 2–4 weeks of neoadjuvant endocrine treatment(5) but is relatively understudied. Intrinsic resistance, as assayed by Ki67, occurs in at least 20% of ER+ HER2− tumors and is an established poor prognosis marker. Unfortunately these tumors also often also fail to respond well when switched to neoadjuvant chemotherapy(6, 7). Consequently, patients diagnosed with intrinsically endocrine resistant ER+HER2− primary breast cancer suffer high rates of relapse and death.
The recently noted correlation between high mutation load and poor prognosis in ER+ breast cancer suggests that defects in DNA damage repair (DDR) genes may constitute an under-explored driver of endocrine therapy resistance(8). While the best-understood DDR defect in breast cancer concerns homologous recombination (HR) deficiency, due to BRCA1/2 loss(9), this mechanism is less pertinent to ER+ disease, which is largely HR-competent. A few preliminary epidemiological studies have noted possible roles for Base Excision Repair (BER) and Nucleotide Excision Repair (NER) in ER+ disease pathogenesis(10, 11). However, our investigations, reported here, establish a new role for a subset of mismatch repair (MMR) pathway components in regulating intrinsic endocrine therapy resistance in ER+ disease. Using in vitro and in vivo models, we demonstrate that defects in MutL complex genes (MLH1, PMS1, PMS2) directly induce endocrine therapy resistance because an intact MutL complex enables an endocrine therapy activated ATM/CHEK2-dependent cell cycle checkpoint control to suppress CDK4 activity. We also use preclinical and neoadjuvant clinical trial data to demonstrate that deregulated CDK4 in MutL-deficient tumors remains targetable by CDK4/6 inhibition (CDK4/6i), explaining the activity of these agents in a subset of endocrine therapy resistant disease and thereby suggesting a new class of predictive markers for CDK4/6 targeted drugs.
Results
Role of DNA damage repair dysregulation in ER+ breast cancer
Since high mutation load is a marker of poor prognosis in ER+ breast cancer(8), we first assessed correlations between mutation load and incidence of non-silent mutations in pathway-unique genes of the five major DDR pathways: MMR, BER, NER, non-homologous end joining (NHEJ) and HR in ER+ human breast tumors. We chose two clinical datasets with whole exome sequencing data for this analysis: (i) several neoadjuvant aromatase inhibitor (AI) trials, collectively termed NeoAI, (Z1031, a study from the American College of Surgeons Oncology Group, which is now part of the Alliance for Clinical Trials in Oncology(6), and the multi-institutional Preoperative Letrozole Phase 2 study (POL)(12)) and (ii) the TCGA dataset(13). Only mutations in MMR genes were significantly associated with increased tumor mutation load in both clinical datasets (Fig 1a–b). Since MMR genes are known to be dysregulated at the gene expression level, the impact of low MMR gene expression (using TCGA definitions of mean-1.5× standard deviation to avoid threshold training) on mutation load was also assessed in both datasets and a correlation between this expanded set of MMR defective tumors and mutation load was confirmed (Fig S1a–b).
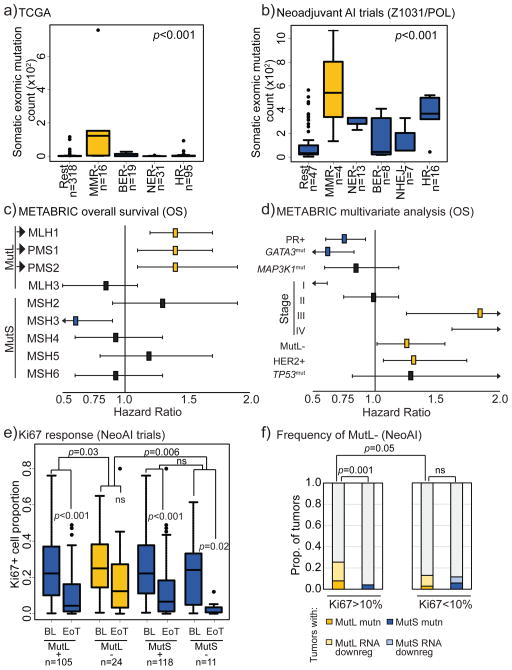
Figure 1. Mismatch repair dysregulation associates with high mutation load and poor clinical outcome in patients with ER+ breast cancer.
a+b) ER+ tumors with non-synonymous mutations in pathway-unique MMR genes from TCGA (a) and NeoAI (b) datasets have increased overall exomic mutation load compared to tumors without. BER, base excision repair; NER, nucleotide excision repair; HR, homologous recombination; NHEJ, non-homologous end joining; Rest, all others. ANOVA with Tukey comparison determined p-values. c+d) Forest plots demonstrating that ER+ tumors with low MutL gene mRNA (<mean-1.5xStDev) in METABRIC associate selectively with poor overall survival (c) and that low MutL mRNA is an independent prognosticator of poor survival in ER+ breast cancer (d). Log rank test was used to determine significance and Cox Regression Proportional Hazards for multivariate analysis. Benjamini-Hochberg correction was applied for multiple comparisons. e) Box plots demonstrating that ER+ tumors from NeoAI (Z1031, POL, NeoPalAna) with mutations in and/or low expression of MutL genes are significantly more likely to be resistant to endocrine therapy as determined by change in Ki67 levels on AI treatment. Friedman Rank Sum test for repeated measures determined p-values for pair-wise comparisons, and Kruskal Wallis test determined p-value for between-group comparisons. f) Stacked column graph depicting frequency of incidence of MutL dysregulation in ER+ tumors from NeoAI. Fisher’s Exact test determined p-values. Accompanying information presented in Figure S1 and Table 1. Horizontal lines indicate mean and error bars denote standard error for box plots.
Mismatch repair dysregulation and endocrine therapy resistance in ER+ breast cancer patients
Next, the effect of low RNA levels of each unique MMR gene on ER+ breast cancer outcome of patients treated with hormone therapy (tamoxifen or AI) was assessed in the METABRIC dataset, chosen for its large sample size and long-term clinical follow-up(14). Analysis in this dataset using the same cut-point for low expression drawn from the TCGA analysis (Fig S1a–b) revealed significant association between low RNA and poor overall (Fig 1c) and disease-free survival (Fig S1c–d) in 3/8 canonical MMR genes: MLH1, PMS1 and PMS2. The poor prognostic effects were independent for each gene and were significant after correction for multiple testing (Table 1).
Table 1.
Proportional hazard calculations for each of the three mismatch repair genes with significant effect on clinical outcome measured by overall survival: MLH1, PMS1 and PMS2. Cox regression was used to calculate p-values.
| Factor | Hazard ratio | Confidence interval | p-value |
|---|---|---|---|
| MLH1high | -- | -- | |
| MLH1low | 1.61 | 1.07–2.43 | 0.02* |
| PR− | -- | -- | |
| PR+ | 0.78 | 0.54–1.13 | 0.19 |
| ERBB2− | -- | -- | |
| ERBB2+ | 1.31 | 0.87–1.99 | 0.19 |
| Stage0 | -- | -- | |
| StageI | 0.47 | 0.26–0.85 | 0.01* |
| StageII | 0.87 | 0.59–1.28 | 0.48 |
| StageIII | 1.46 | 0.68–3.13 | 0.33 |
| StageIV | 2.24 | 0.31–16.48 | 0.43 |
| TP53WT | -- | -- | |
| TP53mut | 1.97 | 1.01–3.85 | 0.04* |
| PMS1high | -- | -- | |
| PMS1low | 1.37 | 1.01–1.86 | 0.04* |
| PR− | -- | -- | |
| PR+ | 0.74 | 0.54–1.02 | 0.07′ |
| ERBB2− | -- | -- | |
| ERBB2+ | 1.46 | 0.2–10.69 | 0.71 |
| Stage0 | -- | -- | |
| StageI | 0.52 | 0.33–0.83 | 0.005* |
| StageII | 0.9 | 0.63–1.29 | 0.58 |
| StageIII | 1.8 | 0.89–3.64 | 0.1′ |
| StageIV | 2.03 | 0.28–14.76 | 0.48 |
| TP53WT | -- | -- | |
| TP53mut | 1.96 | 1.07–3.57 | 0.03* |
| PMS2high | -- | -- | |
| PMS2low | 1.86 | 1.201–2.699 | 0.004* |
| PR− | -- | -- | |
| PR+ | 0.79 | 0.55–1.15 | 0.23 |
| ERBB2− | -- | -- | |
| ERBB2+ | 1.37 | 0.91–2.09 | 0.13 |
| Stage0 | -- | -- | |
| StageI | 0.5 | 0.28–0.92 | 0.03* |
| StageII | 0.94 | 0.63–1.39 | 0.76 |
| StageIII | 1.79 | 0.82–3.91 | 0.14 |
| StageIV | 2.37 | 0.32–17.43 | 0.4 |
| TP53WT | -- | -- | |
| TP53mut | 2.11 | 1.07–4.14 | 0.03* |
p<0.05
p≤0.1
MLH1, PMS1 and PMS2, along with MLH3, constitute the MutL complex of the MMR pathway. Therefore, the finding that only these three genes negatively affect overall and disease-free survival suggested a specific role for the MutL complex in poor outcome ER+ disease. The MutL complex is recruited to DNA mismatches by the MutS complex (MSH genes), whereupon it performs two functions: first, to recruit repair proteins to the mismatched nucleotide and second, to activate ATM/Chk2 signaling in the event of unsuccessful repair(15). The coordinating protein in the MutL complex is MLH1 which, when heterodimerized with PMS1/2, forms a stable complex that is translocated to the nucleus(12). In contrast, MLH3 is largely involved in post-meiotic recombination although it can play a compensatory role in MMR in the absence of MLH1(16), possibly explaining the more equivocal association of MLH3 with clinical outcome in the METABRIC database. Under-expression of any gene serving the MutL complex was used to define a MutL low signature (MutL−) versus a MutL normal group (MutL+). The adverse effects of MutL− status on prognosis was found to be independent of clinical variables (progresterone receptor (PR) positivity, HER2 positivity, tumor stage) as well as mutations known to affect breast cancer prognosis (MAP3K1, GATA3, TP53) (Fig 1d). Interestingly the hazard ratio for MutL− was comparable to the adverse effects of HER2 amplification (Fig 1d+S1e).
Next, MutL selectivity was assessed in the TCGA dataset where ER+ tumors (treated with tamoxifen or AI) with mutations and/or low mRNA levels for any one of MLH1, MLH3, PMS1 or PMS2 (MutL− tumors) also associated with significantly worse survival than tumors without MutL dysregulation (MutL+, Fig S1f). Of note, the association of MutL downregulation with poor overall survival is specific to ER+ breast cancer, with no significant association observed in either METABRIC (p=0.75) or TCGA (p=0.5) datasets in ER− breast cancer cohorts.
To examine whether MutL defects can predict intrinsic response to endocrine therapy (in the form of AI) in primary breast cancer, MutL status was analyzed in the NeoAI dataset (including data from the most recent clinical trial, NeoPalAna(17)) focusing on cases where paired Ki67 data at diagnosis and after 2–4 weeks of AI, as well as exome sequencing and gene expression data, were available. Using the same mutation/expression-based definitions used in the TCGA and METABRIC analyses, MutL− ER+ tumors (n=24) showed no significant fall in Ki67 levels despite AI treatment and less treatment-induced change in Ki67 compared to “MutL+” tumors without MutL under-expression or mutation (p=0.03, Wilcoxon Rank Sum test, Fig 1e). Interestingly, 11 tumors assigned as MutS- by mutation or under-expression of MutS were largely endocrine therapy sensitive with a significant fall in Ki67 (p=0.02) and no difference in Ki67 change compared to the MutS+ group (Fig 1e).
To estimate the relative frequency of MutL− in endocrine therapy resistant ER+ breast cancers, we examined the incidence of either non-synonymous mutations or low mRNA levels in the subset of AI resistant tumors from NeoAI (Ki67>10% on AI treatment). In this setting MutL− status accounted for 27% of poor responders, and MutL, but not MutS dysregulation (4% of poor responders), was enriched in endocrine therapy resistant tumors (Fig 1f). Moreover, MutL dysregulation was enriched in endocrine therapy resistant relative to sensitive tumors in the TCGA dataset as well (p<0.001, MutL− occurring in 25% of tumors from patients who died within 5 years of diagnosis).
MutL complex inactivation causes intrinsic and class-independent endocrine therapy resistance in ER+ breast cancer cells
ER+ breast cancer cell lines were stably transfected with shRNA to suppress expression from each of the three MutL genes most strongly linked to poor clinical outcome: MLH1, PMS1 and PMS2, and one MutS gene, MSH2, to serve as negative control (knockdown validation in Fig S2a–b). As previously published(16, 18, 19), PMS1 silencing also induced loss of stability of MLH1 protein (Fig S2b). Suppression of any one of the three MutL genes in either MCF7 or T47D cells induced resistance to all classes of endocrine interventions: i.e. estrogen deprivation, a surrogate for AI exposure (Fig 2a+S2c), fulvestrant and tamoxifen (Fig 2b+S2d) within a week of administration. In contrast, suppression of MSH2 did not affect response to estrogen stimulation (Fig 2a) or to fulvestrant-mediated ER degradation (Fig S2e). All further experiments were conducted using pharmacologically relevant 100nM doses of fulvestrant and tamoxifen, and largely in shMLH1 (MLH1−) cells because of the central importance of MLH1 to MutL complex formation.
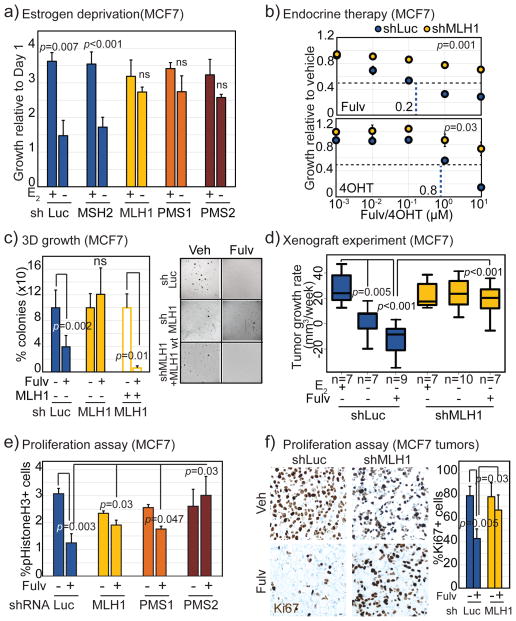
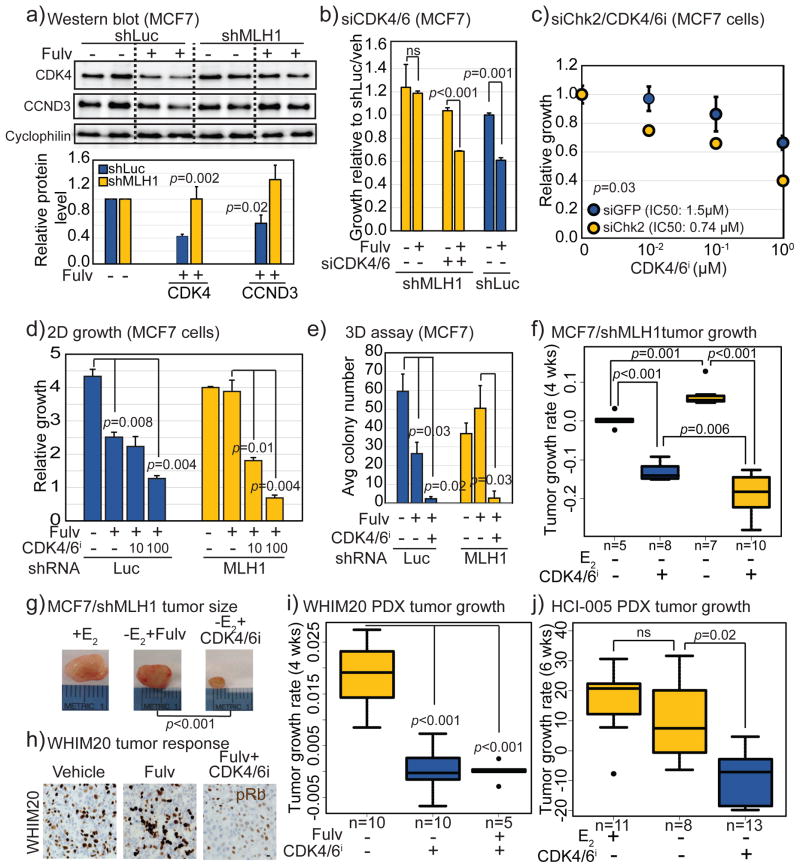
Figure 2. MutL deficiency induces endocrine therapy resistance in ER+ breast cancer cells by preventing proliferative block.
a) Bar graph depicting growth of MCF7 cells stably expressing shRNA against luciferase (control), MSH2, MLH1, PMS1 or PMS2, as indicated, at day 4 relative to day 1 after addition of 10nM beta-estradiol to cells grown in charcoal stripped serum for 4 days. Accompanying knockdown validation in Fig S2a+b, and estradiol dose curve in Fig S2c. b) Dose curves demonstrating increased growth of MCF7 cells stably expressing shRNA against MLH1 in response to increasing doses of 4-hydroxy tamoxifen (4-OHT) and fulvestrant (Fulv) relative to cells expressing shRNA against luciferase control. P-value describes significance of difference between slopes. c) Bar graph depicting percentage of colonies in soft agar (3D growth assay) generated from isotypic MCF7 cells with specified genotype after 4–6 weeks of endocrine treatment with accompanying representative images. Validation of rescue and 2D growth assay in Fig S3a. d) Boxplots depicting differences in tumor growth rate (calculated as slope from growth curve of each tumor) of MutL proficient (shLuc) vs deficient (shMLH1) MCF7 cells, in presence of estrogen (E2+) or placebo (E2−) in drinking water with or without weekly fulvestrant administration as indicated. Differences between the E2+ and E2−, and between the E2+ and E2−Fulv+ groups of shMLH1 tumors were non-significant (p>0.5). e+f) Bar graphs depict fold change in percentage of pHistoneH3+ (pH3, mitotic, e) and Ki67+ (proliferation marker, f) cells in MutL proficient (shLuc) and deficient (shMLH1, shPMS1, shPMS2, as indicated) ER+ breast cancer cells and tumors with representative images. Scale bar=20μm. Associated data for T47D cells in Figs S2+3. For all graphs, error bars describe standard deviation and Student’s t-test determined p-values.
Pooled CRISPR-mediated disruption of each of MLH1, PMS1 and PMS2 genes in both MCF7 and T47D cell lines orthogonally supported a causal association between MutL− and response to fulvestrant (Fig S2f–g) and to estrogen stimulation (Fig S2h). Of note, CRISPR-mediated knockdown of MutL genes was less well tolerated in T47D than in MCF7 cells (Fig S2i), and PMS2 knockdown by shRNA or CRISPR was not well tolerated in either cell line (Fig S2b+i). These data might explain why PMS2 somatic mutations in human breast tumors are characteristically missense as complete loss of function may reduce cell viability, but may also be a reflection of the low baseline levels of PMS2 in MCF7 and T47D cells (approximately 30-fold lower than other MutL genes) rendering the establishment of knockdown efficacy challenging.
MCF7 cells with MLH1 down-regulation demonstrated fulvestrant-resistant growth in soft agar (Fig 2c), and unimpeded xenograft tumor growth after estrogen deprivation, as well as fulvestrant resistance (Fig 2d). Critically, introduction of shRNA-resistant MLH1 cDNA into MLH1− cells (validated in Fig S3a) restored sensitivity to fulvestrant under both 2D (Fig S3a) and 3D growth conditions (Fig 2c).
MutL dysregulation in patient-derived xenograft models
To confirm in vitro findings in a human-in-mouse breast cancer model, the occurrence of MutL gene mutation in patient-derived xenograft (PDX) tumors was examined. A missense mutation (E5K) in the MutL gene, PMS2, previously described in a hypermutator, MMR-deficient colorectal cancer cell line (HT-115) (CoSMiC), was identified in WHIM20, an ER+ PDX(4) with high mutation load (Fig S3d). Notably, WHIM20 exhibits resistance to both fulvestrant treatment (Fig S3b) and estrogen deprivation(4). A second MutL mutant ER+ PDX model, HCI-005(20), was also identified, harboring a frameshift mutation (L160fs) in PMS1 and also associated with high mutation load (Fig S3c). HCI-005 tumors are less estrogen dependent than other ER+ PDX, demonstrating 100% tumor outgrowth in ovariectomized mice(20). These data provide additional support for a role for MutL− in endocrine response as well as model systems to study MutL-deficient breast cancer.
MutL loss decreases Chk2 activation in response to endocrine treatment
Resistance to fulvestrant treatment in MutL− cells was not due to a failure to degrade ER given the comparable levels of ER and ER downstream effectors (PGR and GREB1) at both RNA and protein level before and after fulvestrant treatment in MCF7 shLuc and shMLH1 cells and xenograft tumors (Fig S3e). Additionally, comparable RNA levels for a panel of ER target genes including PGR and GREB1 were observed between MutL− and MutL+ human tumors from the TCGA dataset (Fig S3f) indicating that ER transcriptional activity is not significantly affected by defective MutL function. Since MLH1 re-expression rescued the endocrine therapy sensitive phenotype of MLH1− cells, loss of MutL competence appears to have a direct and causal role in intrinsic endocrine therapy resistance rather than an indirect role through the induction of genome instability and evolution of secondary mutations. This conclusion is supported by the rapidity by which resistance arises upon disruption of MutL function, since clonal selection of resistance mutations usually takes months or even years.
To understand the mechanism whereby MutL gene loss causes intrinsic endocrine therapy resistance, cell cycle activity after fulvestrant treatment was determined by immunofluorescence for pHistoneH3 (mitotic marker) staining in vitro (Fig 2e) and immunohistochemistry for Ki67 (proliferative marker) in xenograft tumors (Fig 2f). Although basal cell cycle profiles were comparable between shLuc and shMLH1 cells (Fig S3g), fulvestrant significantly inhibited proliferation of shLuc but not of shMLH1, shPMS1 or shPMS2 MCF7 and T47D cells (Fig 2e+S3h). Similarly, MCF7 shMLH1 xenograft tumors demonstrated no measurable inhibition of proliferation after fulvestrant treatment, unlike their shLuc counterparts (Fig 2f). These data suggest that loss of any member of the MutL complex prevents the anti-proliferative effects of ER blockade in ER+ breast cancer cells.
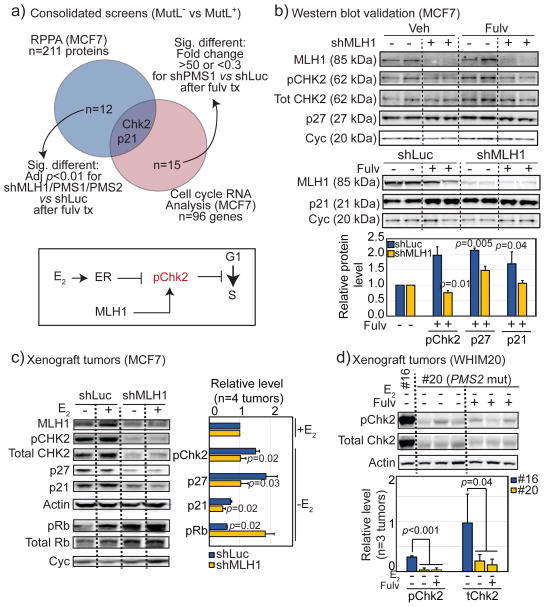
Two screens were performed to explore underlying mechanisms. The first approach used reverse phase protein array (RPPA) analysis and the second compared gene expression of cell cycle related genes in MCF7 MutL+ and MutL− cells. For both screens, cells were treated with either control or fulvestrant for 48 hours before harvest. Analysis of results identified that of 211 proteins screened in the RPPA array, the levels of 12 proteins were significantly different (depicted in Fig S4a), and of the 96 gene RNA levels assayed by qRT-PCR, 15 were significantly differentially regulated (shown in Fig S4b) by fulvestrant treatment in MutL− relative to MutL+ cells. Of these factors, Chk2 and p21 were the only two that were significantly down-regulated in both screens in MutL− vs MutL+ cells after fulvestrant treatment (Fig 3a).
Figure 3. MutL deficiency downregulates Chk2.
a) Venn diagram describing the intersection of proteins identified by RPPA as differentially regulated in MutL− (intersection of shMLH1, shPMS1 and shPMS2) vs MutL+ (shLuc) MCF7 cells, and genes with significantly dysregulated expression levels in MutL− (shPMS1) vs MutL+ (shLuc) MCF7 cells, after fulvestrant treatment. Inset of pathway based on significantly dysregulated genes identified. All significant protein level and RNA level changes are depicted in Fig S4a+b. b–d) Western blot validation of pChk2, p21 and p27 levels in MCF7 MutL+ (shLuc) and MutL− (shMLH1) cells grown in vitro (b) and in vivo as xenograft tumors (c), and in patient derived MutL− (PMS2 mutant, WHIM20) and MutL+ (WHIM16) xenografts (d), with accompanying quantification. Additional Western blot validation in MCF7 shPMS2 cells and in other PDX tumors in Fig S4c–e. Quantification performed using ImageLab software. Treatment as indicated. For all graphs, columns represent the mean, error bars describe standard deviation and Student’s t-test determined p-values.
MutL complex genes are known to activate Chk2 during mismatch repair(21). We therefore proceeded to evaluate the role of Chk2 in mediating intrinsic endocrine therapy resistance. Down-regulation of Chk2 in MutL− relative to MutL+ cells after fulvestrant treatment was observed in vitro by assessing levels of pChk2 and its downstream effectors, p21 and p27, in MCF7/shLuc relative to either shMLH1 (Fig 3b) or shPMS2 cells (Fig S4c). In addition to down-regulation of pChk2 and more modestly, of p27/p21, up-regulation of p-Rb were observed in MLH1− versus MLH1+ MCF7 xenograft tumors grown in the absence of estrogen (Fig 3c). In spite of the heterogeneity of PDX tumors(22), WHIM20 tumors (#20, PMS2 E5K) also exhibited an almost complete inhibition of both total and p-Chk2 protein levels after estrogen deprivation, and fulvestrant treatment, in contrast to a MutL+, ER+ PDX tumor, WHIM 16 (Fig 3d). Both RPPA (Fig S4d) and mass spectrometry-based phosphoproteomics(22) (Fig S4e) across PDX lines confirmed that WHIM20 tumors (-E2) have down-regulated p-Chk2, and p-p27 at multiple phosphorylation sites. Finally, pChk2 was significantly downregulated in primary MutL− ER+ breast tumors in TCGA RPPA data (Fig S5a), and mRNA from CDKN1A (p21) was upregulated after AI treatment in MutL+, but not in MutL−, tumors from the NeoAI dataset (Fig S5b).
ATM/Chk2 activation by the MutL complex is required for endocrine therapy response
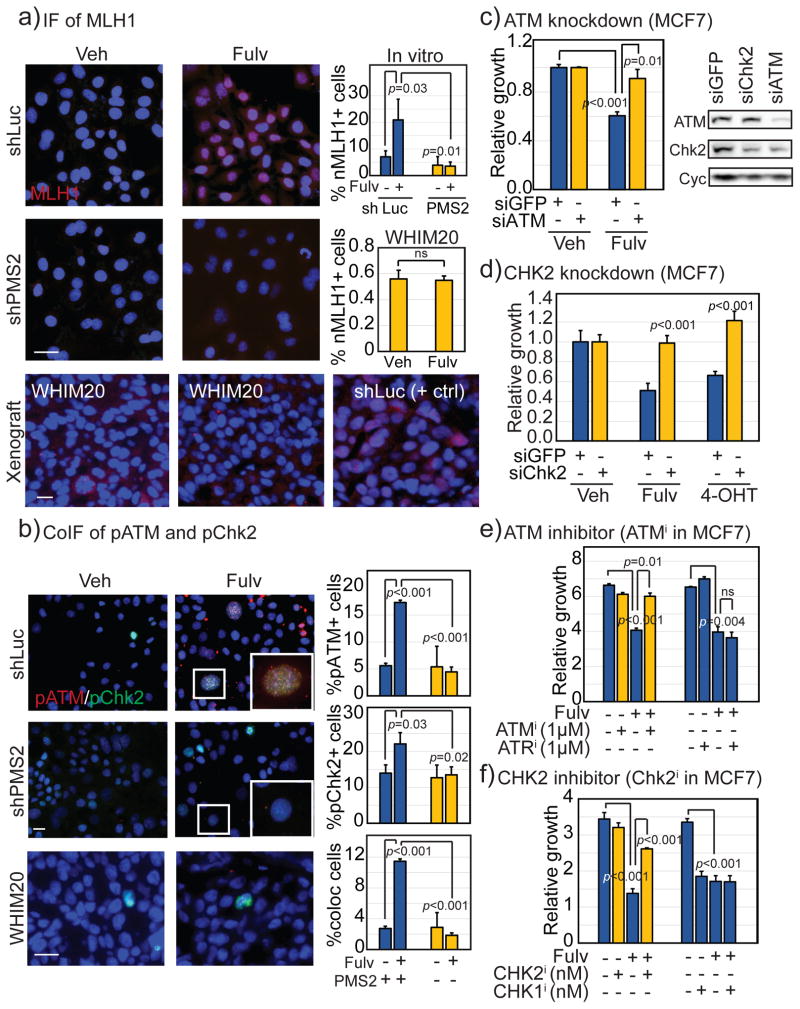
The data presented thus far are consistent with previously reported links between MMR signaling and Chk2 activation(23), and between ER signaling and Chk2(24), presenting the hypothesis that MutL− ER+ breast tumors are less able to phosphorylate Chk2 in response to endocrine therapy thereby decreasing the strength of treatment-associated cell cycle arrest(25). Previously published research suggests that MLH1 acts as a scaffold enabling ATM auto-phosphorylation and subsequent phosphorylation of Chk2(21). To test this association in ER+ breast cancer cells, cellular localization of MLH1 and p-Chk2 were analyzed after fulvestrant treatment in MCF7 MutL+ (shLuc and shMSH2) and MutL− (shMLH1 and shPMS2) cells. In response to fulvestrant treatment, both MutL+ cell lines significantly increased MLH1 translocation to the nucleus where co-localization with p-Chk2 was observed (Fig 4a+S5c). Unsurprisingly, shMLH1 cells did not upregulate MLH1 nuclear translocation (Fig S5c), but neither did shPMS2 cells (Fig 4a+S5c), supporting previously published evidence that the MutL complex requires heterodimerization to facilitate nuclear translocation(18). In corroboration, WHIM20 (PMS2 mutant) tumors had cytoplasmic but no detectable nuclear MLH1 on fulvestrant treatment, in contrast to MCF7 MutL+ xenograft tumors (Fig 4a). Consistent with these data, nuclear co-localization of p-ATM and p-Chk2 increased >4-fold (p<0.001) in MutL+ cells after fulvestrant treatment but demonstrated no perceivable increase in MutL− (0.6-fold) MCF7 cells (Fig 4b). The lack of p-ATM/p-Chk2 foci in response to fulvestrant was also observed in MutL− WHIM20 tumors where pATM was virtually undetectable, although attenuated pChk2 nuclear foci were detected (Fig 4b).
Figure 4. ER+ breast cancer cells require Chk2 activation for sensitivity to endocrine therapy.
a+b) Immunofluorescence depicting relative levels and cellular localization of MLH1 (a), pChk2 and pATM (b) in MutL+ (shLuc), MutL− (shPMS2) cells, and MutL+ (MCF7 shLuc) and MutL− (WHIM20, PMS2 mutant) xenograft tumors grown in the presence of control or fulvestrant with accompanying quantification. Scale bars: 20μm. Accompanying IF of pChk2 in other MutL− cell lines presented in Fig S5c. c–f) Bar graphs depicting change in growth following treatment of specified cells with fulvestrant or tamoxifen as indicated and siGFP, siATM and siChk2 with accompanying Western blot validation (c+d), or ATM and Chk2 inhibitor (KU-55933, Chk2 inhibitor II hydrate, e+f). Growth response to treatment with ATR and Chk1 inhibitors is included (VE-821, LY2606368) as a specificity control. Validation of pharmacological interventions presented in Figure S5f. Accompanying Chk2 activator data presented in Fig S6. Student’s t-test generated all p-values. Columns represent the mean and error bars denote standard deviation.
In support of these functional relationships, pooled siRNA used to transiently down-regulate either ATM or Chk2 in MCF7 parental cells induced resistance to both fulvestrant and tamoxifen within 48 hours (Fig 4c+d), although transient downregulation of ATM, but not Chk2, also inhibited baseline growth of MCF7 cells (Fig S5d). These results were confirmed in T47D cells (Fig S5e), where transient downregulation of neither ATM nor Chk2 significantly affected baseline growth (Fig S5g). Orthogonally, when ATM and Chk2 activation was inhibited pharmacologically(26) (validated in Fig S5f), both MCF7 (Fig 4e+f) and T47D (Fig S6a+b) cells lost the ability to respond with growth inhibition to fulvestrant treatment. This phenotype appeared specific to Chk2, as neither an ATR inhibitor(27) nor a Chk1 inhibitor(28) could induce endocrine treatment resistance (Fig 4e+f, Fig S6a+b). Moreover, ATM/Chk2 activation using the small molecule activator, 3,3′-diindolyl methane (DIM)(29),(30) (validated in Fig S5e), rescued sensitivity of MCF7/MutL− (Fig S6c+e) and T47D/MLH1− (Fig S6d+f) cells to endocrine treatment dose-dependently. These data suggest that Chk2 activation is both necessary and sufficient for MutL− induced endocrine therapy resistance.
MutL/Chk2 dependent inhibition of CDK4 is required for response to endocrine therapy
Several lines of evidence support the hypothesis that the MutL/Chk2 axis regulates induction of a G1/S cell cycle block when ER+ breast cancer cells are exposed to endocrine therapy. Firstly, the two most significantly upregulated cell cycle genes in MCF7 MutL− vs MutL+ cells after fulvestrant treatment were cyclin-dependent kinase, CDK4 and cyclin, CCND3 (Fig S7a+5a), both integral for G1/S cell cycle progression. Secondly, MutL− ER+ patient tumors from the NeoAI studies had increased levels of CDK4 and CCND3 RNA relative to MutL+ tumors and these levels did not significantly decrease upon AI treatment (Fig S7b). Thirdly, and in support of previously published evidence(25, 29, 31, 32), a significant inverse correlation was observed between protein levels of p-Chk2 and CDK4 in cells treated with Chk2 inhibitors and activators (Fig S7d). These lines of evidence are largely indirect, however, and could represent consequences of unchecked proliferation in MutL− ER+ breast tumors treated with endocrine therapy, rather than a causal occurrence. Therefore, we next tested whether the G1/S phase cyclin-dependent kinases, CDK4 and CDK6, were critical for mediating endocrine therapy resistance in ER+, MutL−/Chk2− breast cancer cells.
When pooled siRNA was used to suppress CDK4 and CDK6 in MCF7 MutL+ and MutL− cells (Fig S7c), suppression of either gene singly resulted in partial rescue of endocrine response in the MutL− cells (data not shown) but the combinatorial inhibition of both genes resulted in complete rescue of fulvestrant sensitivity in MCF7/MLH1− cells (Fig 5b). The next step in the proposed genetic pathway, loss of Chk2, was also tested for its role in CDK4/6 regulation in ER+ breast cancer cells. Transient siRNA-induced suppression of Chk2 in MCF7 parental cells resulted in an immediate and significant increase in sensitivity to pharmacological CDK4/6 inhibition (CDK4/6i) (Fig 5c), a result confirmed in T47D/MutL+ cells (Fig S7e). These data suggested that loss of either MutL or Chk2 in ER+ breast cancer cells induces reliance on CDK4/6 upregulation for cell cycle progression, resulting in resistance to endocrine therapy but sensitivity to CDK4/6i. Corroboratively, analysis of the response of 47 breast cancer cell lines to CDK4/6i, palbociclib(33), indicated increased sensitivity to palbociclib in MutL−, but not MutS−, ER+ breast, and not in ER− breast cancer cell lines (Fig S7f).
Figure 5. MutL deficient ER+ breast cancer cells upregulate CDK4 and are sensitive to CDK4/6 inhibitors.
a) Western blot confirming increased levels of CDK4 and CCND3 in shMLH1 MCF7 cells after fulvestrant treatment relative to shLuc cells with accompanying quantification. b–e) Graphs showing relative growth in 2D (b–d) and in soft agar (e), of parental MCF7 (b–c) and MCF7 shLuc and shMLH1 cells (d–e), treated with siCDK4/6 (b, validated in Fig S7d), siChk2 (c, validated in Fig 4c), and increasing doses of Abemaciclib, a CDK4/6 inhibitor, validated in Fig S7g (c–e), in combination with fulvestrant as indicated. f–j) Boxplots showing tumor growth rate of MCF7 shLuc and shMLH1 cells (f) with accompanying representative images of tumor size (g), and WHIM20, PMS2 mutant PDX (i) tumors with accompanying representative images of pRb immunohistochemistry (h), and HCI-005, PMS1 mutant PDX (j) treated as specified. Scale bar = 20μm. Student’s t-test generated all p-values except for (f+i+j) where ANOVA with Tukey comparson was deployed. Columns represent the mean and error bars denote standard deviation. Supporting data in Fig S7.
MutL− ER+ breast tumors are sensitive to CDK4/6 inhibition
To directly test whether MutL− ER+ breast cancer cells lines are sensitive to CDK4/6i, both palbociclib and abemaciclib(34) (validated in Fig S7g) were administered to MCF7 and T47D MutL+ and MutL− cells along with fulvestrant. Both palbociclib (Fig S7h) and abemaciclib (Fig 5d+S7i) profoundly and consistently inhibited 2D growth of MutL− MCF7 and T47D cells. A striking difference in growth response in MutL− vs MutL+ cells was observed when comparing the effect of fulvestrant alone relative to the combinatorial use of CDK4/6i along with fulvestrant treatment. In the case of MCF7 MutL+ cells, the combination of fulvestrant and abemaciclib induced ~50% growth inhibition relative to fulvestrant alone, while in MutL− cells, the combination induced >80% growth inhibition over fulvestrant alone (Fig 5d, p=0.02). This difference was even more noticeable in T47D cells, where the combinatorial growth inhibition relative to fulvestrant alone went from ~25% in MutL+ to >50% in MutL− cells (Fig S7i, p=0.005). Abemaciclib, in combination with fulvestrant, also inhibited 3D growth of MutL− cells (Fig 5e). Of note, because CDK2, another G1/S cyclin-dependent kinase, was also upregulated at RNA levels in the cell cycle screen (Fig S7a) response to CDK2 inhibition in MutL− cells was tested. In contrast to their response to CDK4/6 inhibitors, neither MCF7 nor T47D shMLH1 cells showed increased sensitivity to CDK2 inhibitors; rather both MutL− cell lines trended towards resistance to this inhibitor relative to MutL+ cells. IC50 for MCF7 shLuc cells was 429nM vs 3μM for shMLH1 cells (p=0.05). Similarly, in T47D cells, IC50 for shLuc cells was 7μm vs >50μM for shMLH1 cells (p=0.04). These data suggested a specific role for CDK4/6 in inducing MutL− mediated intrinsic endocrine therapy resistance, and for CDK4/6 inhibitors in targeting MutL− endocrine therapy resistant tumors.
Three xenograft models were utilized for in vivo validation of response to CDK4/6 inhibition. MCF7 MutL− xenograft tumors regressed when treated with a combination of estrogen deprivation and palbociclib but not when treated with estrogen deprivation alone or with fulvestrant (Fig 5f+g). Response of MutL− xenograft tumors to palbociclib was significantly higher than MutL+ MCF7 xenograft tumors (Fig 5f), although the effect size was moderate. CDK4/6i response was also validated in WHIM20 tumors, which demonstrated down-regulation of pRb in response to a combination of fulvestrant and palbociclib treatment but not in response to fulvestrant alone (Fig 5h). WHIM20 tumors also demonstrated significant inhibition of tumor growth in response to either palbociclib alone or to the combination of palbociclib and fulvestrant (Fig 5i). Finally, a second PDX model, HCI-005 with a frameshift mutation in PMS1, also demonstrated endocrine therapy resistance and significant palbociclib sensitivity (Fig 5j). These data together suggested that patients with MutL− ER+ tumors can benefit from CDK4/6i despite intrinsic endocrine therapy resistance.
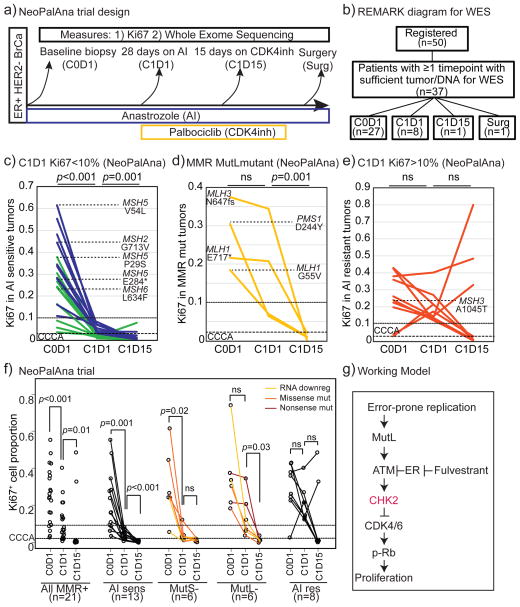
To obtain clinical data to support the postulate that MutL− tumors are sensitive to CDK4/6i, data from the NeoPalAna trial were examined(17). In this study (Fig 6a), 50 patients presenting with clinical stage 2/3 ER+ HER2− breast cancer were treated with an AI, anastrozole, and biopsied after one month (C1D1). Palbociclib was then added to the treatment regimen with a further biopsy after ~2 weeks of combined treatment (C1D15). Patients then completed neoadjuvant treatment with the AI+CDK4/6i combination for approximately 16 weeks before surgery. Whole exome sequencing and RNA expression analysis were conducted on tumors with sufficient material (Fig 6b). The primary endpoint for this study was Complete Cell Cycle Arrest (CCCA) defined as Ki67≤2.7%. More tumors demonstrated CCCA after the combination of AI and CDK4/6i than after AI alone confirming the activity of palbociclib in primary ER+ breast cancer.
Figure 6. MutL deficiency in ER+ breast tumors can predict sensitivity to CDK4 inhibitors.
a+b) Schema of longitudinal clinical trial (a) and REMARK diagram of whole exome sequenced samples (b) in NeoPalAna, testing efficacy of neoadjuvant aromatase inhibitor, anastrozole and CDK4/6 inhibitor, palbociclib combination in patients with ER+ breast cancer. c–e) Line graphs indicating response to anastrozole and palbocilib as measured by on-treatment Ki67 levels in longitudinal biopsies of MutL+, AI-sensitive tumors (Ki67≤10%, c), tumors with mutations in MutL genes (d) and MutL+, AI- resistant tumors (Ki67>10%, e). Green lines indicate tumors that demonstrate complete cell cycle arrest (CCCA, Ki67<2.7%) after 4 weeks of aromatase inhibitor treatment. Tumors with mutations in MutS genes are indicated in c+e. f) Line graphs indicating response to anastrozole and palbocilib as measured by on-treatment Ki67 levels in longitudinal biopsies of all MMR+ tumors (combining AI resistant and Ai sensitive subsets), AI sensitive tumors (Ki67<10%), tumors with mutations in and/or low RNA of MutS and MutL genes, and AI resistant tumors (Ki67>10%). Line color indicates class of mutation or RNA downregulation as indicated in legend. Friedman Rank Sum test for repeated measures determined p-values. g) Working model. Error prone proliferation stimulated by estrogen signaling in ER+ breast cancer cells induces mismatches and activates the mismatch repair pathway, which in turn activates Chk2 resulting in exit from the cell cycle. In the absence of either estrogen signaling or MutL complex activation, there is no/low Chk2 activation and cells do not respond to endocrine therapy.
First, the Ki67 response of tumors based on mutations in MutL and MutS genes alone was analyzed (Fig 6c–e). To serve as reference points in the analysis, the MutL+ cases are divided into AI sensitive and resistant (AI sensitive=Ki67<10% after 4 weeks of AI). When considering mutations alone, six MutS mutant tumors were identified, five of which were AI sensitive, including a tumor with a truncating mutation in MSH5. The majority of the MutS mutant tumors also demonstrated CCCA with AI alone, although the addition of CDK4/6i incrementally increased treatment efficacy (Fig 6c+f). Four MutL mutant tumors were identified (Fig 1e). Consistent with our hypothesis, these tumors exhibited AI resistant proliferation and none demonstrated CCCA with AI alone. However, in keeping with the experimental data presented above, all four MutL-mutant tumors demonstrated significant Ki67 inhibition when palbociclib was added, with all of them achieving CCCA (Fig 6d). Importantly, tumors with inactivating (nonsense or frame shift) mutations in MutL genes demonstrated the strongest endocrine therapy resistance phenotype and appeared most sensitive to CDK4/6i (Fig 6d).
Next, dysregulation of MutL and MutS genes at both DNA and gene expression level (MutL− and MutS− groups) were determined as in previous analyses presented in Fig 1 (described in Fig 6f). Using the combined definition, six MutL− and six MutS− tumors were identified from 37 tumors examined (two combined MutL and MutS deficient were excluded as not fitting a binary definition of deficiency). Again, four of the six MutS− tumors were AI sensitive, even exhibiting CCCA with AI alone (Fig 6f), and only one of the tumors remained AI resistant (Fig 6e+f). In contrast, 3/6 MutL− tumors were AI resistant (Ki67>10%) and 0/6 MutL− tumors demonstrated CCCA on AI alone (Fig 6f). MutL− tumors demonstrated mean Ki67 levels after AI treatment of 13%, comparable to MMR+/AI resistant tumors (19%; p=0.2) and significantly different from both MutS− and MMR+/AI sensitive tumors (4%; p=0.003). However, upon addition of palbociclib all six MutL− tumors achieved CCCA, with mean Ki67 levels falling from 13% on AI treatment to 1.3% on the combination.
Discussion
In this investigation we delineate a pathway involving the MutL complex, along with ATM, Chk2 and CDK4/6 that is required for ER+ HER2− tumors to respond to endocrine therapy (Fig 6g). When components of this pathway are poorly expressed or lost through mutation, feedback control on CDK4/6 is defective. This allows the cell cycle to proceed despite DNA mismatches thereby promoting the growth of high mutation load ER+ breast cancers that are intrinsically resistant to endocrine treatment, but still sensitive to CDK4/6i.
The sequential triple biopsy design of the NeoPalAna trial was executed to determine the effects of CDK4/6i on ER+ HER2− tumors where intrinsic endocrine therapy resistance had been demonstrated. Consistent with our model, CDK4/6i was uniformly effective in suppressing residual proliferation in MutL− ER+ breast cancers, with almost undetectable Ki67 levels after ~2 weeks of palbociclib exposure. In contrast, the degree to which each mutant MutL allele or expression level defect promoted AI-resistant proliferation was variable and likely dependent on the degree of dysfunction induced by the mutation or the degree of under-expression. A comprehensive predictive assay based on MutL gene status would have to combine expression analysis with mutation detection. A combination of somatic mutation detection and expression status is a reasonable proposition for a clinical assay as both approaches have been adapted to routine pathological material. Ultimately, large datasets from randomized trials of adjuvant CDK4/6i therapy that are currently underway will be required to dissect these relationships fully.
The efficacy of CDK4/6i in the advanced ER+ breast cancer setting has been validated in multiple clinical trials(35, 36), but as yet there are no biomarkers to guide treatment. CDK4/6i are expensive, have more severe adverse events than endocrine therapy alone, and have to be administered chronically to control disease. Our findings open up new hypotheses for CDK4/6i predictive test discovery, which to date has not been successful(37). While a weakness of our study concerns the small sample size in the NeoPalAna study, the data are entirely consistent with our extensive preclinical investigation regarding the response of MutL− tumors to CDK4/6i. Clearly, our work merits further correlative studies in the setting of both endocrine monotherapy studies and CDK4/6i combination studies with larger sample sizes and long-term outcomes to determine the prognostic and predictive impact of our findings. A further issue is that the MutL− mechanism clearly does not explain all cases of endocrine therapy resistant yet palbociclib sensitive ER+ breast cancer. Further exploration of the relationship between other DNA repair pathways and this phenotype may be fruitful in this regard.
The endocrine therapy sensitivity of MutS− ER+ breast cancers is an interesting and contrasting phenomenon compared to MutL− tumors. The molecular underpinnings of this gene selectivity is an important area for further research, and is perhaps linked to the canonical association of MLH1 with ATM activation and of MSH2 complex formation with ATR/Chk1 activation(21, 38). Consistent with this hypothesis, in this study, MutS− tumors appear to retain the ability to activate ATM and Chk2 in response to inhibition of ER. A further uncertainty is how ER is linked to ATM function. This has not been entirely delineated but it has been proposed to be through ER up-regulated microRNA-18a and 106a(15). In this model, ER activation drives the microRNA-dependent loss of ATM function, allowing the cell cycle to proceed.
Despite the retention of endocrine therapy sensitivity in MutS− tumors, either MutL− or MutS− may promote secondary mutations though MMR defects that enhance the probability of acquired endocrine resistance. A potential incidence of this phenomenon is observed in WHIM20, where we have previously reported an ESR1 ligand-binding domain mutation (Y537S) that causes endocrine therapy resistance(4). It is possible that the PMS2 E5K mutation in WHIM20 causes only a partial endocrine therapy resistance that is eventually made more profound due to the evolution of an ESR1 gain of function mutation. It is clear from the relationships between MutL status and Ki67 response that partial endocrine resistance is often seen, likely due to incomplete suppression of MutL function. This possibility is illustrated by the HCI005 PDX model where, in presence of wildtype ESR1 but mutant PMS1, there is partial endocrine therapy resistance, and significant palbociclib sensitivity. Even modest levels of persistent KI67 activity despite AI therapy dramatically increases relapse risk(39).
The role of MMR in breast cancer has been underestimated for several reasons. Firstly, there is only a weak relationship between germ-line defects in MMR genes and breast cancer susceptibility although links to increased breast cancer risk have been observed particularly for families with defective MLH1 alleles(40). Secondly, our investigations on ER+/HER2− breast cancer in the neoadjuvant setting illustrated here provide the only published datasets to date that can connect intrinsic endocrine therapy resistance and palbociclib to MMR gene mutation status. Thirdly, an examination of classic markers of MMR dysregulation such as repeat expansion may be unhelpful in the breast cancer context, since MutL alleles are likely frequently hypomorphic, i.e. sufficient to be defective in Chk2 activation but having only relatively weak effects on promoting mutation load, unlike for example, MMRD in classic Lynch syndrome cancers such as endometrial and colorectal cancer(41). Simple immunohistochemistry that is typically used for the routine diagnosis of MMR in colorectal cancer will not identify MutL missense alleles with partially defective function.
Both CHEK2 and ATM are well-established breast cancer susceptibility genes with low to medium penetrance(42). Interestingly, CHEK2 germline variants specifically associate with increased incidence of ER+ tumors(43), suggesting that the role of Chk2 in connecting ER to the cell cycle highlighted in our investigation is a fundamental pathway that contributes to development of luminal tumors. Of concern is that endocrine drugs used for chemoprevention in patients with germ-line mutant MutL, CHEK2, or ATM related breast cancer might be less effective. However, we show that the Chk2 activator and dietary component di-indolylmethane (DIM) could provide an alternative agent in this setting, at least regarding the chemoprevention of MutL and ATM-associated breast cancers.
Methods
Cell lines, mice, CRISPR, si/shRNA transfection and growth assays
Cell lines were obtained from American Type Culture Collection (ATCC, 2015) and maintained and validated as previously reported(44) Mycoplasma tests were performed on parent cell lines and stable cell lines every 6 months (latest test: 05/17) with the Lonza Mycoalert Plus kit (cat# LT07-710) as per manufacturer’s instructions. Cell lines were discarded at <25 passages, and fresh vials were thawed out. Key experiments were repeated with each fresh thaw. shRNA plasmids were purchased from Sigma-Aldrich. CRISPR plasmids were purchased from Santa Cruz. Transfection and validation of CRISPR and shRNA plasmids was conducted as previously23. Transient transfection with siRNA against CHEK2 was conducted as previously(44), and siRNA pools were purchased from Sigma Aldrich. Stable cell lines were established in the presence of specified antibiotics at recommended concentrations. Growth assays were conducted in triplicate and repeated independently as previously using Alamar blue to identify cell viability(44). Growth assay results were plotted as fold change in growth from Day1 to Day7, and normalized to vehicle control where specified. 3D growth assays were conducted over 4–6 weeks with weekly drug treatments as previously(45). Tumor growth assays in vivo were carried out as previously(46) by injecting 2–5×106 MCF7 cells into the L4 mammary fat pad/mouse. Mice for the MCF7 experiments were 4–6 week athymic nu/nu female mice (Envigo) and for the PDX experiments were 6–8 week female SCID/Bg mice, both from Charles River laboratory. Tumor volume was measured twice or thrice weekly using calipers to make two diametric measurements. Tumors were randomized for treatment at 50–150 mm3 volume. Tumors were harvested at <2 cm diameter, and were embedded in paraffin blocks, OCT, and snap frozen as previously(47). Mice that died within 3 weeks of tumor growth rate experiments were excluded from analysis. For all mouse experiments, investigator was blinded to groups and to outcomes. All mouse experiments were performed according to the IACUC rules and regulations (protocol#AN-6934).
Inhibitors and agonists
All drugs were maintained as stock solutions in DMSO, and stock solutions were stored at −80, and working stocks at −20 unless otherwise mentioned. 4-OHT (cat#H7904) and fulvestrant (cat#I4409) were purchased from Sigma-Aldrich and stocks were diluted to 10mM working stocks for all experiments other than dose curves, where specified concentrations were used. For all experiments, cells were treated 24 hours after plating, and thereafter every 48 hours until completion of experiment. For mouse xenograft experiments, fulvestrant concentrations of 250mg/kg body weight were used. Beta-estradiol was purchased from Sigma-Aldrich (cat# E8875), maintained in sterile, nuclease free water, and diluted to obtain 10mM stocks for all experiments. For mouse xenograft experiments, 17 β-estradiol was added to the drinking water twice a week at a final concentration of 8μg/ml (cat#E2758, Sigma). Chk2 Inhibitor II hydrate (cat#C3742, Sigma-Aldrich), Chk1 inhibitor (PF-477736, cat#S2904, SelleckChem) and Chk2 activator (3, 3;-diindolyl methane, cat#sc-204624, Santa Cruz Biotechnology) were used at 10 and 100 nM, and 10 and 100 micromolar concentrations, respectively. Abemaciclib (LY2835219, cat#S7158, SelleckChem) and Palbociclib (PD-0332991, cat#S1116, SelleckChem) were used at 100 nM final concentrations for cell line assays and at 70 mg/kg/day in chow for tumor growth assays as previously(46).
Immunostaining, Comet assay and Microscopy
Immunofluorescence (IF) was performed based on manufacturer instructions. Cells were: washed in PBS; fixed for 20 minutes at RT in 4% PFA; blocked for 1 hour at room temperature in 5% goat serum and 1% Triton X-100 in 1× PBS; incubated with primary antibody overnight at 4 degrees in 1% goat serum and 1% Triton X-100 in 1× PBS antibody diluent; incubated with secondary antibody in diluent for 1 hour at RT; and then mounted with DAPI-containing mounting media (cat#P36935). Primary antibodies used include pHistoneH3 (Cell Signaling; 1:500), pATM (cat#ab36810), pChk2 (cat#21997), MLH1 (cat#WH0004292M2). Cells were treated with fulvestrant for 48 hours before evaluation. Alkaline comet assay was performed as per the manufacturer’s instructions on a CometAssay Electrophoresis SystemII (Trevigen, cat#4250-050-ES). Calculation of nuclei with DNA damage was performed using CASPLab software27 to calculate the ratio of DNA content in tail/head. Cut-offs for categorization were set as 0–0.5, 0.5–1, 1–5 and >5 for No, Low, Med and High DNA damage. Fluorescent images were captured with a Nikon microscope and quantified with ImageJ.
Western blotting, gene expression array, RPPA and phospho-proteomics
Western blotting was conducted as previously described23. All antibodies were purchased from Cell Signaling Technology and used at 1:1000 dilutions unless otherwise specified. Primary antibodies were incubated with the membrane overnight at 4 degrees and included p-Chk2 (cat#2197), total Chk2 (cat#2662), p21 (cat#2947), p27 (cat#2552), MLH1 (1:2000, Sigma-Aldrich, cat# WH0004292M2), PMS1 (1:2000, Sigma-Aldrich, cat#WH0005378M1), PMS2 (1:2000, Sigma-Aldrich, cat#SAB4502223), CDK2 (cat#2546), CDK4 (cat# 12790), and Cyclophilin-A (cat#2175, 1:5000). The gene expression array (Qiagen) was used according to manufacturer’s instructions. Reverse phase protein array (RPPA) assays were carried out as described previously with minor modifications(48). Phospho-proteomics data and methodologies for analysis were performed also as described previously(22).
Statistical Analysis
ANOVA or Student’s T-test was used for independent samples with normal distribution. Where distribution was not normal (assessed using Q-Q plots with the Wilk-Shapiro test of normality), either the Kruskal-Wallis or Wilcoxon Rank Sum test was used. All experiments were conducted in triplicate and each experiment was duplicated independently >2 times. These criteria were formulated to ensure that results from each dataset were calculable within the range of sensitivity of the statistical test used. Databases used for human data mining are from publically available resources: Oncomine, cBio(49) and COSMIC(50). Z1031/POL dataset was used with permission from the Alliance consortium. All patients provided informed consent, studies were conducted according to ethical guidelines and with IRB approval. Lists of DDR genes for initial analyses were obtained from the KEGG database, and list of MMR genes was restricted to MLH1, PMS1, PMS2, MLH3, MSH2, MSH3, MSH4, MSH5, MSH6, PMS2L3, and EXO1. MutL− tumor from METABRIC, TCGA and Z1031/POL datasets were determined in a case list containing all ER+ sample IDs6 based on gene expression less than mean-1.5 standard deviation and/or the presence of non-silent mutations in MLH1, MLH3, PMS1 and PMS2. Mutations identified by next-generation sequencing in the NeoAI trials were validated previously(51) and mutations identified by TCGA whole-exome sequencing have been demonstrated to have ~95% validation efficacy(52). For the multivariate analysis, we analyzed 1415 ER+ tumor samples, extracting mutation data from cBio portal(53), and corresponding clinical data through Oncomine. Only samples with survival metadata were included in the analysis. RPPA, gene expression, and survival data for TCGA samples were downloaded from cBio portal(49). Resistance in the Z1031/POL dataset was determined as previously(6). For combined NeoAI trial data analysis in Fig 1e, a one-sided Wilcoxon Rank Sum test was used to confirm significance of change in Ki67 in MutL− vs MutL+ tumors, and two-sided tests for all other analyses, with correction for multiple comparisons. For NeoPalAna trials, the combined RNA/mutation level analysis in Fig 6f excluded all four tumors that had coincident mutations and/or downregulation of both MutL and MutS family members, including two tumors with MutS gene mutations (E284* and P29S in MSH5). All survival data was analyzed using Kaplan-Meier curves and log-rank tests. Proportional hazards were determined using Cox regression. Sample size for animal experiments was estimated using power calculations in R. P-values were adjusted for multiple comparisons where appropriate using Benjamini-Hochberg. All graphs and statistical analyses were generated either in MS Excel or R(54) and edited in Adobe Photoshop or Illustrator.
Supplementary Material
Statement of Significance.
MutL MMR deficiency in a subset of ER+ primary tumors explains why CDK4/6 inhibition is effective against some de novo endocrine therapy resistant tumors. Therefore, markers of MutL MMR dysregulation could guide CDK4/6 inhibitor use in the adjuvant setting, where the risk benefit ratio for untargeted therapeutic intervention is narrow.
Acknowledgments
Financial support: Research reported in this publication was primarily supported by Susan G. Komen Promise grant (PG12220321 to MJE), Cancer Prevention and Research Institute of Texas (CPRIT) Recruitment of Established Investigators award (RR140033 to MJE), Laura Ziskin award from Stand Up2 Cancer (MJE), and Komen CCR award (CCR16380599 to SMK). Clinical trial data accrual and analysis was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821 and U10CA180882 (to the Alliance for Clinical Trials in Oncology), U10CA077440 (legacy), U10CA180833, and U10CA180858. NeoPalAna trial was supported by Pfizer Pharmaceuticals, the Siteman Cancer Center Grant (P30 CA91842, SCC, Eberlein), NCI Cancer Clinical Investigator Team Leadership Award (3P30 CA091842-12S2, NIH/NIC, Ma), Susan G. Komen Promise Grant (MJE), and the Saint Louis Men’s Group Against Cancer (Ma). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. RPPA experiments were supported in part by funds from the CPRIT Proteomics & Metabolomics Core Facility Support Award (RP120092) (DPE and ShH) and NCI Cancer Center Support Grant to Antibody-based Proteomics Core/Shared Resource (P30CA125123) (DPE and ShH).
The authors would like to acknowledge Drs. Eric Chang, Geoff Wahl, Jeff Rosen and Susan Rosenberg for scientific input, Dr. Alana Welm for generously sharing the HCI-005 PDX model and genomic data, Drs. Kimal Rajapakshe, Cristian Coarfa, and Qianxing Mo for data processing and normalization on RPPA data, and Fuli Jia, and Davis So from the Antibody-based Proteomics Core/Shared Resource for technical assistance in performing RPPA experiments.
Footnotes
ClinicalTrials.gov Identifier: NCT00265759
Conflict of Interest Statement: C. Ma received research funding and ad hoc consulting fees from Pfizer and Novartis. M. Ellis received ad hoc consulting fees from Pfizer, AstraZenica, Celgene, Nanostring, Puma and Novartis. He also reports patents and royalty income for PAM50-based diagnostics to Nanostring/Prosigna. All other authors disclosed no potential conflict of interest.
References
- 1.Kohler BA, Sherman RL, Howlader N, Jemal A, Ryerson AB, Henry KA, et al. Annual Report to the Nation on the Status of Cancer, 1975–2011, Featuring Incidence of Breast Cancer Subtypes by Race/Ethnicity, Poverty, and State. Journal of the National Cancer Institute. 2015;107(6):djv048. doi: 10.1093/jnci/djv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–45. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeRose YS, Gligorich KM, Wang G, Georgelas A, Bowman P, Courdy SJ, et al. Patient-derived models of human breast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr Protoc Pharmacol. 2013;Chapter 14(Unit14):23. doi: 10.1002/0471141755.ph1423s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowsett M, Smith IE, Ebbs SR, Dixon JM, Skene A, A’Hern R, et al. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst. 2007;99(2):167–70. doi: 10.1093/jnci/djk020. [DOI] [PubMed] [Google Scholar]
- 6.Ellis MJ, Suman VJ, Hoog J, Lin L, Snider J, Prat A, et al. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype--ACOSOG Z1031. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2011;29(17):2342–9. doi: 10.1200/JCO.2010.31.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis MJ, Suman VJ, Hoog J, Goncalves R, Sanati S, Creighton CJ, et al. Ki67 Proliferation Index as a Tool for Chemotherapy Decisions During and After Neoadjuvant Aromatase Inhibitor Treatment of Breast Cancer: Results From the American College of Surgeons Oncology Group Z1031 Trial (Alliance) J Clin Oncol. 2017:JCO2016694406. doi: 10.1200/JCO.2016.69.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haricharan S, Bainbridge MN, Scheet P, Brown PH. Somatic mutation load of estrogen receptor-positive breast tumors predicts overall survival: an analysis of genome sequence data. Breast Cancer Research and Treatment. 2014;146(1):211–20. doi: 10.1007/s10549-014-2991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehman FL, Lord CJ, Ashworth A. Synthetic lethal approaches to breast cancer therapy. Nature Reviews Clinical Oncology. 2010;7(12):718–24. doi: 10.1038/nrclinonc.2010.172. [DOI] [PubMed] [Google Scholar]
- 10.Abdel-Fatah TM, Perry C, Arora A, Thompson N, Doherty R, Moseley PM, et al. Is there a role for base excision repair in estrogen/estrogen receptor-driven breast cancers? Antioxid Redox Signal. 2014;21(16):2262–8. doi: 10.1089/ars.2014.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijai J, Topka S, Villano D, Ravichandran V, Maxwell KN, Maria A, et al. A recurrent ERCC3 truncating mutation confers moderate risk for breast cancer. Cancer Discov. 2016 doi: 10.1158/2159-8290.CD-16-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson JA, Jr, Budd GT, Carey LA, Harris LA, Esserman LJ, Fleming GF, et al. Improved surgical outcomes for breast cancer patients receiving neoadjuvant aromatase inhibitor therapy: results from a multicenter phase II trial. J Am Coll Surg. 2009;208(5):906–14. doi: 10.1016/j.jamcollsurg.2009.01.035. discussion 15–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo X, Yang C, Qian X, Lei T, Li Y, Shen H, et al. Estrogen Receptor Alpha regulates ATM expression through miRNAs in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(18):4994–5002. doi: 10.1158/1078-0432.CCR-12-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raschle M, Marra G, Nystrom-Lahti M, Schar P, Jiricny J. Identification of hMutLbeta, a heterodimer of hMLH1 and hPMS1. J Biol Chem. 1999;274(45):32368–75. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- 17.Ma CX, Gao F, Luo J, Northfelt DW, Goetz MP, Forero A, et al. NeoPalAna: Neoadjuvant palbociclib, a cyclin-dependent kinase 4/6 inhibitor, and anastrozole for clinical stage 2 or 3 estrogen receptor positive breast cancer. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-16-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang DK, Ricciardiello L, Goel A, Chang CL, Boland CR. Steady-state regulation of the human DNA mismatch repair system. J Biol Chem. 2000;275(37):29178. [PubMed] [Google Scholar]
- 19.Buermeyer AB, Wilson-Van Patten C, Baker SM, Liskay RM. The human MLH1 cDNA complements DNA mismatch repair defects in Mlh1-deficient mouse embryonic fibroblasts. Cancer Res. 1999;59(3):538–41. [PubMed] [Google Scholar]
- 20.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–20. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown KD, Rathi A, Kamath R, Beardsley DI, Zhan Q, Mannino JL, et al. The mismatch repair system is required for S-phase checkpoint activation. Nat Genet. 2003;33(1):80–4. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 22.Huang KL, Li S, Mertins P, Cao S, Gunawardena HP, Ruggles KV, et al. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat Commun. 2017;8:14864. doi: 10.1038/ncomms14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G-M. Mechanisms and functions of DNA mismatch repair. Cell Research. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 24.Caldon CE. Estrogen Signaling and the DNA Damage Response in Hormone Dependent Breast Cancers. Frontiers in Oncology. 2014:4. doi: 10.3389/fonc.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aliouat-Denis C-M, Dendouga N, Van den Wyngaert I, Goehlmann H, Steller U, van de Weyer I, et al. p53-independent regulation of p21Waf1/Cip1 expression and senescence by Chk2. Mol Cancer Res. 2005;3(11):627–34. doi: 10.1158/1541-7786.MCR-05-0121. [DOI] [PubMed] [Google Scholar]
- 26.Dai B, Zhao XF, Mazan-Mamczarz K, Hagner P, Corl S, Bahassi el M, et al. Functional and molecular interactions between ERK and CHK2 in diffuse large B-cell lymphoma. Nat Commun. 2011;2:402. doi: 10.1038/ncomms1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williamson CT, Miller R, Pemberton HN, Jones SE, Campbell J, Konde A, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun. 2016;7:13837. doi: 10.1038/ncomms13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryant C, Rawlinson R, Massey AJ. Chk1 inhibition as a novel therapeutic strategy for treating triple-negative breast and ovarian cancers. BMC cancer. 2014;14:570. doi: 10.1186/1471-2407-14-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kandala PK, Srivastava SK. Activation of checkpoint kinase 2 by 3,3′-diindolylmethane is required for causing G2/M cell cycle arrest in human ovarian cancer cells. Molecular Pharmacology. 2010;78(2):297–309. doi: 10.1124/mol.110.063750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomson CA, Ho E, Strom MB. Chemopreventive properties of 3,3′-diindolylmethane in breast cancer: evidence from experimental and human studies. Nutr Rev. 2016;74(7):432–43. doi: 10.1093/nutrit/nuw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3(5):421–9. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 32.Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito Si, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21(19):5195–205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast cancer research: BCR. 2009;11(5):R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nature Reviews Clinical Oncology. 2016 doi: 10.1038/nrclinonc.2016.26. advance online publication. [DOI] [PubMed] [Google Scholar]
- 35.Turner NC, Ro J, Andre F, Loi S, Verma S, Iwata H, et al. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. N Engl J Med. 2015;373(3):209–19. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 36.Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N Engl J Med. 2016;375(18):1738–48. doi: 10.1056/NEJMoa1609709. [DOI] [PubMed] [Google Scholar]
- 37.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 38.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 39.Goncalves R, Reinert T, Ellis MJ. Avoidance of Negative Results in Adjuvant Endocrine Therapy Trials for Estrogen Receptor-Positive Breast Cancer. J Clin Oncol. 2017:JCO2017730424. doi: 10.1200/JCO.2017.73.0424. [DOI] [PubMed] [Google Scholar]
- 40.Win AK, Lindor NM, Jenkins MA. Risk of breast cancer in Lynch syndrome: a systematic review. Breast Cancer Res. 2013;15(2):R27. doi: 10.1186/bcr3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donehower LA, Creighton CJ, Schultz N, Shinbrot E, Chang K, Gunaratne PH, et al. MLH1-silenced and non-silenced subgroups of hypermutated colorectal carcinomas have distinct mutational landscapes. J Pathol. 2013;229(1):99–110. doi: 10.1002/path.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tung N, Lin NU, Kidd J, Allen BA, Singh N, Wenstrup RJ, et al. Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients With Breast Cancer. J Clin Oncol. 2016;34(13):1460–8. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Domagala P, Wokolorczyk D, Cybulski C, Huzarski T, Lubinski J, Domagala W. Different CHEK2 germline mutations are associated with distinct immunophenotypic molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;132(3):937–45. doi: 10.1007/s10549-011-1635-7. [DOI] [PubMed] [Google Scholar]
- 44.Haricharan S, Brown P. TLR4 has a TP53-dependent dual role in regulating breast cancer cell growth. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(25):E3216–25. doi: 10.1073/pnas.1420811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discovery. 2013;3(2):224–37. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wardell SE, Ellis MJ, Alley HM, Eisele K, VanArsdale T, Dann SG, et al. Efficacy of SERD/SERM Hybrid-CDK4/6 Inhibitor Combinations in Models of Endocrine Therapy-Resistant Breast Cancer. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2015;21(22):5121–30. doi: 10.1158/1078-0432.CCR-15-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haricharan S, Dong J, Hein S, Reddy JP, Du Z, Toneff M, et al. Mechanism and preclinical prevention of increased breast cancer risk caused by pregnancy. eLife. 2013;2:e00996. doi: 10.7554/eLife.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang C-H, Zhang M, Rajapakshe K, Coarfa C, Edwards D, Huang S, et al. Mammary Stem Cells and Tumor-Initiating Cells Are More Resistant to Apoptosis and Exhibit Increased DNA Repair Activity in Response to DNA Damage. Stem Cell Reports. 2015;5(3):378–91. doi: 10.1016/j.stemcr.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38(Database issue):D652–7. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–60. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. Erratum: The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11908. doi: 10.1038/ncomms11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.